Abstract
The successful growth of hypermutator strains of bacteria contradicts a clear preference for lower mutation rates observed in the microbial world. Whether by general DNA repair deficiency or the inducible action of low-fidelity DNA polymerases, the evolutionary strategies of bacteria include methods of hypermutation. Although both raise mutation rate, general and inducible hypermutation operate through distinct molecular mechanisms and therefore likely impart unique adaptive consequences. Here we compare the influence of general and inducible hypermutation on adaptation in the model organism Pseudomonas aeruginosa PAO1 through experimental evolution. We observed divergent spectra of single base substitutions derived from general and inducible hypermutation by sequencing rpoB in spontaneous rifampicin-resistant (RifR) mutants. Likewise, the pattern of mutation in a draft genome sequence of a derived inducible hypermutator isolate differed from those of general hypermutators reported in the literature. However, following experimental evolution, populations of both mutator types exhibited comparable improvements in fitness across varied conditions that differed from the highly specific adaptation of nonmutators. Our results suggest that despite their unique mutation spectra, general and inducible hypermutation can analogously influence the ecology and adaptation of bacteria, significantly shaping pathogenic populations where hypermutation has been most widely observed.
Keywords: mutS, rulAB
Bacterial evolution requires mutation rates be optimized to permit adaptation through sequence variation while still preserving genome integrity. The broad conservation of multiple DNA error-avoidance and error-repair processes across all domains of life indicates a preference for lower mutation rates to avoid the primarily deleterious nature of mutation (1, 2). However, hypermutator (or mutator) strains of bacteria constitute a pervasive exception to the rule as these bacteria successfully inhabit diverse environments despite up to 1,000-fold increases in spontaneous mutation rates (2, 3).
Hypermutation arises most commonly through disruption of the methyl-directed mismatch repair (MMR) system, and mutS inactivation is the most widespread defect (4). The presence of MMR-deficient strains of the opportunistic pathogen Pseudomonas aeruginosa in chronic cystic fibrosis (CF) respiratory infections stands as the most illustrative example of hypermutator ecology (5). During chronic infection, hypermutation has been credited with facilitating the phenotypic changes and clonal diversification characteristic of P. aeruginosa adaptation to the CF lung environment (6, 7). Furthermore, MMR-deficient hypermutators are overrepresented in populations of various other pathogenic bacteria (3), suggesting that high mutation rates offer advantages in pathogenesis.
Hypermutation need not be constitutive. Mutagenic DNA repair, an inducible source of hypermutation, transiently increases mutation rate through the action of specialized, low-fidelity DNA polymerases as part of the SOS response following exposure to UV radiation (UVR) or other DNA-damaging agents (8). Secondary mutations produced by the Y-family polymerases polIV (Escherichia coli DinB) and polV (E. coli UmuDC) during template-independent synthesis of damaged DNA comprise the majority of sequence alterations derived from UVR exposure. These polymerases also confer UVR tolerance and therefore are critical for epiphytic growth in plant pathogenic bacteria harboring the umuDC-homolog rulAB that reside on leaf surface habitats optimized for solar UVR exposure (9). However, Y-family polymerases are broadly distributed among diverse organisms where their function is not understood, suggesting inducible hypermutation likely influences bacterial adaptation in diverse environments (10, 11).
Hypermutation has been observed in a wide variety of clinical, environmental, and laboratory populations suggesting that evolutionary strategies of bacteria include systems for increasing mutability. We hypothesized that general (mutS-deficient) and inducible (rulAB-mediated) hypermutation likely produce unique mutation spectra, which differentially affect adaptive improvements in fitness. Although hypermutation universally increases mutation rate, the influences of different molecular mechanisms on adaptation are likely not equal but have yet to be compared experimentally. Fortunately, bacteria are well suited for the experimental and genomic study of evolution (12–14). To compare the influence of general and inducible hypermutation on adaptation, we have conducted experimental evolution with the model organism P. aeruginosa PAO1, compared single-base substitution preferences, and sequenced the genome of a derived inducible mutator isolate. We observed comparable fitness improvements in both general and inducible mutator lineages despite divergent point mutation spectra, suggesting that their unique mechanisms do not impose a strong bias on adaptation.
Results
Frequency of Rifampicin-Resistant (RifR) Mutants During Lineage Growth.
