Abstract
Many tumors are characterized by recurrent translocations between a tissue-specific gene and a proto-oncogene. The juxtaposition of the Ig heavy chain gene and Myc in Burkitt’s lymphoma and in murine plasmacytoma is a classic example. Regulatory elements within the heavy chain constant region locus are required for Myc translocation and/or deregulation. However, many genes are regulated by cis-acting elements at distances up to 1,000 kb outside the locus. Such putative distal elements have not been examined for the heavy chain locus, particularly in the context of Myc translocations. We demonstrate that a transgene containing the Ig heavy chain constant region locus, inserted into five different chromosomal locations, can undergo translocations involving Myc. Furthermore, these translocations are able to generate plasmacytomas in each transgenic line. We conclude that the heavy chain constant region locus itself includes all of the elements necessary for both the translocation and the deregulation of the proto-oncogene.
Many tumor types, including most leukemias and lymphomas, are characterized by reciprocal translocations of the same two chromosomal loci in independent tumors. A number of B lineage lymphomas harbor recurrent translocations that involve the Ig locus, whereas the T-cell receptor locus is involved in most T lineage lymphoma translocations (1, 2). As a result of these recurrent translocations, promoters or enhancers in one translocation partner often change the regulation of the proto-oncogene in the other partner. A classic example is the translocation of the Myc proto-oncogene on human chromosome (Chr) 8 (mouse Chr 15) to the Ig heavy chain locus (Igh) on human Chr 14 (mouse Chr 12) (3–6). This translocation results in Myc being placed under the strong transcriptional control of Igh. The Myc:Igh translocation occurs in more than 85% of human Burkitt’s lymphoma and mouse plasmacytoma, and it is one of the earliest events in tumorigenesis, indicating it to be the driving force of these tumors. Molecularly, the translocation junction occurs most frequently in the first (noncoding) exon or first intron of the Myc gene, joining the “tail” (second and third exons) of Myc to the tail (3′ or constant region) end of Igh. Because it is retained in the majority of these translocations, enhancer elements in the tail (3′ regulatory region) of the Igh locus presumably deregulate Myc expression, a primary event in the tumorigenesis (7). In part, the prevalence of this recurrent translocation is due to strong selection for deregulated Myc expression (3–6).
Chromosomal location also has a role in recurrent translocations. Although the Myc and N-myc genes are very similar, Myc, but not N-myc, is found as a translocation partner in murine pro–B-cell lymphomas that arise in mice deficient in the DNA repair factors p53 and Ligase 4. Gostissa et al. tested whether this cell-type specific use of Myc in translocations was due to selection for specific activities of the protein encoded by the Myc gene, by replacing the Myc coding exons with the N-myc coding exons (8). They found translocations in pro–B-cell lymphomas now joined heavy chain genes to the N-myc gene in the Myc location. These investigators concluded that, at least for this pair of genes in this genetic background, selection for the activities of the specific protein is less important than cis-acting elements in the Myc locus that target translocations with some degree of cell-type preference (8). Apparently, preferential targeting of specific loci can vary depending on the cell type and on DNA repair pathways used; the N-myc locus is a target for chromosomal rearrangements in other genetic backgrounds (9, 10).
Myc:Igh translocations in plasmacytoma are thought to result from aberrant heavy chain class switch recombination (1, 2). Normal switch recombination occurs through double-stranded breaks that are introduced into 2- to 8-kb switch (S) regions that precede the Igh constant region genes (11). S regions are characterized by multiple copies of simple sequences, some of which are preferred sites for action by the activation-induced cytidine deaminase (AID), the enzyme that initiates class switching (12, 13). Recombination joins double-stranded breaks in two S regions, bringing the exon encoding the Igh variable region into physical and functional association with a new heavy chain constant region with different effector functions (11). Like class switch recombination, translocations to Myc usually involve S regions (1, 4–6) and depend on AID (14–16).
The known regulatory elements contained within the Igh locus have been examined for a role in Myc translocation and deregulation. The Igh intronic enhancer (Eμ) is not physically associated with the Myc coding sequences after translocation and is, therefore, unlikely to be important for Myc deregulation (3–6). Gostissa et al. demonstrated that elements in the Igh 3′ regulatory region are required for Myc translocation and/or deregulation (17). The potential for additional cis-acting elements outside the Igh constant region locus has not been investigated. In this study, we addressed a fundamental question: Are DNA sequences flanking the Igh constant region locus in its normal chromosomal location required for tumorigenic translocations? Or, are the sequences within the constant region locus sufficient? We used an Igh transgene in five genomic locations and determined that all five different chromosomal locations are permissive for translocations with Myc that result in plasmacytoma.
Results
Igh Transgene Is Able to Undergo Translocations with Myc at Multiple Chromosomal Locations.
