The process by which a linear sequence of amino acids folds into a discrete and functional three-dimensional protein is one on which life depends. Although the code that governs folding remains a mystery, we do know that the primary sequence is subject to evolutionary pressure to adjust folding rate and product stability according to physiological needs. The failure of a protein to fold correctly leads to a functional deficit, which can have serious consequences, as in cystic fibrosis. However, there is an emerging class of late-onset, slow-progressing diseases that appear to result instead from a gain of function associated with the abnormally folded form of the protein. These diseases, which are characterized by ordered, fibrillar aggregates comprising different proteins, include common neurodegenerative diseases such as Alzheimer’s disease (AD) and Parkinson’s disease (PD) as well as many rare systemic diseases such as familial amyloid polyneuropathy (1, 2).
What do the fibrillizing proteins have in common? Although all of the fibrils share some morphological features (3–6), examination of the primary sequences of the constituent proteins does not reveal a conserved motif. In fact, the ability to form fibrils in vitro is not limited to the disease-associated proteins, as illustrated by the paper of Chiti et al. in this issue of the Proceedings (7) and an earlier paper from the same group (8). Thus two globular proteins with no homology to the disease-associated proteins or to each other have the ability to form fibrils that resemble those extracted from diseased tissue. In both cases, the globular proteins are prone to fibril formation under conditions that stabilize a partially folded structured form of the protein. Most proteins appear to have evolved to fold without populating such an intermediate (9). The rationale for this phenomenon and its possible implications for degenerative disease will be discussed.
A Group of Age-Associated Degenerative Diseases Are Characterized by Abnormally Folded, Fibrillar Proteins
Extracellular fibrillar protein deposits, or amyloid plaques, are characteristic of a group of degenerative diseases that affect the brain, pancreas, heart, kidneys, and other tissues (1). The “native” properties of the constituent amyloid proteins vary: some are soluble oligomers in vivo (transthyretin in familial amyloid polyneuropathy), whereas others are flexible peptides (Aβ in AD). AD, the most prevalent of the amyloid diseases, is diagnosed based on the presence of extracellular amyloid plaques together with cytoplasmic neuronal inclusions (neurofibrillary tangles) in the affected regions of the brain (10). Other neurodegenerative diseases, PD being the most prevalent, are characterized by the presence of cytoplasmic protein inclusions in the affected regions of the brain (e.g., Lewy bodies comprising fibrillar α-synuclein in PD, neurofibrillary tangles comprising fibrillar tau in frontotemporal dementia, nuclear inclusions comprising fibrillar huntingtin in Huntington’s disease) (2, 11).
Mutant Genes Linked to Familial Disease Effect Changes That Accelerate in Vitro Fibrillization by Several Mechanisms, Suggesting a Causal Role for Fibrillogenesis
AD and PD exist in two forms, the common sporadic form and a rare early-onset form, transmitted by autosomal dominant inheritance (2). Early-onset PD was linked to two point mutations in the gene encoding α-synuclein (12, 13), the primary component of Lewy body-derived fibrils (11). One of these mutations promotes in vitro fibril formation by α-synuclein (6). Similarly, all of the early-onset AD mutations, located to three genes, influence the properties of Aβ or the production of Aβ congeners in a manner that accelerates in vitro fibrillization, consistent with acceleration of fibril formation also being the pathogenic mechanism (10, 14). Factors responsible for fibrillization in sporadic AD and PD have yet to be determined. The mutant gene products associated with the polyglutamine expansion diseases (e.g., Huntington’s disease) are also constituents of the characteristic inclusions. Polyglutamine-containing proteins form amyloid fibrils in vitro (15). Finally, some familial systemic amyloid diseases result from mutations in globular proteins that destabilize their folded structures; transthyretin mutants associated with familial amyloid polyneuropathy have been shown to be more prone to unfolding under acidic conditions (16), and lysozyme mutants linked to systemic amyloid disease have reduced stability (17). The recurring correlation between fibrillar pathologies, disease-associated mutations, and predisposition to fibrillization strongly suggests that the process of fibril formation initiates the pathogenic cascade.
The Key to the Underlying Etiology of These Diseases May Lie in a Careful Examination of the Differences Between Normal Folding and Pathogenic Fibrillization
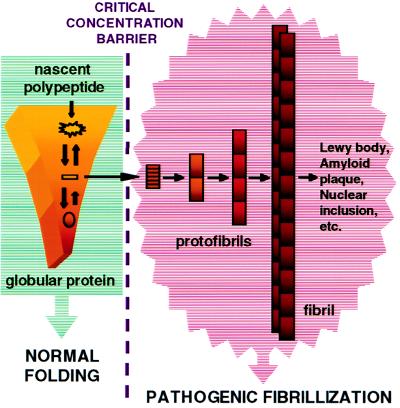
Fig. 1 offers a simplified depiction of some critical differences between the two processes discussed below:
Figure 1.
