Abstract
In polymicrobial infections, microbes can interact with both the host immune system and one another through direct contact or the secretion of metabolites, affecting disease progression and treatment options. The thick mucus in the lungs of patients with cystic fibrosis is highly susceptible to polymicrobial infections by opportunistic pathogens, including the bacterium Pseudomonas aeruginosa and the fungus Aspergillus fumigatus. Unravelling the hidden molecular interactions within such polymicrobial communities and their metabolic exchange processes will require effective enabling technologies applied to model systems. In the present study, MALDI-TOF and MALDI-FT-ICR imaging mass spectrometry (MALDI-IMS) combined with MS/MS networking were used to provide insight into the interkingdom interaction between P. aeruginosa and A. fumigatus at the molecular level. The combination of these technologies enabled the visualization and identification of metabolites secreted by these microorganisms grown on agar. A complex molecular interplay was revealed involving suppression, increased production, and biotransformation of a range of metabolites. Of particular interest is the observation that P. aeruginosa phenazine metabolites were converted by A. fumigatus into other chemical entities with alternative properties, including enhanced toxicities and the ability to induce fungal siderophores. This work highlights the capabilities of MALDI-IMS and MS/MS network analysis to study interkingdom interactions and provides insight into the complex nature of polymicrobial metabolic exchange and biotransformations.
Microbes that colonize mammalian hosts can form polymicrobial communities, such as biofilms, where they establish commensual, mutualistic, competitive, or antagonistic interactions with one another and with the host. In microbial disease, this complex interplay can affect the outcome of antimicrobial therapy (1). Therefore, it is important to understand polymicrobial populations and their interactions at the molecular level.
In persons with cystic fibrosis (CF), the lungs are lined with a viscous mucus layer susceptible to polymicrobial infections (2). Pseudomonas aeruginosa, a Gram-negative bacterial opportunistic pathogen, is the most prevalent and persistent microorganism (3) isolated from the sputum of CF lungs and leading cause of mortality in CF patients (4). Within the CF lung, P. aeruginosa exists in biofilm-like macrocolonies (5) and is refractory to antimicrobial agents and the host immune response (6). Aspergillus fumigatus, an opportunistic fungal pathogen, is the second-most persistent microbe in the CF lung, with a 10–57% prevalence rate (3), and is capable of causing allergic bronchopulmonary aspergillosis (7).
Superinfection with both P. aeruginosa and A. fumigatus in CF patients leads to decreased pulmonary function compared with monoinfection with either microbe (8). Interestingly, however, in a pulmonary mouse model, mice coinfected with P. aeruginosa and A. fumigatus had a higher survival rate than mice infected by A. fumigatus alone (9). Additional in vitro studies have suggested that P. aeruginosa has an inhibitory effect on filamentation and biofilm formation of A. fumigatus through both direct contact and secreted molecules (10). The coexistence of P. aeruginosa and A. fumigatus in the CF lung, species composition, spatial orientation, and molecular interaction remain to be elucidated, however.
Understanding these interkingdom interactions requires a combination of innovative enabling technologies and in vitro model systems. MALDI imaging mass spectrometry (MALDI-IMS) is a powerful technology (11) capable of simultaneously visualizing the spatial and temporal distribution of hundreds of metabolites secreted by microorganisms directly on agar, rather than focusing on single molecules or pathways (12). The objective of the present study was to use MALDI-IMS to identify key metabolic exchange factors in interactions between P. aeruginosa and A. fumigatus and to uncover roles for these metabolites in the regulation of polymicrobial systems.
Identification of metabolites by MALDI-TOF IMS in combination with high accuracy (<10 ppm) MALDI FT-ICR IMS was facilitated by the recently developed MS/MS network analysis on microbial extracts. This computational methodology uses similarities in MS fragmentation data to associate structurally similar metabolites, including novel analogs (13). This multipronged approach revealed a complex assortment of secreted metabolites and pointed toward previously unknown metabolic interactions between P. aeruginosa and A. fumigatus grown in close proximity on agar.
Of the metabolite classes described herein, phenazines produced by P. aeruginosa play important roles in electron shuttling, generation of toxic superoxides, and biofilm development through signaling and redox chemistry (14, 15). In addition, the phenazines pyocyanin (PYO; 1) and 1-hydroxyphenazine (1-HP; 2) are reported inhibitors of A. fumigatus (16). (Details of the numbered structures here and below are provided in SI Appendix, Fig. S1). We observed that phenazine metabolites produced by P. aeruginosa were converted by the fungus into unique products with alternative biological functions. These biotransformations included conversion of phenazine-1-carboxylic acid (PCA; 3) into 1-HP (2), 1-methoxyphenazine (1-MP; 4), and phenazine-1-sulfate (5). Both 1-HP (2) and 1-MP (4) inhibited fungal growth, while the phenazine-1-sulfate (5) did not. 1-HP induced up-regulation of the extracellular fungal siderophores triacetylfusarinine C (6, 7) and fusarinine C (8). A. fumigatus also converted the P. aeruginosa metabolites PCA (3) and PYO (1) into phenazine dimers (9, 10), potentially in defense against P. aeruginosa and its elaborate system of virulence and signaling factors. This work demonstrates the application of MALDI-IMS in identifying microbial bioconversion metabolites, opens up opportunities to study the effects of these metabolites on both the producing organism and the competing bacterium, and could ultimately lead to alternative therapeutic interventions for infections by these and other microbial pathogens.
Results and Discussion
Interaction of P. aeruginosa and A. fumigatus.
