Abstract
Calmodulin (CaM) is a ubiquitous intracellular calcium sensor that directly binds to and modulates a wide variety of ion channels. Despite the large repository of high-resolution structures of CaM bound to peptide fragments derived from ion channels, there is no structural information about CaM bound to a fully folded ion channel at the plasma membrane. To determine the location of CaM docked to a functioning KCNQ K+ channel, we developed an intracellular tethered blocker approach to measure distances between CaM residues and the ion-conducting pathway. Combining these distance restraints with structural bioinformatics, we generated an archetypal quaternary structural model of an ion channel–CaM complex in the open state. These models place CaM close to the cytoplasmic gate, where it is well positioned to modulate channel function.
Keywords: KCNQ2, KCNQ3, M-current, calmodulation, tetraethylammonium
Voltage-gated ion channels respond to fluctuations in Ca2+ concentration in order to regulate membrane excitability, cardiac rhythm, and synaptic transmission between neurons. Although many ion channels are regulated by intracellular calcium, most voltage-gated channels do not directly bind calcium; instead, they employ the ubiquitous Ca2+-binding protein, calmodulin (CaM). CaM consists of two globular domains (N- and C-lobes), each of which contains two calcium-binding EF hand structures permitting the binding of up to four Ca2+ ions. CaM communicates changes in intracellular Ca2+ levels by binding to consensus sites known as CaM-binding motifs (e.g., IQ, 1-5-10, 1-8-14) in both the presence and absence of Ca2+, leading to changes in channel function (calmodulation) (1–4). Furthermore, calcification of either the N- or C-lobe of CaM allows for the modular regulation of channel assembly and gating through lobe-specific interactions of CaM with voltage-gated ion channels (5, 6).
For the KCNQ family of potassium channels [Q1–Q5; voltage-gated K+ channel (Kv)7.1–Kv7.5], CaM binds to the intracellular C-terminus to regulate channel assembly (7, 8), trafficking (9, 10), and function (7, 11, 12). The KCNQ C-terminus contains two CaM-binding motifs: a Ca2+ independent (IQ-like) and Ca2+ dependent (1-5-10), which are separated by approximately 135 residues (13). CaM binds to KCNQ channels in the absence or presence of calcium; however, CaM binding requires both motifs to be intact, consistent with in vitro studies that indicate a 1∶1 stoichiometry for the CaM∶KCNQ C-terminus protein–protein interaction (8, 11, 14). Fittingly, mutations that disrupt CaM binding to either of these motifs in Q1 and Q2 channels are associated with long QT syndrome (LQTS) and benign familial neonatal convulsions (BFNC), respectively (7, 9–11). Taken together, these studies indicate that one CaM molecule will simultaneously interact with both CaM-binding motifs in the KCNQ C-terminus.
Current structural information for ion channel–CaM complexes is limited to structures of CaM bound to peptides derived from the cytoplasmic parts of ion channels. In many of these high-resolution structures, CaM wraps around a single α-helical peptide (15, 16). However, for KCNQ subunits harboring two CaM-binding motifs, CaM is predicted to wrap around multiple cytoplasmic KCNQ helices similar to structures of CaM bound to peptides derived from either the SK Ca2+-activated K+ channel or the pre-IQ domain of the Cav1.2 voltage-gated Ca2+ channel (17–19). Although these published structures (and the inevitable structure of CaM bound to a KCNQ-derived peptide) are expected to faithfully mirror the structure of CaM when bound to a functioning channel, these isolated structures do not provide any information about the quaternary structure of the full-length ion channel–CaM complex. To determine where CaM resides on a functioning KCNQ channel, we developed an intracellular tethered blocker approach to measure distances between CaM residues and the crystallographically known quaternary ammonium-blocking site on voltage-gated K+ channels (20). Using these distance restraints, we generated 3D structural models of the Q2/Q3 channel–CaM complex in the open state using the rKv1.2 and CaM–Cav1.2 crystal structures (21, 22). These models place CaM close to the cytoplasmic gate, where it is well positioned to communicate changes in intracellular calcium to a functioning Q2/Q3 channel.
Results
To convert CaM binding into a structural reporter for Q2/Q3 channels, we chemically derivatized calmodulin into a “tethered blocker” (Fig. 1A) when bound to Q2/Q3 channels. In this intracellular variant, current inhibition requires CaM binding to the channel instead of covalent modification of an extracellular target cysteine residue (23). Similar to the extracellular version, inhibition of channel function is dependent on tether length; thus, distances between CaM and the blocker’s binding site can be determined by comparing the magnitude of current inhibition with a panel of CaM proteins chemically derivatized with different-length tethers. For this intracellular tether blocker approach to work as cartooned, three requirements must be met: (i) The binding site of the blocker is known; (ii) The blocker binds to Q2/Q3 channels with low affinity such that the freely diffusible tethered blocker does not measurably contribute to channel inhibition; and (iii) inhibition of Q2/Q3 function depends on CaM binding to increase the local concentration of the tethered blocker for its binding site.
