Abstract
Human centromeres are defined by megabases of homogenous alpha-satellite DNA arrays that are packaged into specialized chromatin marked by the centromeric histone variant, centromeric protein A (CENP-A). Although most human chromosomes have a single higher-order repeat (HOR) array of alpha satellites, several chromosomes have more than one HOR array. Homo sapiens chromosome 17 (HSA17) has two juxtaposed HOR arrays, D17Z1 and D17Z1-B. Only D17Z1 has been linked to CENP-A chromatin assembly. Here, we use human artificial chromosome assembly assays to show that both D17Z1 and D17Z1-B can support de novo centromere assembly independently. We extend these in vitro studies and demonstrate, using immunostaining and chromatin analyses, that in human cells the centromere can be assembled at D17Z1 or D17Z1-B. Intriguingly, some humans are functional heterozygotes, meaning that CENP-A is located at a different HOR array on the two HSA17 homologs. The site of CENP-A assembly on HSA17 is stable and is transmitted through meiosis, as evidenced by inheritance of CENP-A location through multigenerational families. Differences in histone modifications are not linked clearly with active and inactive D17Z1 and D17Z1-B arrays; however, we detect a correlation between the presence of variant repeat units of D17Z1 and CENP-A assembly at the opposite array, D17Z1-B. Our studies reveal the presence of centromeric epialleles on an endogenous human chromosome and suggest genomic complexities underlying the mechanisms that determine centromere identity in humans.
Keywords: kinetochore, dicentric, epigenetics, polymorphism, heterochromatin
The centromere is an essential locus required for chromosomal attachment to the spindle and chromosomal movement during cell division. Several lines of evidence support epigenetic mechanisms of eukaryotic centromere specification. First, DNA sequences are different among centromeres within the same and different organisms. On dicentric chromosomes that contain two regions of centromeric DNA, one centromere often is inactivated, suggesting that DNA sequence alone is insufficient for centromere function. An important epigenetic determinant of centromere identity is the centromeric histone, centromeric protein A (CENP-A)/centromeric histone 3 (CENH3), a histone H3 variant that is enriched within centromeric nucleosomes at endogenous centromeres (1–3) but is absent from inactive centromeres of dicentric chromosomes (4, 5). CENP-A/CENH3 distinguishes the centromere from the rest of the genome by contributing to uniquely organized chromatin that exists as alternating domains of CENP-A/CENH3 and H3-containing nucleosomes (6, 7). CENP-A/CENH3 is thought to establish epigenetic memory so that at each cell cycle, centromeric nucleosomes containing CENP-A/CENH3 are replenished, presumably by the CENP-A/CENH3 chaperone Holliday junction-recognition protein (HJURP) (8, 9). CENP-A chromatin serves as a platform for recruitment of other centromere proteins (10).
Endogenous human centromeres are composed of sequence alpha-satellite DNA, a 171-bp monomeric repeat that is present in thousands of copies on each chromosome. A genomic basis for centromere specification stems from the ability of alpha-satellite DNA to assemble de novo centromeres when introduced into cultured human cells (11, 12). At each centromere, alpha-satellite monomers are arranged tandemly. A defined number of monomers comprise a higher-order repeat (HOR) unit that then is repeated hundreds to thousands of times, producing highly homogenous arrays that are 97–100% identical. Most Homo sapiens chromosomes (HSA) are thought to have a single homogeneous alpha-satellite array. However, some chromosomes, such as HSA1, HSA5, HSA7, and HSA15, have two or more distinct arrays that are each defined by different HORs (13–15). On HSA5 and HSA7, the two alpha-satellite arrays are separated by up to a megabase. HSA17 also has two directly adjacent alpha-satellite arrays, D17Z1 and D17Z1-B. D17Z1-B is positioned toward the short arm (16, 17). D17Z1 is accepted as the functional centromere on HSA17, based on the demonstration that synthetic and cloned D17Z1 sequences efficiently seed de novo centromere assembly (11, 18).
However, the functional significance of D17Z1-B, the second HOR array on HSA17, is currently unknown. To better understand this second higher-order alpha-satellite array on HSA17, we tested for centromere function associated with D17Z1-B in various biological contexts. We show that D17Z1-B is able to support CENP-A assembly independently in vitro and in vivo and that, on a given HSA17, CENP-A assembly at D17Z1 or D17Z1-B is mutually exclusive. We also explored the stability and heritability of centromere assembly at D17Z1 or D17Z1-B by tracking the CENP-A position on HSA17 through multigenerational families.
Results
Two Distinctive Higher-Order Alpha-Satellite Arrays on HSA17.
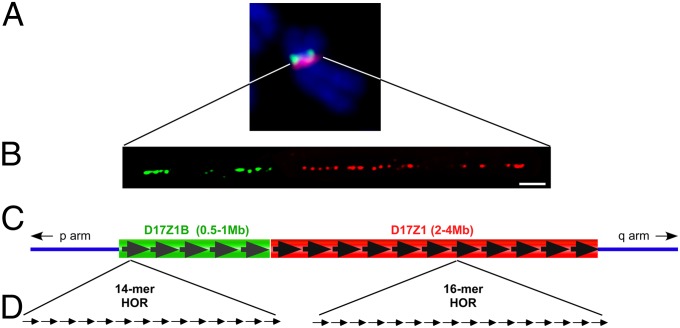
D17Z1 is the larger, extensively studied array on HSA17. It is comprised of a 16-monomer (16-mer) HOR unit, whereas D17Z1-B is composed of a 14-mer HOR (16, 19) (Fig. 1). D17Z1 and D17Z1-B share only 92% sequence identity (16) and can be distinguished by FISH, Southern blotting, and PCR under high-stringency conditions. FISH on metaphase chromosomes indicated that D17Z1 and D17Z1-B are arranged contiguously (Fig. 1). Because of the proximity of D17Z1 and D17Z1-B, we also used extended chromatin fibers to evaluate their long-range organization on individual HSA17s from several cell lines (Fig. 1, Fig. S1A, and Table S1). We confirmed that within the HSA17 centromere, D17Z1 and D17Z1-B are distinct and juxtaposed, with D17Z1-B oriented toward the short arm.
