Abstract
The generation of induced pluripotent stem (iPS) cells is an important tool for regenerative medicine. However, the main restriction is the risk of tumor development. In this study we found that during the early stages of somatic cell reprogramming toward a pluripotent state, specific gene expression patterns are altered. Therefore, we developed a method to generate partial-iPS (PiPS) cells by transferring four reprogramming factors (OCT4, SOX2, KLF4, and c-MYC) to human fibroblasts for 4 d. PiPS cells did not form tumors in vivo and clearly displayed the potential to differentiate into endothelial cells (ECs) in response to defined media and culture conditions. To clarify the mechanism of PiPS cell differentiation into ECs, SET translocation (myeloid leukemia-associated) (SET) similar protein (SETSIP) was indentified to be induced during somatic cell reprogramming. Importantly, when PiPS cells were treated with VEGF, SETSIP was translocated to the cell nucleus, directly bound to the VE-cadherin promoter, increasing vascular endothelial-cadherin (VE-cadherin) expression levels and EC differentiation. Functionally, PiPS-ECs improved neovascularization and blood flow recovery in a hindlimb ischemic model. Furthermore, PiPS-ECs displayed good attachment, stabilization, patency, and typical vascular structure when seeded on decellularized vessel scaffolds. These findings indicate that reprogramming of fibroblasts into ECs via SETSIP and VEGF has a potential clinical application.
Keywords: shear stress, stem cell therapy, vascular tissue engineering
An interesting aspect of research today is focused on the generation of functional cells to be used for regenerative medicine. For example, the damaged endothelial cells (ECs) on the vessel wall could be replaced by using EC-based therapy. The discovery of reprogramming induced pluripotent stem (iPS) cells from somatic cells (1–3) could pave the way for major advances in regenerative medicine. In comparison with embryonic stem cells, they could offer a tool for clinical application that does not raise either ethical or alloimmune concerns. iPS cell generation is mainly based on the sufficient delivery of a combination of key transcription factors, such as c-Myc, Klf4, Oct4, and Sox2 (2), or Lin28 and Nanog (4), which initiates the reprogramming of somatic cells into a pluripotent state. Methods have now been developed to drive the reprogramming factors in a way that overcomes the lentiviral vectors that stably integrated into the host cell genome (2, 5, 6). These methods include nonintegrating adenoviral vectors (7) and plasmids (8, 9), or delivery of the reprogramming factors as purified recombinant proteins (10) and modified RNA molecules (11–13).
iPS cells have displayed the potential to differentiate into a number of cell lineages, such as CD34+ progenitor cells (14), cardiomyocytes (15, 16), and ECs (17). However, the main limitation for iPS cell application is the risk of tumor development, because these cells are reprogrammed to a fully pluripotent state. On the basis of the fact that cell reprogramming is a process with low efficiency, it is also possible that different stages of cell reprogramming may regulate signal pathways able to direct the differentiation of reprogrammed cells before the pluripotent state. Therefore, “skipping pluripotency” is a way to convert a somatic cell from one type to another. In this study we have established a method to generate partially induced pluripotent stem (PiPS) cells. This method includes transferring of the genes encoding the four transcription factors (OCT4, SOX2, KLF4, and c-MYC) to human fibroblasts, and culture in reprogramming media for 4 d. PiPS cells did not form tumors in vivo and had the potential to differentiate into ECs in response to defined media and culture conditions. We demonstrated that these PiPS cell-derived ECs are functional in angiogenesis in infarcted tissues in ischemic limb and in reendothelialization in tissue-engineered vessels ex vivo.
Results
Alterations of Gene Expression During Fibroblast Cell Reprogramming as Early as Day 4.
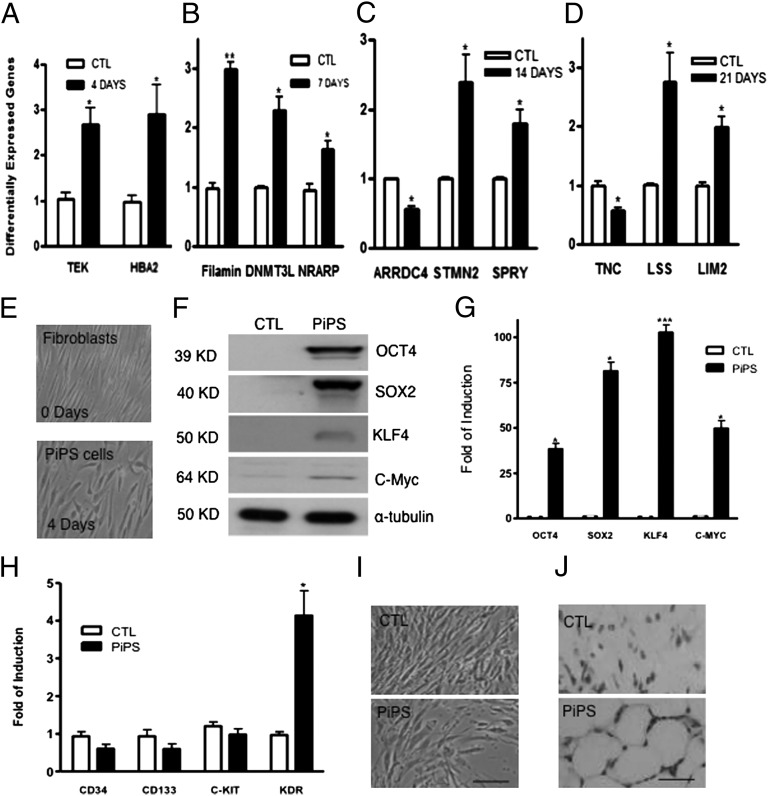
Human fibroblasts were virally transduced with genes encoding the four transcription factors OCT4, SOX2, KLF4, and c-MYC, cultured in reprogramming media for 4, 7, 14, and 21 d, and subjected to microarray analysis. The results revealed that 198 genes were altered at day 4, 107 genes at day 7, 97 genes at day 14, and 131 genes at day 21 compared with day 0. Interestingly, when functional classification of the differentially expressed genes was performed using Ingenuity Systems software, significant differences observed in the expression of genes involved in cellular movement, cell death, cellular growth, proliferation, and development demonstrated that a high number of changes in the expression profile occur during early stages of reprogramming (4 d) compared with days 7, 14, and 21 (SI Appendix, Fig. S1, and Tables S2–S5). Importantly, a high number of genes mainly associated with vascular lineages, such as TEK, NRARP, STMN2, and filamin were also identified as differentially expressed in the microarray and confirmed by real-time PCR (Fig. 1 A–D). These results demonstrate that during somatic cell reprogramming toward a pluripotent state, specific gene expression patterns are altered as early as day 4.