Fig. 1.
Tethered blocker strategy for detecting CaM bound to functioning KCNQ channels. (A) CaM protein chemically derivatized with an internal channel blocker inhibits KCNQ function depending on tether length: If the tether is long enough, the blocker will reach its binding site and block K+ conduction (Left), whereas shorter tethers prevent the blocker from effectively inhibiting channel function (Right). Because the effective concentration of the blocker is dependent on tether length, the magnitude of inhibition provides a distance between a CaM residue and the internal blocker site. (B) Structures of the maleimido–QA linkers: n, number of glycines in the linker; d, extended length of linkers rounded to the nearest angstrom from the center of the quaternary ammonium (shown in red) to the olefinic carbons on the maleimide.
For the blocker, we chose a quaternary ammonium, tetraethylammonium (TEA), because its intracellular binding site on voltage-gated K+ channels is well established (20, 24) and TEA internally blocks Q2/Q3 channels at millimolar concentrations in excised patches (25). We chose to perform the tethered blocker experiments in Xenopus oocytes to ensure that the chemically derivatized CaM proteins would coassemble with KCNQ subunits during biogenesis (7–10). Therefore, we first determined the IC50 value for internal block of Q2/Q3 channels in Xenopus oocytes by recording current from individual oocytes before and after injection with various concentrations of TEA. Assuming a cytoplasmic volume of 500 nL (26), the IC50 for internal TEA block of Q2/Q3 channels was 2 ± 1 mM (Fig. S1), which, in contrast to Shaker K+ channels (27), was essentially independent of voltage.
With a suitable blocker in hand, we generated a panel of maleimido-quaternary ammoniums (QAs) with various length tethers (32–66 Å) by varying the number of glycines between the two moieties using solid-phase peptide synthesis (23) (Fig. 1B). To transform CaM into a tethered blocker, we engineered individual cysteines into the N- and C-lobes of CaM, which does not contain any native cysteines, and labeled the purified mutants with approximately 10-fold molar excess of a maleimido-QA (Materials and Methods). The chemically derivatized CaM proteins were purified by HPLC and the presence of the modification was confirmed by electrospray mass spectrometry (Table S1). Chemical modification of CaM did not disrupt Ca2+ binding, because the QA-derivatized CaM proteins bound phenyl-Sepharose only in the presence of Ca2+ (Fig. S2A).
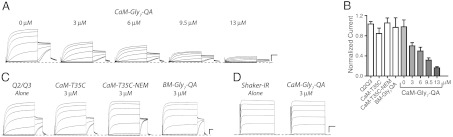
We first determined whether CaM binding to K+ channels could be exploited in an intracellular tethered blocker approach by examining CaM derivatized at position T35C (N-lobe) (Fig. 2). To allow the differently derivatized CaM proteins to compete with endogenous CaM (6–15 μM) (21, 28) during Q2/Q3 channel biogenesis (7–10), we coinjected CaM protein with channel mRNA. This experimental design required us to compare oocytes injected with derivatized CaM protein to water-injected controls. Oocytes were coinjected with KCNQ channel mRNA and CaM–T35C protein, and families of Q2/Q3 currents were measured 3 d after coinjection. Oocytes injected with CaM–Gly7–QA (3 μM in ovo) resulted in 43 ± 4% decrease in current, as compared to water-injected controls (0 μM) (Fig. 2 A and B). Inhibition was dependent on the amount of CaM–Gly7–QA injected (Fig. 2A) and at 13 μM was comparable to maximal block with 10 mM TEA (Fig. S1), consistent with the injected CaM protein competing with endogenous CaM for binding to Q2/Q3 channels. Inhibition of Q2/Q3 current required the presence of the blocker (Fig. 2C) because neither unlabeled CaM–T35C nor N-ethylmaleimide (NEM)-capped version (CaM–T35C–NEM) reduced the current (Fig. 2B).
Fig. 2.
Chemically derivatized CaM proteins behave as intracellular tethered blockers. (A) Families of Q2/Q3 currents recorded from oocytes injected with channel mRNA or coinjected with T35C–Gly7–QA protein. Currents were elicited by 1-s test potentials from -100 to +40 mV in 20-mV increments from a holding potential of -80 mV followed by a tail pulse to -30 mV. Dashed line indicates zero current. Scale bars, 0.5 μA and 0.2 s. (B) Quantification of current levels at 40 mV from oocytes co-injected with different CaM proteins. Values are normalized to oocytes injected with only channel mRNA. Data are presented as the mean ± SEM from 2–4 batches of oocytes. (C) Families of Q2/Q3 currents recorded from oocytes injected with channel mRNA or co-injected with CaM protein. Currents were elicited by 4 s test potentials from -100 to +40 mV in 20 mV increments from a holding potential of -80 mV followed by a tail pulse to -30 mV. Dashed line indicates zero current. Scale bars represent 0.5 μA and 0.5 s. (D) Families of Shaker–IR currents were elicited by test potentials from -100 to +60 mV in 20-mV increments from a holding potential of -80 mV. Scale bars, 1 μA and 10 ms.