Fig. 1.
Genomic organization of HSA17 centromere. (A) HSA17 contains two HOR alpha-satellite arrays, D17Z1 (red) and D17Z1-B (green), that can be identified by high-stringency FISH. D17Z1-B is located on the short (p) arm side of the centromere. (B) Stretched chromatin fibers hybridized with D17Z1 (red) or D17Z1-B (green) FISH probes confirm that the two arrays are juxtaposed. (C and D) D17Z1 primarily contains reiterated HOR units composed of 16 individual 171-bp alpha-satellite monomers that are tandemly arranged. Overall, the array is estimated to cover 2–4 Mb. The HOR of D17Z1-B contains 14 monomers, and the entire array is estimated to be one-fourth the size of D17Z1. (Scale bar: 10 microns.)
D17Z1 and D17Z1-B Support de novo Centromere Assembly Independently.
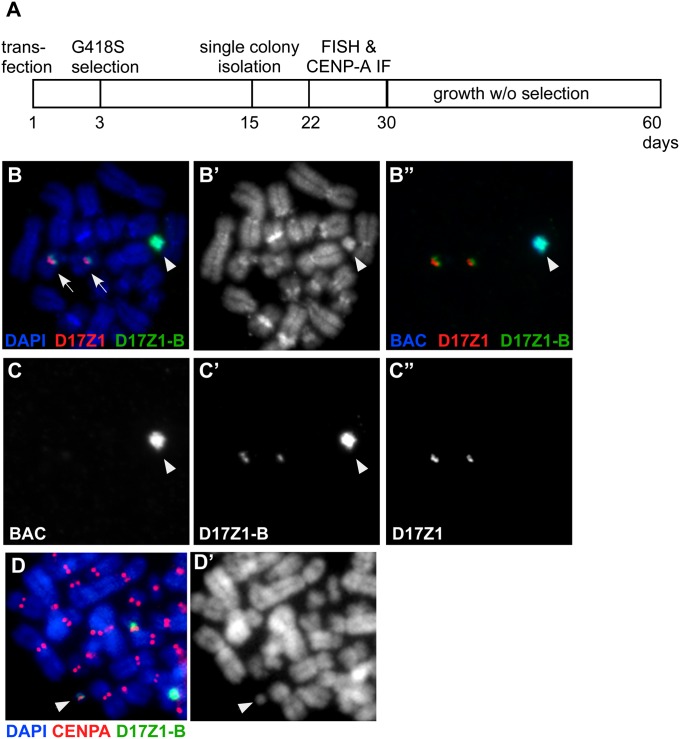
When introduced into human cells, cloned or synthetic arrays of D17Z1 are efficient substrates for de novo centromere assembly (11, 20). However, it was not known if D17Z1-B also could be an efficient centromeric substrate. We used human artificial chromosome (HAC) assays to test the ability of D17Z1-B to form de novo centromeres independently. We identified two BACs, RP11-285M22 (158 kb) and RP11-458D13 (149 kb), containing 20 kb of D17Z1-B and ∼130 kb of monomeric alpha-satellite DNA (16). RP11-285M22 was subcloned further to produce a 60-kb construct, RP11-285M22PmlI, containing ∼19 kb of D17Z1-B (eight HOR units) and 31 kb of monomeric alpha-satellite. The three BACs were transfected independently into HT1080 cells. RP11-354P13, a BAC containing D17Z1, and RP11-971O21SwaI, containing 55 kb of DXZ1 (HSAX alpha-satellite), were used as controls for HAC formation (Table S2) (21). D17Z1 and DXZ1 constructs formed HACs at previously reported frequencies (i.e., >20% of cell lines contained HACs) (Table S2) (11, 21). Of the D17Z1-B transfected lines, 58 were analyzed by FISH for the presence of extrachromosomal structures that were DAPI positive and hybridized with the BAC backbone and D17Z1-B probes but not with D17Z1. A HAC was observed in 28 of the 58 lines (48%) (Fig. 2 and Tables S2 and S3). In 18 of the 28 lines (30%), a HAC was present in ≥20% of the spreads, suggesting these were the most stable de novo chromosomes. Notably, HAC formation was most efficient using the construct containing the least amount of monomeric alpha-satellite. Several lines also were analyzed by CENP-A immunostaining-FISH (Fig. 2); CENP-A colocalized with the alpha-satellite probe on the HAC in >90% of spreads. HACs also were stable for ≥30 d when grown in the absence of selection. These results indicate that D17Z1-B independently can form HACs that contain functional centromeres, recruit centromere proteins, and segregate faithfully in mitosis.
Fig. 2.
HAC assays confirm that D17Z1-B can form de novo centromeres. (A) Schematic of HAC assay. BAC constructs containing large D17Z1-B (or D17Z1 control) sequence inserts and a neomycin-selectable marker were introduced into proliferating human HT1080 cells. After 2–3 d, cells were grown in medium containing G418 sulfate (G418S). By day 15, drug-resistant colonies were isolated and expanded to evaluate centromere assembly. (B) FISH with probes to BAC vector, D17Z1, and D17Z1-B shows that the HAC (arrowhead) hybridizes only with D17Z1-B and BAC probes. Endogenous HSA17s are denoted by arrows. B shows a merged image; B′ shows DAPI staining only; B′′ shows BAC (blue), D17Z1 (red), and D17Z1-B (green). (C) Single-channel images of B showing that only the HAC contains BAC and D17Z1-B sequences. C shows BAC; C′ shows D17Z1-B; C′′ shows D17Z1. (D) CENP-A immunostaining and FISH showing that the D17Z1-B–derived HAC (green) assembles a kinetochore, as denoted by CENP-A (red). D′ is DAPI staining.
CENP-A Assembly on HSA17 Reveals Functional Epialleles.