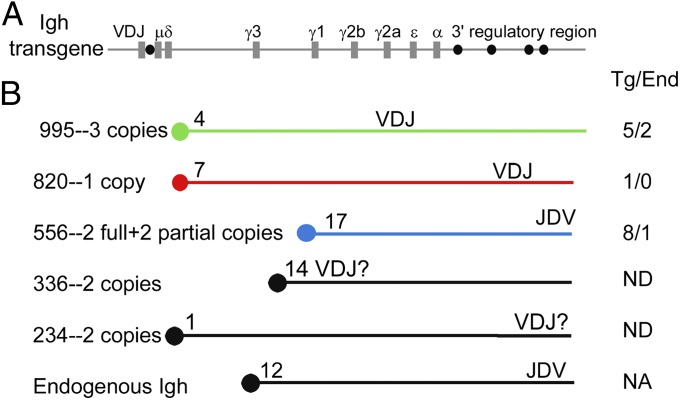
To determine whether the chromosomal environment impacts the development of Myc:Igh translocations, we analyzed these events in transgenic mouse models expressing Igh from five different chromosomes. The transgene consists of a 230-kb bacterial artificial chromosome (BAC) carrying a prerearranged VDJ exon and the entire heavy chain constant domain, including the 28-kb 3′ regulatory region (7) and an additional 15 kb (Fig. 1A) (18). This heavy chain transgene undergoes normal class switch recombination that is qualitatively and quantitatively comparable to that of the endogenous locus, regardless of the chromosomal insertion site (18). To determine whether this construct can participate in Myc translocations, transgenic B cells were induced ex vivo to undergo class switching. To examine a reasonable number of translocation events, we increased the frequency of them by overexpression of AID from a retrovirus. Three days after activation, Myc:Igh translocations were cloned by using a PCR-based approach (19, 20) and characterized by DNA sequencing. Three independent transgenic lines were studied. Line 995 contains three complete copies of the transgene on chromosome 4. The variable region-encoding exon of at least one transgene copy in line 995 is proximal to the centromere and inverted compared with the endogenous locus (Fig. 1B). Line 820 contains a single copy of the Igh transgene inserted into chromosome 7 and inverted compared with the endogenous locus. Line 556 contains two complete and two partial copies of the transgene inserted near the telomere of chromosome 17 in the same orientation as the endogenous Igh. We found that the Igh transgenes were a frequent partner for Myc translocations in tissue culture; 79% of translocations were to the transgene (Fig. 1B). The translocations to the transgene were indistinguishable from translocations to the endogenous locus, in that they were distributed over a 2-kb region, including the 5′-most Myc exon and intron, and the translocations were widely distributed in S regions (Table S1). The interpretation of these experiments includes the assumption that the ratio of translocations involving the transgene to translocations involving the endogenous genes will not be changed significantly by AID overexpression. Even considering this assumption, these results established that the Igh transgene, in three different chromosomal locations, can undergo recombination with the Myc proto-oncogene. Recombination of Myc with the transgene is at least as efficient as recombination with the endogenous Igh genes.
Fig. 1.
Myc:Igh translocations. (A) Structure of the 230-kb Igh transgene. Coding exons are depicted as gray rectangles, and regulatory elements are depicted as black circles. The transgene is drawn approximately to scale. (B) Five transgenic lines used in this study are shown, with copy number, a schematic of chromosome location, and transgene orientation relative to the centromere depicted by the orientation of the VDJ exon. The location of the endogenous Igh locus on Chr 12 is shown for comparison. To the right of the chromosome schematics is shown an enumeration of Myc:Igh translocation sites cloned from in vitro cultures. Translocation to the transgene or to the endogenous genes (number of sequences of each type shown, separated by a slash) was determined by evaluating three to 52 polymorphisms in the various S regions (see SI Materials and Methods for sequence data and methods).
Myc:Igh Translocations to the Transgene Are Found in Plasmacytoma.
