Abstract
Cell surface receptors play ubiquitous roles in cell signaling and communication and their expression levels are important biomarkers for many diseases. Expression levels are, however, only one factor that determines the physiological activity of a receptor. For some surface receptors their distribution on the cell surface, especially their clustering, provides additional mechanisms for regulation. To access this spatial information robust assays are required that provide detailed insight into the organization of cell surface receptors on nanometer length scales. In this manuscript we demonstrate through combination of scattering spectroscopy, electron microscopy, and generalized multiple particle Mie theory (GMT) simulations that the density- and morphology-dependent spectral response of Au nanoparticle (NP) immunolabels bound to the epidermal growth factor receptors ErbB1 and ErbB2 encodes quantitative information of both the cell surface expression and spatial clustering of the two receptors in different unliganded in vitro cancer cell lines (SKBR3, MCF7, A431). A systematic characterization of the collective spectral responses of NPs targeted at ErbB1 and ErbB2 at various NP concentrations indicates differences in the large-scale organization of ErbB1 and ErbB2 in cell lines that overexpress these receptors. Validation experiments in the scanning electron microscope (SEM) confirm that NPs targeted at ErbB1 on A431 are more strongly clustered than NPs bound to ErbB2 on SKBR3 or MCF7 at overall comparable NP surface densities. This finding is consistent with the existence of larger receptor clusters for ErbB1 than for ErbB2 in the plasma membranes of the respective cells.
Keywords: Cell Signaling, Epidermal Growth Factor Receptor, Immunophenotyping, Molecular Imaging, Plasmon Hybridization, Receptor Clustering
The transmembrane proteins ErbB1 and ErbB2 are members of the epidermal growth factor receptor (ErbB) family that play prominent roles in the development and progression of various malignancies.1–4 Although overexpression of ErbB1 and ErbB2 can provide useful diagnostic and prognostic cues, it is not trivial to discern pathologic dysregulations from fluctuations that lie within the natural spread of biological activity based on expression levels alone. Consequently, synergistic information, for instance, about the spatial receptor organization that shed additional light on the physiological state of the receptor would be highly beneficial for optimizing treatment strategies. There is growing evidence that the segregation of ErbB family members into micro- or nanodomains, in the following simply referred to as “clusters”, plays an important role in regulating the signal transduction through the plasma membrane.5–9 The characterization of the clustering of cell surface receptors goes, however, beyond the capabilities of classical immunophenotyping methods (e.g. flow cytometry) and, instead requires approaches that can probe the spatial organization of cell surface receptors on sub-diffraction limit length scales. Methods that have been applied for this purpose in the case of ErbB family members include electron microscopy10 and different fluorescence based approaches, such as fluorescence resonance energy transfer (FRET),11–14 near-field scanning optical microscopy (NSOM),15, 16 fluorescence brightness analysis,17, 18 and superresolution fluorescence microscopies.19 We have recently shown that the density-dependent spectral response of 40 nm Au nanoparticle (NP) immunolabels targeted at the respective receptors provides an alternative, non-fluorescence based optical approach for mapping the spatial distribution of receptor densities on cellular surfaces.20, 21 The optical properties of NPs are determined by coherent collective electron oscillations, the plasmons. If two or more NPs approach each other to within approx. one particle diameter on the cell surface, the oscillating E-fields surrounding the NPs interfere and induce a hybridization of the plasmon modes of the individual NPs.22, 23 Although plasmon hybridization is an intrinsic near-field response, the accompanying spectral shift of the plasmon resonance can be conveniently detected in the far-field using conventional optical microscopy and spectroscopy.24–31 This unique feature of NP labels has enabled new bio-imaging approaches for monitoring the structure and dynamics of biological systems on sub-diffraction limit length scales.20, 21, 32–43
In this manuscript we demonstrate that plasmon coupling between NP immunolabels targeted at a specific cell surface receptor facilitates a quantitative characterization not only of its surface expression but also of its spatial organization into clusters. Our approach is based on the fact that at constant ambient refractive index the scattering spectra of NP immunolabels targeted to specific cell surface receptors depend both on the average separation and the geometric arrangement of the NPs on the cell surface and, therefore, on factors which are determined by the underlying spatial distribution of the targeted receptors (Figure 1a). Since plasmon coupling enables to probe distances on the tens of nanometer length scale (and thus significantly beyond typical spatial FRET barriers),25, 26 the technique is particularly suited for characterizing the “large-scale”13 association of ErbBs into clusters on cellular surfaces, provided that scattering spectra can be acquired with sufficient signal-to-noise.