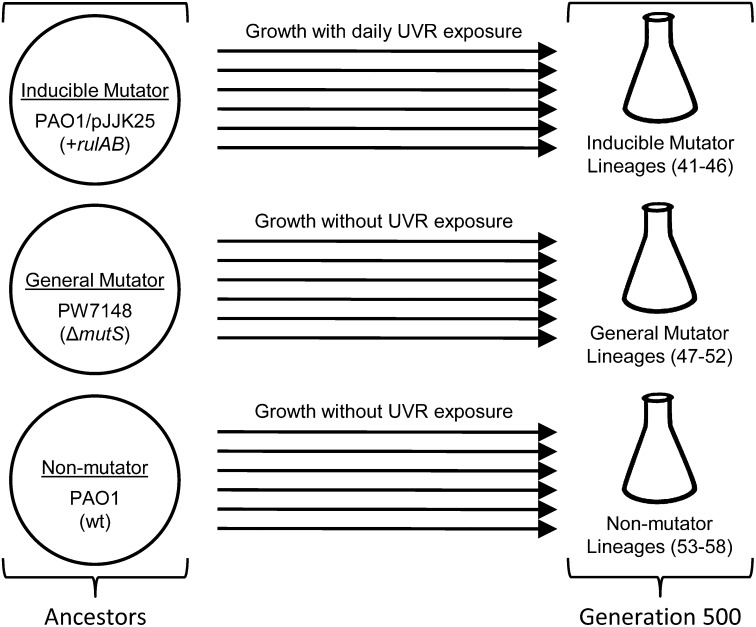
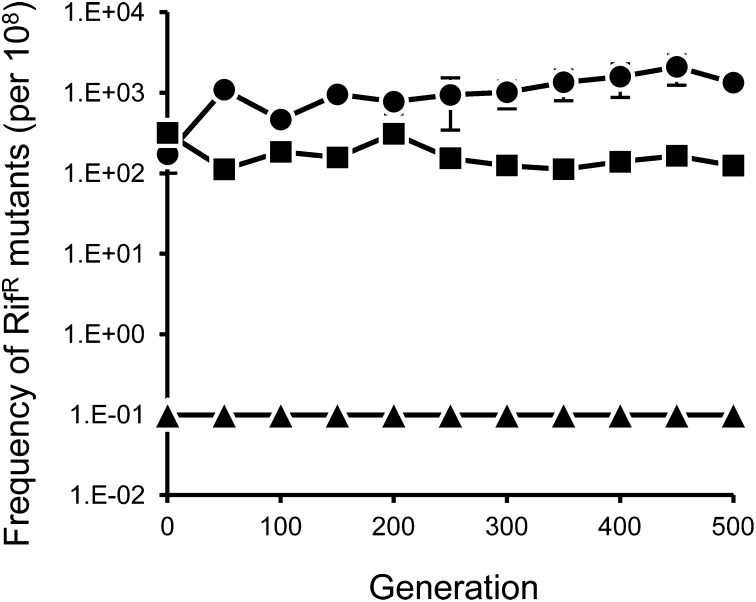
Eighteen parallel lineages of P. aeruginosa were serially propagated for ∼500 generations in a manner similar to another study conducted previously (Fig. 1 and ref. 15). Every 24 h, expression of rulAB was activated in inducible mutator lineages of PAO1/pJJK25 by exposure to UVR at a dosage optimized to closely match the rate of spontaneous RifR mutants generated in general mutator lineages of PW7148. In this manner, the mutability rates of PAO1/pJJK25 (1.7 × 102) and PW7148 (3.2 × 102) were standardized at the beginning of the evolution experiment. Within the first 50 generations of the experiment, however, inducible mutator lineages established and maintained a 10-fold greater average frequency of RifR mutants (Fig. 2). Despite the difference in overall frequency of RifR mutants in each population, the average frequency of RifR mutants in inducible and general mutator lineages largely remained stable throughout lineage growth (Fig. 2). We expect that rates of RifR mutability remained constant throughout lineage propagation and that the disparity in RifR mutant frequency does not reflect divergent supply rates. Therefore, the observed difference in RifR mutant frequency between inducible and general mutator lineages implies that rulAB-mediated and mutS-deficient hypermutation are not equivalent at the rpoB locus. In contrast, the frequency of RifR mutants in nonmutator lineages was effectively zero and only a few resistant colonies were ever recovered (Fig. 2), consistent with the comparatively low mutation rate of P. aeruginosa nonmutators (16).
Fig. 1.
Relationship of ancestral P. aeruginosa PAO1 genotypes and their derived experimental lineages. Six parallel lineages of each ancestral genotype were derived from a single colony and propagated by serial transfer for 500 generations.
Fig. 2.
Average frequency of RifR mutants in lineages of (●) inducible mutators, (■) general mutators, and (▲) nonmutators during experimental evolution. Error bars represent SEM and if not visible it is because they are smaller than the size of the marker.
Spectra of Single Base Substitutions in RifR Mutants.
The distribution and frequency of mutations in RifR colonies of the ancestral PAO1/pJJK25 and PW7148 strains were compared with identify differences in point mutation spectra associated with rulAB-mediated and mutS-deficient hypermutation. Mutations responsible for RifR map exclusively to rpoB and produce amino acid substitutions in three primary regions of the β subunit of RNA polymerase (17). These regions, termed clusters I–III, are highly conserved among bacteria (18) and well characterized in P. aeruginosa (19). Furthermore, the 38 diverse mutational possibilities in rpoB available to achieve resistance include all transition and transversion substitutions, providing a suitable system for analyzing point mutation spectra.
We sequenced rpoB clusters I–III from RifR mutants of PAO1/pJJK25 and PW7148 and observed point mutations at 18 unique nucleotide positions in clusters I and II (Table S1). All sequenced rpoB fragments from RifR mutants contained only one point mutation each. Both general and inducible hypermutation favored specific base substitutions at one of a few sites with little overlap observed between mechanisms. Tabulating all substitution types revealed that PAO1/pJJK25 and PW7148 produced significantly divergent spectra of mutations in rpoB (P = 0.0005, Table 1). The CG→TA transition was observed most frequently in RifR mutants of PAO1/pJJK25, whereas the opposing TA→CG transition was most abundant in RifR mutants of PW7148 (Table 1). These results demonstrate that general and inducible hypermutation generate divergent point mutation spectra at the rpoB locus, consistent with our hypothesis.