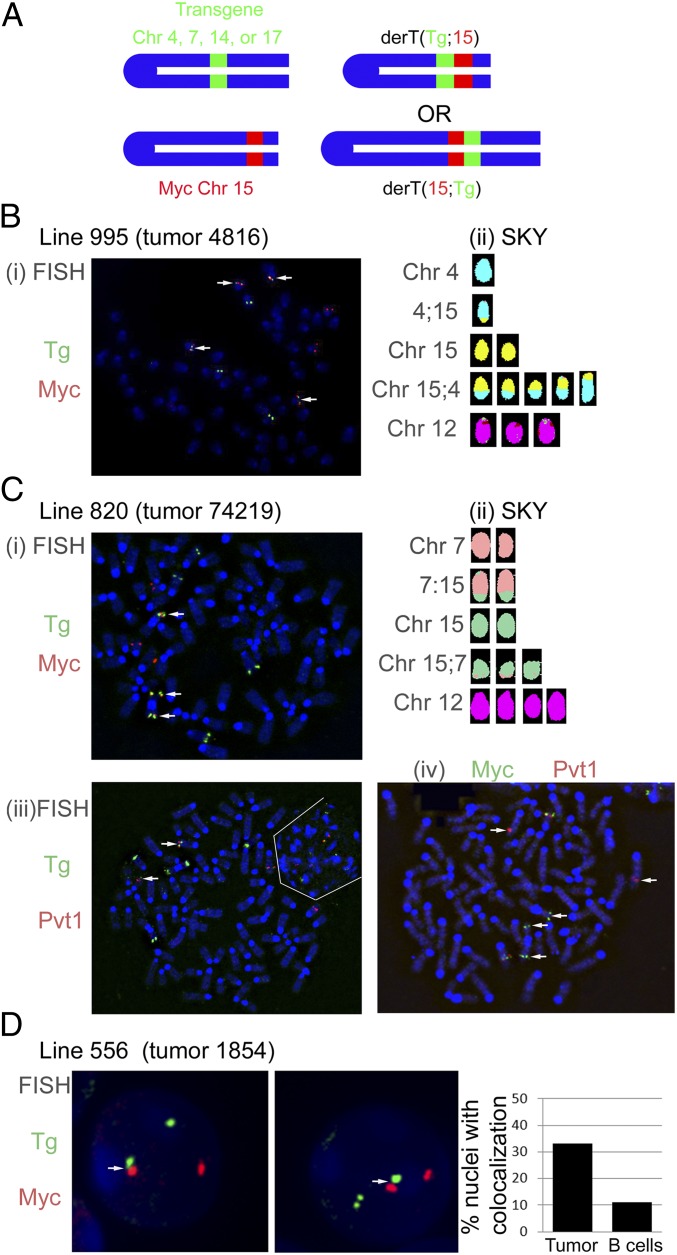
To determine whether these translocations involving the Igh transgene could promote tumor development, mice from each of the three transgenic lines, and two additional transgenic lines, designated line 336 (two copies integrated near the centromere of Chr 14) and 234 (two copies integrated near the telomere of Chr 1), were injected with pristane to induce plasmacytoma. Many of the mice developed abundant ascites containing cells with the characteristic cytology of malignant plasma cells. Primary tumor cells, tumor cells adapted to tissue culture, or highly homogenous tumor cells passaged in SCID or nude mice were characterized for chromosomal translocations by two-color fluorescent in-situ hybridization (FISH) and whole genome spectral karyotyping (SKY) (21). For FISH, metaphase or interphase chromosome spreads were hybridized with a BAC probe spanning the Myc gene on Chr 15. The same chromosome spreads were also hybridized with a second BAC (labeled with a different fluorochrome) that spans the insertion site of the transgene for the given line (Fig. 2A). This second probe lacked any Igh sequences, containing only sequences from Chr 1, 4, 7, 14, or 17. Translocation between the Igh transgene and Myc were defined by colocalization of the two BAC probes. Representative two-color FISH data are shown in Fig. 2 B–D. For example, the tumor from line 995 (Fig. 2B, i) has four Chr with colocalization of the Myc and Igh transgene probes. SKY analysis confirmed the presence of both der (4, 15) and der (15, 4) chromosomes within this tumor (Fig. 2B, ii). Notably, Chr 12 carrying the endogenous Igh was not rearranged. Seven tumors with translocations between Myc and the Igh transgene in line 820 had no rearrangements involving Chr 12 (Table 1). In a single tumor with a T(7;15) translocation, Chr 12 was involved in another translocation (Table 1). More than 50% of the plasmacytomas demonstrated colocalization of the Myc probe and the Igh transgene probe and, therefore, contained translocations between Myc and the Igh transgene (Table 1 and Figs. S2--S5). As expected, the plasmacytomas that lacked a translocation between Myc and the Igh transgene had translocations involving the endogenous Igh locus and Myc (Table 1). Of the five chromosomal locations we tested, all were permissive for translocations to Myc. Apparently, the chromosomal location for line 234 (telomeric on Chr 1) is relatively inefficient for translocations with Myc that lead to plasmacytoma.
Fig. 2.