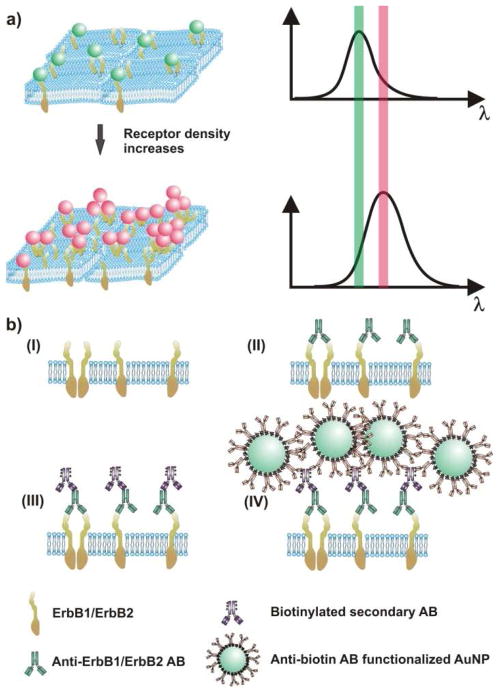
Figure 1. The spectral response of NP immunolabels depends on the NP density and clustering on the cell surface.
a) As the average interparticle separation between the NP decreases (left) the area-averaged plasmon spectrum red-shifts and increase in scattering intensity (right). b) Labeling strategy for cell-surface ErbB1 and ErbB2: (I) ErbB receptors embedded in the plasma membrane are (II) labeled with primary antibodies (ABs). (III) Biotinylated secondary ABs are tethered to the primary antibodies before, in the final step (IV), anti-biotin AB-functionalized NPs are targeted to biotin binding sites.
We validate in the following the applicability of plasmon coupling microscopy20, 21, 32 to differentiate ErbB1 and ErbB2 cell surface expression levels in three different cancer model cell lines (SKBR3, MCF7, and A431) using established surface profiling approaches, such as fluorescence flow cytometry and electron microscopy. We then examine the density- and morphology-dependence of the spectral response of NP immunolabels targeted at cell surface receptors using state-of-the-art electromagnetic simulations. A concise understanding of the combined effect of average NP density and spatial organization on the spectral response of the bound NP immunolabels facilitates a systematic experimental investigation of the ErbB1 and ErbB2 clustering in the investigated cell/receptor systems in the last part of this manuscript.
ErbB1 and ErbB2 Labeling for an Optical Characterization of Relative Expression Levels
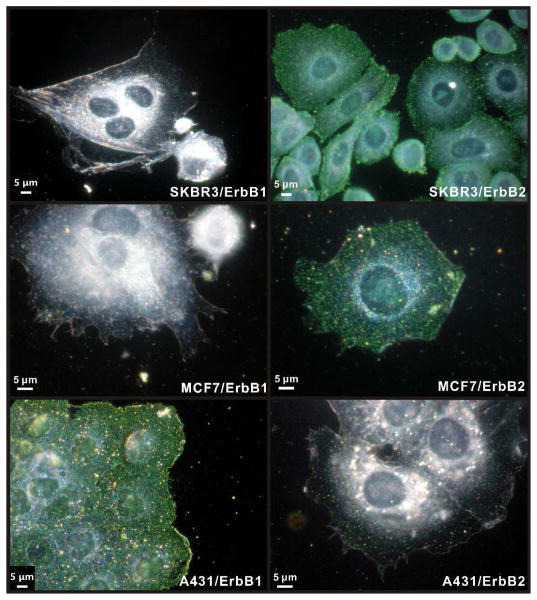
Our labeling strategy for ErbB1 and ErbB2 follows established procedures21 and is outlined in Figure 1b. In the first labeling step the formaldehyde-fixed cells are incubated with primary antibodies targeted at the receptor of interest. Then, biotinylated secondary antibodies are tethered to the primary antibodies. Finally, anti-biotin antibody functionalized 40 nm diameter Au NPs are bound to the generated biotin binding sites on the cell surface. The NPs used in this work were functionalized with 3.4 kDa long polyethylene glycols (PEGs) that carry a terminal azide group to facilitate a cross-linking with alkine functionalized anti-biotin antibodies (see Methods in the Supporting Information) through a Cu(I) catalyzed 1,3-cycloaddition reaction.44 Figure 2 shows optical images of MCF7, SKBR3, and A431 cells with labeled ErbB1 and ErbB2 receptors taken with a conventional digital camera mounted via an eye-piece adaptor to a darkfield microscope. The immunolabel concentration used in these experiments was cAu ≅ 0.8 nM, and the cell samples were incubated with NP immunolabel solution for 3 hours. Already an inspection by eye shows clear differences between the investigated cell/receptor combinations. MCF7/ErbB2, SKBR3/ErbB2, and A431/ErbB1 all show strong labeling. In the case of A431/ErbB2 and MCF7/ErbB1 the labeling efficiency is significantly lower but individual “green spots” indicative of bound NP labels are still detectable, whereas for SKBR3/ErbB1 no labels are detectable in the optical microscope. These trends are in good agreement with conventional flow cytometry results obtained for these cell/receptor combinations (Figure S1). Although the optical images of the NP labeled cells provide a quick survey over the relative expression levels, they don’t lend themselves to an accurate quantification required for a systematic comparison of ErbB1 and ErbB2 expression levels on different cell surfaces. For instance, it is difficult to determine differences in receptor expression for those cell/receptor combinations that show a high degree of labeling (MCF7/ErbB2, SKBR3/ErbB2, A431/ErbB1) based on the optical images, alone.
Figure 2. Optical immunophenotyping with Au NP labels.
The left column shows optical images of SKBR3, MCF7, and A431 cells (top to bottom) after labeling ErbB1 with 40 nm Au NPs. The right column shows the same cells after labeling ErbB2.