Fig. 1.
Different expression of genes during reprogramming and characterization of 4-day PiPS cells. Differential expression profile of genes altered according to microarray analysis during reprogramming was confirmed by real-time PCR assays on day 4 (A), day 7 (B), day 14 (C), and day 21 (D) [data are means ± SEM (n = 3); *P < 0.05, **P < 0.01]. Human fibroblasts were nucleofected with a linearized pCAG2LMKOSimO plasmid encoding the four reprogramming genes (OCT4, SOX2, KLF4, and C-MYC) or an empty vector. Images show the morphology of fibroblasts, 4-day PiPS cells (E). PiPS cells expressed the four reprogramming factors at protein (F) and mRNA levels (G) [data are means ± SEM (n = 3); *P < 0.05, ***P < 0.001]. (H) Real-time PCR assays for progenitor markers CD34, CD133, c-Kit, and KDR (VEGFR2) [data are means ± SEM (n = 3); *P < 0.05]. (I) PiPS cells were negative for alkaline phosphatase, whereas they formed capillary-like structures in in vivo Matrigel plug assays, when injected to SCID mice for 2 wk (J). (Scale bar, 50 μm.)
Characterization of 4-d PiPS Cells.
Human fibroblasts were reprogrammed for 4 d by nucleofecting with a linearized pCAG2LMKOSimO plasmid encoding the four genes defined as PiPS cells. PiPS cells displayed an alternate morphology distinct from the fibroblasts and did not form colonies in this early stage of reprogramming (Fig. 1E). PiPS cells expressed the four reprogramming factors at the protein (Fig. 1F) and mRNA levels (Fig. 1G). They showed an induced expression of VEGF receptor 2 (VEGFR2), kinase insert domain receptor (KDR) compared with the control, but not for the progenitor markers such as CD34, CD133, and c-Kit (Fig. 1H). PiPS cells were negative for alkaline phosphatase (Fig. 1I) and pluripotent markers such as SSEA-1 and TRA 1-81 and did not form tumors in vivo 2 mo after s.c. injection in SCID mice (SI Appendix, Fig. S2). In parallel, fully reprogrammed iPS cells were s.c. injected in SCID mice, where tumors were observed (SI Appendix, Fig. S2). Importantly, PiPS cells formed capillary-like structures in in vivo Matrigel plug assays in SCID mice, as revealed by H&E staining (Fig. 1J).
PiPS Cells Display the Potential to Differentiate into ECs.
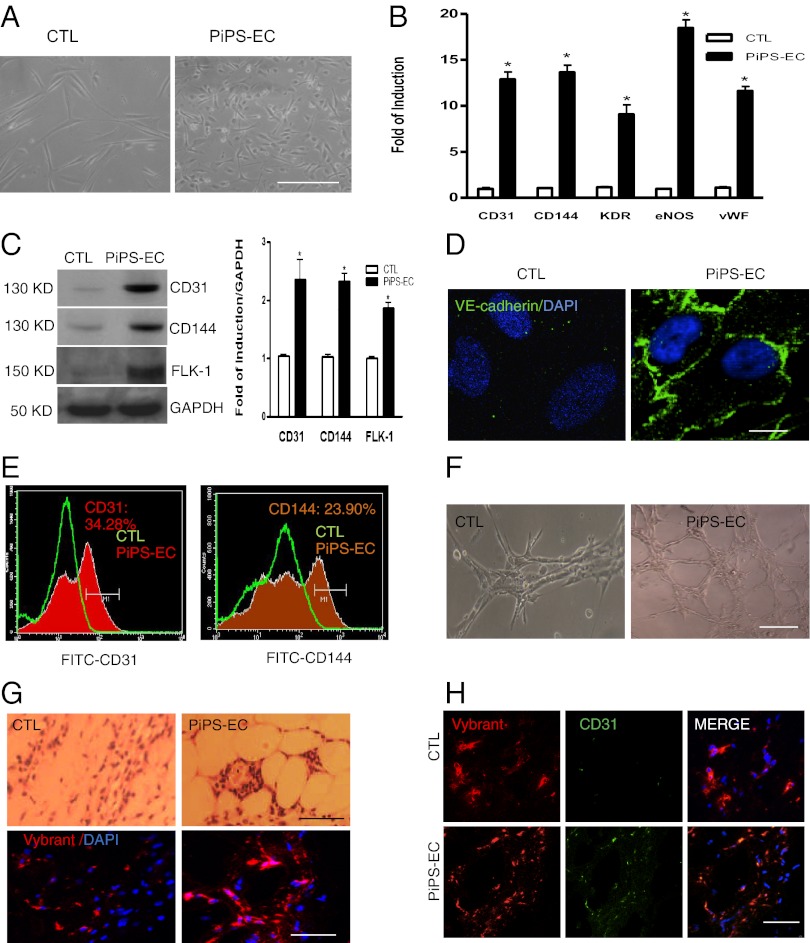
Because PiPS cells expressed VEGFR2, we wondered whether they can differentiate into vascular lineages and specifically into ECs. We found that PiPS cells from day 3 under the culture conditions start expressing EC markers, whereas high expression was detected up to day 9 (SI Appendix, Fig. S3). PiPS cells displayed an endothelial-like morphology in comparison with control cells on day 6 (Fig. 2A) and expressed endothelial-specific markers such as CD31, CD144, VEGFR2, eNOS, and vWF at mRNA (Fig. 2B) and protein levels (Fig. 2C). Immunofluorescence staining also revealed a typical endothelial staining for CD144 (Fig. 2D). FACS analysis confirmed expression of CD31 and CD144 (Fig. 2E) for PiPS-ECs, whereas undifferentiated PiPS cells were negative for CD31 (SI Appendix, Fig. S4A). PiPS cells were also tested for pluripotent markers, which were not expressed as FACS and real-time PCR analyses revealed (SI Appendix, Fig. S4 B and C). Moreover, the differentiated ECs were able to form vascular-like tubes in vitro (Fig. 2F) and in vivo (Fig. 2G, Upper) in Matrigel plugs. To distinguish from the endogenous ECs, the differentiated cells were labeled with Vybrant (red) before s.c. injection to SCID mice. As shown in Fig. 2G (Lower), Vybrant staining confirmed the contribution of the exogenous human cells to form tube-like structures in SCID mice. Further experiments indicated double staining of the cells with Vybrant and CD31 (Fig. 2H). These results suggest that PiPS cells can differentiate into ECs. We defined these cells as PiPS-ECs.