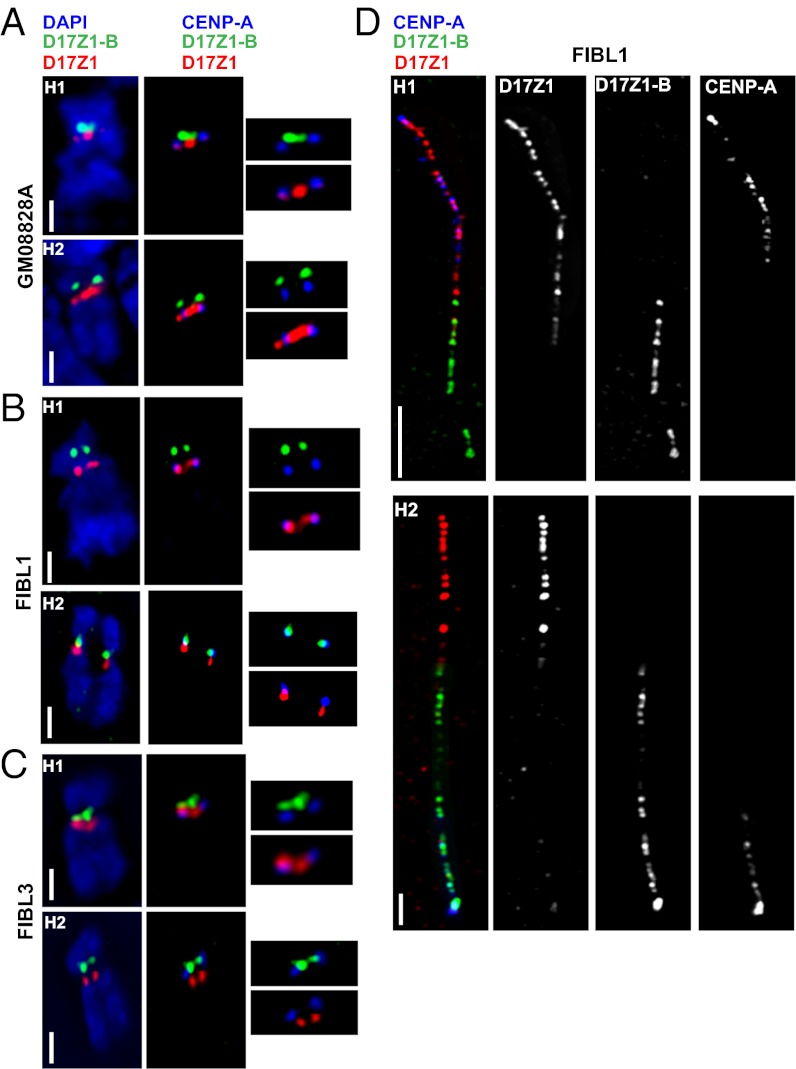
The HAC experiments indicated that in vitro both D17Z1 and D17Z1-B independently support centromere assembly. However, it was unclear if these functional properties exist in vivo, so we determined the location of CENP-A on HSA17 in 31 cell lines (Table S1). The site of CENP-A was assigned using CENP-A immunostaining-FISH (Fig. 3). To ensure that D17Z1 and D17Z1-B could be distinguished easily on condensed metaphase chromosomes, proliferating cell cultures were treated with ethidium bromide or were centrifuged at a lower cell concentration to obtain extended metaphases and mechanically stretched chromosomes. After CENP-A immunostaining and cohybridizing with FISH probes specific for D17Z1 and D17Z1-B, metaphase images were assessed visually and analyzed using a custom chromosome analysis script (22) that plots fluorescence intensities along the chromosome. Overlap of the CENP-A curve with the D17Z1 or D17Z1-B curves was recorded (Fig. S1B). Most HSA17s in the cell lines studied showed localization of CENP-A at D17Z1 (Fig. 3A). Surprisingly, we observed that ∼25% of individuals or cell lines were functionally heterozygous for the CENP-A location on HSA17 (Fig. 3 B and C and Table S1). [A functional heterozygote was defined as a diploid line in which CENP-A was located at D17Z1 (CENP-AZ1) on one HSA17 homolog and at D17Z1-B (CENP-AZ1-B) on the other HSA17.] The remaining 75% of individuals studied were functionally homozygous for CENP-AZ1. We have not yet identified an individual or cell line in which CENP-A was located at D17Z1-B on both HSA17 homologs.
Fig. 3.
Identification of centromeric epialleles on HSA17. (A) FISH for D17Z1 (red) and D17Z1-B (green) was combined with CENP-A immunostaining to determine the CENP-A location in the diploid line GM08828. CENP-A was located at D17Z1 on both homologs (H1 and H2). (B and C) CENP-A immunostaining and D17Z1/D17Z1-B FISH were performed on line FIBL1 (B) and line FIBL3 (C). In both lines, CENP-A was located at D17Z1 (red) on H1 but at D17Z1-B (green) on H2. (Scale bars: 5 microns in A–C.) (D) Extended chromatin fibers from line FIBL1 confirmed the CENP-A location (blue) at D17Z1 (red) on H1 and at D17Z1-B (green) on H2. (Scale bars: 15 microns for H1; 5 microns for H2).