This work used P. aeruginosa PA14 (17) and A. fumigatus Af293 (18) as model strains to study this interkingdom interaction at the metabolic level. The two strains were grown in a side-by-side interaction on ISP2 agar using high-cell-density spot inoculants as a model for this microbial encounter. P. aeruginosa can persist in high densities (108–1010 cfu/g) in the airways of CF patients (3), and high-cell-density spot inoculants have been used previously to model colony biofilms (19).
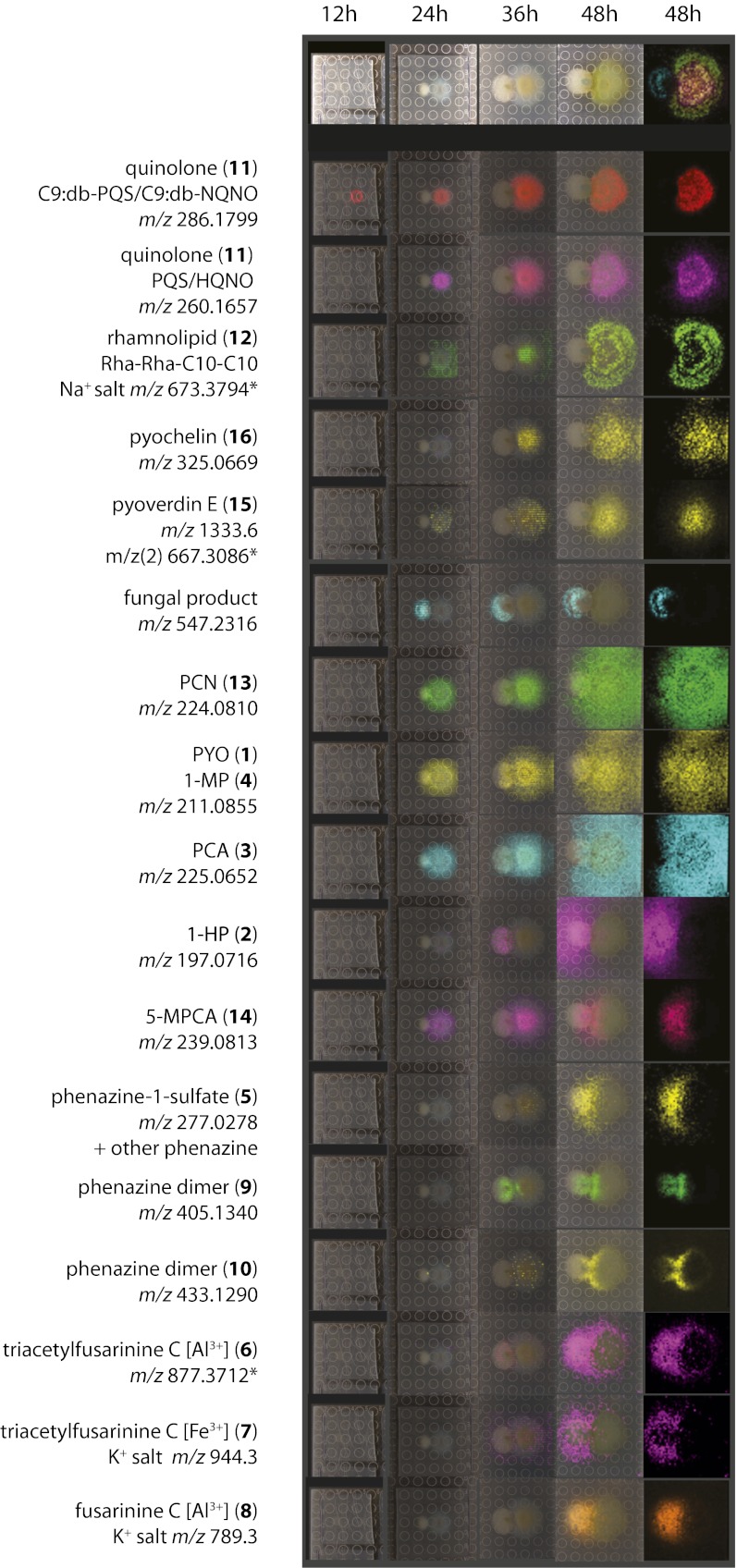
Time-dependent metabolic exchange between P. aeruginosa and A. fumigatus was studied at 30 °C and analyzed at 12 h, 24 h, 36 h, and 48 h. Significant fungal inhibition was observed at 36 h and 48 h. The P. aeruginosa colony also appeared inhibited and exhibited yellow pigmentation at the interface at 48 h (Fig. 1 and SI Appendix, Fig. S2).
Fig. 1.
(A) MALDI-IMS images of selected metabolites for A. fumigatus (Left) and P. aeruginosa (Right) grown for 12 h, 24 h, 36 h, and 48 h on ISP2. Optical images are displayed in the top row. Overlays of m/z distributions with the optical images are shown in all of the other images except those in column 5, which show the m/z distribution only. Column 5, top panel, shows an overlay of 543, 260, and 673 m/z. Representative quinolones (11) observed in the interaction (m/z 286 and 260 Da) are displayed. db, double bond. At least 10 quinolones were identified with a similar distribution showing limited diffusion due to their hydrophobic nature. A representative example for the rhamnolipid family (12) (m/z 673 Da, Na+ form) is shown. At least 10 rhamnolipids were identified, including monorhamnolipids and di-rhamno lipid congeners, which are widely secreted into the media. P. aeruginosa siderophores pyoverdin E (15) and pyochelin (16) displayed a similar distribution. Various phenazines were identified, including previously unreported phenazine-1 sulfate (5) and phenazine dimers (9, 10). Fungal fusarinine-derived siderophores (6, H+ form; 7, K+ form; 8, K+ form shown) were induced and were predominantly present in the aluminum-complexed form. Accurate masses are derived from a replicate MALDI FT-ICR IMS (SI Appendix, Fig. S4) or FT-ICR from an extract (indicated by an asterisk). MALDI-IMS spatial resolution was 400–600 μm, with a detection range of 100–3,000 Da.