Table 1.
Spectra of single base substitutions observed in rpoB from RifR mutants of P. aeruginosa PAO1/pJJK25 and PW7148 compared to those in the genome of MRW44.1
| PAO1/pJJK25 |
PW7148 |
MRW44.1 |
||||
| Substitution | Number | Frequency | Number | Frequency | Number | Frequency |
| TA→CG | 1 | 0.011 | 75 | 0.872 | 22 | 0.067 |
| CG→TA | 67 | 0.744 | 10 | 0.116 | 197 | 0.604 |
| TA→GC | 0 | 0.000 | 0 | 0.000 | 14 | 0.043 |
| TA→AT | 13 | 0.144 | 0 | 0.000 | 43 | 0.132 |
| CG→AT | 8 | 0.089 | 1 | 0.012 | 34 | 0.104 |
| CG→GC | 1 | 0.011 | 0 | 0.000 | 16 | 0.049 |
| Total | 90 | 86 | 326 | |||
Relative Fitness of Derived Lineages.
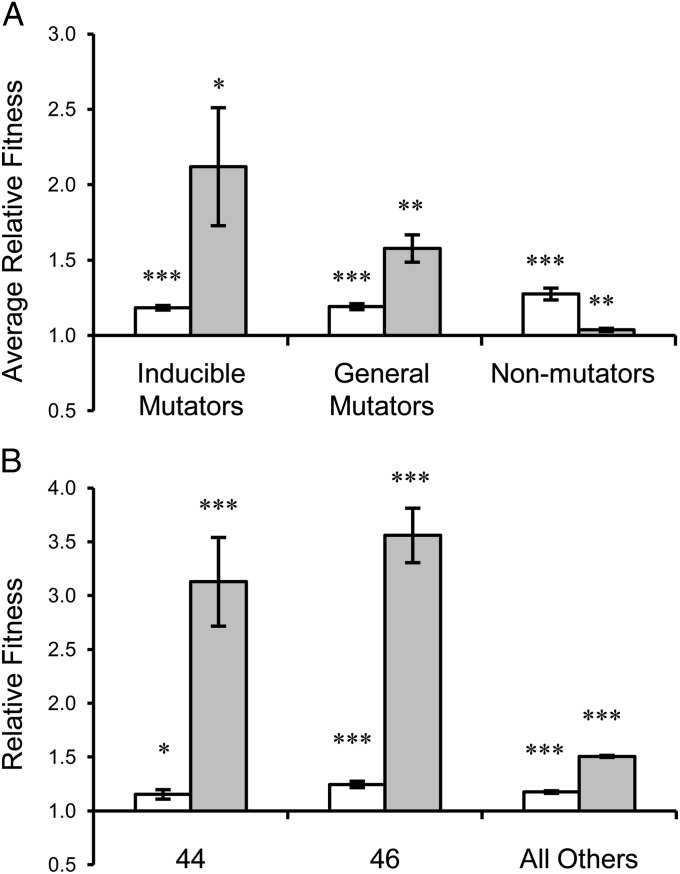
To evaluate adaptation during our evolution experiment, the relative fitness of each derived lineage was determined by direct competition with its respective ancestor (Fig. 1) under both UVR and non-UVR conditions. Many of the inducible and general mutator lineages diversified into mixtures of distinct colony morphologies and so the relative fitness of lineage population samples, which comprised a subset of the entire population, was evaluated to maintain the assemblage of sympatric genotypes. All derived lineages of inducible mutators, general mutators, and nonmutators exhibited improvements in fitness under their native conditions (Fig. 3A and Fig. S1). When assayed under non-UVR conditions, inducible mutator lineages exhibited an 18% average improvement in fitness (all P < 0.02, Fig. 3A). Inducible mutator lineages exhibited significantly higher fitness under UVR conditions compared with non-UVR conditions (all P < 0.004, Fig. 3B), and inducible mutator lineages 44 and 46 displayed an exceptional 234% average improvement in relative fitness under UVR conditions (all P < 0.004, Fig. 3B), whereas the remaining inducible mutator lineages exhibited a 51% average improvement (all P < 0.007, Fig. 3B). This pattern of specificity suggests that inducible mutator populations acquired a combination of adaptive improvements specialized for growth after exposure to UVR along with general improvements that also contributed to fitness under non-UVR conditions.
Fig. 3.
Relative fitness of derived lineages under non-UVR (open bars) and UVR (shaded bars) conditions. (A) Average relative fitness of all derived lineages of inducible, general, and nonmutators. (B) Exceptional relative fitness of lineages 44 and 46 compared with the average of all other inducible mutator lineages. Values are means of six independent replicates and error bars represent SEM. Values are significant by two-tailed independent t test where indicated (*P < 0.05, **P < 0.01, ***P < 0.001).