These results with CaM–Gly7–QA indicated that comparing batches of oocytes injected with derivatized CaM protein to water-injected controls is a reliable approach for determining intracellular block. To confirm this experimental approach, we also injected CaM–Gly7–QA into oocytes already expressing Q2/Q3 currents (Fig. S2B). Families of Q2/Q3 currents from individual oocytes were recorded before and after injection of CaM–Gly7–QA (3 μM in ovo). Injection of CaM–Gly7–QA resulted in 39 ± 9% reduction in current, which was comparable to the reduction observed from oocytes coinjected with mRNA and CaM protein (43 ± 4%) (Fig. S2C). The similar reduction of current observed for both approaches validated the batch comparison method and demonstrated that the magnitude of inhibition does not critically depend on when the chemically derivatized CaM is injected.
Although Q2/Q3 inhibition with CaM–Gly7–QA was approximately 1,000-fold more potent than internal block by TEA (3 μM compared to 2 mM), this increased potency did not directly demonstrate that the observed inhibition with CaM–Gly7–QA was caused by CaM binding to Q2/Q3 channels. To determine whether CaM binding was required for inhibition, we first tested CaM–Gly7–QA on Shaker–IR, which is a TEA-sensitive K+ channel that does not directly bind CaM (29). Coinjection of CaM–Gly7–QA with Shaker–IR mRNA did not reduce current levels (Fig. 2D), consistent with the notion that CaM binding is required for inhibition with CaM–Gly7–QA. We also derivatized a truncated CaM mutant with maleimido–Gly7–QA to generate a tethered blocker (BM–Gly7–QA) that cannot bind Q2/Q3 channels (13). Similar to the Shaker–IR control, BM–Gly7–QA did not inhibit Q2/Q3 currents (Fig. 2 B and C). Together, these results confirmed that CaM–Gly7–QA requires binding for inhibition, fully satisfying the requirements of a tethered blocker.
We first used the tethered blockers to determine a distance between the N-lobe of CaM and the TEA-binding site on Q2/Q3 channels. At the N-lobe residue, T35C, six different tether lengths were tested at 3 μM final oocyte concentration (Fig. 3). As expected for a bona fide tether blocker, the magnitude of channel inhibition was dependent on tether length: The shortest tether, CaM–Gly3–QA, showed little to no inhibition; intermediate-length tethers showed partial inhibition; and all tethers that contained more than six glycine residues showed maximal inhibition (approximately 50%). Plotting inhibition (normalized to maximal inhibition) as a function of fully extended tether length (Fig. 3) resulted in a monophasic curve, indicating that the tethered blockers are reporting from a single CaM-binding site on a KCNQ subunit. To calculate a distance, previous tethered blocker studies on the Shaker K+ channel have used the fully extended linker length of the tethered blocker that results in the first sign of inhibition (23). However, this metric for tether length yields distances that are systematically shorter than the atomic distances in the subsequently published rKv1.2 structure (30). A reexamination (31) of these data revealed that a better metric for calculating a tethered blocker distance is the end-to-end tether length of the linker that results in half-maximal inhibition (d1/2). Therefore, we fitted the data to a Boltzmann equation to objectively obtain a d1/2 of 40 ± 1 Å between the N-lobe residue, T35C, and the Q2/Q3 TEA-binding site.
Fig. 3.
Distance measurements for CaM residues: T35, T45, and T111. Superimposed currents recorded from oocytes injected with 3 μM T35C–Glyn–QA, T111C–Glyn–QA, or T45C–Glyn–QA protein are shown for each residue (Left). Currents were elicited with 4-s test potentials to +40 mV from a holding potential of -80 mV followed by a tail pulse to -30 mV. Dashed line indicates zero current. (Right) Normalized inhibition values plotted as a function of end-to-end linker length. Data are presented as the mean ± SEM from two to four batches of oocytes. The data were fitted to a Boltzmann equation to generate the midpoint of inhibition (d1/2): T35C, 40 ± 1 Å; T111C, 50 ± 1 Å; T45C, 49 ± 1 Å. Values are reported as mean ± the error of the fit.