Quantification of NP Immunolabel Densities through Inspection in the Scanning Electron Microscope (SEM)
To obtain quantitative information about the level of immunolabeling, we inspected cell samples that were labeled under identical conditions as the ones shown in Figure 2 in the SEM and quantified the average surface densities (ρ) of the NP immunolabels bound to the peripheral, stretched areas of the plasma membrane (see Methods in the Supporting Information). We limited our analysis herein on the relatively even peripheral cell regions where the optical spectra of NPs can be acquired with best signal-to-noise and where the NP density is highest (Table S1). For each cell/receptor combination we averaged over multiple cells and considered a total number of at least 10,000 NPs in our calculation of ρ to account for cell-to-cell fluctuations.
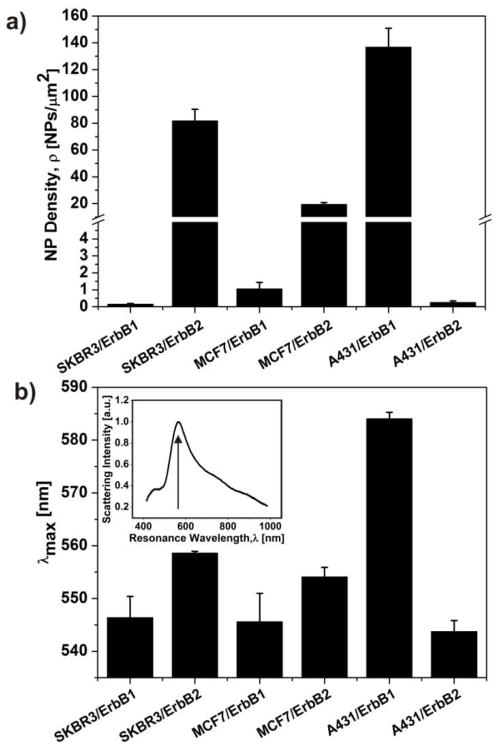
The resulting ρ values are summarized in Figure 3a. The average NP density is highest for A431/ErbB1 (136.7±14.1 NPs/μm2), followed by SKBR3/ErbB2 (81.7±8.7 NPs/μm2), and MCF7/ErbB2 (19.4±1.4 NPs/μm2). MCF7/ErbB1 (1.05±0.38 NPs/μm2), A431/ErbB2 (0.25±0.01 NPs/μm2), and SKBR3/ErbB1 (0.15±0.04 NPs/μm2) show significant lower labeling. The average NP density for the different cell/receptor systems is a quantitative metric that enables a differentiated analysis of the expression levels of the two growth factor receptors on one cell line and with some caution also between different cell lines. For comparison between different cell lines, one has to consider that the NP binding efficiencies can be slightly modified by differences in the cell surface. The cell surface is a complex hybrid material that is covered with a glycocalyx coat and differences in the thickness and composition of this coat can affect NP binding at a given NP concentration. Since all of the investigated cells are epithelial cells, we assume that this effect will be small for the performed studies.
Figure 3. Multimodal NP immunolabels facilitate the quantification of relative surface expression levels through SEM (a) or plasmon coupling (b).
The histogram in (a) gives an overview of the average nanoparticle densities (ρ) for the investigated cell/receptor systems. The ρ-values were measured after incubating the cell samples with a 0.8 nM Au NP immunolabel solution for 3 hours. The histogram in (b) shows the measured resonance wavelength (λmax) obtained under saturation binding conditions for the investigated cell/receptor systems. The inset shows a representative scattering spectrum of an Au NP labeled cell surface, and the peak wavelength of the spectrum is indicated by an arrow. Error bars in (a) and (b) are standard deviations of three independent experiments.
The measured ρ values are consistent with a strong overexpression of ErbB1 on A431 and ErbB2 on SKBR3, whereas the expression of ErbB1 on both MCF7 and SKBR3 as well as of ErbB2 on A431 is very low. The measured ρ values confirm the impression from the optical images in Figure 2 that the expression of ErbB2 on MCF7 is higher than for ErbB1 on MCF7 and SKBR3 and for ErbB2 on A431, but they clarify that the expression is still low when compared with A431/ErbB1 or SKBR3/ErbB2. Due to the large optical cross-sections of the NP labels, the optical images of NP labeled MCF7/ErbB2 are somewhat deceiving since they create a vivid optical labeling already for relatively moderate NP densities.
Quantification of NP Immunolabel Densities through Plasmon Coupling Microscopy
Although ρ provides an accurate differentiation of ErbB1 and ErbB2 expression levels, its determination in the SEM is very time-consuming and requires special equipment. The interparticle separation 10, 25, 45, 46 (and thus ρ) 20, 21 dependent spectral response of noble metal NPs provides new opportunities for an all-optical quantification of relative receptor expression levels using NP immunolabels. To determine the ideal concentration of the Au NP immunolabel solution for the assay, we first determined the resonance wavelength, λ, as function of the NP concentration (cAu) (Figure S2). To that end, we incubated the samples with increasing NP concentrations for 3 h at room temperature before we measured the scattering spectra of the cell surfaces (see Methods in the Supporting Information). The spectra were measured in the peripheral regions of well-stretched cells where the scattering background from the nucleus was negligible. In all investigated cell/receptor pairs, the resonance wavelength was found to red-shift with increasing NP concentration until at high concentrations it converged against an asymptotic limit (λmax) and did no longer red-shift when the NP concentration was further increased. The observed increase in λ with growing cAu results from an increasing electromagnetic coupling between the NPs when the average NP density (ρ) rises.21 The number of NP labels bound to the cell surface after a constant incubation time depends on the NP concentration in solution, the number of receptors on the cell surface, and the binding affinity of the NP labels. The NP concentrations at which λ converged differed for the investigated cell/surface receptor combinations, but we found that for cAu = 0.8 nM λmax had been obtained in all cases.