General mutator lineages displayed a 19% average improvement in relative fitness under their native, non-UVR conditions (all P < 0.002, Fig. 3A) and a 57% average improvement in fitness under UVR conditions (all P < 0.002, Fig. 3A). Surprisingly, the average relative fitness of general mutators was significantly higher under UVR conditions (P = 0.012, Fig. 3A), despite a lack of previous exposure to UVR, suggesting that bacteria in these lineages acquired generalized adaptive improvements useful under both conditions. Conversely, the adaptation of nonmutator lineages was highly specific to the non-UVR conditions under which they were propagated (Fig. 3A). Nonmutator lineages displayed a 27% average improvement in relative fitness under non-UVR conditions (all P < 0.008, Fig. 3A). Under UVR conditions, nonmutator lineages exhibited a 4% average improvement in relative fitness (Fig. 3A) but only nonmutator lineage 54 exhibited a significant improvement in fitness (P = 0.028, Fig. S1C).
Draft Genome Sequence Analysis of a Derived Inducible Mutator.
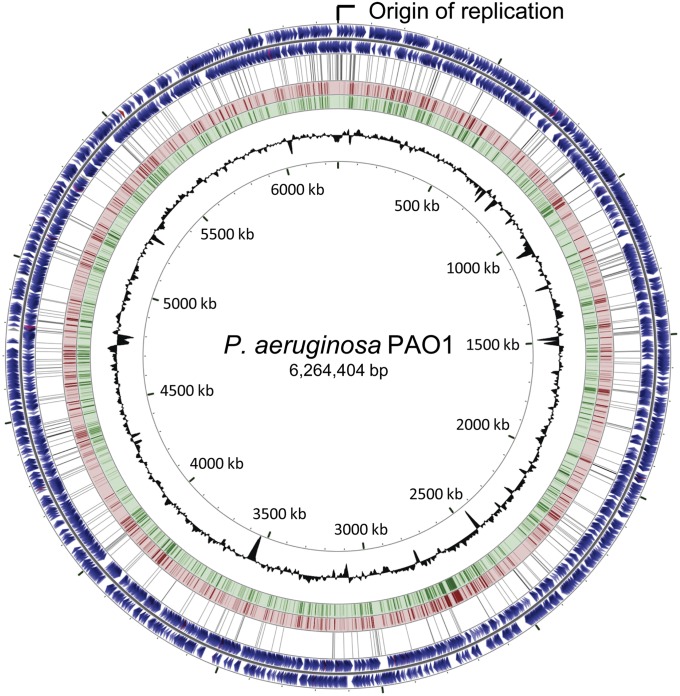
We sequenced the genome of MRW44.1, an isolate from inducible mutator lineage 44, to determine the complete list of mutational variations relative to the published PAO1 reference sequence (GenBank accession no. AE004091; ref. 20). Inducible mutator lineage 44 was chosen because of its pronounced relative fitness under UVR conditions (Fig. 3B) and MRW44.1 showed similarly high fitness (Fig. S2). Sequencing of the shotgun library generated 383,834 total passed filter reads that, when mapped to the published PAO1 reference sequence, produced 54 contigs totaling 6,227,769 bp in length and covering 99.4% of the reference sequence. This strategy identified 403 high-confidence sequence variations that included single base substitutions, multiple base substitutions, insertions, and deletions. Eighteen mutational differences have been previously shown to separate the ancestral PAO1 strain used in this study from the published PAO1 reference sequence (21) and all were present in the draft genome sequence of MRW44.1. The remaining 385 new mutations (Table S2 and Dataset S1) included nonsense mutations in 12 genes (Table S3) and missense mutations in 145 genes from various functional classes (Dataset S2) that were distributed throughout the chromosome but appeared particularly dense near the origin of replication (Fig. 4).
Fig. 4.
Genetic map of the chromosome of P. aeruginosa PAO1 based on the published reference sequence (AE004091, ref. 20). Starting at the outside, rings one and two are protein-coding sequences (blue), tRNA (red), rRNA (purple), and other (gray) ORFs on the forward and reverse strands, respectively. Ring three (black) indicates the position of the 385 variations identified in the derived inducible mutator isolate MRW44.1. Rings four and five map pairwise Blastn alignments of the PAO1 reference sequence (red) and the de novo assembled contigs (green), respectively. Ring six shows G + C content (deviation from average) and ring seven indicates scale (in kilobase pairs). The origin of replication is also indicated. The map was generated using the CGView Server (22).
The draft genome sequence of MRW44.1 revealed 326 single base substitutions. To determine the genome-wide mutation bias of rulAB-mediated hypermutation in this strain, we compared the spectra of single base substitutions with a simple random distribution based on the PAO1 genome composition that assumes an equal likelihood of all substitutions (23). A comparison of the mutation spectra observed in MRW44.1 with the expected frequencies of all possible nucleotide substitutions using a binomial test indicated a strong bias toward CG→TA transitions and possibly TA→AT transversions (Table S4). The observed frequency of all other substitutions was significantly lower than expected. This pattern closely matches the frequency distribution of single base substitutions observed in RifR mutants of PAO1/pJJK25 (Table 1) and together they suggest that rulAB-mediated hypermutation has a clear preference for CG→TA substitutions.