To determine the precision of the measured distance, we also measured the distance from T35C and the Q2/Q3 TEA-binding site at different depolarizing potentials and with higher in ovo concentrations of derivatized CaM. Repeating the analysis at different test potentials consistently resulted in a d1/2 of 40 ± 1 Å (Table S2), demonstrating that the measured distance is not dependent on the depolarizing pulse. To determine whether the calculated distances were also independent of the amount of injected CaM protein, we repeated the experiments at an in ovo concentration of approximately 15 μM (Fig. S2D). As expected, the higher concentration of injected CaM–Glyn–QA that contained more than six glycine residues (n = 7 or 8) resulted in maximal inhibition (approximately 85%) that was greater than that observed with 3 μM (approximately 50%). In addition, this increased inhibition was not caused by disrupting CaM homeostasis—as has been observed with CaM overexpression (32)—because CaM protein derivatized with shorter tethers did not inhibit Q2/Q3 channel function (Fig. S2D). Moreover, fitting the 15-μM data to a Boltzmann yielded a d1/2 of 43 ± 1 Å, demonstrating that the number of tethered blockers bound to functioning Q2/Q3 channels does not appreciably affect the distance measured between CaM and the TEA-binding site. In total, the consistency of the calculated distance at different voltages and in ovo concentrations of derivatized CaM bolstered our confidence in both the methodology’s robustness and the precision of the distance measured with intracellular tethered blockers.
We next switched to the C-lobe of CaM and tested residue T111C (Fig. 3). Compared to T35C, T111C is further away from the TEA-binding site because maximal inhibition required tethers with at least 10 glycine residues. Fitting the data to a Boltzmann equation afforded a d1/2 of 50 ± 1 Å. Lastly, to triangulate the position of CaM bound to Q2/Q3 channels, we picked an additional CaM residue, T45C, to generate a third distance restraint (Fig. 3). Although this residue is in the N-lobe of CaM, its d1/2 (49 ± 1 Å) was comparable to the C-lobe residue, T111C (Fig. 3).
Because no high-resolution structural information exists for the Q2/Q3–CaM complex, we modeled a quaternary structure using ion channel domains with known structures and our distance constraints. For Q2/Q3, we used the membrane-embedded portion of rKv1.2 [Protein Data Bank (PDB) 2A79] (30) to model the S1–S6 segments and transplanted the crystallographically determined TEA-binding site from the KcsA–tetrabutylantimony structure (PDB 2HJF) (20). For CaM, we used the crystal structure of CaM bound to the Cav1.2 pre-IQ domain (PDB 3G43) (17) for three reasons: (i) Both CaM-binding motifs must be intact for the KCNQ–CaM protein–protein interaction, suggesting that CaM wraps around two helices (14); (ii) hydrophobic residues in the pre-IQ domain that make contact with CaM are also present in the KCNQ–CaM–binding motifs (Fig. S3A); and (iii) CaM binds to both KCNQ and Cav channels in the absence and presence of calcium (18, 33, 34). To apply the radial distance restraints to these high-resolution structures, we approximated the trajectory taken by the tethered blocker from CaM to the quaternary ammonium-binding site (Fig. 4, large purple sphere). Therefore, the first 20 Å was modeled as a straight line until it emerged from the Kv inner vestibule (Fig. 4, blue sphere) where it was allowed to splay off at an angle collinear with the S6 helix that enabled the fourfold arrangement of CaM proteins. Four CaM molecules (sans Cav1.2 peptides) were modeled because in vitro data indicate a 1∶1 KCNQ∶CaM stoichiometry (8, 11). Two KCNQ–CaM quaternary models satisfied the distance restraints without observable van der Waals (VDW) clashes (Fig. 4). The lack of a single KCNQ–CaM model arises from the degeneracy with which CaM binds to its targets; thus, Fig. 4A depicts one structural model where T111C faces the channel, whereas in Fig. 4B the CaM subunits are essentially inverted (note the position of T111C). In both models, the modeled CaMs are close to the KCNQ channel gate (Fig. 4, Left).
Fig. 4.
Structural models of the Q2/Q3–CaM complex. (A) Side and cytoplasmic views of the complex, showing the channel subunits colored grey and the four CaM molecules in different pastels. In the side view, only three subunits are shown for clarity. Residues T35C (red), T45C (green), and T111C (blue) are shown in space fill. Red spheres, potassium ions; purple sphere, TEA-binding site; dark blue sphere, dummy ion to simulate trajectory of the tethered blocker. (B) Membrane and cytoplasmic view of the complex with CaM subunits inverted. Colors are the same as in A.
We also generated quaternary structural models (Fig. S3 B and C) of the complex using the crystal structure of CaM bound to a peptide from the SK2 channel (PDB 1G4Y) (19) because functional data suggest that CaM binds to a continuous KCNQ peptide (35). Using the SK2–CaM structure as a model of the KCNQ–CaM protein–protein interaction, however, did not fit the distance restraints and required the systematic addition of 5 Å to yield quaternary structures that did not contain nonnative contacts. Thus, our experimentally determined distance restraints indicate the protein fold of CaM in the CaV 1.2 pre-IQ domain structure (17) may better represent CaM when it is bound to full-length Q2/Q3 channels.