Plasmacytomas harbor translocations to the Igh transgene. (A) Colocalization of the Igh transgene with Myc or Pvt1. Colocalization of BACs spanning the Igh transgene insertion site on Chr 4 (line 995), 7 (line 820), or 17 (line 556) and either Myc or Pvt1 reveal translocations involving the two loci. White arrows note separation of probes (Civ) or colocalization (C i–iii). (B) Two-color FISH (i) and SKY (ii) on metaphase spreads from tumor no. 4816, line 995. The color coding for Chr 15 for the SKY analysis of these metaphases was changed to yellow to better contrast with the aqua color coding for Chr 4. (C) Two-color FISH and SKY analysis of tumor no. 74219, line 820. In Ci, the Igh transgene is shown to colocalize with Myc. In Ciii, the Igh transgene is shown to colocalize with Pvt1. The white lines delimit a second, interphase cell that lies next to the metaphase. In Civ, the Myc and Pvt1 probes are rearranged onto different chromosomes. (D) Interphase two-color FISH analysis of tumor no. 1854, line 556 (two representative cells shown). Myc and the Igh transgene were colocalized in 33% of the interphases from tumor no. 1854 ascites, and in 11% of the interphases from normal B-cell controls (P < 0.001, Fisher’s exact test).
Table 1.
Myc:Igh translocations
| Transgenic line | Primary tumor | Genetic background | Translocations | Other |
| 995 Chr 4 at 98.3 of 154.3 Mb VDJ centromere proximal | 4816 | C57BL/6, Bcl-xL | 4;15 | Myc-Sμ (cloning), secretes IgA |
| 1022 | C57BL/6, Bcl-xL | 4;15 | ||
| 820 Chr 7 at 122 of 145 Mb VDJ centromere proximal | 74149 | Mixed, Bcl-xL* | 7;15 | |
| 74150 | Mixed, Bcl-xL | 7;15 | ||
| 74160 | Mixed, Bcl-xL | 7;15 and 12;18 | Secretes IgA | |
| 74162 | Mixed, Bcl-xL | 12;15 | ||
| 74163 | Mixed, Bcl-xL | 7;15 | Secretes IgA, Pvt1-Sα (cloning) | |
| 74219 | Mixed, Bcl-xL | 7;15 and 6;13 | ||
| 74220 | Mixed, Bcl-xL | 12;15 | ||
| 74223 | Mixed, Bcl-xL | 7;15 | ||
| 74997 | Mixed, Bcl-xL | 12;15 | ||
| 75000 | Mixed, Bcl-xL | 12;15 | ||
| 75001 | Mixed, Bcl-xL | 7;15 | Chr 4 insertion into 7;15 | |
| 75002† | Mixed, Bcl-xL | 7;15 | Secretes IgA and IgG2a | |
| 75002† | Mixed, Bcl-xL | 12;15 | ||
| 75004 | Mixed, Bcl-xL | 12;15 | ||
| 336 Chr 14 near centromere orientation unknown | 78286 | Mixed, Bcl-xL* | 14:15 | |
| 79130 | Mixed, Bcl-xL | 14?;15‡, 15;18, and 1;4 | Myc-Sγ2a (cloning) | |
| 79134 | Mixed, Bcl-xL | 14;15 | ||
| 79710 | Mixed, Bcl-xL | 14;15 | ||
| 79711 | Mixed, Bcl-xL | 14;15 | ||
| 79716 | Mixed, Bcl-xL | 14;15 and 9;16 | ||
| 80635 | Mixed, Bcl-xL | 14;15 | ||
| 80639 | Mixed, Bcl-xL | 12;15 | ||
| 80648 | Mixed, Bcl-xL | 14;15 | ||
| 82028 | Mixed, Bcl-xL | 12;15 | ||
| 82043 | Mixed, Bcl-xL | 14;15 | ||
| 82048 | Mixed, Bcl-xL | 12;15 | ||
| 82181 | Mixed, Bcl-xL | 14;15 | ||
| 83353 | Mixed, Bcl-xL | 12;15 | ||
| 556 Chr 17 at 89.6 of 95.2 Mb VDJ centromere distal | 1854 | BALB/cAn | 15;17 | Interphase, Myc-Sα (cloning) |
| 2731 | C57BL/6, Bcl-xL | not 15;17 | Interphase | |
| 234 Chr 1 near telomere | 78709 | Mixed, bclxL* | 12;15 | |
| 79145 | Mixed, bclxL | 12;15 | ||
| 80616 | Mixed, Bcl-xL | 1;15 and 6;13 | ||
| 80621 | Mixed, Bcl-xL | 12;15 | ||
| 83366 | Mixed, Bcl-xL | 12;15 |
A concern of the transgenic approach is that unusual transcription rates or chromosomal structure of the Igh transgenes may create a nonphysiologic target for Myc translocations. It is possible that multiple copies of large Igh transgenes, with their strong regulatory elements, result in nonphysiologic chromatin structures. Such a criticism is difficult to rule out in the absence of any experimental data for or against this possibility. To equalize the Igh transgenic and endogenous loci to the extent possible, we “knocked-in” an assembled VH region segment, with its physiologic promoter, into one of the endogenous loci in the single-copy line 820. In line 820, where the transcriptional activity of the transgene and the endogenous genes should be similar, the frequency of translocation to the transgene and endogenous Igh genes was virtually identical (Table 1). Analogous results were obtained with the two-copy line 336 (Table 1).