A section of the agar containing the side-by-side interactions was cut out, treated with matrix, and subjected to MALDI-TOF IMS (SI Appendix, Fig. S3) to examine the spatial and temporal distribution of the secreted metabolites (12). At 48 h, more than 90 ions were observed localized on, around, and between both P. aeruginosa and A. fumigatus, indicating the complex chemical and dynamic metabolic exchange between these organisms. Examples of these observed m/z signals corresponding to the metabolites reported in this paper are shown in Fig. 1. To facilitate identification of the molecules observed on IMS, a duplicate plate with an incubation time of 48 h was subjected to MALDI FT-ICR-IMS, which allowed accurate mass determination of metabolites with <10 ppm error (Fig. 1 and SI Appendix, Fig. S4). In addition, extracts of the interaction zone between A. fumigatus and P. aeruginosa were analyzed by HPLC, FT-ICR, and ion-trap MS. This enabled the construction of MS/MS networks (13) of the P. aeruginosa–A. fumigatus interaction site, displaying metabolite ions (m/z) as nodes (SI Appendix, SI Methods). Nodes with a high MS/MS spectral analogy are clustered together and often belong to the same chemical class. Clusters of the different metabolite classes can be correlated to the ions observed in the MALDI image of the P. aeruginosa colony grown in proximity to A. fumigatus, to facilitate their annotation (Fig. 1 and SI Appendix, Fig. S5).
Metabolite Classes Involved in P. aeruginosa–A. fumigatus Interaction.
P. aeruginosa and A. fumigatus are known to be prolific producers of a wide variety of secondary metabolites (20, 21). In the interaction between these organisms, several fungal metabolites were suppressed compared with control fungus; for example, fungal metabolite with m/z 547 was not observed at the interaction site and was present only on the outer edge of the fungal colony, whereas the mycotoxin fumigaclavines A and C were nearly undetectable (Fig. 1 and SI Appendix, Fig. S6). From these observations, it is apparent that P. aeruginosa has a significant influence on the metabolic arsenal of A. fumigatus. This influence is reminiscent of the way in which Bacillus subtilis can suppress the defensive arsenal of Streptomyces coelicolor (22). The identification of this phenomenon in an interkingdom interaction raises the intriguing possibility that such alterations of metabolic factors may affect disease progression.
In contrast to the fungal metabolites, many of the P. aeruginosa-secreted products did not appear to be affected by the interaction with A. fumigatus, as exemplified by two metabolite classes, the quinolones (11) and the rhamnolipids (12). As evaluated by MALDI-IMS and MS/MS networks, more than 10 members of the 4(1H)-quinolone (11) family, including (2-heptyl-3-hydroxy-4-hydroxyquinoline (PQS), were observed localized to P. aeruginosa at 48h (Fig. 1 and SI Appendix, Figs. S5 and S6). Several quinolones have been identified as quorum-sensing (QS) signaling molecules (PQS and 2-heptyl-4-hydroxyquinoline), iron chelators (PQS), and antimicrobials (2-heptyl-4-hydroxyquinoline N-oxide), but most quinolones remain uncharacterized (23, 24). More than 10 rhamnolipids were produced by P. aeruginosa and secreted extensively in the media, as exemplified by m/z 673 Da (12) (Fig. 1 and SI Appendix, Figs. S5 and S6). These biosurfactants have been reported to promote uptake and biodegradation of insoluble compounds, affect swarming and biofilm formation, act as virulence factors, and inhibit a variety of microorganisms, including fungi (25).
The most surprising results were from our analysis of the phenazines, a major class of P. aeruginosa metabolites, which had strikingly unique distributions. Although some abundant members of the phenazines completely engulfed the fungal colony, others were detected only at the interface or had unexpected distributions under the fungal colony. Phenazine-1-carboxamide (PCN; 13) and PCA (3) were both so profusely secreted by P. aeruginosa that they diffused throughout the fungal colony (Fig. 1). PCN (13), which has reported antifungal activity against Fusarium species (26), did not lead to a clearance zone in a disk diffusion assay (up to 0.20 μmol/disk) on an A. fumigatus lawn on ISP2 agar; however, PCN (13) did exhibit a zone of discoloration starting at 0.08 μmol/disk. PCA (3) showed no observable phenotypic effect under these conditions (SI Appendix, Fig. S7).
An ion with m/z 211 Da was localized well into the fungal territory and of similar intensity to PCA (3) and PCN (13). This IMS signal originated from two phenazines, PYO (1) and 1-MP (4). Whereas PYO is a well-studied P. aeruginosa virulence factor and QS signaling molecule (27) with reported antifungal activity, 1-MP (4) is not a previously described P. aeruginosa PA14 metabolite. When the two compounds were tested in an antifungal disk diffusion assay, PYO (1) showed no antifungal effect up to 0.20 μmol/disk, whereas 1-MP (4) demonstrated robust inhibition of A. fumigatus starting at 0.08 μmol/disk (SI Appendix, Fig. S7).
Unexpectedly, 1-HP (2; m/z 197 Da), a known P. aeruginosa phenazine metabolite, exhibited an unusual distribution at much higher concentrations in and around the A. fumigatus colony than surrounding the P. aeruginosa colony. This finding was confirmed by comparing extracts from the outer edge of the fungal colony, the interaction site, and the outer edge of the P. aeruginosa colony with a 1-HP (2) standard (SI Appendix, Fig. S8), using HPLC and MS analysis. Because P. aeruginosa did not grow around the fungal colony, this distribution allowed us to hypothesize that the fungus might play a role in the production of this metabolite. 1-HP (2) showed fungal inhibition in the lawn assay starting at 0.16 μmol, and discoloration was observed at 0.08 μmol/disk (SI Appendix, Fig. S7) This antifungal activity is in accordance with previously reported findings (16).