Discussion
Motivated by the plethora of high-resolution structures of isolated ion channel domains, we developed an intracellular tethered blocker approach to generate quaternary structural models of ion channel–CaM complexes. Using a panel of intracellular tethered blockers to generate distance restraints between CaM and the Kv channel TEA-binding site, we generated quaternary models of the Q2/Q3–CaM complex in the open state. In these models, CaM is very close to the cytoplasmic gate of Q2/Q3 channels where it is well positioned to modulate Q2/Q3 channel gating (calmodulation). The juxtaposition of the “CaM ring” to the Q2/Q3 channel gate results in a cytoplasmic vestibule that is continuous with the Q2/Q3 pore domain, which is in contrast to the hanging gondola structure observed in classic Kv channels (Kv1–4). This elongated permeation pathway predicts that the potassium ions will enter and exit the Q2/Q3–CaM complex through the center of the CaM ring. Given that the spacing between the first CaM-binding motif and the cytoplasmic “bundle crossing” (22) is conserved in the KCNQ family, our quaternary models serve as structural scaffolds for all KCNQ–CaM complexes.
While developing the intracellular tethered blocker approach, we were also able to glean some information about the dynamics of the KCNQ–CaM protein–protein interaction at the plasma membrane (Fig. S2B). Injecting CaM–Gly7–QA into oocytes already expressing Q2/Q3 currents resulted in a similar magnitude of inhibition compared to coinjection with channel mRNA (Fig. S2C). The rapid onset (approximately 2 h) indicates that the chemically derivatized CaM proteins need not coassemble with Q2 and Q3 channel subunits during biogenesis and can bind to functioning channels at the plasma membrane. These results suggest that at least one CaM-binding site is free or the injected CaM rapidly exchanges with KCNQ-bound CaM proteins or other intracellular scaffolding proteins (e.g., syntaxin 1A and AKAP79/150) (35–37). Both scenarios illustrate the dynamic nature of the KCNQ–CaM complex.
Although the KCNQ–CaM protein–protein interaction is dynamic, the monophasic inhibition curves indicate that the tethered blockers are reporting from a single location (within approximately 66 Å) on the KCNQ C-terminus and are not oscillating between the two CaM-binding motifs. A single CaM on the KCNQ C-terminus is also consistent with previous functional data that suggest the syntaxin 1A and AKAP79/150 abrogate CaM binding by partially masking only one CaM-binding motif (35–37). In spite of this competition for the KCNQ C-terminus, the distance restraints did not depend on the amount of injected tethered blocker (Fig. 3 and Fig. S2D), suggesting that KCNQ need not be fully occupied with QA-derivatized CaM molecules to determine the location of CaM bound to KCNQ channels.
As with all structural determinations that rely on functional measurements, there are some limitations to the intracellular tethered blocker approach. First, the models depict the Q2/Q3–CaM complex in the open state. Second, the distances measured by tethered blockers are radial; thus, the absolute position of CaM with respect to a particular KCNQ subunit cannot be determined. To approximate the position of each CaM molecule in the quaternary structures, we have positioned the KCNQ subunit such that the S6 pore helix bisects the center of each CaM molecule. Given the short distance between the bundle crossing and the first CaM-binding domain (approximately 15 aa), this is a good approximation of the absolute position of CaM in the full-length complex. Third, the approach does not speak to how the KCNQ C-terminus is threaded through the lobes of CaM. With a high-resolution structure of a CaM–KCNQ peptide complex, a single Q2/Q3–CaM quaternary model could be generated using tethered blockers; however, the molecular details of the protein–protein interaction inherent to the full-length complex, such as whether a single CaM binds to one KCNQ C-terminus or to two adjacent C-termini simultaneously, will not be resolvable. Lastly, we do not know whether the measured distances using wild-type protein are from fully calcified, half-calcified, uncalcified, or an amalgam of differently calcified CaM proteins. Because injecting micromolar amounts of purified CaM protein does not affect Q2/Q3 function (Fig. 2C), our current structural models are most likely representative of the Q2/Q3–CaM complex under endogenous calcium concentrations. Future studies that utilize CaM calcium-binding mutants (CaM12, CaM34, etc.) (38) will provide insight into how calcification of the different CaM lobes affects orientation on the full-length channel.
Given that the number of high-resolution structures of CaM bound to peptide fragments from ion channels is steadily increasing, the need to connect these structures to their respective ion-conducting domains is vital for unraveling the mechanisms of calmodulation. By chemically derivatizing CaM with different channel blockers, the intracellular blocker approach can be expanded to determine the location of CaM on a wide variety of ion channels. Moreover, the approach is not limited to CaM, and can be applied to other cytoplasmic regulatory and scaffolding proteins that are essential for ion channel modulation.
Materials and Methods
Molecular Biology, CaM Expression, and Purification.
For mRNA synthesis, DNA constructs were linearized with the appropriate restriction enzyme and cRNA was synthesized using in vitro runoff transcription with SP6 or T7 polymerase. CaM DNA was obtained from D. Yue (Johns Hopkins, Baltimore, MD) and subcloned into the pET–DUET1 vector for protein expression and purification. Cysteine point mutants were introduced by Quikchange site-directed mutagenesis (Strategene); the CaM-binding mutant truncation (BM) was generated by cassette mutagenesis. All CaM mutations were confirmed by DNA sequencing of the entire gene. Wild-type and mutant CaM proteins were expressed and purified as described previously (39, 40), with some modifications (SI Material and Methods).