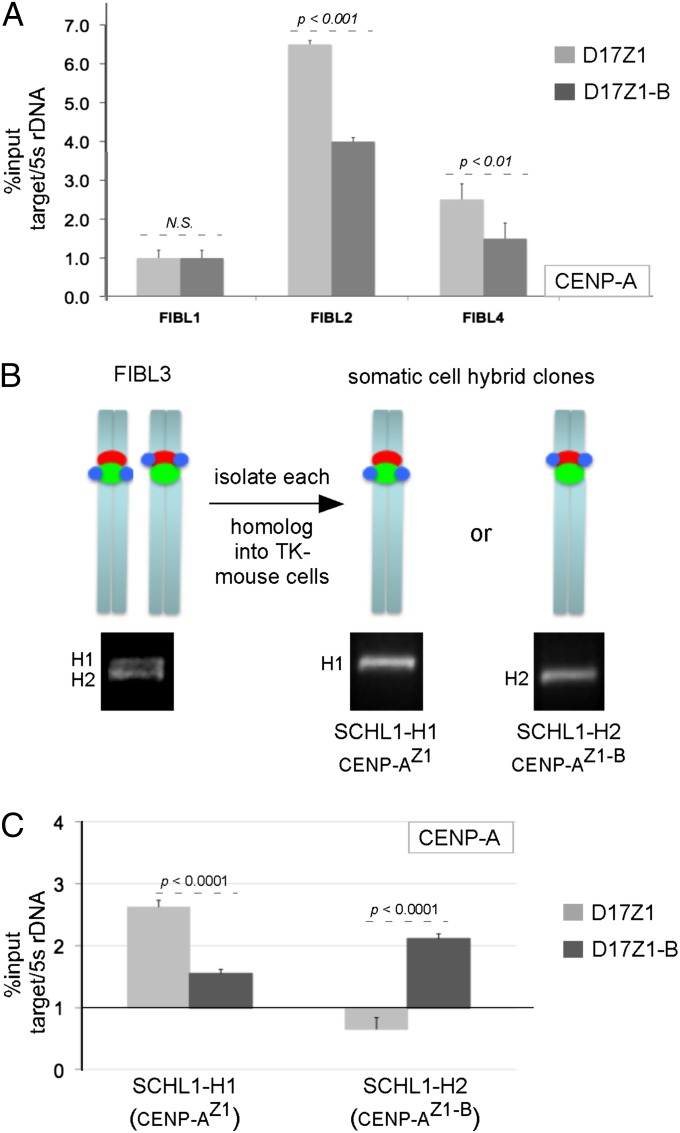
To support the cytological studies, we also used ChIP with CENP-A antibodies and primers specific to D17Z1 and D17Z1-B (Fig. 4 and Fig. S2). Five diploid cell lines were studied (FIBL1, FIBL2, FIBL4, CEPH1345-11, and CEPH1345-13). Of these, CENP-A was enriched at D17Z1 in the two lines FIBL2 and CEPH1345-13 (Fig. 4A and Fig. S2B). However, ChIP showed nearly equal enrichment of CENP-A on both arrays in lines FIBL1 and CEPH1345-11 (Fig. 4A and Fig. S2B). In FIBL4, CENP-A appeared to be more enriched on D17Z1 than on D17Z1-B, but immunostaining revealed this enrichment was misleading because the line is hypertetraploid. It contains three copies of HSA17 that have CENP-A at D17Z1 and one copy of HSA17 that has CENP-A at D17Z1-B. Analysis of FIBL3, the diploid parental line of FIBL4 before it became tetraploid, showed that this line originally existed with CENP-A at D17Z1 on one HSA17 and at D17Z1-B on the other homolog (Fig. 3C and Table S1). In several instances the ChIP results would have been confusing to interpret in the absence of metaphase immunostaining. However, using both methods, we were able to determine that enrichment of CENP-A at D17Z1 and D17Z1-B in diploid lines represented the presence of distinct functional alleles on the two HSA17 homologs: CENP-AZ1 on one homolog and CENP-AZ1-B on the other. These studies reveal that CENP-A assembly in normal individuals can occur naturally at either D17Z1 or D17Z1-B on HSA17. Collectively, the HAC and in vivo CENP-A assignment experiments support the existence of functional centromeric polymorphisms, or epialleles, at an endogenous human centromere.
Fig. 4.
Confirmation of functional heterozygotes for CENP-A location. (A) ChIP analysis of diploid lines FIBL1, FIBL2, and FIBL4. In FIBL1, CENP-A was enriched at both D17Z1 and D17Z1-B, indicating functional heterozygosity. CENP-A was enriched at D17Z1 in FIBL2, suggesting functional homozygosity. In FIBL4, CENP-A appeared more enriched on D17Z1, but immunostaining of FIBL4 revealed tetraploidy for HSA17. Immunostaining of FIBL3, the diploid version of FIBL4, showed one HSA17 with CENP-A at D17Z1 and the other with CENP-A at D17Z1-B (Fig. 3C). (B) Homologs (H) of FIBL3 were separated into mouse–human somatic cell hybrids. H1 and H2 were molecularly distinguished from one another using PCR for a polymorphic 17q locus. (C) CENP-A location was confirmed by ChIP on each HSA17 homolog. CENP-A was maintained stably at D17Z1 on H1 and at D17Z1-B on H2. Data represent three independent experiments. CENP-A enrichment at the target site is reported as percent of input shown relative to the CENP-A–negative site mouse 5s rDNA. Error bars represent SEM.
CENP-A Assembly at D17Z1 or D17Z1-B Is Stably Inherited.
The observation that some cell lines or individuals showed opposing patterns of CENP-A location on HSA17 in the same nucleus raised questions regarding the stability and inheritance of CENP-A location and centromere function on HSA17. Although several of these cell lines were genetically unrelated (Table S1), many represented individuals from Centre du Etude Polymorphisme Humain (CEPH) families spanning three generations. These individuals were included to evaluate inheritance of CENP-A location on HSA17 through multiple meioses. We considered the possibility that CENP-A might shift between D17Z1 and D17Z1-B on one or both HSA17 homologs, particularly in functionally heterozygous individuals. To test this notion, we focused on CEPH family 1345 (CEPH1345) that contained functionally heterozygous individuals in each generation (Fig. S2A). The CENP-AZ1-B epiallele was passed from the paternal grandmother to her son and at least two grandchildren, thus suggesting that CENP-A epialleles on HSA17 are stably inherited through meiosis. However, these cell lines are diploid, and it was formally possible that in these lines CENP-A location alternated on both HSA17 homologs. To address more precisely the stability of CENP-A location in functionally heterozygous lines, we separated the HSA17 homologs from lines FIBL1 and FIBL3. The diploid lines were fused with mouse cells to create somatic cell hybrid lines containing only a single HSA17. Human cell lines were fused to a mouse cell line that lacks thymidine kinase (tk). When grown on hypoxanthine-aminopterin-thymidine (HAT) medium, the human TK gene located on HSA17 complements the mouse mutation. Genotyping of the diploid cell lines for a polymorphic marker (Table S4) allowed us to verify that each HSA17 homolog had been transferred to distinct somatic cell hybrid lines (Fig. 4B). Mouse Cenp-A immunostaining and ChIP confirmed that each HSA17 homolog maintained the location of Cenp-A (D17Z1 vs. D17Z1-B) originally identified in the parental diploid line. Continued growth of cells for more than 20 cell divisions and repeated immunostaining and FISH confirmed that the position of Cenp-A did not move over time. We conclude that assembly of CENP-A can occur at either D17Z1 or D17Z1-B and, once established, is stable over time in both human cells and in a nonhuman genetic background.