We explored the evolutionary forces behind sequence variation in MRW44.1 by examining the proportion of synonymous mutations. Significantly more synonymous point mutations were observed within coding regions (103 or 38.0%) than expected by random chance (68 or 25.2%) based on the composition of the PAO1 genome. Single base substitutions in MRW44.1 were highly skewed toward CG→TA transitions, which have a high probability of producing synonymous changes (46.1%), and TA→AT transversions, which have a very low probably of producing synonymous changes (5.6%). To reflect this bias, we separated point mutations into two categories: rulAB (either CG→TA or TA→AT) or non-rulAB (all other substitutions) and these categories included 86 and 17 synonymous substitutions, respectively. The sum of the individual binomial distributions of the CG→TA and TA→AT substitutions indicated that the number of observed synonymous mutations in the composite rulAB category did not differ significantly from random (P = 0.144). These results point to random genetic drift as the dominant process behind single base substitution during the experimental evolution of MRW44.1.
Discussion
Our results suggest that despite generating unique mutation spectra, general and inducible hypermutation may not differentially affect adaptation. We demonstrated the analogous influence of unique mechanisms of hypermutation on the ecology and adaptation of bacterial populations by investigating the point mutation spectra of antibiotic-resistant clones, the genome of a derived inducible mutator, and the relative fitness of derived populations. Thus, distinct strategies for increasing mutation rate may significantly shape bacterial populations, especially in pathogens where hypermutation has been most widely observed.
We tracked the frequency of RifR mutants in each lineage to monitor population structure dynamics through fluctuations in a readily measurable phenotype associated with a known genetic determinant. The average frequency of RifR mutants never rose above 2.1 × 10−5 in our mutator lineages, suggesting that mutations in rpoB derived from general and inducible hypermutation were deleterious. These results are consistent with reports of reduced fitness and RNA polymerase function in RifR mutants of E. coli and P. aeruginosa grown in the absence of rifampicin (24, 25). A balance between the supply of new RifR mutations, provided by hypermutation, and the loss of mutants due to fitness costs linked to defects in rpoB, could explain the observed stable average frequency of RifR mutants in our lineages.
General and inducible mutator lineages exhibited different RifR mutant frequencies, but given the diverse mutational possibilities available to achieve resistance, this disparity likely did not result from a bias toward the mutation spectra of either hypermutation mechanism. Rather, not all point mutations in rpoB are equal with respect to their pleiotropic fitness costs (24, 25), and general and inducible hypermutation clearly favor different nucleotide substitutions at the rpoB locus. The point mutation spectra of PW7148 may favor rpoB mutations that carry greater fitness defects and thus produced the observed lower frequency of RifR mutants in these lineages. Indeed a recent investigation of fitness costs associated with specific rpoB mutations in PAO1 showed that mutants harboring H531Y, the predominant rulAB-derived substitution observed here, always exhibited higher fitness than mutants with D521G, the most abundant mutS-derived substitution observed in this study, in the same background (25). Furthermore, mutants with the H531R substitution (only observed in mutS lineages here) displayed the lowest fitness of all strains tested in the prior study (25).
The spectra of point mutations present in the derived inducible mutator isolate MRW44.1 bore a striking resemblance to that observed in rpoB from RifR mutants of the parental strain PAO1/pJJK2. Although the overlap between the two spectra was only weakly significant (P = 0.019), both were heavily biased toward CG→TA transitions. These results confirm that RifR frequency serves as a reasonable estimate for single base substitution rates throughout the genome as previously reported for E. coli (17). Therefore, the point mutations observed in rpoB from RifR mutants of PW7148 also likely represent the genome-wide single base substitution spectra associated with mutS-deficient hypermutation. However, although widely used to evaluate hypermutator status (26), RifR mutant frequency does not provide a comprehensive assessment of mutagenesis. Because rpoB is essential, measurement of RifR ignores insertions, deletions, and other sequence polymorphisms that eliminate RNA polymerase function. Both general and inducible hypermutation have been shown to produce these additional polymorphisms (27, 28) and the genome of MRW44.1 included 59 such mutations. We suspect that these hypermutation mechanisms produce varied spectra with respect to all forms of mutation but can only conclude here that mutS-deficient and rulAB-mediated hypermutation differ in their production of single base substitutions. However, the predominant mutations reported here in MRW44.1 and previously in general mutators from similar studies (16, 23) were single base substitutions, indicating their significant role in genome evolution driven by hypermutation.
To our knowledge, we present a unique sequence of the genome of an inducible mutator following experimental evolution. Sequences of general mutators present in the literature (16, 23) allowed us to compare how these mechanisms influence genome evolution. We observed 385 mutations in MRW44.1 after 500 generations of laboratory evolution with daily activation of rulAB-mediated hypermutation. Following 20,000 generations of growth in a similar laboratory evolution experiment, the genome sequence of a mutT-deficient E. coli hypermutator included 653 new mutations highly skewed toward AT→CG transversions (23). A longitudinal study of CF lung-infecting P. aeruginosa found primarily transitions among 959 variations in a mutL-deficient hypermutator isolated 250 mo postinfection (16). In contrast, genome sequences of derived nonmutators from both laboratory and clinical studies bore considerably fewer mutations (13, 16, 23).