Other ions assigned to phenazine metabolites were observed with a specific distribution at the A. fumigatus–P. aeruginosa interface. This type of distribution is often indicative of a specific chemical response, including defense, syntropism, signaling, or related to nutrient deficiency (28–30). One of these ions (m/z 239 Da) was annotated as 5-N-methylated PCA (5-MPCA; 14), a phenazine recently reported to induce fungal cell death in P. aeruginosa–Candida albicans interactions (31). This metabolite was more directed toward A. fumigatus at 48 h than at 12 h–36 h, suggesting a comparable defensive role. A phenazine ion with m/z of 277 Da had a similar distribution to 5-MPCA at the microbial interface at 48 h. This metabolite was abundant in an extract of the P. aeruginosa–A. fumigatus interaction and was characterized as phenazine-1-sulfate (5) (SI Appendix, SI Spectral Analyses). The phenazine-1-sulfate showed no fungal inhibition in the disk diffusion lawn assay up to 0.20 μmol/disk (SI Appendix, Fig. S7). Although the formation of a sulfated phenazine metabolite by P. aeruginosa through interaction with a fungal strain is unprecedented, sulfonation is a known metabolic process used by a variety of fungi to solubilize and detoxify xenobiotics (32–34). Therefore, we hypothesized that this metabolite is a fungal biotransformation product.
Finally, two additional induced phenazine ions (m/z 405 and 433 Da) with comparable distributions were detected at the interface of the P. aeruginosa and A. fumigatus colonies. The distribution of m/z 405 Da at various time points provides clues as to its origin. Specifically, when comparing the 36 h and 48 h time points, the signal seems to emerge from the fungus. A phenazine dimer with this molecular formula was previously isolated from P. aureofaciens (35), but its structure does not fit our MSn fragmentation data, which indicate the presence of a different crosslink between the phenazine moieties (9) (SI Appendix, SI Spectral Analyses). The MSn spectrum of ion m/z 433 Da could not be fully annotated, but is suggestive of a phenazine dimer containing a PCA unit (10) (SI Appendix, SI Spectral Analyses). Both of these molecules appeared to be fungal biotransformation products, but neither could be isolated in sufficient quantity for further NMR characterization.
Biotransformation of P. aeruginosa Phenazine Metabolites by A. fumigatus.
We were intrigued by the unexpected distribution of 1-HP (2) in the P. aeruginosa– A. fumigatus MALDI-IMS, indicating a higher concentration under A. fumigatus than under P. aeruginosa. Whereas P. aeruginosa is known to produce low levels of 1-HP (2) compared with PCA (3) (36), A. fumigatus does not have the biosynthetic machinery to make phenazines. Of additional interest was the spatial distribution of both phenazine-1-sulfate (5) and the dimeric phenazine (9, 10) compounds, because it was unclear whether these metabolites originated from P. aeruginosa or the fungal colony (Fig. 1).
To gain insight into why the majority of 1-HP (2) was accumulated under the fungal colony and which organism was responsible for the formation of the induced phenazine metabolites, we investigated the effect of the major phenazine metabolites observed in the interaction [PCN (13), PYO (1), PCA (3), 1-HP(2), and 1-MP (4)] with A. fumigatus (Fig. 2) using MALDI-IMS. With this method, changes in metabolic output are monitored directly on the agar with minimal sample manipulation, and the spatial distribution of multiple divergent metabolites can be observed in a single experiment. This provides a large-scale picture of metabolic transformation without bias based on extraction procedures.
Fig. 2.
MALDI-IMS of A. fumigatus treated with various phenazines demonstrating the formation of phenazine biotransformation products and up-regulation of siderophores. The top row identifies the fungal phenotype after treatment of a pregrown (for 24 h) fungal colony with a phenazine-impregnated paper disk for 24 h on ISP2. The second row identifies which phenazine is applied to the agar. The metabolites detected are shown on the left. MALDI-IMS spatial resolution was 400–600 μm, with a detection range of 100–2,000 Da.
For analysis of the biotransformation of phenazines by A. fumigatus, HPLC-purified commercial phenazines applied to paper disks (3 mm radius) were placed next to a 24-h colony of A. fumigatus on ISP2 and incubated at 30 °C for 24 h. After the paper disk was removed, the fungal colony was subjected to MALDI-IMS as described above and monitored for the ions previously observed in the P. aeruginosa–A. fumigatus interaction (Fig. 1).
We did not find any changes in the metabolite profile of A. fumigatus treated with PCN (13) that correlated with the ions observed in the interaction of A. fumigatus and P. aeruginosa (Fig. 2). There was no evidence of the formation of 1-HP (2), 1-MP (4), phenazine sulfate (5), or the dimeric phenazines (9, 10). These findings were confirmed by extraction and subsequent HPLC with diode array UV detection coupled to offline MS analysis (SI Appendix, Fig. S9). Similar experiments with PYO (1) provided no further insight into the formation of these metabolites observed at the interface of the P. aeruginosa–A. fumigatus interaction.