Chromatin Organization of CENP-A–Associated and –Depleted HOR Arrays on HSA17.
Histone H3 methylated at lysine 4 (K4) and lysine 36 (K36) generally is present within the same chromatin domain as CENP-A (7, 23). Chromatin containing H3K9me3 usually is located outside the CENP-A chromatin core (7). We assayed histone modifications at D17Z1 and D17Z1-B when each was the active or inactive centromere. HSA17 homologs that had been transferred to mouse–human somatic cell hybrids were studied by ChIP for two euchromatic (H3K4me2 and H3K36me2) and two heterochromatic (H3K9me3 and H3K27me3) modifications (Figs. S3 and S4). All somatic cell hybrids have the same rodent background; lines differ only in the HSA17 each contains. Mouse minor and major satellite DNAs were used as controls for Cenp-A and histone modifications (Fig. S3) (6, 24). Neither D17Z1 nor D17Z1-B was depleted completely or enriched for histone modifications in either the active or inactive state, but some modest differences were observed. When D17Z1 was the site of centromere assembly (i.e., lines SCHL1-H1, SCHL3-H1, Table S1), it had slightly more H3K4me2 and H3K36me2 (Fig. S4 A and B). However, when D17Z1-B was the functional centromere, H3K4me2 and H3K36me2 appeared equivalently enriched at D17Z1 and D17Z1-B (Fig. S4 A and B). H3K9me3 and H3K27me3 also were present on both arrays (Fig. S4 C and D). H3K9me3 appeared more enriched on active and inactive D17Z1 (Fig. S4C). These results indicate that, with the exception of CENP-A chromatin, obvious differences in chromatin organization are not observed between D17Z1 and D17Z1-B.
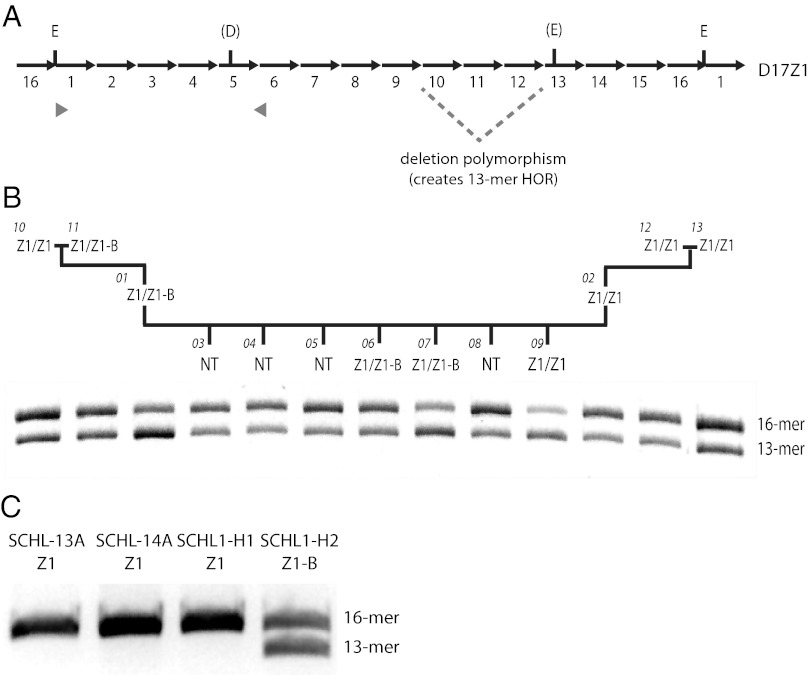
D17Z1 Polymorphisms and CENP-A Location on HSA17.
D17Z1 contains several well-characterized polymorphisms, including sequence variants and total number of monomers within the HOR (25). The prevalent repeat unit is a 2.7-kb HOR, defined by EcoRI restriction sites and consisting of 16-mers (Fig. 5A). HOR size polymorphisms range from 11-mer to 15-mer repeat units. Two major D17Z1 haplotypes are present in humans. Haplotype I, defined primarily as 16-mers with some 15- and 14-mers, is present in ∼65% of HSA17s; haplotype II, containing additional 13-mers, is present in ∼35% of HSA17s (26). Haplotypes I and II are distinguished molecularly using PCR-restriction digestion, because the 13-mer variant is in linkage disequilibrium with the restriction site polymorphism for DraI. We noted that the distributions of haplotype I and haplotype II were similar to the frequencies of CENP-A at D17Z1 (∼70%) vs. D17Z1-B (∼30%) in the individuals in our study (Table S1), so we tested if specific D17Z1 haplotypes correlate with CENP-A location on HSA17. An 850-bp portion of D17Z1 was PCR amplified and digested with DraI. The presence of the polymorphism/restriction site reduces the 850-bp product to 750 bp, and band intensities indicate relative amounts of each repeat variant (27). In the CEPH1345 family, the genotypes of all individuals were 16/13 heterozygotes, that is, their HSA17s have both 16-mer and 13-mer forms of the HOR unit. Individuals in which CENP-A was located at D17Z1 on both HSA17s (Z1/Z1) generally exhibited more intense 850-bp bands (Fig. 5B), whereas Z1/Z1-B individuals tended to have a more intense lower 750-bp band. These results suggested that in functional homozygotes, CENP-A is located at the D17Z1 arrays with a greater proportion of 16-mers. We infer from the genotypes of Z1/Z1-B functional heterozygotes that the location of CENP-A at D17Z1-B may be linked to the smaller D17Z1 HOR variant. However, all the genotyped CEPH individuals were diploid for HSA17, so it was not possible to discriminate between amounts and types of variants associated with a particular HSA17. Therefore, D17Z1 haplotypes of HSA17s that had been separated into somatic cell hybrids were determined (Table S1). Three different HSA17s in which CENP-A was located at D17Z1 showed a predominant genotype that included only the 16-mer HOR. However, the HSA17 that had CENP-A at D17Z1-B exhibited both 16-mer and 13-mer forms of the HOR. Taken together, the polymorphism data suggest that the presence of sequence and size variants of D17Z1 may influence where CENP-A is assembled on HSA17.