In Figure 3b we histogram the λmax values obtained for ErbB1 and ErbB2 on MCF7, SKBR3, and A431 cell lines. For SKBR3/ErbB2 and, especially, A431/ErbB1, the measured λmax values of 559 ± 0.23 nm and 584 ± 0.85 nm, respectively, indicate short average NP separations due to a strong receptor overexpression. For MCF7/ErbB2 we obtained λmax = 554 ± 1.26 nm, which is still slightly red-shifted compared with λmax = 544 ± 1.49 nm for A431/ErbB2, 546 ± 2.84 nm for SKBR3/ErbB1 and 545 ± 3.81 nm for MCF7/ErbB1. The sequence of the measured λmax values (A431/ErbB1 > SKBR3/ErbB2 > MCF7/ErbB2 > SKBR3/ErbB1 > MCF7/ErbB1 > A431/ErbB2) reproduces the overall trend observed for ρ very well. One noticeable difference is that λmax for SKBR3/ErbB1 is higher than for MCF7/ErbB1, although the measured ρ values are in reverse order. This difference can be rationalized if one considers that the exact value of λmax does not only depend on ρ (we averaged over a relatively large area of ~ 12×12 μm2 in our calculations of ρ) but also on its spatial organization within that area. A preferential clustering of NP labels due to a heterogenous receptor distribution will lead to a λmax value that is significantly red-shifted when compared to a homogenous random NP distribution, even if the area-averaged ρ value is identical. The dependence of λmax on both ρ and the heterogeneity of the NP distribution provides unique opportunities for deriving detailed insights into the spatial organization of the labeled receptors, provided the interplay between ρ, NP clustering, and the spectral response is sufficiently understood. Since in the limit of low ρ it is possible to record scattering spectra of individual NPs and clusters, the accuracy of the quantitative spectroscopic characterization of low NP densities can in the future be further improved by replacing the areaaveraged spectroscopic analysis through a statistical analysis of the spectra recorded from individual scatterers.
Examining the Influence of NP Clustering on the Measured Spectral Response
To elucidate the influence of the spatial NP organization on the λ(ρ) relationship, we simulated the polarization-averaged darkfield scattering spectra of different 2-dimensional NP distributions for various ρ values through Generalized Multiple Particle Mie Theory (GMT) calculations.47 In these calculations we considered four different clustering configurations (random, 50% dimers, 50% trigonal trimers, 50% random clusters) for NP densities between ρ = 0 (individual NPs) and ρ = 130 NPs/μm2. The interparticle separation for the dimers and trimers was kept constant at 5 nm, as was the minimum inter-particle separation in random arrays. This minimum separation is justified by the PEG and antibody coatings on the individual NPs. The 50% dimer (trimer) configurations were generated by randomly distributing individual NPs (constituting 50% of the total number of NPs) and randomly-oriented dimer (trimer) clusters in an area of 2 × 2 μm2. The 50% random cluster configurations were generated by arbitrarily positioning 1/4 of the total number of NPs in the vicinity of any of the already randomly distributed 3/4 of NPs with a minimum separation of 5 nm. Further details about the GMT simulations are provided in the Methods section in the Supporting Information.
Selected NP distributions for ρ = ~25, ~45, ~90 NPs/μm2 are shown in Figure 4a-d, and Figure 4e compares the λ(ρ) relationships obtained for the three different clustering configurations and random NP distribution across the entire investigated ρ range. The graphs confirm that clustering leads to a significant steepening of the λ(ρ) relationship, which results in a red-shift of the peak resonance wavelength of the clustered NPs relative to that of the random NP distribution at a given ρ. For the clustered NP configurations (50% dimers, 50% trimers, 50% clustered) the simulated λ(ρ) relationships show a rapid convergence of the measured peak resonance wavelength against an asymptotic λmax value, since the spectral response of the simulated clusters are dominated by nearest-neighbor NP interactions.23, 48–51 In contrast, the λ(ρ) relationship of the random NP distribution shows initially a much shallower slope (due to the absence of significant near-field coupling), which increases - as expected - when the average interparticle separation decreases as ρ grows. Within the ρ range considered in our simulations, the random NP distribution does not yet show a convergence against an asymptotic λmax.
Figure 4. The resonance wavelength (λ) depends on the level of NP clustering for a given NP density (ρ).
Typical spatial distributions of 40nm Au NPs in a) random, b) 50% dimers, c) 50% trimers, and d) 50% clustered NP assemblies with varying particle density. e) Peak resonance wavelengths of the ensemble-averaged scattering spectra of the NP assemblies shown in (a–d) as a function of NP density.