However, when we grew A. fumigatus adjacent to PCA (3; m/z 225 Da), MALDI-IMS revealed products with m/z 197 and 211 Da that were not detected in either the control fungus or the control agar treated with PCA (Fig. 2). After extraction and HPLC analysis, not only PCA (3) itself, but also three major additional phenazines were detected based on their characteristic UV absorbance (SI Appendix, Fig. S9) (37). Comparison with commercial standards identified 1-HP (2; m/z 197 Da) and 1-MP (4; m/z 211 Da), along with phenazine-1-sulfate (5; m/z 277 Da) previously identified at the P. aeruginosa–A. fumigatus interaction site. These findings indicate that A. fumigatus is responsible for the biotransformation of the P. aeruginosa metabolite PCA (3) into 1-HP (2), 1-MP (4), and phenazine-1-sulfate (5). This is a remarkable result, because although phenazines have been studied for decades and are toxic to various organisms, few studies have detailed bioconversion or sequestering of these metabolites by other species. One relevant example is the reported conversion of PCN (13) and PCA (3) to a partially characterized phenazine by A. sclerotiorum (38). More recently, fungal addition products to 5-MPCA were observed in the interaction of C. albicans with P. aeruginosa, resulting in increased fungal toxicity (31, 39).
Aspergilli are known to metabolize a wide variety of aromatic compounds using an extensive repertoire of chemical transformations including decarboxylation, oxidation, methylation, sulfonation, and aromatic acid reduction (32–34, 40). The capability of fungi to metabolize a variety of molecules has been exploited in studies using fungal–bacterial cocultures to detoxifiy and mineralize environmentally persistent compounds (41). In contrast, studies on the fungal bioconversion of metabolites produced by pathogenic microbes in the context of polymicrobial disease are limited.
To test whether 1-MP (4) and phenazine-1-sulfate (5) were derived from the 1-HP (2) intermediate, we investigated the influence of 1-HP (2) on the fungal colony using MALDI-IMS. Indeed, 1-HP (2) applied in proximity to A. fumigatus was converted by the fungus to 1-MP (4), as illustrated by formation of the m/z 211 product in the MALDI-IMS, which was further confirmed by HPLC (SI Appendix, Fig. S9) and MS analysis of the extract. Further HPLC and FT-ICR/ion-trap analysis identified the presence of phenazine-1-sulfate as well. Surprisingly, the fungal siderophores triacetylfusarinine C and fusarinine C were also detected in this MALDI-IMS dataset and corresponding extracts. Triacetylfusarinine C was present in the unbound (m/z 853) and iron-complexed form (m/z 906 Da; 7), as well as in the aluminum-chelated state (m/z 877 Da; 6), whereas fusarinine C was detected predominantly as the aluminum-bound complex (K+ salt m/z 789 Da shown; 8). These siderophores were previously detected at the A. fumigatus –P. aeruginosa interaction site, where they localized around A. fumigatus, reaching around P. aeruginosa in the MALDI-IMS (Fig. 1). In the interaction with P. aeruginosa, significantly more Al3+-bound triacetylfusarinine C and fusarinine C were detected compared with the Fe3+-chelated forms. Neither the metal-complexed nor the des-metal siderophores were detected in the MALDI-IMS or the extract of the control fungus. The chemical identification of these aluminum-complexed siderophores was aided by MS/MS network analysis of the extract (SI Appendix, SI Methods and Fig. S5). Because no exogenous aluminum was added to the media, the observation of aluminum-complexed fungal fusarinines was unexpected. These siderophores are known to facilitate the uptake of iron required for a variety of cellular processes (42). Because of its similar size and charge as iron, aluminum can also complex siderophores (43). Aluminum is toxic to a variety of organisms (44) and, like iron, can often stimulate siderophore production (45). Thus, the fusarinines may also function as protective agents by trapping aluminum extracellularly. It has been proposed that unlike Fe3+, Al3+ chelated to siderophores is not reduced intracellularly and remains bound to the siderophore, which is hydrolyzed and secreted (42).
After identifying 1-MP (4) as a fungal biotransformation product, we then examined whether 1-MP was responsible for additional phenazine biotransformation products, such as the phenazine dimers, through both MALDI-IMS and HPLC analysis of the extract (Fig. 2). We detected no additional phenazine-derived products, however. Given that PCA (3) is converted by A. fumigatus to 1-HP (2), we hypothesized that a reaction intermediate of this transformation process might also react with other phenazines, such as PYO (1), and thus may be responsible for phenazine dimer formation. When PYO (1) and PCA (3) were applied simultaneously near A. fumigatus, both phenazine dimers m/z 405 and 433 Da (9, 10) were observed in MALDI-IMS (Fig. 2).
The biotransformation of P. aeruginosa metabolite PCA (3) by A. fumigatus produces multiple modified phenazines with diverse potential properties through decarboxylation/hydroxylation, methylation, and sulfonation (Fig. 3). Conversion of PCA (3) to 1-HP (2) and subsequently to phenazine-1-sulfate (5) appears to follow the paradigm of detoxification (32–34). In contrast, transformation of PCA (3) to 1-HP (2) and 1-MP (4) leads to more potent fungal inhibition than PCA itself and does not appear to be a detoxification biotransformation. Similarly, in the Lentinus fungal species, methylation of phenolic hydroxyl groups was found to increase toxicity to fungal growth, and it was suggested that methylation might serve other functions, including prevention of repolymerization or facilitation of further biotransformations (46).
Fig. 3.
Observed biotransformation of P. aeruginosa metabolites by A. fumigatus. P. aeruginosa produces PCA (3) and PYO (1), which diffuse in agar and flood the A. fumigatus colony. It also produces 1-HP (2), albeit at relatively low levels. A. fumigatus converts PCA (3) into 1-HP (2), resulting in the up-regulation of the siderophore triacetylfusarinine C (6, 7) and fusarinine C (8). 1-HP (2) is subsequently further metabolized by A. fumigatus to 1-MP (4) and phenazine-1 sulfate (5). PCA (3) not only undergoes a biotransformation by A. fumigatus as a single species, but is also converted by the fungus in the presence of PYO (1) to mixed phenazine dimers (9, 10).