A simplified mechanistic scenario highlighting the differences between normal folding and pathogenic fibrillization. The approximate free energy of each species is encoded by color; from the least stable in yellow to the most stable in red. Normal folding occurs by an intermediate-free pathway involving a descent through a funnel-like energy surface to the low-energy folded structure. The fibrillogenic intermediate depicted as an orange rectangle will not accumulate under normal circumstances. However, point mutations in the protein sequence could stabilize this species relative to the folded state, for example, by increasing the rate of unfolding of the native state. Fibril formation will not occur, however, unless a critical concentration of this intermediate accumulates. With respect to pathogenic fibrillization, structured intermediates, designated protofibrils, are populated during the process, an important distinction between the pathways. The intermediate protofibrils grow slowly, then are rapidly converted to large amyloid fibrils (4).
Folding and Unfolding Are Typically Concentration Independent, While Fibrillization Is Likely, Under in Vivo Conditions, To Be Highly Concentration Dependent.
With few exceptions, globular proteins have evolved to fold as monomers under conditions of “infinite dilution.” Fibril nucleaton, in contrast, may require the cooperative ordered aggregation of many monomers. Thus, fibril formation requires a critical concentration of protein to be reached to occur at all (Fig. 1) (18, 19). Because the critical concentration can depend on the environment, slight alterations in lysosomal pH, for example, could promote fibrillization (16). Once the concentration creeps above the critical concentration, nucleation is likely to be highly concentration dependent, thus small changes in local protein concentration could abruptly shift the protein population from nearly 100% monomeric and soluble to mostly oligomeric and fibrillar. Chiti et al. (7) note the importance of protein concentration for the production of acylphosphatase amyloid fibrils in vitro, by analogy to crystal growth (19). To prevent the critical concentration from being exceeded in vivo, the cell has evolved the chaperone system to enforce “infinite dilution” by binding to nascent polypeptide chains and preventing their aggregation or sequestering them so that folding to the globular form has a chance to occur. Furthermore, unfolded proteins that escape the chaperone system are degraded by the proteasome. Gradual decreases in the efficiency of these systems with aging (many of the components are ATP dependent) could lead to late-onset disease. However, the fact that only a select group of proteins are found in disease-associated fibrils suggests that these proteins are subject to episodes of abnormally high expression such that the chaperone/proteasome system is temporarily overwhelmed.
Folding and Fibrillization Both Reduce a Protein’s Entropy, But in the Highly Concentrated Cytoplasmic Environment, Fibril Formation Can Increase the Total Protein Entropy.
Most in vitro experiments are done in dilute solutions with a single protein component. However, the typical cytoplasm is a nonideal medium with a total protein concentration approaching 1 M. Thus fibril formation could be induced by increasing the concentration of “innocent bystander” globular proteins that can increase their own translational entropy by removing fibrillogenic proteins from the medium. This phenomenon, dubbed “molecular crowding” (20), could be a factor in increasing flux of protein down the fibrillogenic pathway by effectively reducing the critical concentration. For example, one could imagine that a burst of protein production associated with a neuronal repair mechanism could induce fibrillization of an unrelated protein that is approaching its critical concentration.
Though Fibril-Forming Proteins Often Contain Little β-Sheet Structure in Their Monomeric Forms, the Fibrillar Forms Are Always Enriched in β-Sheet Secondary Structure.
Fibrillizing proteins include some largely unstructured, “natively unfolded” proteins (Aβ and tau in AD, ref. 21; α-synuclein in PD, ref. 22), some that are primarily β-sheet (transthyretin in familial amyloid polyneuropathy, ref. 16), but most, like acyl phosphatase (7), are typical globular proteins with helical structure as well as β-sheet structure, most of which is located in the protein interior. All amyloid fibrils contain β-sheet structure in which the peptide strands are aligned orthogonal to the direction of fibril growth (1). This “cross-β” motif may be responsible for the similar widths of most fibrils; the typical β-strand length in globular proteins is ca. 8–10 residues because of strand twisting, which corresponds to a β-sheet with a width of 28–35 Å, similar to the width/diameter of filaments that seem to be structural subunits of most fibrils (3, 5, 23, 24). Although sequence is presumed to play some role in the conversion from a multiconformational globular protein to the predominantly β-sheet fibril, a striking sequence dependence would not be expected, because the energetic differences associated with assumption of β-strand structure between all the amino acids except proline are small (25).
The Sequences of Globular Proteins Have Responded to Evolutionary Pressures to Fold Rapidly, Without Populating Structured Intermediates.