Quantitative Characterization of ErbB1 and ErbB2 Clustering
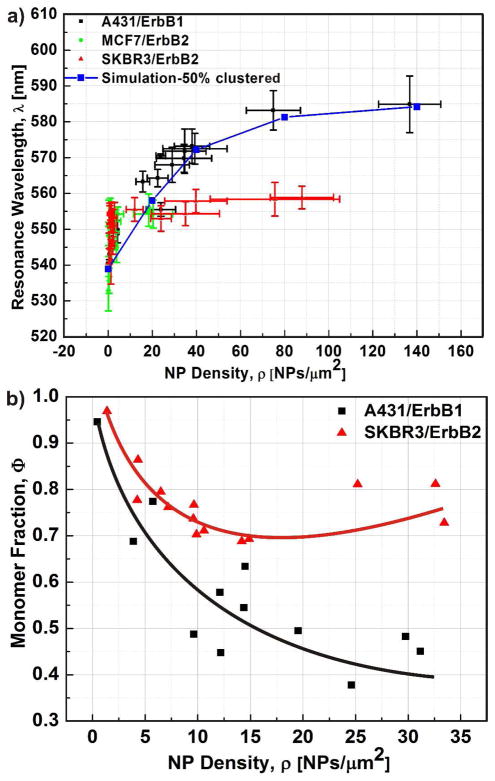
The simulated λ(ρ) relationships are a valuable guide for the analysis of the collective spectral response of NP labeled cell surfaces. The experimentally obtained λ(ρ) relationships for the three cell/receptor combinations, which exhibit significant binding and reached NP densities of ρ ≥ 20 NPs/μm2, are shown in Figure 5a. The samples were prepared by incubating the cells with increasing concentrations of NP immunolabels (cAu) for 3h. In all three cases (SKBR3/ErbB2, MCF7/ErbB2, A431/ErbB1) we observe a steep initial increase of λ as function of NP density for ρ < 10 NPs/μm2. This behavior is consistent with a strong clustering of the NP binding sites, which preferentially arranges the NPs in close vicinity to each other so that their plasmons can efficiently couple. This clustering at low NP densities was also confirmed through a Hopkins test performed with the NP locations as obtained by SEM (Figure S3). For larger ρ values significant differences between ErbB1 and ErbB2 appear. Whereas for SKBR3/ErbB2 and MCF7/ErbB2 λ converges against λmax at ρ ≥ 10 NPs/μm2, in the case of A431/ErbB1 λ continues to increase and does not converge against λmax until at ρ = 100 NPs/μm2. We included the simulated λ(ρ) curve for the 50% random cluster configuration as solid blue line in Figure 5a. The simulation reproduces the experimental A431/ErbB1 relationship very closely and confirms that a continuous increase in cluster size with growing ρ leads to the formation of larger clusters containing more strongly coupled NPs. The systematic blue-shift of the λ(ρ) curves for SKBR3/ErbB2 and MCF7/ErbB2 for ρ ≥ 10 NPs/μm2 indicates less strongly coupled NPs than in A431/ErbB1. To validate this conclusion we inspected the NP labeled cell surfaces in the SEM and performed a statistical analysis of the cluster size distributions as function of average ρ for A431/ErbB1 and SKBR3/ErbB2. In Figure 5b we plot the monomer fraction (Φ) defined as the number of monomers divided by the total number of NPs in clusters (i.e. dimers and higher order oligomers) for the two cell/receptor combinations. The continuous decrease of Φ as function of ρ in the case of A431/ErbB1 is consistent with an increasing number and/or size of NP clusters across the entire investigated ρ-range. In contrast, for SKBR3/ErbB2, Φ does not continue to decrease below Φ ≅ 0.7 for ρ > 15 NPs/Sm2. Instead, Φ slightly increases again for growing ρ. This behavior indicates that the fraction of clustered NPs becomes maximum at ρ ≅ 15 NPs/Sm2 and then decreases again as more and more NPs bind to the cell surface.
Figure 5. Differences in the spectral response and NP clustering indicate non-identical clustering propensities of ErbB1 and ErbB2.
a) Peak resonance wavelength (λ) as function of NP density (ρ) for A431/ErbB1 (black), MCF7/ErbB2 (green), and SKBR3/ErbB2 (red). The simulated λ(ρ)-curve for 50% clustered is included in blue. The A431/ErbB1 curve shows the data reported in ref. 21. b) Monomer fraction (Φ) as function of ρ as determined by SEM. Φ is defined as the ratio of the number of individual NPs divided by the total number of NPs in clusters. A cluster was defined as a conglomeration of NPs that lie within a separation of 15 nm for this analysis. For A431/ErbB1 Φ is observed to continuously decrease with ρ, while for SKBR3/ErbB2 Φ does not drop below 70%. A total number of at least 1000 NPs were evaluated for data points with ρ ≥ 5 NPs/μm2. For data points with ρ < 5 NPs/μm2 at least 500 NPs were considered. Overall 5 independent labeling experiments were performed.