Linker Synthesis, CaM Labeling, and Purification.
The panel of maleimido-quaternary ammoniums were synthesized and purified as previously described (23). To chemically derivatize CaM with a maleimido-quaternary ammonium or N-ethylmalemide, a CaM cysteine mutant was dissolved in 1 mL of PBS and reduced with 10-fold excess of tris(2-carboxyethyl)phosphine (TCEP) for 10 min at room temperature (pH = 7.6). The glycine linker or NEM (12-fold molar excess) was dissolved in 1 mL of PBS and added dropwise to the CaM protein solution. After 2 h, the reaction mixtures were transferred to a 3-mL Slide-A-Lyzer (3500 MWCO; Thermo Scientific) and dialyzed against 3 L of PBS at room temperature. The labeled CaM proteins were purified by using a C4 HPLC column (10 × 250 mm, 5-μm particles), eluting with solvent A (95% water, 5% acetonitrile, and 0.1% TFA) and solvent B (95% acetonitrile, 5% water, and 0.1% TFA), forming a gradient of 35–50% B over 38 min. Fractions containing strong absorption at 214, 260, and 280 nm were collected and lyophilized. Labeling was confirmed by electrospray ionization mass spectrometry (Table S1).
Electrophysiology.
Oocytes were surgically removed from Xenopus laevis, defolliculated, and stored as previously described (41). Oocytes were microinjected with 15.2 ng of mRNA and 27–130 ng purified CaM protein 8–24 h after surgery. For Q2/Q3 heteromeric channels, an equal amount of mRNA (7.6 ng) for each subunit was injected. The final CaM concentration inside the oocyte (3–18 μM) was calculated by assuming a volume of 500 nL for each oocyte (26). Currents were recorded 66–74 h (24 h for Shaker) after injection. Inhibition was determined by batch comparison of oocytes injected with channel mRNA alone versus channel mRNA with CaM protein. The percent inhibition obtained from each batch was normalized to the maximal inhibition value. Normalized values were plotted as a function of extended tether length for each modified CaM. Distance curves were fit to a Boltzmann equation to generate a midpoint distance (d1/2).
KCNQ2/3–CaM Model Generation.
The rKv1.2 potassium channel (PDB 2A79) (30) and CaM–Cav1.2 peptide (PDB 3G43) (17) crystal structure coordinates were obtained from the Protein Data Bank (http://www.rcsb.org) and processed using PyMOL (Schrodinger LLC) to isolate putatively extracellular and intramembranous regions of the rKv1.2 channel and a CaM chain from respective PDB coordinates. Using PyMOL, a dummy atom was placed 20 Å below the quaternary ammonium-binding site (20, 24) to indicate the inflection point of a flexible chemical linker as it would angle into the channel pore. Threonine residues (residues 34, 44, 110) were mutated to cysteine using the mutagenesis wizard within PyMOL, selecting backbone-restrained rotamers with minimal VDW clashes as indicated in the PyMOL graphical user interface. The mutated CaM subunit was positioned at the intracellular end of the isolated potassium channel structure to satisfy the distance restraints between the gamma sulfur atoms of the mutated cysteine residues and the quaternary ammonium-binding site (20, 24). VDW clashes between the channel and CaM were visualized using PyMOL (http://pymolwiki.org/index.php/Show_bumps). Once a CaM orientation was determined that satisfied the measured distance restraints without VDW clashes, the remaining three CaM molecules were generated using basic symmetry operators within PyMOL and confirmed to satisfy the distances without any CaM–channel or CaM–CaM VDW clashes as described above.
Supplementary Material
ACKNOWLEDGMENTS.
This work was supported by National Institutes of Health Grant GM-070650 (to W.R.K.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207606109/-/DCSupplemental.