Fig. 5.
CENP-A location on HSA17 and D17Z1 HOR polymorphisms. (A) The D17Z1 HOR contains 16 monomeric repeats (16-mer) and is defined by EcoRI restriction sites (E). A 3-monomer deletion generates a 13-mer from the 16-mer. Some HORs also contain sequence polymorphisms that create DraI (D) and additional EcoRI restriction sites. Part of the HOR (gray arrowheads) was amplified by PCR and digested with DraI. If the DraI site is present, the 850-bp PCR product is reduced to 750-bp. Each HOR variant can be correlated with the 850-bp (16-mer) or 750-bp (13-mer) bands, and the intensity of bands indicates relative amounts of each repeat form. (B) In the CEPH1345 family, 16-mer and 13-mer D17Z1 variants were present in each individual. Functional homozygotes (Z1/Z1, with CENP-A located at D17Z1 on both HSA17s), such as individual 13, had darker upper bands, indicating a greater proportion of 16-mers. Functional heterozygotes (Z1/Z1-B), such as individual 07, typically had darker lower bands or equivalently dark upper and lower bands. NT, not tested. Because CEPH lines were diploid, proportions of each variant could not be assigned to specific HSA17s. Thus, single HSA17s in somatic cell hybrids were genotyped. (C) In SCHL-13A, SCHL-14A, and SCHL1-H1 that have CENP-A at D17Z1, only the 16-mer was detected. In SCHL1-H2, which shows CENP-A at D17Z1-B, the 13-mer and 16-mer were equally present.
Discussion
Our studies reveal the existence of functional epigenetic polymorphisms, or epialleles, associated with centromere function on HSA17 and suggest that some endogenous human chromosomes are arranged as structurally dicentric chromosomes. Epialleles are influenced by environmental, stochastic, or genetic factors that affect the plasticity by which a genotype is observed. Metastable epialleles can affect the expression of developmental genes, causing cell-to-cell variation in gene expression in a specific tissue and causing visible phenotypic variations, such as mouse coat color or plant petal color (28, 29). Obligatory epialleles arise as a consequence of DNA polymorphisms that affect epigenetic modifications, whereas nonobligatory, or pure, epialleles occur independently of genetic variation and are affected more profoundly by environment or other external signals. In the fission yeast Schizosaccharomyces pombe, centromeric epialleles were described on minichromosomes containing central core and outer repeat sequences. Minichromosomes lacking CENP-A switched functional states in the presence of heterochromatin (30). It is not known if CENP-A assembly at D17Z1 and D17Z1-B is controlled by deterministic or stochastic factors. Transgenerational inheritance of CENP-A position strengthens the argument that functional epialleles exist at the centromere of HSA17, and the Mendelian inheritance of centromere function suggests deterministic factors. Without sequence information for individual HOR arrays, it will be difficult to determine the mechanistic nature of the epialleles.
Dicentric chromosomes typically are considered abnormal chromosomes associated with cancers and birth defects, but HSA17 is a clear example of a normal human chromosome that is structurally arranged as a dicentric chromosome in an entire species and behaves as a functionally monocentric chromosome. Other human chromosomes, such as HSA5 and HSA7, also have two HOR arrays. However, unlike HSA17 in which either HOR array can support centromere formation, only one HOR array on HSA5 and HSA7 is always the site of centromere assembly (14). The existence on the same chromosome of two genomically similar but functionally distinct alpha-satellite arrays, either of which can be the site of centromere assembly, raises the question of what determines the location of CENP-A on HSA17. Total array size may influence CENP-A assembly, so that larger arrays recruit CENP-A and other assembly factors more efficiently. This hypothesis may be supported by our observations that CENP-A was located at what visually appeared to be a much larger D17Z1 array in SCHL1-H1 and SCHL3-H1 as compared with the D17Z1-B on the same chromosome (Figs. 1B and 3D). Conversely, on the HSA17 in SCHL1-H2 in which CENP-A was located at D17Z1-B, the D17Z1 and D17Z1-B arrays visually appeared equivalent in size (FIBL1 H2) (Fig. 3C). If D17Z1-B is equivalent to or larger in size than D17Z1, CENP-A chromatin may be more likely to be assembled at this array. At human centromeres, unordered monomeric alpha-satellite DNA is located between the HOR alpha-satellite array and chromosome arms. The amount of monomeric alpha-satellite separating D17Z1 from the long arm and D17Z1-B from the short arm also may influence where CENP-A is located on HSA17.
Chromatin environment is an important factor in centromere assembly and function. Previous studies using synthetic HACs have shown that heterochromatin, and to a lesser extent euchromatin, is incompatible with CENP-A assembly and maintenance of centromere function (31, 32). These studies suggest that inactive centromeres, or HOR arrays that do not associate with CENP-A, would be much more heterochromatic. More recent findings indicate that there can be equal enrichment of heterochromatic marks at active and inactive alpha-satellite arrays on HSA5 and HSA7 in diploid primary human cells (14). Thus, it may be necessary to consider both total array size and the proportion of each array that contributes to heterochromatin and centromeric chromatin. CENP-A is assembled on a portion of any alpha-satellite array, and the rest of the array is assembled into heterochromatin (33). If D17Z1 is larger than D17Z1-B, even when it is the site of CENP-A assembly, it may appear more enriched for H3K9me3 even though only a portion of the array is assembled into heterochromatin. It will be necessary to develop methods to combine long-range optical mapping of protein binding and accurate determination of the size of the alpha-satellite array to define more precisely the relationship of histone modifications to centromere function and suppression at this endogenous centromere.