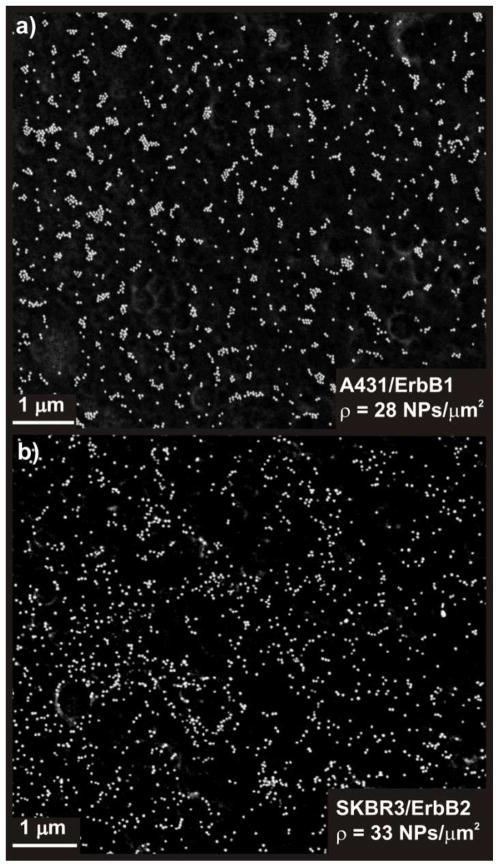
We rationalize the observed differences in the saturation behavior of the λ(ρ) relationships for ErbB2 and ErbB1 and the observed trends for Φ with the fact that MCF7 and SKBR3 exhibit less and smaller high-affinity binding sites (i.e. receptor clusters) for NP immunolabels targeted at ErbB2 than A431 for NPs targeted at ErbB1. Preferential binding of the NPs to these high-affinity binding sites leads to the observed initial spectral shifts, but once these sites are occupied, the immunolabels bind to areas with lower ErbB2 receptor concentration, resulting in a more random NP distribution and an increase in ρ without a further red-shift of λ. In contrast, for A431/ErbB1 the NPs keep assembling into larger clusters. This hypothesis is supported by the SEM images in Figure 6 where we compare the spatial distribution of NP labels for A431/ErbB1 and SKBR3/ErbB2 for similar (high) ρ values (28 NPs/μm2 vs. 33 NPs/μm2). In the case of A431/ErbB1 the NP immunolabels are organized into larger clusters than for SKBR3/ErbB2, for which the NP distribution appears more stochastic.
Figure 6. Optical inspection of NP distribution in the SEM confirms differences in the spatial clustering.
a) SEM micrograph of NP labeled A431/ErbB1 (ρ = 28 NPs/μm2) and b) of SKBR3/ErbB2 (ρ = 33 NPs/μm2). The NPs in (a) show a higher propensity to cluster than in (b).
Future studies will have to show if this behavior is specific to ErbB1 on A431 cells, which are known to dramatically overexpress this receptors, or whether the formation of larger clusters is a general characteristic of ErbB1. Experimental support for the latter is provided by recent NSOM and superresolution fluorescence microscopic studies that have shown that ErbB1 forms clusters with an average diameter of ~160 nm in HeLa cells,16 which compares with average cluster sizes for ErbB2 of ~70 nm in Cal-51 and SKBR3 cells.19 In the context of this work, it is sufficient to conclude that our combined experimental and theoretical studies confirm that the resonance wavelength of NP immunolabels targeted at cell surface receptors provide information not only about relative expression levels (if λ is evaluated at a fixed NP concentration) but also about the spatial organization of the receptors on the cell surface into clusters (if the slope of λ is evaluated as function of the NP concentration). The differences in the spectral shift of the collective plasmon resonance wavelength as function of NP concentration for the investigated cell/receptor combinations are consistent with a stronger clustering for ErbB1 in A431 than for ErbB2 in SKBR3 and MCF7.