Either Orientation of the Igh Transgene Is Able to Undergo Translocation.
In approximately 85% of mouse plasmacytomas and human Burkitt lymphomas, the Igh locus is translocated 5′ of Myc coding exons (3–6). Conversely, the remaining 15% of plasmacytomas juxtapose Igκ or Igλ sequences 3′ of Myc, to a region known as plasmacytoma variant translocation locus or Pvt1 (3, 22, 23). This bias is best explained by the orientation of heavy and light chain loci relative to their respective centromeres. Light chain gene rearrangements 5′ of Myc are thought to be precluded because they would generate dicentric chromosomes that are incompatible with cell viability. Because the orientation of the transgene in line 820 is the same as Igκ and Igλ (Fig. 1B), we speculated that translocations involving the Igh transgene in this line would deregulate Myc primarily through Pvt1 rearrangments. Indeed, two-color FISH analysis showed colocalization of Pvt1 and the Igh transgene in tumors from this line (Fig. 2C, iii). Furthermore, two-color FISH revealed that Myc and part of the Pvt1 probe are rearranged to separate chromosomes, as would be expected if the Pvt1 locus was used in the translocation (Fig. 2C, iv). SKY confirmed the presence of a T(7;15) translocation (Fig. 2C, ii and Fig. S4).
Translocations of Myc to the Transgene and to the Endogenous Igh Are Similar at the Molecular Level.
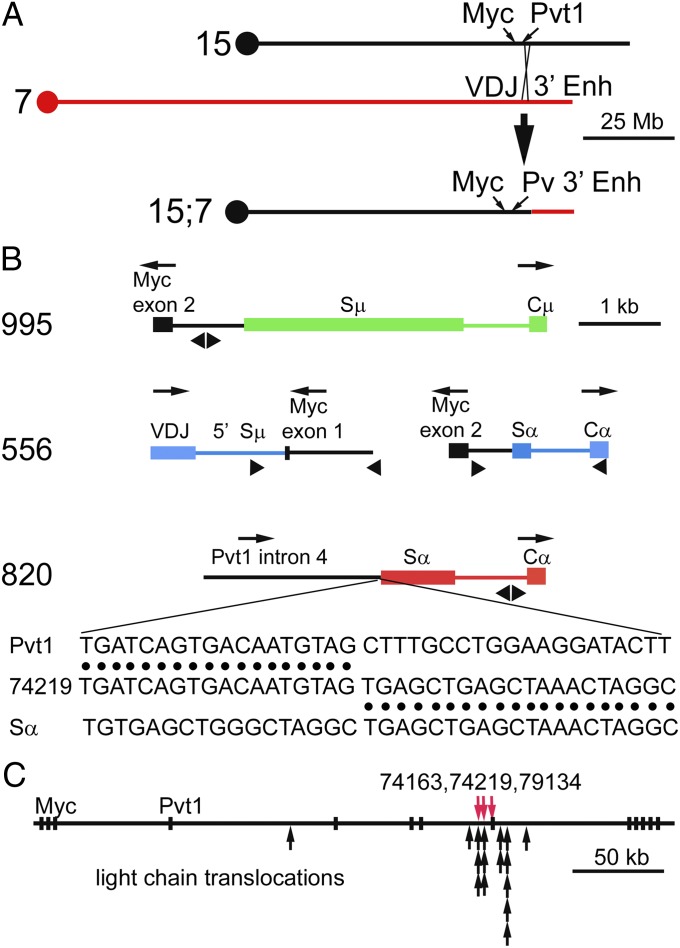
To characterize the translocations at the molecular level, the translocation site from at least one tumor from each of four transgenic lines was cloned and sequenced (Fig. 3). The translocation event from a line 995 tumor was typical of Myc:Igh translocations, joining exons 2 and 3 of Myc to the 3′ end of Igh, in the opposite transcriptional orientation (Fig. 3B and Table S2). The recombination event occurred in the first intron of Myc and in the Sμ region of the Igh transgene. Recombination sites in two tumors from line 336 were also typical Myc:Igh translocations. Recombination in the first intron of Myc and in Igh Sγ2 joined exons 2 and 3 of Myc to the 3′ end of the Igh locus (Table S2). Both products of the reciprocal translocation from a tumor from line 556 were cloned and sequenced. Both recombination products were similar to Myc:Igh translocations involving the endogenous Igh in terms of translocation sites in Myc, Sμ, and Sα, and in terms of orientation (Fig. 3B). Molecular cloning of the translocation site in a tumor from line 820 was consistent with the results of FISH and SKY analysis—sequences in the fourth intron of the Pvt1 gene were joined to Sα from the transgene in a 3′ to 5′ orientation (Fig. 3B and Table S2). Molecular cloning revealed that the translocation sites in a second line 820 tumor and in a line 336 tumor were also in the fourth intron/fifth exon of Pvt1 (Table S2). The fourth intron/fifth exon/fifth intron of the Pvt1 gene may be unusually susceptible to dsDNA breaks in B cells, because it is a site of RNA polymerase initiation or pausing and of AID binding (24). Most Pvt1:kappa light chain translocations are located in this part of the Pvt1 gene (Fig. 3C) (25). Frequent translocation by the Igh transgene in line 820 suggests that the orientation of the Igh locus is more or less irrelevant for oncogenic rearrangements. Apparently, the cell population examines all possible translocations (26, 27), and those translocations that allow cell viability and neoplastic transformation are selected.