An observed role for the transient biotransformation product 1-HP (2) is the induction of fusarinine-based fungal siderophores. This appears to be a well-regulated process, because 1-MP (4), the bioconversion product from 1-HP (2), does not up-regulate these fungal siderophores. These results contrast with the role of phenazines in producing bacteria, where they repress genes responsible for siderophore biosynthesis and transport (47). Unexpectedly, we found fungal siderophores predominantly complexed with aluminum and, to a lesser degree, iron at the A. fumigatus–P. aeruginosa. interface. It is possible that P. aeruginosa siderophores, such as pyoverdin E (15) and pyochelin (16), already depleted iron at the interaction site between these organisms. In addition, fungal siderophores may play an active role in sequestering extracellular aluminum, a potentially toxic metal, without release in the cell (Fig. 2) (42, 48).
The metabolism of PCA (3) by A. fumigatus also may disrupt the siderophore-independent pathway used by P. aeruginosa for iron acquisition. P. aeruginosa uses PCA (3) to mediate the reduction of extracellular Fe(III) to Fe(II) important for biofilm formation (49). Metabolism of PCA (3) by A. fumigatus yields many phenazine products with potentially modified redox potentials that may interfere with this process. Through additional fungal biotransformation processes, phenazines PYO (1) and PCA (3) produced by P. aeruginosa are converted into phenazine dimers. PYO (1) is a P. aeruginosa QS signaling molecule that influences transcription regulation through SoxR and helps coordinate biofilm formation (27). Bioconversion of PYO (1) by A. fumigatus might lower its concentration and thus possibly affect the QS mechanisms of P. aeruginosa and subsequent physiological responses. This hypothesized process is akin to a strategy used by root-associated fungi and other species that degrade QS homoserine lactones to attenuate bacterial virulence (50). The role of these previously unreported phenazine dimers and their effect on both A. fumigatus and P. aeruginosa will require isolation for additional studies. Whether the biotransformations and molecular interactions observed in our model system between A. fumigatus and P. aeruginosa also play roles in the complex in vivo environment of the CF lung will remain a topic for future studies.
In summary, this paper highlights the capability of MALDI-IMS combined with MS/MS networking to capture the multiplexed nature of polymicrobial interactions. Contrary to the common perception, MALDI-IMS is not limited to the detection of large peptidic metabolites. In a single experiment, we surveyed structurally divergent metabolites ranging from small nonpeptide phenazines and their biotransformations (MW <300) to quinolones (MW <350), rhamnolipids (MW 500–800), and larger siderophores (MW 800–1400). The pairwise interaction of P. aeruginosa and A. fumigatus revealed suppression and increased production of metabolites and multiple biotransformations within a single microbial encounter. A. fumigatus converted the P. aeruginosa metabolite PCA (3) to 1-HP (2), 1-MP (4), and phenazine-1-sulfate (5). 1-HP (2) itself was transformed by the fungus to 1-MP (4) and phenazine-1-sulfate (5). The 1-HP (2) and 1-MP (4) exhibited increased fungal inhibitory activity compared with PCA (3), whereas phenazine-1-sulfate (5) showed no antifungal activity. These metabolites could be intermediates in the detoxification process or serve other, as-yet unexplored roles, as exemplified by 1-HP (2), which turned on the production of fungal siderophores. MALDI-IMS also revealed that A. fumigatus converted the P. aeruginosa metabolites PCA (3) and PYO (1) into dimeric phenazines (9, 10). The identification of these unique fungal bioconversion metabolites provides opportunities to study their effects on both the producing organism and competing bacterium.
As exemplified by these experiments, the nature of metabolic exchange within mixed-species interactions is highly complex. We have demonstrated the application of MALDI-IMS to capture metabolic exchange and bioconversions of P. aeruginosa and A. fumigatus in a model system. Our findings further suggest that metabolite biotransformations by neighboring microorganisms may play a role in the progression of CF and other polymicrobial diseases. We anticipate this will be an important area for future studies. Therefore, it is imperative to continue the development of enabling technologies such as MALDI-IMS to visualize the hidden molecular world of microbial encounters.
Methods
Detailed information on strains, culture conditions, bioassays, extractions, purification of metabolites, HPLC conditions, MALDI-TOF IMS, MALDI-FT-ICR IMS, MS analysis including MS/MS networking, and NMR analysis are provided in SI Appendix, SI Methods. Enlarged pictures of the A. fumigatus–P. aeruginosa interactions; the workflow for MALDI imaging; MALDI-FT-ICR imaging data; MALDI-IMS data on fungal products, quinolones, and rhamnolipids; MS/MS networks of P. aeruginosa–A. fumigatus extracts; fungal inhibition assays; and HPLC chromatograms are provided in SI Appendix. MS spectral annotations and NMR analyses are reported in SI Appendix, SI Spectral Analyses.
Supplementary Material
Acknowledgments
We thank Ashlee Dravis (J. Craig Venter Institute) for providing the Aspergillus fumigatus Af293 strain, and Jane Yang and Dr. Steven Bark for critically reviewing the manuscript. D.S.C. thanks Prof. Richard Caprioli at the Vanderbilt University Mass Spectrometry Research Center for use of the Bruker MALDI FT-ICR device. This work was supported by AI095125 and S10RR029121.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1206855109/-/DCSupplemental.