In conclusion, we have demonstrated that the superb optical properties of NP immunolabels in combination with efficient labeling procedures enable the differentiation of relative expression levels of cell surface receptors (in this work ErbB1 and ErbB2) “by eye”. We further quantified the differences in the NP labeling between the investigated cell/receptor systems through spectral analysis of the collective plasmon resonance wavelength of the NP labels on the cell surface. A systematic characterization of the electromagnetic interactions between NPs immunolabels confined to a 2-dimensional cell surface revealed that the measured collective resonance wavelength depends both on the average NP density as well as on the level of NP clustering on the cell surface. We showed that different levels of NP clustering lead to different functional dependencies of the collective NP plasmon resonance wavelength (λ) on the average NP density (ρ), which can be conveniently varied through control of the NP concentration (cAu). We mapped the λ(ρ) relationships for A431/ErbB1, SKBR3/ErbB2, and MCF7/ErbB2 and found that λ for SKBR3/ErbB2 and MCF7/ErbB2 converged against an asymptotic λmax value for significantly smaller ρ values than for A431/ErbB1. The differences in the respective λ(ρ) relationships indicate a higher degree of clustering for ErbB1 on A431 than for ErbB2 on SKBR3 or MCF7. Considering the potential relevance of receptor clustering in modulating and controlling the cell signaling activity, plasmon coupling based expression profiling modalities that provide synergistic information about both biomarker expression and clustering on the single cell level represent useful augmentations to conventional immunophentyping approaches. The resulting gain in information could contribute to enhance the prognostic and/or predictive value of cell surface biomarkers in the near future.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (NIH/NCI) through grant 5R01CA138509-04.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Table S1, Figure S1–S4 and Methods. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Bublil EM, Yarden Y. Current Opinion in Cell Biology. 2007;19(2):124–134. doi: 10.1016/j.ceb.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 2.Citri A, Yarden Y. Nature Reviews Molecular Cell Biology. 2006;7(7):505–516. doi: 10.1038/nrm1962. [DOI] [PubMed] [Google Scholar]
- 3.Yarden Y, Sliwkowski MX. Nature Reviews Molecular Cell Biology. 2001;2(2):127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 4.Leth-Larsen R, Lund RR, Ditzel HJ. Molecular & Cellular Proteomics. 2010;9(7):1369–1382. doi: 10.1074/mcp.R900006-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ariotti N, Liang H, Xu YF, Zhang YQ, Yonekubo Y, Inder K, Du GW, Parton RG, Hancock JF, Plowman SJ. Molecular and Cellular Biology. 2010;30(15):3795–3804. doi: 10.1128/MCB.01615-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsieh MY, Yang S, Raymond-Stinz MA, Steinberg S, Vlachos DG, Shu W, Wilson B, Edwards JS. Iet Systems Biology. 2008;2(5):256–272. doi: 10.1049/iet-syb:20070073. [DOI] [PubMed] [Google Scholar]
- 7.Mayawala K, Vlachos DG, Edwards JS. FEBS Letters. 2005;579(14):3043–3047. doi: 10.1016/j.febslet.2005.04.059. [DOI] [PubMed] [Google Scholar]
- 8.Bray D, Levin MD, Morton-Firth CJ. Nature. 1998;393(6680):85–88. doi: 10.1038/30018. [DOI] [PubMed] [Google Scholar]
- 9.Bethani I, Skanland SS, Dikic I, Acker-Palmer A. EMBO Journal. 2010;29(16):2677–2688. doi: 10.1038/emboj.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang S, Raymond-Stintz MA, Ying W, Zhang J, Lidke DS, Steinberg SL, Williams L, Oliver JM, Wilson BS. Journal of Cell Science. 2007;120(16):2763–2773. doi: 10.1242/jcs.007658. [DOI] [PubMed] [Google Scholar]
- 11.Clayton AHA, Tavarnesi ML, Johns TG. Biochemistry. 2007;46(15):4589–4597. doi: 10.1021/bi700002b. [DOI] [PubMed] [Google Scholar]
- 12.Clayton AHA, Walker F, Orchard SG, Henderson C, Fuchs D, Rothacker J, Nice EC, Burgess AW. Journal of Biological Chemistry. 2005;280(34):30392–30399. doi: 10.1074/jbc.M504770200. [DOI] [PubMed] [Google Scholar]
- 13.Szabo A, Horvath G, Szollosi J, Nagy P. Biophysical Journal. 2008;95(4):2086–2096. doi: 10.1529/biophysj.108.133371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Webb SED, Roberts SK, Needham SR, Tynan CJ, Rolfe DJ, Winn MD, Clarke DT, Barraclough R, Martin-Fernandez ML. Biophysical Journal. 2008;94(3):803–819. doi: 10.1529/biophysj.107.112623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagy P, Jenei A, Kirsch AK, Szollosi J, Damjanovich S, Jovin TM. Journal of Cell Science. 1999;112(11):1733–1741. doi: 10.1242/jcs.112.11.1733. [DOI] [PubMed] [Google Scholar]
- 16.Abulrob A, Lu ZF, Baumann E, Vobornik D, Taylor R, Stanimirovic D, Johnston LJ. Journal of Biological Chemistry. 2010;285(5):3145–3156. doi: 10.1074/jbc.M109.073338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saffarian S, Li Y, Elson EL, Pike LJ. Biophysical Journal. 2007;93(3):1021–1031. doi: 10.1529/biophysj.107.105494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagy P, Claus J, Jovin TM, Arndt-Jovin DJ. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(38):16524–16529. doi: 10.1073/pnas.1002642107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaufmann R, Muller P, Hildenbrand G, Hausmann M, Cremer C. Journal of Microscopy. 2011;242(1):46–54. doi: 10.1111/j.1365-2818.2010.03436.x. [DOI] [PubMed] [Google Scholar]