The mutations observed in MRW44.1 mapped to the PAO1 chromosome with high density near the origin of replication. By comparison, mutations in the derived genomes of mutT-deficient E. coli (23) and mutL-deficient P. aeruginosa (16) hypermutators were evenly distributed throughout their respective genomes. Because UVR exposure occurred before the transfer to fresh media in our experiment, inducible hypermutation should have been actively resolving lesions only during the first round of genome replication. However, multiple rounds of replication can occur simultaneously during growth, effectively overrepresenting genes near the origin of replication (29). Active polV competes for replication forks of undamaged DNA (30) and could displace polIII in the early stages of a subsequent replication. This additional mutagenesis of undamaged DNA preferentially near the origin of replication could explain the biased distribution of mutations observed in MRW44.1. Because MMR-deficient hypermutation is not constrained by the same replication-linked temporal mechanics, mutations would not be expected to exhibit higher density at the origin of replication and this is true of the mutations present in the genomes of general hypermutators reported elsewhere (16, 23).
To compare the effects of the divergent mutS-deficient and rulAB-mediated mutation spectra on adaptation, we measured changes in fitness relative to each lineage’s respective ancestor. All derived lineage populations exhibited similar gains in fitness under non-UVR conditions, signifying that each adapted comparably to the shared culture media, independent of mutation supply rate or mechanism. Inducible mutator lineages received regular UVR exposure during propagation and thus exhibited significantly higher fitness under UVR conditions similar to a previous evolution experiment with the plant-associated inducible mutator Pseudomonas cichorii 302959 (15). The exceptional fitness gains exhibited by lineages 44 and 46 under UVR conditions likely reflect additional adaptive improvements not yet realized by the other lineages.
We sequenced the genome of an isolate from one lineage to inventory all mutations that arose during adaptation, and the improved fitness of MRW44.1 indicates that some of these mutations are beneficial. However, our analysis of synonymous, neutral substitutions suggests that the dominant process driving single base substitution during experimental evolution was random genetic drift. Therefore, most of the 168 nonsynonymous mutations are also likely neutral or nearly so. The signals of rare beneficial mutations are overwhelmed by the more abundant random mutations, preventing their identification for further analysis. A similar analysis with mutT-deficient E. coli (23) revealed the same random pattern, suggesting that both inducible and general hypermutation influence genome evolution primarily through a simple neutral accumulation derived from an elevated mutation rate.
Surprisingly, fitness improvements in general mutator lineages were also significantly higher under UVR conditions despite an absence of previous exposure to UVR. The genome of PAO1, and consequently PW7148, lacks a umuDC homolog (31) and so we added a rulAB-encoding plasmid to generate the UVR-inducible hypermutator PAO1/pJJK25 used in this study. Addition of rulAB to PAO1 also increases UVR tolerance (9). In microorganisms, UVR tolerance is linked to direct and indirect mechanisms of DNA maintenance (32). General mutator lineages may have acquired compensatory improvements to mitigate the mutagenic effects of mutS inactivation and ensure genome stability. Such mutations could carry added fitness benefits following UVR exposure by better equipping cells to handle additional DNA damage. Therefore, although both mutator types achieved comparable fitness gains under UVR conditions, inducible mutators likely did so through specialized adaptive changes, whereas general mutators relied on less specific improvements. This distinction reduces any specific bias imposed by the mutation spectra of either mutS-deficient or rulAB-mediated hypermutation, allowing both to reach a common phenotypic outcome through unique mutational routes.
Unlike mutator lineages, the adaptation of nonmutator lineages was specific to their native, non-UVR conditions, and no improvements in fitness under the alternate, UVR conditions were observed. This specificity could simply reflect the lower mutation supply rate of nonmutators that may have limited access to additional beneficial mutations in the timeframe of our experiment. However, the exclusive ability for mutator lineages to exhibit additional fitness improvements across varied conditions could highlight the capacity for hypermutation to explore genotypic space in pursuit of general adaptive improvements. Hypermutation also stimulated population diversity and increased fitness under varied conditions could reflect beneficial mutations present in specific subpopulations. Both explanations are consistent with the observed prevalence of hypermutators in chronic CF infection and their contribution to adaptive phenotypic shifts and clonal expansion that promote long-term survival (5–7). Our findings expand the current paradigm of the role hypermutation plays in the ecology and adaptation of bacterial populations to include additional mechanisms responsible for increasing mutation rates.
The results of our evolution experiment with the model organism P. aeruginosa PAO1 suggest that both general and inducible hypermutation analogously encourage adaptation and population diversification despite their divergent mutation spectra. Characteristic adaptive changes and clonal expansion observed in populations of P. aeruginosa during chronic CF infection have been attributed to MMR-deficient hypermutation. Our results suggest that inducible hypermutation could serve the same function, and indeed DNA polIV can promote mucoid conversion (33), raising the question of why inducible hypermutation has received little attention in CF-infecting populations. The source of inoculum for respiratory infection likely includes numerous environmental pools of P. aeruginosa (34, 35) where plasmid-encoded umuDC homologs have been reported (36). Furthermore, polV is common among enteric bacteria (37) and the CF lung environment includes potential DNA-damaging compounds that induce the SOS response (10). On the basis of this reasoning and the data presented here, we suspect that inducible hypermutation likely plays a larger role in chronic respiratory infection than currently thought. Inducible and MMR-deficient hypermutation are not mutually exclusive mechanisms and may well function concurrently during infection, warranting further studies to build on this initial comparison.