References
- 1.Jabra-Rizk M. Pathogenesis of polymicrobial biofilms. Open Mycol J. 2011;5:39–43. [Google Scholar]
- 2.Rowe SM, Miller S, Sorscher EJ. Cystic fibrosis. N Engl J Med. 2005;352:1992–2001. doi: 10.1056/NEJMra043184. [DOI] [PubMed] [Google Scholar]
- 3.Bauernfeind A, et al. Qualitative and quantitative microbiological analysis of sputa of 102 patients with cystic fibrosis. Infection. 1987;15:270–277. doi: 10.1007/BF01644137. [DOI] [PubMed] [Google Scholar]
- 4.Finnan S, Morrissey JP, O’Gara F, Boyd EF. Genome diversity of Pseudomonas aeruginosa isolates from cystic fibrosis patients and the hospital environment. J Clin Microbiol. 2004;42:5783–5792. doi: 10.1128/JCM.42.12.5783-5792.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh PK, et al. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 2000;407:762–764. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- 6.Lambert PA. Mechanisms of antibiotic resistance in Pseudomonas aeruginosa. J R Soc Med. 2002;95(Suppl 41):22–26. [PMC free article] [PubMed] [Google Scholar]
- 7.Latgé J-P. Aspergillus fumigatus and aspergillosis. Clin Microbiol Rev. 1999;12:310–350. doi: 10.1128/cmr.12.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amin R, Dupuis A, Aaron SD, Ratjen F. The effect of chronic infection with Aspergillus fumigatus on lung function and hospitalization in patients with cystic fibrosis. Chest. 2010;137:171–176. doi: 10.1378/chest.09-1103. [DOI] [PubMed] [Google Scholar]
- 9.Yonezawa M, et al. A new model of pulmonary superinfection with Aspergillus fumigatus and Pseudomonas aeruginosa in mice. J Infect Chemother. 2000;6:155–161. doi: 10.1007/s101560070015. [DOI] [PubMed] [Google Scholar]
- 10.Mowat E, et al. Pseudomonas aeruginosa and their small diffusible extracellular molecules inhibit Aspergillus fumigatus biofilm formation. FEMS Microbiol Lett. 2010;313:96–102. doi: 10.1111/j.1574-6968.2010.02130.x. [DOI] [PubMed] [Google Scholar]
- 11.Cornett DS, Reyzer ML, Chaurand P, Caprioli RM. MALDI imaging mass spectrometry: Molecular snapshots of biochemical systems. Nat Methods. 2007;4:828–833. doi: 10.1038/nmeth1094. [DOI] [PubMed] [Google Scholar]
- 12.Yang YL, Xu Y, Straight P, Dorrestein PC. Translating metabolic exchange with imaging mass spectrometry. Nat Chem Biol. 2009;5:885–887. doi: 10.1038/nchembio.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watrous J, et al. Mass spectral molecular networking of living microbial colonies. Proc Natl Acad Sci USA. 2012;109:E1743–E1752. doi: 10.1073/pnas.1203689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Price-Whelan A, Dietrich LEP, Newman DK. Rethinking “secondary” metabolism: physiological roles for phenazine antibiotics. Nat Chem Biol. 2006;2:71–78. doi: 10.1038/nchembio764. [DOI] [PubMed] [Google Scholar]
- 15.Pierson LS, 3rd, Pierson EA. Metabolism and function of phenazines in bacteria: Impacts on the behavior of bacteria in the environment and biotechnological processes. Appl Microbiol Biotechnol. 2010;86:1659–1670. doi: 10.1007/s00253-010-2509-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kerr JR, et al. Pseudomonas aeruginosa pyocyanin and 1-hydroxyphenazine inhibit fungal growth. J Clin Pathol. 1999;52:385–387. doi: 10.1136/jcp.52.5.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rahme LG, et al. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 18.Pain A, et al. Insight into the genome of Aspergillus fumigatus: Analysis of a 922-kb region encompassing the nitrate assimilation gene cluster. Fungal Genet Biol. 2004;41:443–453. doi: 10.1016/j.fgb.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 19.Ramos I, Dietrich LEP, Price-Whelan A, Newman DK. Phenazines affect biofilm formation by Pseudomonas aeruginosa in similar ways at various scales. Res Microbiol. 2010;161:187–191. doi: 10.1016/j.resmic.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Budzikiewicz H. Secondary metabolites from fluorescent pseudomonads. FEMS Microbiol Rev. 1993;10:209–228. doi: 10.1111/j.1574-6968.1993.tb05868.x. [DOI] [PubMed] [Google Scholar]
- 21.Frisvad JC, Rank C, Nielsen KF, Larsen TO. Metabolomics of Aspergillus fumigatus. Med Mycol. 2008;47(Suppl 1):S1–S4. doi: 10.1080/13693780802307720. [DOI] [PubMed] [Google Scholar]
- 22.Straight PD, Willey JM, Kolter R. Interactions between Streptomyces coelicolor and Bacillus subtilis: Role of surfactants in raising aerial structures. J Bacteriol. 2006;188:4918–4925. doi: 10.1128/JB.00162-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huse H, Whiteley M. 4-Quinolones: Smart phones of the microbial world. Chem Rev. 2011;111:152–159. doi: 10.1021/cr100063u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heeb S, et al. Quinolones: From antibiotics to autoinducers. FEMS Microbiol Rev. 2011;35:247–274. doi: 10.1111/j.1574-6976.2010.00247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soberón-Chávez G, Lépine F, Déziel E. Production of rhamnolipids by Pseudomonas aeruginosa. Appl Microbiol Biotechnol. 2005;68:718–725. doi: 10.1007/s00253-005-0150-3. [DOI] [PubMed] [Google Scholar]
- 26.Chin-A-Woeng TFC, Bloemberg GV, van der Bij AJ. Biocontrol by phenazine-1-carboxamide–producing Pseudomonas chlororaphis PCL 1391 of tomato root rot caused by Fusarium oxysporum f. sp. radicis-lycopersici. Mol Plant Microbe Interact. 1998;11:1069–1077. [Google Scholar]