Fig. 3.
Molecular characteristics of translocations between the Igh transgene and Myc. (A) Schematic diagrams of Chr 15 with the approximate location of the Myc and Pvt1 genes, Chr 7 with the Igh transgene, and the resulting derivative Chr 15 after translocation. (B) Molecular clones of translocations to the Igh transgenes. These molecular clones are from primary tumor no. 4816 (line 995), from primary tumor no. 1854 [line 556, both der (15) and der (17) translocation products], and from tumor no. 74219 (line 820). Igh transgenic sequences at the translocation site are color coded according to the chromosome on which the transgene resides. Myc- or Pvt1-associated sequences (Chr 15) are shown in black. The transcriptional direction of each gene is shown above each schematic. Triangles below each schematic approximate the location of PCR primers used to clone these translocation sites. The sequence data corresponding to the structure shown in the line 820 schematic is presented below the schematic. Sequence data for other translocation sites in tumors are shown in Table S2. (C) Structure of the Myc and Pvt1 loci on Chr 15. Exons are depicted as black rectangles. The Myc gene is encoded by the three exons on the left of the map; the Pvt1 transcripts are encoded by the next exon designated “Pvt1” and the remaining nine exons (and additional exons not included in this figure). The translocation sites in tumors 74163, 74219, and 79134 are shown as red arrows. Translocation sites to light chain genes in plasmacytomas (25) are shown below as black arrows.
Discussion
We sought to determine whether the recurrent Igh:Myc translocations in human Burkitt lymphoma and in murine plasmacytoma depend on the particular chromosomal location of Igh and its orientation relative to the centromere. To determine the role of chromosomal location, we used a 230-kb Igh transgene inserted into five different chromosomal locations and in both orientations relative to the centromere. Each of the five Igh transgenes is capable of class switch recombination at near physiologic levels (18, 28), a likely prerequisite for translocation (1, 4–6). We found that the Myc proto-oncogene can translocate to a 230-kb Igh locus inserted into any of five different chromosomal locations (Fig. 2 and Table 1). Not only was the transgene able to undergo translocations with Myc, but the frequency of translocation to the transgenic Igh was at least equal to the frequency of translocation to the endogenous Igh. At the molecular level, Myc translocations to the Igh transgene are similar, if not identical, to translocations to the endogenous Igh (Fig. 3, Table S1, and Table S2).
Lymphoid cells expressing the RAG endonuclease or AID can suffer any one of thousands of different translocations that are spread throughout the genome (26, 27, 29, 30). Presumably it is selection for the Igh:Myc translocation, and its resulting deregulated Myc expression, that leads to the recurrence of this translocation in human lymphoma and murine plasmacytoma. However, Igh:Myc translocations are overrepresented in the primary pool of translocations (26, 27, 29, 31). Several factors have been hypothesized to increase the frequency of Igh:Myc translocation, including gene proximity due to chromosome territories (32, 33), gene proximity perhaps due to shared transcription factors (34, 35), high frequency of DNA breaks after AID activity (29), gene proximity after DNA breaks perhaps due to sharing of repair machinery (32), and failure to repair after DNA breaks (16, 36). Chromosome environment is likely to play a role in some or all of these factors. The Igh transgene therefore represents a valuable model for future studies on the effect of different chromosome environments on translocation and each of the potential factors mentioned above.
The 230-kb transgene is sufficient not only for the recombination event with Myc, but it also includes all of the regulatory elements necessary for oncogenic deregulation of Myc (Table 1). Because the Igh 3′ regulatory region is required for the genesis of plasmacytomas (17, 37–39), it is likely that the 3′ regulatory region in the Igh transgene both enhances Myc recombination to the transgene and deregulates Myc expression, resulting in tumor development. Thus, putative regulatory elements outside of the 230-kb constant region gene region play only a minor role for oncogenic translocation and for Myc deregulation.
Materials and Methods
Identification of Transgene Insertion Sites into the Mouse Genome.