References
- 1.Rhoads AR, Friedberg F. Sequence motifs for calmodulin recognition. FASEB J. 1997;11:331–340. doi: 10.1096/fasebj.11.5.9141499. [DOI] [PubMed] [Google Scholar]
- 2.Schonherr R, Lober K, Heinemann SH. Inhibition of human ether a go-go potassium channels by Ca(2+)/calmodulin. EMBO J. 2000;19:3263–3271. doi: 10.1093/emboj/19.13.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirschberg B, Maylie J, Adelman JP, Marrion NV. Gating of recombinant small-conductance Ca-activated K+ channels by calcium. J Gen Physiol. 1998;111:565–581. doi: 10.1085/jgp.111.4.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black DJ, et al. Calmodulin interactions with IQ peptides from voltage-dependent calcium channels. Am J Physiol Cell Physiol. 2005;288:C669–C676. doi: 10.1152/ajpcell.00191.2004. [DOI] [PubMed] [Google Scholar]
- 5.Maylie J, Bond CT, Herson PS, Lee WS, Adelman JP. Small conductance Ca2+-activated K+ channels and calmodulin. J Physiol. 2004;554:255–261. doi: 10.1113/jphysiol.2003.049072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeMaria CD, Soong TW, Alseikhan BA, Alvania RS, Yue DT. Calmodulin bifurcates the local Ca2+ signal that modulates P/Q-type Ca2+ channels. Nature. 2001;411:484–489. doi: 10.1038/35078091. [DOI] [PubMed] [Google Scholar]
- 7.Shamgar L, et al. Calmodulin is essential for cardiac IKS channel gating and assembly: Impaired function in long-QT mutations. Circ Res. 2006;98:1055–1063. doi: 10.1161/01.RES.0000218979.40770.69. [DOI] [PubMed] [Google Scholar]
- 8.Wiener R, et al. The KCNQ1 (Kv7.1) COOH terminus, a multitiered scaffold for subunit assembly and protein interaction. J Biol Chem. 2008;283:5815–5830. doi: 10.1074/jbc.M707541200. [DOI] [PubMed] [Google Scholar]
- 9.Alaimo A, et al. Calmodulin activation limits the rate of KCNQ2 K+ channel exit from the endoplasmic reticulum. J Biol Chem. 2009;284:20668–20675. doi: 10.1074/jbc.M109.019539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Etxeberria A, et al. Calmodulin regulates the trafficking of KCNQ2 potassium channels. FASEB J. 2008;22:1135–1143. doi: 10.1096/fj.07-9712com. [DOI] [PubMed] [Google Scholar]
- 11.Ghosh S, Nunziato DA, Pitt GS. KCNQ1 assembly and function is blocked by long-QT syndrome mutations that disrupt interaction with calmodulin. Circ Res. 2006;98:1048–1054. doi: 10.1161/01.RES.0000218863.44140.f2. [DOI] [PubMed] [Google Scholar]
- 12.Gamper N, Shapiro MS. Calmodulin mediates Ca2+-dependent modulation of M-type K+ channels. J Gen Physiol. 2003;122:17–31. doi: 10.1085/jgp.200208783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yus-Najera E, Santana-Castro I, Villarroel A. The identification and characterization of a noncontinuous calmodulin-binding site in noninactivating voltage-dependent KCNQ potassium channels. J Biol Chem. 2002;277:28545–28553. doi: 10.1074/jbc.M204130200. [DOI] [PubMed] [Google Scholar]
- 14.Gomez-Posada JC, et al. Kv7 channels can function without constitutive calmodulin tethering. PLoS One. 2011;6:e25508. doi: 10.1371/journal.pone.0025508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mori MX, Vander Kooi CW, Leahy DJ, Yue DT. Crystal structure of the CaV2 IQ domain in complex with Ca2+/calmodulin: High-resolution mechanistic implications for channel regulation by Ca2+ Structure. 2008;16:607–620. doi: 10.1016/j.str.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Contessa GM, et al. Structure of calmodulin complexed with an olfactory CNG channel fragment and role of the central linker: Residual dipolar couplings to evaluate calmodulin binding modes outside the kinase family. J Biomol NMR. 2005;31:185–199. doi: 10.1007/s10858-005-0165-1. [DOI] [PubMed] [Google Scholar]
- 17.Fallon JL, et al. Crystal structure of dimeric cardiac L-type calcium channel regulatory domains bridged by Ca2+* calmodulins. Proc Natl Acad Sci USA. 2009;106:5135–5140. doi: 10.1073/pnas.0807487106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim EY, et al. Multiple C-terminal tail Ca(2+)/CaMs regulate Ca(V)1.2 function but do not mediate channel dimerization. EMBO J. 2010;29:3924–3938. doi: 10.1038/emboj.2010.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schumacher MA, Rivard AF, Bachinger HP, Adelman JP. Structure of the gating domain of a Ca2+-activated K+ channel complexed with Ca2+/calmodulin. Nature. 2001;410:1120–1124. doi: 10.1038/35074145. [DOI] [PubMed] [Google Scholar]
- 20.Zhou M, Morais-Cabral JH, Mann S, MacKinnon R. Potassium channel receptor site for the inactivation gate and quaternary amine inhibitors. Nature. 2001;411:657–661. doi: 10.1038/35079500. [DOI] [PubMed] [Google Scholar]