D17Z1 and D17Z1-B are 92% identical by sequence. Although D17Z1-B exists only as a 14-mer HOR (16), D17Z1 HORs often are polymorphic in size, ranging from 16-mers to as few as 11-mers (26, 34). Such variable repeat unit lengths are the products of the homologous unequal crossing-over that contributes to homogenization of the long arrays of tandem repeats (26). The results of D17Z1 haplotyping from the CEPH family and the individual HSA17s isolated into somatic cell hybrids suggest that CENP-A assembly at D17Z1-B correlates with the presence of D17Z1 haplotype II, i.e., the presence of 13-mer HORs. This finding implies that D17Z1 arrays containing the shorter HOR may be less favorable for CENP-A assembly. However, the number of HOR types varies, so some haplotype I individuals may have more 13-mers than 16-mers, but other haplotype I individuals may have more 16-mers and only a few 13-mers. Our studies in the multigenerational family emphasize this variability. Because the cell lines were diploid, the proportion of each type of HOR remains undetermined. Future experiments will require analyzing individual HSA17s that have been isolated into somatic cell hybrids to define the proportion of 16-mers to 13-mers.
Why might an array of D17Z1 with more 13-mers be less favorable for CENP-A assembly? In individuals exhibiting haplotype II (13-mers), a fraction of D17Z1 HORs are smaller than D17Z1-B HORs (14-mers). These differences could affect the number and location of CENP-B boxes, the binding site for the centromeric protein CENP-B. The alpha satellite-binding protein CENP-B is important for de novo CENP-A assembly and phasing of centromeric nucleosomes (32, 35). The 16-mer HOR unit of D17Z1 has six CENP-B boxes, and the 14-mer HOR of D17Z1-B has only three CENP-B boxes. Thus, CENP-B may be important for positioning centromeric nucleosomes, so that D17Z1 16-mer arrays with greater numbers of CENP-B boxes may be more amenable for CENP-A assembly. An array of D17Z1 that contains more 13-mers than 16-mers naturally would have fewer CENP-B boxes. Therefore the 14-mer of D17Z1-B, with three or four CENP-B boxes per HOR, may be a better substrate for centromeric chromatin assembly. Our ongoing studies to sequence long arrays of D17Z1 and D17Z1-B should allow us to identify organizational properties of the two alpha-satellite arrays that impact centromere assembly on HSA17.
From these studies, we conclude that centromere identity on HSA17 involves synergy between genomic and epigenetic processes. This work also establishes the existence in the normal human karyotype of endogenous chromosomes that are structurally dicentric. This finding raises questions about centromere inactivation/repression and long-term stability of universal dicentrics. HSA17 is particularly interesting in this regard, because it is one of the most unstable and rearranged chromosomes in cancers. The establishment of HSA17 as a model for dicentric behavior will allow further investigation into how DNA sequence and epigenetic processes impact centromere function and repression in the context of a normal chromosome.
Materials and Methods
Native Chromatin Isolation, ChIP, and Quantitative PCR.
Native chromatin was incubated with protein A/G beads preconjugated to commercial human CENP-A antibodies (ab13939; Abcam) or custom mouse Cenp-A antibodies (AP601) using the MAGnify ChIP kit (Invitrogen). Samples were amplified using the GenomePlex Complete Whole Genome Amplification Kit (Sigma). Quantitative PCR (qPCR) was performed using EXPRESS SYBR GreenER qPCR Universal Supermix and Mx3000P Q-PCR System (Stratagene) with custom primers for D17Z1, D17Z1-B, and 5s rDNA (control) (Table S4) (32). ChIPs were done at least three times, and PCRs were performed in triplicate. Analysis and quantification were performed using MXPro software (Stratagene). For all ChIPs, relative enrichment was calculated using the Percent Input Method [100*2^(Adjusted input − Ct (IP)]. Significant enrichment was determined using a Student’s t test (GraphPad, http://www.graphpad.com/quickcalcs/).
HSA17 and D17Z1 PCR-Restriction Digestion Genotyping.
Individual HSA17s were distinguished by PCR analysis of the polymorphic SLC6A4 locus located on 17q (Table S4). Genomic DNA was amplified according to previous methods (36). PCR of the DraI polymorphic region of D17Z1 was performed on human genomic DNA (gDNA) or somatic cell hybrid gDNA using primers 17α1 and 17α2 (Table S4) exactly as described (27). Ethanol-precipitated PCR product was digested using 80 U of DraI (R0129; New England BioLabs).
Supplementary Material
Acknowledgments
We thank Hunt Willard for DNA probes and human and mouse lines and Nakano Megumi and Hiroshi Masumoto (Kazusa Reseach Institute, Japan) for advice on minor and major satellite qPCR. This work was supported by National Institutes of Health Grant GM069514 (to B.A.S.), March of Dimes Foundation Grants 6-FY06-377 and 6-FY10-294 (to B.A.S.), and the Duke School of Medicine Dean’s Office.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1203126109/-/DCSupplemental.