Fig. 2.
PiPS cells differentiate into ECs. PiPS or control cells were seeded on collagen IV-coated plates and cultured with endothelial cell growth medium-2 (EGM-2) for 6 d. (A) Images show an endothelial-like morphology for PiPS-ECs in comparison with control cells, and expressed endothelial-specific cell markers, such as CD31, CD144, KDR, eNOS, and vWF, at the mRNA [data are means ± SEM (n = 3); *P < 0.05] (B) and protein levels and quantification (C). (D) Immunoflurorescence staining showed a typical endothelial staining for CD144, and DAPI was used and stained the cell nucleus. (Scale bar, 25 μm.) (E) FACS analysis confirmed expression of CD31 and CD144. Representative images show vascular-like tubes in vitro (F) and in vivo (G, Upper) in Matrigel plug assays. PiPS-ECs labeled with Vybrant (red) before the s.c. injection in SCID mice (G, Lower) confirmed the presence of labeled human cells 2 wk later. The PiPS-ECs stained positive for Vybrant and CD31, whereas the control cells were positive only for the Vybrant but not for CD31 (H). (Scale bar, 50 μm.)
SET Translocation (Myeloid Leukemia-Associated) (SET) Similar Protein (SETSIP) Is Involved in Differentiation of PiPS-ECs.
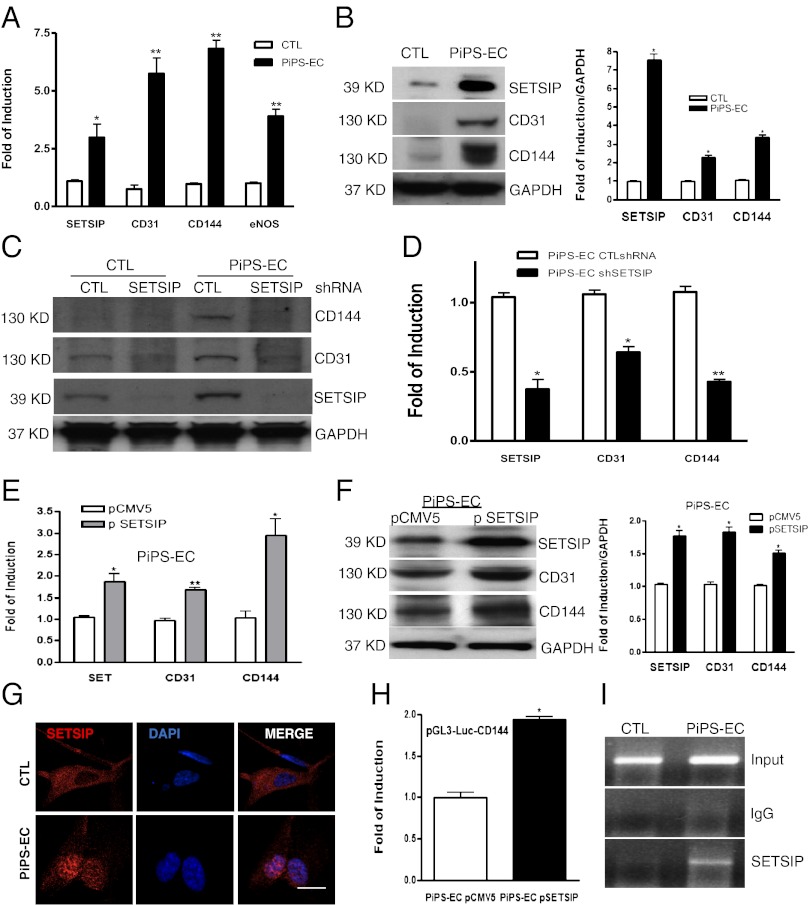
To shed light on the mechanisms involved in PiPS-ECs differentiation, a number of genes indentified from microarray analysis were screened. A gene defined as “similar to SET translocation protein” (SETSIP) was studied. SETSIP gives rise to a protein containing 10 additional amino acids in the N terminus (SI Appendix, Fig. S5A) in comparison with the known SET protein (SI Appendix, Fig. S5B). In our PiPS cell model, we found that SETSIP was expressed in parallel with endothelial markers (Fig. 3A) at mRNA and protein levels (Fig. 3B, Left, and quantification, Right). Additional experiments also indicated that VEGF further induced SETSIP expression at the protein level (SI Appendix, Fig. S6). Interestingly, down-regulation of SETSIP by shRNA in control and PIPS-ECs resulted in suppression of endothelial marker expression at protein (Fig. 3C) and mRNA levels on day 6 (Fig. 3D). Moreover, SETSIP overexpression in PiPS-ECs induced EC marker expression in mRNA (Fig. 3E) and proteins (Fig. 3F, Left, and quantification, Right). Importantly, SETSIP was translocated to the cell nucleus during PiPS-EC differentiation (Fig. 3G). To elucidate the underlying mechanism of the EC regulation by SETSIP, luciferase assays demonstrated that SETSIP induced the promoter activity of vascular endothelial-cadherin (VE-cadherin) (Fig. 3H). Moreover, ChIP assays confirmed these findings by showing a direct binding of SETSIP to the VE-cadherin gene promoter at region −864 to −1152 nt upstream of the transcription initiation site (Fig. 3I). These results indicate that SETSIP is important in PiPS-ECs differentiation. To answer questions such as how SETSIP is activated during cell reprogramming, examination of the SETSIP promoter binding sites predicts a possible binding with OCT1. To verify whether OCT1 is involved in the OCT4 signal pathway, further experiments revealed that OCT1 expression is induced during the early stages of reprogramming (SI Appendix, Fig. S7A), whereas overexpression of OCT4 in fibroblasts resulted in OCT1 and SETSIP activation (SI Appendix, Fig. S7B).