- 21.Cartaud A, Ozon R, Walsh MP, Haiech J, Demaille JG. Xenopus laevis oocyte calmodulin in the process of meiotic maturation. J Biol Chem. 1980;255:9404–9408. [PubMed] [Google Scholar]
- 22.Holmgren M, Shin KS, Yellen G. The activation gate of a voltage-gated K+ channel can be trapped in the open state by an intersubunit metal bridge. Neuron. 1998;21:617–621. doi: 10.1016/s0896-6273(00)80571-1. [DOI] [PubMed] [Google Scholar]
- 23.Blaustein RO, Cole PA, Williams C, Miller C. Tethered blockers as molecular “tape measures” for a voltage-gated K+ channel. Nat Struct Biol. 2000;7:309–311. doi: 10.1038/74076. [DOI] [PubMed] [Google Scholar]
- 24.Yellen G, Jurman ME, Abramson T, MacKinnon R. Mutations affecting internal TEA blockade identify the probable pore-forming region of a K+ channel. Science. 1991;251:939–942. doi: 10.1126/science.2000494. [DOI] [PubMed] [Google Scholar]
- 25.Suh BC, Hille B. Electrostatic interaction of internal Mg2+ with membrane PIP2 Seen with KCNQ K+ channels. J Gen Physiol. 2007;130:241–256. doi: 10.1085/jgp.200709821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cicirelli MF, Robinson KR, Smith LD. Internal pH of Xenopus oocytes: A study of the mechanism and role of pH changes during meiotic maturation. Dev Biol. 1983;100:133–146. doi: 10.1016/0012-1606(83)90204-x. [DOI] [PubMed] [Google Scholar]
- 27.Thompson J, Begenisich T. Affinity and location of an internal K+ ion binding sitein Shaker K channels. J Gen Physiol. 2001;117:373–384. doi: 10.1085/jgp.117.5.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cicirelli MF, Smith LD. Calmodulin synthesis and accumulation during oogenesis and maturation of Xenopus laevis oocytes. Dev Biol. 1986;113:174–181. doi: 10.1016/0012-1606(86)90120-x. [DOI] [PubMed] [Google Scholar]
- 29.Cukovic D, Lu GW, Wible B, Steele DF, Fedida D. A discrete amino terminal domain of Kv1.5 and Kv1.4 potassium channels interacts with the spectrin repeats of alpha-actinin-2. FEBS Lett. 2001;498:87–92. doi: 10.1016/s0014-5793(01)02505-4. [DOI] [PubMed] [Google Scholar]
- 30.Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 31.Morin TJ, Kobertz WR. Tethering chemistry and K+ channels. J Biol Chem. 2008;283:25105–25109. doi: 10.1074/jbc.R800033200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gamper N, Li Y, Shapiro MS. Structural requirements for differential sensitivity of KCNQ K+ channels to modulation by Ca2+/calmodulin. Mol Biol Cell. 2005;16:3538–3551. doi: 10.1091/mbc.E04-09-0849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wen H, Levitan IB. Calmodulin is an auxiliary subunit of KCNQ2/3 potassium channels. J Neurosci. 2002;22:7991–8001. doi: 10.1523/JNEUROSCI.22-18-07991.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Erickson MG, Liang H, Mori MX, Yue DT. FRET two-hybrid mapping reveals function and location of L-type Ca2+ channel CaM preassociation. Neuron. 2003;39:97–107. doi: 10.1016/s0896-6273(03)00395-7. [DOI] [PubMed] [Google Scholar]
- 35.Etzioni A, et al. Regulation of neuronal M-channel gating in an isoform-specific manner: Functional interplay between calmodulin and syntaxin 1A. J Neurosci. 2011;31:14158–14171. doi: 10.1523/JNEUROSCI.2666-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Regev N, et al. Selective interaction of syntaxin 1A with KCNQ2: Possible implications for specific modulation of presynaptic activity. PLoS One. 2009;4:e6586. doi: 10.1371/journal.pone.0006586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bal M, Zhang J, Hernandez CC, Zaika O, Shapiro MS. Ca2+/calmodulin disrupts AKAP79/150 interactions with KCNQ (M-Type) K+ channels. J Neurosci. 2010;30:2311–2323. doi: 10.1523/JNEUROSCI.5175-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klevit RE, Dalgarno DC, Levine BA, Williams RJ. 1H-NMR studies of calmodulin. The nature of the Ca2+-dependent conformational change. Eur J Biochem. 1984;139:109–114. doi: 10.1111/j.1432-1033.1984.tb07983.x. [DOI] [PubMed] [Google Scholar]
- 39.Gopalakrishna R, Anderson WB. Ca2+-induced hydrophobic site on calmodulin: Application for purification of calmodulin by phenyl-Sepharose affinity chromatography. Biochem Biophys Res Commun. 1982;104:830–836. doi: 10.1016/0006-291x(82)90712-4. [DOI] [PubMed] [Google Scholar]
- 40.Allen MW, et al. Fluorescence labeling, purification, and immobilization of a double cysteine mutant calmodulin fusion protein for single-molecule experiments. Anal Biochem. 2004;325:273–284. doi: 10.1016/j.ab.2003.10.045. [DOI] [PubMed] [Google Scholar]
- 41.O’Connell D, Mruk K, Rocheleau JM, Kobertz WR. Xenopus laevis oocytes infected with multi-drug resistant bacteria: Implications for electrical recordings. J Gen Physiol. 2011;138:271–277. doi: 10.1085/jgp.201110661. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.