Materials and Methods
The bacterial strains and plasmids used in this study and detailed methods for strain construction are described in SI Materials and Methods. Methods for our evolution experiments with general and inducible mutator lineages and with control lineages and relative fitness assays are found in SI Materials and Methods. Methods for DNA sequence analysis to identify RifR mutations and for determination of a draft genome sequence of P. aeruginosa MRW44.1 are also in SI Materials and Methods.
Supplementary Material
Acknowledgments
The authors thank Rich Lenski, Tom Schmidt, and Barb Sears for helpful comments; Jeff Barrick for help with genome substitution calculations; Kevin Carr for help with genome sequence analysis; Paul Stothard for genome map customization with the CGview server; and Vinh Tran and Bryan Lenneman for technical assistance. This work was supported by the US Department of Agriculture and Michigan State University AgBioResearch. The development and maintenance of strains in the University of Washington P. aeruginosa PAO1 two-allele transposon mutant library was funded by National Institutes of Health Grant P30 DK089507.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession no. ALBW01000000).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205357109/-/DCSupplemental.
References
- 1.Sniegowski PD, Gerrish PJ, Johnson T, Shaver A. The evolution of mutation rates: Separating causes from consequences. Bioessays. 2000;22:1057–1066. doi: 10.1002/1521-1878(200012)22:12<1057::AID-BIES3>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 2.de Visser JAGM. The fate of microbial mutators. Microbiology. 2002;148:1247–1252. doi: 10.1099/00221287-148-5-1247. [DOI] [PubMed] [Google Scholar]
- 3.Sundin GW, Weigand MR. The microbiology of mutability. FEMS Microbiol Lett. 2007;277:11–20. doi: 10.1111/j.1574-6968.2007.00901.x. [DOI] [PubMed] [Google Scholar]
- 4.Hall LMC, Henderson-Begg SK. Hypermutable bacteria isolated from humans—a critical analysis. Microbiology. 2006;152:2505–2514. doi: 10.1099/mic.0.29079-0. [DOI] [PubMed] [Google Scholar]
- 5.Oliver A, Mena A. Bacterial hypermutation in cystic fibrosis, not only for antibiotic resistance. Clin Microbiol Infect. 2010;16:798–808. doi: 10.1111/j.1469-0691.2010.03250.x. [DOI] [PubMed] [Google Scholar]
- 6.Hogardt M, et al. Stage-specific adaptation of hypermutable Pseudomonas aeruginosa isolates during chronic pulmonary infection in patients with cystic fibrosis. J Infect Dis. 2007;195:70–80. doi: 10.1086/509821. [DOI] [PubMed] [Google Scholar]
- 7.Mena A, et al. Genetic adaptation of Pseudomonas aeruginosa to the airways of cystic fibrosis patients is catalyzed by hypermutation. J Bacteriol. 2008;190:7910–7917. doi: 10.1128/JB.01147-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schlacher K, Goodman MF. Lessons from 50 years of SOS DNA-damage-induced mutagenesis. Nat Rev Mol Cell Biol. 2007;8:587–594. doi: 10.1038/nrm2198. [DOI] [PubMed] [Google Scholar]
- 9.Kim JJ, Sundin GW. Regulation of the rulAB mutagenic DNA repair operon of Pseudomonas syringae by UV-B (290 to 320 nanometers) radiation and analysis of rulAB-mediated mutability in vitro and in planta. J Bacteriol. 2000;182:6137–6144. doi: 10.1128/jb.182.21.6137-6144.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erill I, Campoy S, Barbé J. Aeons of distress: An evolutionary perspective on the bacterial SOS response. FEMS Microbiol Rev. 2007;31:637–656. doi: 10.1111/j.1574-6976.2007.00082.x. [DOI] [PubMed] [Google Scholar]
- 11.Andersson DI, Koskiniemi S, Hughes D. Biological roles of translesion synthesis DNA polymerases in eubacteria. Mol Microbiol. 2010;77:540–548. doi: 10.1111/j.1365-2958.2010.07260.x. [DOI] [PubMed] [Google Scholar]
- 12.Elena SF, Lenski RE. Evolution experiments with microorganisms: The dynamics and genetic bases of adaptation. Nat Rev Genet. 2003;4:457–469. doi: 10.1038/nrg1088. [DOI] [PubMed] [Google Scholar]
- 13.Herring CD, et al. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat Genet. 2006;38:1406–1412. doi: 10.1038/ng1906. [DOI] [PubMed] [Google Scholar]
- 14.Hegreness M, Kishony R. Analysis of genetic systems using experimental evolution and whole-genome sequencing. Genome Biol. 2007;8:201. doi: 10.1186/gb-2007-8-1-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weigand MR, Sundin GW. Long-term effects of inducible mutagenic DNA repair on relative fitness and phenotypic diversification in Pseudomonas cichorii 302959. Genetics. 2009;181:199–208. doi: 10.1534/genetics.108.096131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cramer N, et al. Microevolution of the major common Pseudomonas aeruginosa clones C and PA14 in cystic fibrosis lungs. Environ Microbiol. 2011;13:1690–1704. doi: 10.1111/j.1462-2920.2011.02483.x. [DOI] [PubMed] [Google Scholar]