Fig. 3.
SETSIP is involved in differentiation of PiPS-ECs. Real-time PCR shows SETSIP parallel expression with EC markers at the mRNA [A; data are means ± SEM (n = 3); *P < 0.05, **P < 0.01] and protein (B) levels and quantification (Right), in PiPS-ECs at day 6 of differentiation. SETSIP was knocked down by shRNA in CTL and PiPS-ECs at day 3 of differentiation, showing a suppression in endothelial markers assessed on day 6 at protein (C) and mRNA levels [D; data are means ± SEM (n = 3); *P < 0.05, **P < 0.01]. SETSIP overexpression using pCMV5-SETSIP construct (p SETSIP) or empty vector pCMV5 in PiPS-ECs during EC differentiation at day 4, showing further induction in EC marker expression at mRNA [E; data are means ± SEM (n = 3); *P < 0.05, **P < 0.01] and protein levels, and quantification (F), when assessed at day 6. Immunostaining shows translocation of SETSIP to the cell nucleus during PiPS-ECs differentiation at day 6; DAPI was used and stained the cell nucleus (G). (Scale bar, 50 μm.) Luciferase assays were performed at day 4 during PiPS-ECs differentiation, showing an increased promoter activity for VE-cadherin in the presence of the SETSIP at day 6 of differentiation (H; data are means ± SEM (n = 3); *P < 0.05]. ChIP assays revealed that SETSIP binds directly to the VE-cadherin gene promoter at region (−864 to −1152) nt upstream of the transcription initiation site in 6-day differentiated PiPS-ECs (I).
Establishing PiPS Cell Lines That Differentiate into EC Cells.
To improve the purity of PiPS cells, human fibroblasts were nucleofected with the pCAG2LMKOSimO plasmid, which contains a neomycin resistant gene and an mOrange marker. PiPS cells were selected by neomycin treatment for 4 d, and a pure population of PiPS cells was obtained expressing the mOrange marker (SI Appendix, Fig. S8A) and the four genes (SI Appendix, Fig. S8B). The pure population of PiPS cells was subjected to differentiation and confirmed the potential of these cells to differentiate into the EC lineage, expressing endothelial markers (SI Appendix, Fig. S8C). Moreover, PiPS-EC–derived cells displayed the capacity of low-density lipoprotein uptake (SI Appendix, Fig. S8D). Importantly, luciferase assays demonstrated that PiPS-ECs derived from a pure population of PiPS cells had an increased promoter activity of VE-cadherin compared with control cells (SI Appendix, Fig. S8E). Additional experiments revealed that knockdown of SETSIP by shRNA in PiPS-ECs after selection resulted in suppression of VE-cadherin at the protein level (SI Appendix, Fig. S8F). Luciferase assays also showed that knockdown of SETSIP decreased the VE-cadherin promoter activity in PiPS-ECs (SI Appendix, Fig. S8G). Furthermore, knockdown of SETSIP in PiPS-ECs by shRNA abolished the formation of vascular-like tubes in vivo in Matrigel plugs (SI Appendix, Fig. S8H). These results support the notion that SETSIP has an important role in PiPS-EC differentiation derived from a pure population of PiPS cells.