Transgenic DNA was digested with Sau3A and ligated to form circles. Primers were used from the 3′ end of the Igh transgene that would amplify around the circle (including the adjacent chromosomal insertion sequences—see Fig. S1 and Table S3). The resulting PCR product was cloned and sequenced as described in detail in SI Materials and Methods.
Plasmacytoma induction and analysis.
Plasmacytomas were induced with pristane in mice expressing a Bcl-xL transgene or on a BALB/cAN background (SI Materials and Methods). All mouse studies were approved by the institutional animal use committees at the University of Michigan or the National Cancer Institute. FISH and SKY analysis of tumor cells used standard reagents and approaches (SI Materials and Methods and Table S4). Molecular clones of tumor translocation sites were obtained by PCR-based approaches as described in detail in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. Michael Potter, Seigfried Janz, and Konrad Huppi for advice; Jian Shi for technical assistance; and Dr. Gary Huffnagel for the use of his labotatory’s cytospin centrifuge and reagents. This work was supported by National Institutes of Health (NIH) Grants AI068749 and AI076057 (to W.D.), HL079118 (to D.O.F.), and T32-AI007413 (to E.S.) and, in part, by the Intramural Research Program of the NIH, National Institute of Allergy and Infectious Diseases, National Cancer Institute, and National Institute of Arthritis and Musculoskeletal and Skin Diseases.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. JX080033–JX080057).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1202882109/-/DCSupplemental.
References
- 1.Nussenzweig A, Nussenzweig MC. Origin of chromosomal translocations in lymphoid cancer. Cell. 2010;141:27–38. doi: 10.1016/j.cell.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gostissa M, Alt FW, Chiarle R. Mechanisms that promote and suppress chromosomal translocations in lymphocytes. Annu Rev Immunol. 2011;29:319–350. doi: 10.1146/annurev-immunol-031210-101329. [DOI] [PubMed] [Google Scholar]
- 3.Cory S. Activation of cellular oncogenes in hemopoietic cells by chromosome translocation. Adv Cancer Res. 1986;47:189–234. doi: 10.1016/s0065-230x(08)60200-6. [DOI] [PubMed] [Google Scholar]
- 4.Potter M. Neoplastic development in plasma cells. Immunol Rev. 2003;194:177–195. doi: 10.1034/j.1600-065x.2003.00061.x. [DOI] [PubMed] [Google Scholar]
- 5.Janz S. Myc translocations in B cell and plasma cell neoplasms. DNA Repair (Amst) 2006;5:1213–1224. doi: 10.1016/j.dnarep.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 6.Küppers R, Dalla-Favera R. Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene. 2001;20:5580–5594. doi: 10.1038/sj.onc.1204640. [DOI] [PubMed] [Google Scholar]
- 7.Cogne M, Birshtein BK. In: Molecular Biology of B Cells. Honjo T, Alt FW, Neuberger MS, editors. London: Academic; 2004. pp. 289–305. [Google Scholar]
- 8.Gostissa M, Ranganath S, Bianco JM, Alt FW. Chromosomal location targets different MYC family gene members for oncogenic translocations. Proc Natl Acad Sci USA. 2009;106:2265–2270. doi: 10.1073/pnas.0812763106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kovalchuk AL, et al. AID-deficient Bcl-xL transgenic mice develop delayed atypical plasma cell tumors with unusual Ig/Myc chromosomal rearrangements. J Exp Med. 2007;204:2989–3001. doi: 10.1084/jem.20070882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rooney S, et al. Artemis and p53 cooperate to suppress oncogenic N-myc amplification in progenitor B cells. Proc Natl Acad Sci USA. 2004;101:2410–2415. doi: 10.1073/pnas.0308757101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manis JP, Tian M, Alt FW. Mechanism and control of class-switch recombination. Trends Immunol. 2002;23:31–39. doi: 10.1016/s1471-4906(01)02111-1. [DOI] [PubMed] [Google Scholar]
- 12.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 13.Revy P, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 14.Ramiro AR, et al. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 2004;118:431–438. doi: 10.1016/j.cell.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 15.Dorsett Y, et al. A role for AID in chromosome translocations between c-myc and the IgH variable region. J Exp Med. 2007;204:2225–2232. doi: 10.1084/jem.20070884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robbiani DF, et al. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135:1028–1038. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gostissa M, et al. Long-range oncogenic activation of Igh-c-myc translocations by the Igh 3′ regulatory region. Nature. 2009;462:803–807. doi: 10.1038/nature08633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunnick WA, et al. Switch recombination and somatic hypermutation are controlled by the heavy chain 3′ enhancer region. J Exp Med. 2009;206:2613–2623. doi: 10.1084/jem.20091280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janz S, Müller J, Shaughnessy J, Potter M. Detection of recombinations between c-myc and immunoglobulin switch alpha in murine plasma cell tumors and preneoplastic lesions by polymerase chain reaction. Proc Natl Acad Sci USA. 1993;90:7361–7365. doi: 10.1073/pnas.90.15.7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramiro AR, et al. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 2006;440:105–109. doi: 10.1038/nature04495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liyanage M, et al. Multicolor spectral karyotyping of mouse chromosomes. Nat Genet. 2004;14:312–315. doi: 10.1038/ng1196-312. [DOI] [PubMed] [Google Scholar]