- 27.Dietrich LEP, Price-Whelan A, Petersen A, Whiteley M, Newman DK. The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol Microbiol. 2006;61:1308–1321. doi: 10.1111/j.1365-2958.2006.05306.x. [DOI] [PubMed] [Google Scholar]
- 28.Liu W-T, et al. Imaging mass spectrometry of intraspecies metabolic exchange revealed the cannibalistic factors of Bacillus subtilis. Proc Natl Acad Sci USA. 2010;107:16286–16290. doi: 10.1073/pnas.1008368107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Y-L, et al. Connecting chemotypes and phenotypes of cultured marine microbial assemblages by imaging mass spectrometry. Angew Chem Int Ed Engl. 2011;50:5839–5842. doi: 10.1002/anie.201101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gonzalez DJ, et al. Microbial competition between Bacillus subtilis and Staphylococcus aureus monitored by imaging mass spectrometry. Microbiology. 2011;157:2485–2492. doi: 10.1099/mic.0.048736-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gibson J, Sood A, Hogan DA. Pseudomonas aeruginosa–Candida albicans interactions: Localization and fungal toxicity of a phenazine derivative. Appl Environ Microbiol. 2009;75:504–513. doi: 10.1128/AEM.01037-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cerniglia CE, Freeman JP, Mitchum RK. Glucuronide and sulfate conjugation in the fungal metabolism of aromatic hydrocarbons. Appl Environ Microbiol. 1982;43:1070–1075. doi: 10.1128/aem.43.5.1070-1075.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sack U, et al. Novel metabolites in phenanthrene and pyrene transformation by Aspergillus niger. Appl Environ Microbiol. 1997;63:2906–2909. doi: 10.1128/aem.63.7.2906-2909.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Capotorti G, Cesti P, Lombardi A, Guglielmetti G. Formation of sulfate conjugates metabolites in the degradation of phenanthrene, anthracene, pyrene and benzo[A]pyrene by the ascomycete Aspergillus terreus. Polycycl Aromat Comp. 2005;25:197–213. [Google Scholar]
- 35.Neuenhaus W, Roemer A, Budzikiewicz H, Korth H, Pulverer G. Bis(2-hydroxy-1-phenazinyl)methane: A compound of new structure from Pseudomonas aureofaciens. Z Naturforsch B. 1980;35B:385–388. [Google Scholar]
- 36.Denning GM, et al. Phenazine-1-carboxylic acid, a secondary metabolite of Pseudomonas aeruginosa, alters expression of immunomodulatory proteins by human airway epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2003;285:L584–L592. doi: 10.1152/ajplung.00086.2003. [DOI] [PubMed] [Google Scholar]
- 37.Fernandez RO, Pizarro RA. High-performance liquid chromatographic analysis of Pseudomonas aeruginosa phenazines. J Chromatogr A. 1997;771:99–104. [Google Scholar]
- 38.Hill JC, Johnson GT. Microbial transformation of phenazines by Aspergillus sclerotiorum. Mycologia. 1969;61:452–467. [PubMed] [Google Scholar]
- 39.Morales DK, et al. Antifungal mechanisms by which a novel Pseudomonas aeruginosa phenazine toxin kills Candida albicans in biofilms. Mol Microbiol. 2010;78:1379–1392. doi: 10.1111/j.1365-2958.2010.07414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Milstein O, et al. Metabolism of lignin related aromatic compounds by Aspergillus japonicas. Arch Microbiol. 1983;135:147–154. [Google Scholar]
- 41.Boonchan S, Britz ML, Stanley GA. Degradation and mineralization of high-molecular-weight polycyclic aromatic hydrocarbons by defined fungal-bacterial cocultures. Appl Environ Microbiol. 2000;66:1007–1019. doi: 10.1128/aem.66.3.1007-1019.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Renshaw JC, et al. Fungal siderophores: Structures, functions and applications. Mycol Res. 2002;106:1123–1142. [Google Scholar]
- 43.Evers ARD, Hancock RD, Martell AE, Motekaitis RJ. Metal ion recognition in ligands with negatively charged oxygen donor groups: Complexation of Fe(III), Ga(III), In(III), Al(III) and other highly charged metal ions. Inorg Chem. 1989;28:2189–2195. [Google Scholar]
- 44.Burrows WD. Aquatic aluminum: Chemistry, toxicology, and environmental prevalence. Crit Rev Environ Control. 1977;7:167–216. [Google Scholar]
- 45.Hu X, Boyer GL. Siderophore-mediated aluminum uptake by Bacillus megaterium ATCC 19213. Appl Environ Microbiol. 1996;62:4044–4048. doi: 10.1128/aem.62.11.4044-4048.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shuen SK, Buswell JA. Effect of lignin-derived phenols and their methylated derivatives on the growth of Lentinus spp. Lett Appl Microbiol. 1992;15:12–14. [Google Scholar]
- 47.Dietrich LEP, Teal TK, Price-Whelan A, Newman DK. Redox-active antibiotics control gene expression and community behavior in divergent bacteria. Science. 2008;321:1203–1206. doi: 10.1126/science.1160619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schalk IJ, Hannauer M, Braud A. New roles for bacterial siderophores in metal transport and tolerance. Environ Microbiol. 2011;13:2844–2854. doi: 10.1111/j.1462-2920.2011.02556.x. [DOI] [PubMed] [Google Scholar]
- 49.Wang Y, et al. Phenazine-1-carboxylic acid promotes bacterial biofilm development via ferrous iron acquisition. J Bacteriol. 2011;193:3606–3617. doi: 10.1128/JB.00396-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amara N, Krom BP, Kaufmann GF, Meijler MM. Macromolecular inhibition of quorum sensing: Enzymes, antibodies, and beyond. Chem Rev. 2011;111:195–208. doi: 10.1021/cr100101c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.