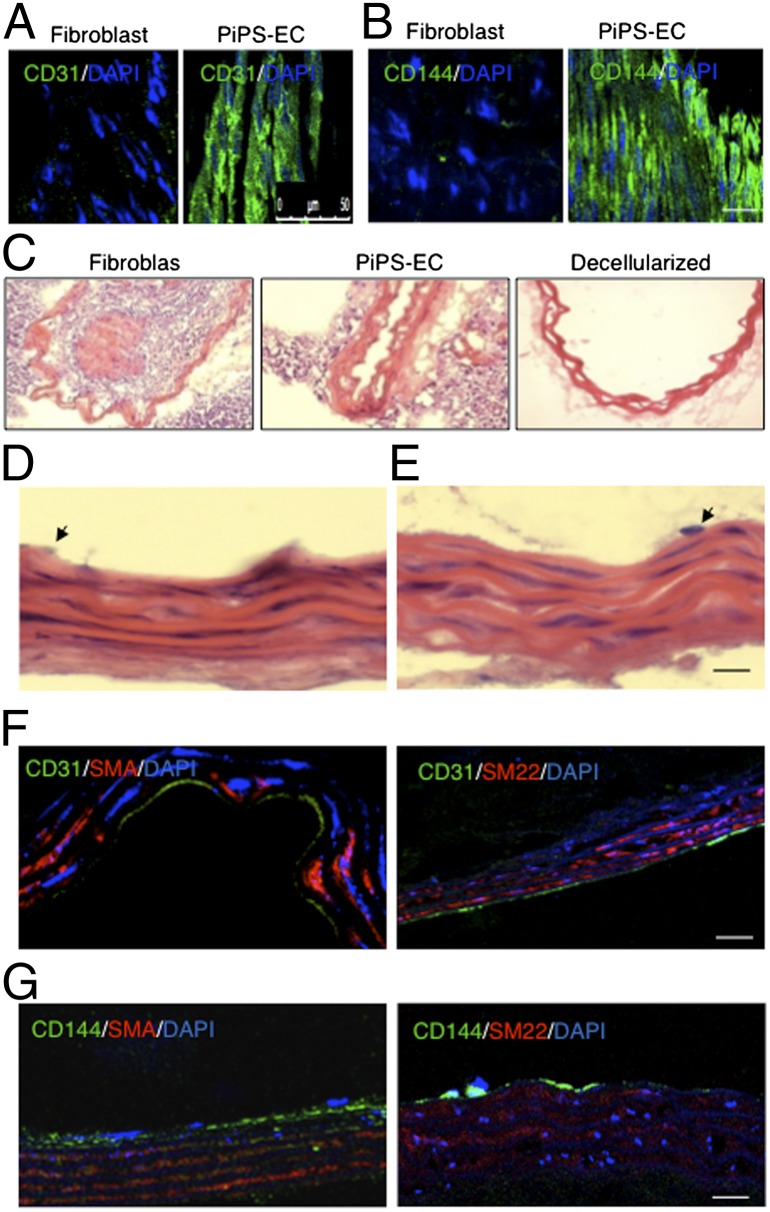
PiPS-ECs Display Endothelial Properties in Vivo.
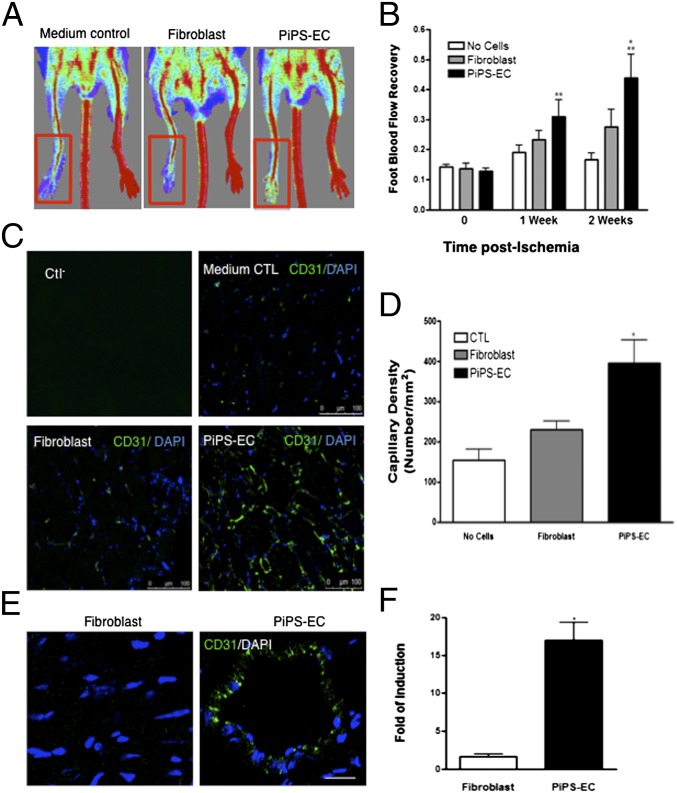
Functionally, PiPS-ECs promoted significantly higher blood flow compared with fibroblasts when injected i.m. into an ischemic model of SCID mice (Fig. 4A). They showed significantly increased blood flow compared with control (no cells) and fibroblasts (Fig. 4B). PiPS-ECs also displayed significantly higher capillary numbers in comparison with fibroblasts (Fig. 4 C and D) when staining with CD31 antibody. Engrafted PiPS-ECs displayed a typical vascular architecture, whereas in the control experiments the injected fibroblasts were present in a random pattern, and no vascular structures were observed (Fig. 4E). Finally, when PiPS-ECs or fibroblasts were stained and quantified with a human specific CD31 antibody, PiPS-ECs displayed an enhanced engraftment ability (Fig. 4 E and F). In addition, PiPS-ECs showed the ability to participate in tissue regeneration when they were seeded on decellularized vessel scaffolds in a specially constructed bioreactor and harvested on day 5. Ex vivo the PiPS-ECs seeded vessels were fixed and stained positive for endothelial markers CD31 (Fig. 5A) and CD144 (Fig. 5B), demonstrating a highly elongated and oriented pattern. Such staining was not obtained for control fibroblasts (Fig. 5 A and B). Importantly, in the ex vivo bioreactor, the PiPS-ECs seeded scaffolds displayed normal vessel morphology, whereas the lumen was almost blocked in some of the fibroblast-seeded vessels (Fig. 5C). These results indicate that PiPS-ECs display endothelial functions when tested ex vivo. To test whether PiPS cells have an ability to directly differentiate into vascular lineage such as smooth-muscle cells and EC ex vivo, the reendothelialization potential of PiPS cells in tissue-engineered vessels was assessed. The vessels were sectioned and stained with H&E (Fig. 5D). The double-seeded scaffolds with PiPS cells showed a native vessel architecture with multiple layers of smooth-muscle cells while an EC layer was present (Fig. 5 D and E). The multiple layers of cells increased vessel wall stability and integrity to a high level, matching results of native nondecellularized aortic grafts (Fig. 5D). The double-seeded PiPS cell-derived vessels were fixed and stained positive for endothelial markers such as CD31 (Fig. 5F) and CD144 (Fig. 5G) and for smooth-muscle markers such as SMA (Fig. 5 F and G, Left panel) and SM22 (Fig. 5 F and G, Right).
Fig. 4.
PiPS-ECs improved neovascularization and blood flow recovery in a hindlimb ischemic model. PiPS-ECs, fibroblasts, or medium control (no cells) were injected i.m. into adductors of an ischemic model of SCID mice. (A) Representative color laser Doppler images of superficial blood flow (BF) in lower limbs taken 2 wk after ischemia induction. (B) Line graph shows the time course of postischemic foot BF recovery (calculated as the ratio between ischemic foot BF and contralateral foot BF) in mice given medium as control, fibroblasts, and PiPS-ECs. Statistical analysis showed significantly higher foot