- 22.Webb E, Adams JM, Cory S. Variant (6; 15) translocation in a murine plasmacytoma occurs near an immunoglobulin κ gene but far from the myc oncogene. Nature. 1984;312:777–779. doi: 10.1038/312777a0. [DOI] [PubMed] [Google Scholar]
- 23.Siwarski D, et al. Structure and expression of the c-Myc/Pvt 1 megagene locus. Curr Top Microbiol Immunol. 1997;224:67–72. doi: 10.1007/978-3-642-60801-8_6. [DOI] [PubMed] [Google Scholar]
- 24.Yamane A, et al. Deep-sequencing identification of the genomic targets of the cytidine deaminase AID and its cofactor RPA in B lymphocytes. Nat Immunol. 2011;12:62–69. doi: 10.1038/ni.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huppi K, Pitt J, Wahlberg B, Caplen NJ. Genomic instability and mouse microRNAs. Toxicol Mech Methods. 2011;21:325–333. doi: 10.3109/15376516.2011.562759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klein IA, et al. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell. 2011;147:95–106. doi: 10.1016/j.cell.2011.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiarle R, et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell. 2011;147:107–119. doi: 10.1016/j.cell.2011.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dunnick WA, Shi J, Graves KA, Collins JT. Germline transcription and switch recombination of a transgene containing the entire H chain constant region locus: Effect of a mutation in a STAT6 binding site in the γ 1 promoter. J Immunol. 2004;173:5531–5539. doi: 10.4049/jimmunol.173.9.5531. [DOI] [PubMed] [Google Scholar]
- 29.Hakim O, et al. DNA damage defines sites of recurrent chromosomal translocations in B lymphocytes. Nature. 2012;484:69–74. doi: 10.1038/nature10909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hasham MG, et al. Widespread genomic breaks generated by activation-induced cytidine deaminase are prevented by homologous recombination. Nat Immunol. 2010;11:820–826. doi: 10.1038/ni.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, et al. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148:908–921. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meaburn KJ, Misteli T, Soutoglou E. Spatial genome organization in the formation of chromosomal translocations. Semin Cancer Biol. 2007;17:80–90. doi: 10.1016/j.semcancer.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parada LA, McQueen PG, Munson PJ, Misteli T. Conservation of relative chromosome positioning in normal and cancer cells. Curr Biol. 2002;12:1692–1697. doi: 10.1016/s0960-9822(02)01166-1. [DOI] [PubMed] [Google Scholar]
- 34.Roix JJ, McQueen PG, Munson PJ, Parada LA, Misteli T. Spatial proximity of translocation-prone gene loci in human lymphomas. Nat Genet. 2003;34:287–291. doi: 10.1038/ng1177. [DOI] [PubMed] [Google Scholar]
- 35.Osborne CS, et al. Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biol. 2007;5:e192. doi: 10.1371/journal.pbio.0050192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu M, et al. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- 37.Madisen L, Groudine M. Identification of a locus control region in the immunoglobulin heavy-chain locus that deregulates c-myc expression in plasmacytoma and Burkitt’s lymphoma cells. Genes Dev. 1994;8:2212–2226. doi: 10.1101/gad.8.18.2212. [DOI] [PubMed] [Google Scholar]
- 38.Truffinet V, et al. The 3′ IgH locus control region is sufficient to deregulate a c-myc transgene and promote mature B cell malignancies with a predominant Burkitt-like phenotype. J Immunol. 2007;179:6033–6042. doi: 10.4049/jimmunol.179.9.6033. [DOI] [PubMed] [Google Scholar]
- 39.Duan H, Heckman CA, Boxer LM. The immunoglobulin heavy-chain gene 3′ enhancers deregulate bcl-2 promoter usage in t(14;18) lymphoma cells. Oncogene. 2007;26:2635–2641. doi: 10.1038/sj.onc.1210061. [DOI] [PubMed] [Google Scholar]
- 40.Pelanda R, et al. Receptor editing in a transgenic mouse model: Site, efficiency, and role in B cell tolerance and antibody diversification. Immunity. 1997;7:765–775. doi: 10.1016/s1074-7613(00)80395-7. [DOI] [PubMed] [Google Scholar]
- 41.Grillot DAM, et al. bcl-x exhibits regulated expression during B cell development and activation and modulates lymphocyte survival in transgenic mice. J Exp Med. 1996;183:381–391. doi: 10.1084/jem.183.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.