- 17.Garibyan L, et al. Use of the rpoB gene to determine the specificity of base substitution mutations on the Escherichia coli chromosome. DNA Repair (Amst) 2003;2:593–608. doi: 10.1016/s1568-7864(03)00024-7. [DOI] [PubMed] [Google Scholar]
- 18.Campbell EA, et al. Structural mechanism for rifampicin inhibition of bacterial rna polymerase. Cell. 2001;104:901–912. doi: 10.1016/s0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- 19.Jatsenko T, Tover A, Tegova R, Kivisaar M. Molecular characterization of Rif(r) mutations in Pseudomonas aeruginosa and Pseudomonas putida. Mutat Res. 2010;683:106–114. doi: 10.1016/j.mrfmmm.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 20.Stover CK, et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 21.Klockgether J, et al. Genome diversity of Pseudomonas aeruginosa PAO1 laboratory strains. J Bacteriol. 2010;192:1113–1121. doi: 10.1128/JB.01515-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grant JR, Stothard P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008;36(Web Server issue):W181–4. doi: 10.1093/nar/gkn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrick JE, et al. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461:1243–1247. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- 24.Barrick JE, Kauth MR, Strelioff CC, Lenski RE. Escherichia coli rpoB mutants have increased evolvability in proportion to their fitness defects. Mol Biol Evol. 2010;27:1338–1347. doi: 10.1093/molbev/msq024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hall AR, Iles JC, MacLean RC. The fitness cost of rifampicin resistance in Pseudomonas aeruginosa depends on demand for RNA polymerase. Genetics. 2011;187:817–822. doi: 10.1534/genetics.110.124628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliver A, Cantón R, Campo P, Baquero F, Blázquez J. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science. 2000;288:1251–1254. doi: 10.1126/science.288.5469.1251. [DOI] [PubMed] [Google Scholar]
- 27.Schaaper RM, Dunn RL. Spectra of spontaneous mutations in Escherichia coli strains defective in mismatch correction: The nature of in vivo DNA replication errors. Proc Natl Acad Sci USA. 1987;84:6220–6224. doi: 10.1073/pnas.84.17.6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smania AM, et al. Emergence of phenotypic variants upon mismatch repair disruption in Pseudomonas aeruginosa. Microbiology. 2004;150:1327–1338. doi: 10.1099/mic.0.26751-0. [DOI] [PubMed] [Google Scholar]
- 29.Chandler MG, Pritchard RH. The effect of gene concentration and relative gene dosage on gene output in Escherichia coli. Mol Gen Genet. 1975;138:127–141. doi: 10.1007/BF02428117. [DOI] [PubMed] [Google Scholar]
- 30.Fijalkowska IJ, Dunn RL, Schaaper RM. Genetic requirements and mutational specificity of the Escherichia coli SOS mutator activity. J Bacteriol. 1997;179:7435–7445. doi: 10.1128/jb.179.23.7435-7445.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simonson CS, Kokjohn TA, Miller RV. Inducible UV repair potential of Pseudomonas aeruginosa PAO. J Gen Microbiol. 1990;136:1241–1249. doi: 10.1099/00221287-136-7-1241. [DOI] [PubMed] [Google Scholar]
- 32.Jacobs JL, Carroll TL, Sundin GW. The role of pigmentation, ultraviolet radiation tolerance, and leaf colonization strategies in the epiphytic survival of phyllosphere bacteria. Microb Ecol. 2005;49:104–113. doi: 10.1007/s00248-003-1061-4. [DOI] [PubMed] [Google Scholar]
- 33.Moyano AJ, Luján AM, Argaraña CE, Smania AM. MutS deficiency and activity of the error-prone DNA polymerase IV are crucial for determining mucA as the main target for mucoid conversion in Pseudomonas aeruginosa. Mol Microbiol. 2007;64:547–559. doi: 10.1111/j.1365-2958.2007.05675.x. [DOI] [PubMed] [Google Scholar]
- 34.Gibson RL, Burns JL, Ramsey BW. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am J Respir Crit Care Med. 2003;168:918–951. doi: 10.1164/rccm.200304-505SO. [DOI] [PubMed] [Google Scholar]
- 35.Schelstraete P, et al. Pseudomonas aeruginosa in the home environment of newly infected cystic fibrosis patients. Eur Respir J. 2008;31:822–829. doi: 10.1183/09031936.00088907. [DOI] [PubMed] [Google Scholar]
- 36.Stokes HW, Krishnapillai V. Prevalence of Pseudomonas aeruginosa FP plasmids which enhance spontaneous and uv-induced mutagenesis. Mutat Res. 1978;50:19–28. doi: 10.1016/0027-5107(78)90056-8. [DOI] [PubMed] [Google Scholar]
- 37.Ohmori H, et al. The Y-family of DNA polymerases. Mol Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.