BF recovery for PiPS-ECs in comparison with both “no cells” control and fibroblasts at weeks 1 and 2 [data are means ± SEM (n = 6)]. Week 1: **P < 0.01, PiPS-EC vs. “no cells” control; **P < 0.01, PiPS-EC vs. fibroblasts. Week 2: **P < 0.01, PiPS-EC vs “no cell” control; *P < 0.05 PiPS-EC vs. fibroblasts. No significant differences were detected when fibroblasts were compared with “no cells” control. (C) Sections of adductors muscles were stained with CD31 antibody, and capillary density was expressed as the capillary number per mm2 [D; data are means ± SEM (n = 3); *P < 0.05]. (Scale bar, 100 μm.) (E) PiPS-ECs displayed an enhanced engraftment ability compared with fibroblasts when stained and quantified with a human-specific CD31 antibody at six randomly selected microscopic fields (at ×100) [F; data are means ± SEM (n = 3); *P < 0.05]. (Scale bar, 50 μm.)
Fig. 5.
PiPS-ECs displayed endothelial properties ex vivo. PiPS-ECs or fibroblasts were seeded on decellularized vessel scaffolds in a specially constructed bioreactor and harvested on day 5. (A and B) En face positive staining for endothelial markers CD31 and CD144 is shown in the PiPS-EC. (Scale bar, 50 μm.) (C) H&E staining showed that fibroblast-seeded vessels were almost occluded, whereas PiPS-ECs-seeded scaffolds presented with a normal vessel morphology. (D and E) Cross-sections with H&E staining of the double-seeded scaffolds with selected PiPS cells, which were induced to be differentiated into smooth-muscle cells and ECs, showing a native-like vessel architecture with multiple layers of smooth-muscle cells and a monolayer of ECs. Arrows indicate an endothelial-like cell. (F and G) Double-seeded PiPS cell-derived vessels were fixed and stained positive for EC markers, such as CD31 and CD144, and for smooth-muscle markers, such as SMA and SM22. (Scale bar, 50 μm.)
Discussion
iPS cell generation is a fascinating tool for regenerative medicine that provides the potential to create patient-specific cells to be used in cell-based therapies. Although important technical progress has been made in iPS cell generation (3, 8, 11, 13, 18) from a number of somatic cell sources, iPS cell research is still in its initial stages. Various issues need to be addressed regarding iPS safety and reprogramming mechanisms. In this study, we developed a method to generate a population of PiPS cells from human fibroblasts through short-term reprogramming and selection. We found that during somatic cell reprogramming the expression of genes involved in cell differentiation into specific cell lineages was altered as early as day 4. PiPS cells displayed the ability to differentiate into ECs through defined media and culture conditions by “skipping pluripotency” without tumor risk. In line with our findings, recent studies have reported the direct conversion of human fibroblasts to blood progenitors (19), germ cells into neurons (20), conversion of mouse and human fibroblasts into functional spinal motor neurons (21), and conversion of mouse fibroblasts into cardiomyocytes by defined factors (22) or using a direct reprogramming strategy (23).