References
- 1.Yoda K, et al. Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proc Natl Acad Sci USA. 2000;97:7266–7271. doi: 10.1073/pnas.130189697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sullivan KF, Hechenberger M, Masri K. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J Cell Biol. 1994;127:581–592. doi: 10.1083/jcb.127.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palmer DK, O’Day K, Margolis RL. Biochemical analysis of CENP-A, a centromeric protein with histone-like properties. Prog Clin Biol Res. 1989;318:61–72. [PubMed] [Google Scholar]
- 4.Stimpson KM, et al. Telomere disruption results in non-random formation of de novo dicentric chromosomes involving acrocentric human chromosomes. PLoS Genet. 2010;6:e1001061. doi: 10.1371/journal.pgen.1001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Warburton PE, et al. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr Biol. 1997;7:901–904. doi: 10.1016/s0960-9822(06)00382-4. [DOI] [PubMed] [Google Scholar]
- 6.Greaves IK, Rangasamy D, Ridgway P, Tremethick DJ. H2A.Z contributes to the unique 3D structure of the centromere. Proc Natl Acad Sci USA. 2007;104:525–530. doi: 10.1073/pnas.0607870104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sullivan BA, Karpen GH. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol. 2004;11:1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunleavy EM, et al. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- 9.Foltz DR, et al. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perpelescu M, Fukagawa T. The ABCs of CENPs. Chromosoma. 2011;120:425–446. doi: 10.1007/s00412-011-0330-0. [DOI] [PubMed] [Google Scholar]
- 11.Grimes BR, Rhoades AA, Willard HF. Alpha-satellite DNA and vector composition influence rates of human artificial chromosome formation. Mol Ther. 2002;5:798–805. doi: 10.1006/mthe.2002.0612. [DOI] [PubMed] [Google Scholar]
- 12.Ikeno M, et al. Construction of YAC-based mammalian artificial chromosomes. Nat Biotechnol. 1998;16:431–439. doi: 10.1038/nbt0598-431. [DOI] [PubMed] [Google Scholar]
- 13.Choo KH, Earle E, Vissel B, Filby RG. Identification of two distinct subfamilies of alpha satellite DNA that are highly specific for human chromosome 15. Genomics. 1990;7:143–151. doi: 10.1016/0888-7543(90)90534-2. [DOI] [PubMed] [Google Scholar]
- 14.Slee RB, et al. Cancer-associated alteration of pericentromeric heterochromatin may contribute to chromosome instability. Oncogene. 2012;31:3244–3253. doi: 10.1038/onc.2011.502. [DOI] [PubMed] [Google Scholar]
- 15.Wevrick R, Willard HF. Physical map of the centromeric region of human chromosome 7: Relationship between two distinct alpha satellite arrays. Nucleic Acids Res. 1991;19:2295–2301. doi: 10.1093/nar/19.9.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rudd MK, Willard HF. Analysis of the centromeric regions of the human genome assembly. Trends Genet. 2004;20:529–533. doi: 10.1016/j.tig.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 17.Waye JS, Willard HF. Structure, organization, and sequence of alpha satellite DNA from human chromosome 17: Evidence for evolution by unequal crossing-over and an ancestral pentamer repeat shared with the human X chromosome. Mol Cell Biol. 1986;6:3156–3165. doi: 10.1128/mcb.6.9.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrington JJ, Van Bokkelen G, Mays RW, Gustashaw K, Willard HF. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat Genet. 1997;15:345–355. doi: 10.1038/ng0497-345. [DOI] [PubMed] [Google Scholar]
- 19.Warburton PE, Willard HF. Genomic analysis of sequence variation in tandemly repeated DNA. Evidence for localized homogeneous sequence domains within arrays of alpha-satellite DNA. J Mol Biol. 1990;216:3–16. doi: 10.1016/s0022-2836(05)80056-7. [DOI] [PubMed] [Google Scholar]
- 20.Grimes BR, Babcock J, Rudd MK, Chadwick B, Willard HF. Assembly and characterization of heterochromatin and euchromatin on human artificial chromosomes. Genome Biol. 2004;5:R89. doi: 10.1186/gb-2004-5-11-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rudd MK, Mays RW, Schwartz S, Willard HF. Human artificial chromosomes with alpha satellite-based de novo centromeres show increased frequency of nondisjunction and anaphase lag. Mol Cell Biol. 2003;23:7689–7697. doi: 10.1128/MCB.23.21.7689-7697.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blower MD, Sullivan BA, Karpen GH. Conserved organization of centromeric chromatin in flies and humans. Dev Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bergmann JH, et al. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 2011;30:328–340. doi: 10.1038/emboj.2010.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferri F, Bouzinba-Segard H, Velasco G, Hubé F, Francastel C. Non-coding murine centromeric transcripts associate with and potentiate Aurora B kinase. Nucleic Acids Res. 2009;37:5071–5080. doi: 10.1093/nar/gkp529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willard HF, Greig GM, Powers VE, Waye JS. Molecular organization and haplotype analysis of centromeric DNA from human chromosome 17: Implications for linkage in neurofibromatosis. Genomics. 1987;1:368–373. doi: 10.1016/0888-7543(87)90041-3. [DOI] [PubMed] [Google Scholar]
- 26.Warburton PE, Willard HF. Interhomologue sequence variation of alpha satellite DNA from human chromosome 17: Evidence for concerted evolution along haplotypic lineages. J Mol Evol. 1995;41:1006–1015. doi: 10.1007/BF00173182. [DOI] [PubMed] [Google Scholar]
- 27.Warburton PE, Greig GM, Haaf T, Willard HF. PCR amplification of chromosome-specific alpha satellite DNA: Definition of centromeric STS markers and polymorphic analysis. Genomics. 1991;11:324–333. doi: 10.1016/0888-7543(91)90139-6. [DOI] [PubMed] [Google Scholar]
- 28.Finer S, Holland ML, Nanty L, Rakyan VK. The hunt for the epiallele. Environ Mol Mutagen. 2011;52:1–11. doi: 10.1002/em.20590. [DOI] [PubMed] [Google Scholar]
- 29.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 30.Folco HD, Pidoux AL, Urano T, Allshire RC. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science. 2008;319:94–97. doi: 10.1126/science.1150944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakano M, et al. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev Cell. 2008;14:507–522. doi: 10.1016/j.devcel.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okada T, et al. CENP-B controls centromere formation depending on the chromatin context. Cell. 2007;131:1287–1300. doi: 10.1016/j.cell.2007.10.045. [DOI] [PubMed] [Google Scholar]
- 33.Sullivan LL, Boivin CD, Mravinac B, Song IY, Sullivan BA. Genomic size of CENP-A domain is proportional to total alpha satellite array size at human centromeres and expands in cancer cells. Chromosome Res. 2011;19:457–470. doi: 10.1007/s10577-011-9208-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warburton PE, Waye JS, Willard HF. Nonrandom localization of recombination events in human alpha satellite repeat unit variants: Implications for higher-order structural characteristics within centromeric heterochromatin. Mol Cell Biol. 1993;13:6520–6529. doi: 10.1128/mcb.13.10.6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoda K, Ando S, Okuda A, Kikuchi A, Okazaki T. In vitro assembly of the CENP-B/alpha-satellite DNA/core histone complex: CENP-B causes nucleosome positioning. Genes Cells. 1998;3:533–548. doi: 10.1046/j.1365-2443.1998.00210.x. [DOI] [PubMed] [Google Scholar]
- 36.Wendland JR, Martin BJ, Kruse MR, Lesch KP, Murphy DL. Simultaneous genotyping of four functional loci of human SLC6A4, with a reappraisal of 5-HTTLPR and rs25531. Mol Psychiatry. 2006;11:224–226. doi: 10.1038/sj.mp.4001789. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.