- 20.Wang HY, Rong GX, Yan B, Yang LL, Reinhard BM. Nano Letters. 2011;11(2):498–504. doi: 10.1021/nl103315t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, Boriskina SV, Wang HY, Reinhard BM. ACS Nano. 2011;5(8):6619–6628. doi: 10.1021/nn202055b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prodan E, Radloff C, Halas NJ, Nordlander P. Science. 2003;302(5644):419–422. doi: 10.1126/science.1089171. [DOI] [PubMed] [Google Scholar]
- 23.Halas NJ, Lal S, Chang WS, Link S, Nordlander P. Chemical Reviews. 2011;111(6):3913–3961. doi: 10.1021/cr200061k. [DOI] [PubMed] [Google Scholar]
- 24.Schultz S, Smith DR, Mock JJ, Schultz DA. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(3):996–1001. doi: 10.1073/pnas.97.3.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reinhard BM, Siu M, Agarwal H, Alivisatos AP, Liphardt J. Nano Letters. 2005;5(11):2246–2252. doi: 10.1021/nl051592s. [DOI] [PubMed] [Google Scholar]
- 26.Jain PK, Huang WY, El-Sayed MA. Nano Letters. 2007;7(7):2080–2088. [Google Scholar]
- 27.Wei QH, Su KH, Durant S, Zhang X. Nano Letters. 2004;4(6):1067–1071. [Google Scholar]
- 28.Maier SA, Brongersma ML, Kik PG, Atwater HA. Physical Review B. 2002;65(19) [Google Scholar]
- 29.Romo-Herrera JM, Alvarez-Puebla RA, Liz-Marzan LM. Nanoscale. 2011;3(4):1304–1315. doi: 10.1039/c0nr00804d. [DOI] [PubMed] [Google Scholar]
- 30.EI-Kouedi M, Keating CD. Biofunctionalized Nanoparticles for Surface-Enhanced Raman Scattering and Surface Plasmon Resonance. In: Niemeyer CM, Mirkin CA, editors. Nanobiotechnology: Concepts, Applications and Perspectives. Wiley-VCH Verlag GmbH & Co; Weinheim: 2005. pp. 429–443. [Google Scholar]
- 31.Weber ML, Willets KA. Journal of Physical Chemistry Letters. 2011;2(14):1766–1770. [Google Scholar]
- 32.Rong GX, Wang HY, Skewis LR, Reinhard BM. Nano Letters. 2008;8(10):3386–3393. doi: 10.1021/nl802058q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rong GX, Wang HY, Reinhard BM. Nano Letters. 2010;10(1):230–238. doi: 10.1021/nl903350f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rong GX, Reinhard BM. PLoS ONE. 2012;7(3):e34175. doi: 10.1371/journal.pone.0034175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crow MJ, Grant G, Provenzale JM, Wax A. American Journal of Roentgenology. 2009;192(4):1021–1028. doi: 10.2214/AJR.07.3535. [DOI] [PubMed] [Google Scholar]
- 36.Aaron J, Travis K, Harrison N, Sokolov K. Nano Letters. 2009;9(10):3612–3618. doi: 10.1021/nl9018275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Austin LA, Kang B, Yen CW, El-Sayed MA. Journal of the American Chemical Society. 2011;133(44):17594–17597. doi: 10.1021/ja207807t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qian W, Huang XH, Kang B, El-Sayed MA. Journal of Biomedical Optics. 2010;15(4):046025. doi: 10.1117/1.3477179. [DOI] [PubMed] [Google Scholar]
- 39.Seekell K, Crow MJ, Marinakos S, Ostrander J, Chilkoti A, Wax A. Journal of Biomedical Optics. 2011;16(11):116003. doi: 10.1117/1.3646529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crow MJ, Seekell K, Ostrander JH, Wax A. ACS Nano. 2011;5(11):8532–8540. doi: 10.1021/nn201451c. [DOI] [PubMed] [Google Scholar]
- 41.Ma LL, Tam JO, Willsey BW, Rigdon D, Ramesh R, Sokolov K, Johnston KP. Langmuir. 2011;27(12):7681–7690. doi: 10.1021/la200659z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kennedy DC, Tay LL, Lyn RK, Rouleau Y, Hulse J, Pezacki JP. ACS Nano. 2009;3(8):2329–2339. doi: 10.1021/nn900488u. [DOI] [PubMed] [Google Scholar]
- 43.Hodges MD, Kelly JG, Bentley AJ, Fogarty S, Patel II, Martin FL, Fullwood NJ. ACS Nano. 2011;5(12):9535–9541. doi: 10.1021/nn202652h. [DOI] [PubMed] [Google Scholar]
- 44.Kolb HC, Finn MG, Sharpless KB. Angewandte Chemie-International Edition. 2001;40(11):2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 45.Yang LL, Wang HY, Yan B, Reinhard BM. Journal of Physical Chemistry C. 2010;114(11):4901–4908. doi: 10.1021/jp911858v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rechberger W, Hohenau A, Leitner A, Krenn JR, Lamprecht B, Aussenegg FR. Optics Communications. 2003;220(1–3):137–141. [Google Scholar]
- 47.Mackowski DW. Journal of the Optical Society of America a-Optics Image Science and Vision. 1994;11(11):2851–2861. doi: 10.1364/josaa.19.000881. [DOI] [PubMed] [Google Scholar]
- 48.Yan B, Boriskina SV, Reinhard BM. Journal of Physical Chemistry C. 2011;115(50):24437–24453. doi: 10.1021/jp207821t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan B, Boriskina SV, Reinhard BM. Journal of Physical Chemistry C. 2011;115(11):4578–4583. doi: 10.1021/jp112146d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang J, Yang LL, Boriskina S, Yan B, Reinhard BM. Analytical Chemistry. 2011;83(6):2243–2249. doi: 10.1021/ac103123r. [DOI] [PubMed] [Google Scholar]
- 51.Ye J, Wen FF, Sobhan H, Lassiter JB, Van Dorpe P, Nordlander P, Halas NJ. Nano Letters. 2012;12(3):1660–1667. doi: 10.1021/nl3000453. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.