During reprogramming the genome of a somatic cell is entering a process to reach pluripotency. Interestingly, microarray analysis revealed that during the early stages of reprogramming, a number of signal pathways involved in cell-specific lineage differentiation were changed. For example, the expression of genes related to differentiation of connective tissue, muscle cell lines, adipocytes, bone cell, mononuclear cells, osteoclast-like cells, or even adhesion of ECs were altered as early as day 4. Moreover, the induction of genes such as DNA (cytosine-5-)-methyltransferase 3-like (DNMT3L) from day 7 of cell reprogramming, sustains the notion that the above signal pathways may be supported by genome-wide adjustments of repressive and active epigenetic features such as DNA methylation and histone modifications, which change the chromatin remodeling complexes (24–26). Additionally, during the later stages of reprogramming, such as days 14 and 21, more signal pathways linked with cell lineage differentiation were identified to display differential gene expression profiles. Therefore, PiPS cells are in a transient state that is able to undergo direct differentiation in response to specific stimulus, such as the expression of lineage-specific factors.
VEGF has been shown to be the first secreted molecule with specificity for the endothelium during development (27). Cells that respond to VEGF must first express its receptors (VEGFR1 and VEGFR2 or KDR). KDR, which forms a complex with VE-cadherin, β-catenin, and PI3-K (28), is the earliest marker for the endothelial lineage during development. In our PiPS cell model high KDR expression was detected, suggesting that these cells may be responsive to VEGF stimulation and thus capable of differentiation to ECs. Furthermore, SET (29) is a multitasking protein that is involved in apoptosis, transcription, differentiation, nucleosome assembly, and histone binding. In our study we found that when reprogrammed cells were stimulated with VEGF, EC differentiation was induced, and SETSIP was translocated to the cell nucleus. SETSIP was binding to the VE-cadherin gene promoter, resulting in increased VE-cadherin gene expression. Further experiments have shown that SETSIP is expressed in mature ECs in comparable levels to PiPS-ECs (SI Appendix, Fig. S9 A and B). Interestingly, in both mature ECs and PiPS-ECs SETSIP is localized in the nucleus (SI Appendix, Fig. S9A). Therefore, SETSIP through regulation of VE-cadherin has an important role in the functional properties of ECs. Our results demonstrate that EC differentiation is induced through the OCT4-SETSIP and VEGF pathway and link the cell reprogramming of fibroblasts with the directed differentiation to EC cells.
The present findings of PiPS cells-derived ECs that expressed a panel of endothelial markers at mRNA and protein levels and formed vascular-like tubes in vitro and in vivo have several implications. First, the time from reprogramming to obtaining ECs is less than 2 wk, which makes this protocol feasible for treatment of patients with a personalized cell therapy (e.g., leg ischemia). In this study three human fibroblast cell lines have been reprogrammed and differentiated to ECs, achieving comparable results. However, we should point out that it has not been tested with fibroblasts from patients. Second, one of the main concerns of stem cell therapy, tumor formation, is eliminated,and the purity of the autologous cells used is no longer an issue because PiPS cells do not develop teratomas in SCID mice. In addition, even if the efficiency of human fibroblast transfected with the four reprogramming factors (using a single plasmid by nucleofection) was ∼30%, the purity of PiPS cells is not an issue because a pure population of PiPS cells can be selected and further differentiated into ECs. Importantly, PiPS-ECs functionally displayed good attachment, stabilization, patency, and typical endothelial structure [e.g., formation of adhesion junction (CD144) in animal models]. When they were used for preparation of tissue-engineered vessels, the vessels displayed functional properties and showed better recovery of tissue blood flow when injected i.m. into ischemic legs in comparison with control fibroblasts. Finally, when a pure population of PiPS cells is used, tissue-engineered vessels show a native-vessel architecture, with multiple layers of smooth-muscle cells and an EC layer. Therefore, PiPS cells could be a valuable source for treatment of ischemic tissues and for generation of tissue-engineered blood vessels.
In summary, PiPS cells have the potential to differentiate into ECs in response to defined media and culture conditions. SETSIP was identified to be translocated to the cell nucleus and directly bound to the VE-cadherin promoter, inducing EC differentiation. Thus, we developed a method to generate PiPS cells from human fibroblasts through short-term reprogramming that can differentiate into ECs without tumor risk, via the OCT4-SETSIP and VEGF pathway (SI Appendix, Fig. S10). PiPS cells can be a useful cell source for regenerating damaged tissue and vascular tissue engineering ex vivo.
Materials and Methods
Cell Reprogramming.
Fibroblasts were nucleofected with the four reprogrammed factors or control empty vector and cultured with reprogramming media composed of Knockout DMEM (Invitrogen, SKU-10829-018), 20% Knockout Serum Replacement (Invitrogen, SKU 10828-028), 10 ng/mL basic fibroblast growth factor (Miltenyi Biotec, 130-093-837), 0.1 mM β-mercaptoethanol, and 0.1 mM MEM nonessential amino acids. The media were changed every day. Procedure details are in SI Appendix, SI Materials and Methods.
SETSIP Cloning and Nucleofection.
Full-length human SETSIP cDNA fragment was obtained by RT-PCR from PiPS-ECs with the primer set shown in SI Appendix, Table S2 and cloned into Kpn1 and Xba1 sites of a pCMV5 plasmid defined as pCMV5-SETSIP.
Selection of PiPS Cells.
Fibroblasts were nucleofected with a polycistronic plasmid containing all four factors, OCT4, SOX2, KLF4, and C-MYC (OSKM), as shown above. Neomycin selection was started 1 d after the nucleofection up to day 4, when a pure population of PiPS cells has been obtained expressing an mOrange marker and the four reprogrammed genes.
Cell Seeding and Vascular Bioreactor.
PiPS-ECs or fibroblasts were seeded on aortic grafts, which were previously decellularized (with SDS at 0.075% and washed in PBS), in a special constructed bioreactor, and shear stress was applied. Procedure details are in SI Appendix, SI Materials and Methods.
En Face Preparation and Immunofluoresence Staining and Frozen Section Staining.
The procedure for en face preparation is similar to that described elsewhere (30).
Statistical Analysis.
Data were analysis using GraphPad Prism Software. Data expressed as means ± SEM were analyzed with a two-tailed Student t test for two groups. A value of P < 0.05 was considered significant.
Supplementary Material
Acknowledgments
This work was supported by grants from the British Heart Foundation, the Oak Foundation, Jubilaeumsstiftung Basel, Stem Cell Leading Project XDA01010303, and by Wellcome Trust Grant 075491/Z/04.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205526109/-/DCSupplemental.
References
- 1.Takahashi K, Okita K, Nakagawa M, Yamanaka S. Induction of pluripotent stem cells from fibroblast cultures. Nat Protoc. 2007;2:3081–3089. doi: 10.1038/nprot.2007.418. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 3.Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 4.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 5.Maherali N, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 6.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 7.Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science. 2008;322:945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang CW, et al. Polycistronic lentiviral vector for “hit and run” reprogramming of adult skin fibroblasts to induced pluripotent stem cells. Stem Cells. 2009;27:1042–1049. doi: 10.1002/stem.39. [DOI] [PubMed] [Google Scholar]
- 9.Sommer CA, et al. Excision of reprogramming transgenes improves the differentiation potential of iPS cells generated with a single excisable vector. Stem Cells. 2010;28:64–74. doi: 10.1002/stem.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou H, et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warren L, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desponts C, Ding S. Using small molecules to improve generation of induced pluripotent stem cells from somatic cells. Methods Mol Biol. 2010;636:207–218. doi: 10.1007/978-1-60761-691-7_13. [DOI] [PubMed] [Google Scholar]
- 13.Li W, Ding S. Small molecules that modulate embryonic stem cell fate and somatic cell reprogramming. Trends Pharmacol Sci. 2010;31:36–45. doi: 10.1016/j.tips.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 14.Park SW, et al. Efficient differentiation of human pluripotent stem cells into functional CD34+ progenitor cells by combined modulation of the MEK/ERK and BMP4 signaling pathways. Blood. 2010;116:5762–5772. doi: 10.1182/blood-2010-04-280719. [DOI] [PubMed] [Google Scholar]
- 15.Dambrot C, Passier R, Atsma D, Mummery CL. Cardiomyocyte differentiation of pluripotent stem cells and their use as cardiac disease models. Biochem J. 2011;434:25–35. doi: 10.1042/BJ20101707. [DOI] [PubMed] [Google Scholar]
- 16.Kattman SJ, et al. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 17.Li Z, Hu S, Ghosh Z, Han Z, Wu JC. Functional characterization and expression profiling of human induced pluripotent stem cell- and embryonic stem cell-derived endothelial cells. Stem Cells Dev. 2011;20:1701–1710. doi: 10.1089/scd.2010.0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaji K, et al. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szabo E, et al. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature. 2010;468:521–526. doi: 10.1038/nature09591. [DOI] [PubMed] [Google Scholar]
- 20.Tursun B, Patel T, Kratsios P, Hobert O. Direct conversion of C. elegans germ cells into specific neuron types. Science. 2011;331:304–308. doi: 10.1126/science.1199082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Son EY, et al. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell. 2011;9:205–218. doi: 10.1016/j.stem.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ieda M, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Efe JA, et al. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat Cell Biol. 2011;13:215–222. doi: 10.1038/ncb2164. [DOI] [PubMed] [Google Scholar]
- 24.Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Q, Melton DA. Extreme makeover: converting one cell into another. Cell Stem Cell. 2008;3:382–388. doi: 10.1016/j.stem.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 26.Welstead GG, Schorderet P, Boyer LA. The reprogramming language of pluripotency. Curr Opin Genet Dev. 2008;18:123–129. doi: 10.1016/j.gde.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 27.Carmeliet P, et al. Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nature. 1996;380:435–439. doi: 10.1038/380435a0. [DOI] [PubMed] [Google Scholar]
- 28.Carmeliet P, et al. Targeted deficiency or cytosolic truncation of the VE-cadherin gene in mice impairs VEGF-mediated endothelial survival and angiogenesis. Cell. 1999;98:147–157. doi: 10.1016/s0092-8674(00)81010-7. [DOI] [PubMed] [Google Scholar]
- 29.Fan Z, Beresford PJ, Oh DY, Zhang D, Lieberman J. Tumor suppressor NM23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nucleosome assembly protein SET is its inhibitor. Cell. 2003;112:659–672. doi: 10.1016/s0092-8674(03)00150-8. [DOI] [PubMed] [Google Scholar]
- 30.Zeng L, et al. Sustained activation of XBP1 splicing leads to endothelial apoptosis and atherosclerosis development in response to disturbed flow. Proc Natl Acad Sci USA. 2009;106:8326–8331. doi: 10.1073/pnas.0903197106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.