The sequences of globular proteins have been subject to evolutionary pressure to adjust the kinetic and thermodynamic stability of the folded structure and to optimize the efficiency of the folding pathway. Optimal folding involves the avoidance of structured intermediates that can act as kinetic traps, delaying arrival at the correct folded structure (9). Thus, the process can be thought of as an energy funnel (see Fig. 1), where there are many pathways, all of which lead energetically downhill to the final folded structure. The first step involves rapid collapse of the polypeptide to a “molten globule” form in which the hydrophobic residues are removed from aqueous solvent. Subsequently, secondary and tertiary structure rapidly and simultaneously evolve, without population of intermediate states. To stabilize and structurally elucidate potential kinetic intermediates, the Dobson research group at Oxford (7, 8), and others, utilize unusual solvent conditions, often involving low pH media or organic cosolvents. These are the precise conditions that give rise to amyloid fibrils in vitro, when the protein concentration exceeds the critical concentration. Stable structured intermediates, especially those containing exposed β-sheet hydrogen-bonding edges, pose a risk of self-association or high-affinity interactions with other proteins because they have prepaid the entropic cost of β-sheet formation and may have exposed hydrophobic surfaces of the type normally sequestered in globular proteins (26). In contrast, the sequences of fibrillizing proteins are not subject to evolutionary pressure to optimize the fibril or its assembly, but merely to avoid sequences that would lead to juvenile-onset diseases that could interfere with reproduction. Thus, only mutations that promote very rapid fibrillization and possibly, juvenile-onset disease, would be selected against.
Many in Vitro Fibrillization Processes Involve Structured Prefibrillar Oligomeric Intermediates.
Normal (e.g., flagella and microtubule formation, refs. 19 and 27) and disease-associated (sickle-cell fibril formation, ref. 28) nucleation-dependent polymerization processes proceed without the population of intermediate species. In contrast, the in vitro fibrillogenesis processes discussed herein, although they also require a critical protein concentration and are susceptible to seeding with preformed fibrils, involve a discrete, structured intermediate. Such an intermediate first was identified in the case of the Aβ protein of AD and designated the Aβ amyloid protofibril (4, 29) (see Fig. 1). Aβ protofibrils contain β-sheet structure (D. Walsh and D. Teplow, personal communication) and can be rapidly converted to fibrils in the presence of a small amount of preformed Aβ fibril seeds (5). Transthyretin, α-synuclein, and the type II diabetes-linked islet amyloid polypeptide all form morphologically similar species (6, 16, 30). Strikingly, the fibrillization of acylphosphatase also proceeds via a protofibrillar intermediate (7).
Is a Structured Prefibrillar Intermediate Pathogenic?
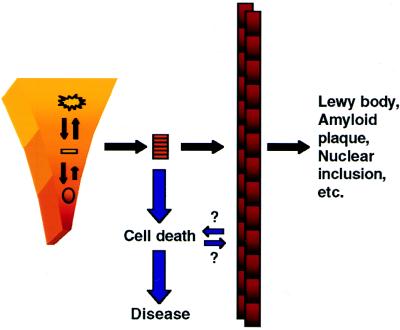
The shared general features among all in vitro fibrillization processes suggest that structured intermediates may be responsible for the biological consequences of the in vivo processes. Like the monomeric folding intermediates discussed above, a β-sheet-containing oligomeric species could bind tightly to any number of cellular targets, triggering, for example, a cytotoxic cascade (see Fig. 2). The smallest intermediate in which the binding site (possibly a β-sheet) is stable would have the greatest specific activity (moles binding site per mole protein). Thus, polymerization would compete with binding and specific activity would decrease as polymerization continued. In fact, the fibril itself actually may be protective; fibrillization would be an efficient way for the cell to sequester potentially toxic protofibrils. This proposal has ramifications for the design of screens for discovery of candidate therapeutic agents, because it suggests that some of the compounds that inhibit fibril formation actually could produce a deleterious effect by causing accumulation of a prefibrillar toxic species.
Figure 2.
A “toxic intermediate” proposal for the underlying etiology of degenerative disease, motivated by consideration of the evolved energetics of protein folding, as discussed in the text. The small arrows topped by questions marks are meant to indicate uncertainty regarding the potential importance of slight biological activity of the fibril and the possibility that cell death could trigger the protofibril-to-fibril transition.
Although there is no direct evidence for protofibrillar intermediates in vivo, their existence can explain many observations that are inconsistent with the fibril itself being the toxic species. First, several in vitro biological activities that have been ascribed to the Aβ amyloid fibril seem to peak early in the aggregation process and then diminish, consistent with a prefibrillar intermediate being most active (33). A nonfibrillar Aβ aggregate with dimensions indistinguishable from the early Aβ protofibril (4), has been reported to have a spectrum of potentially relevant biological activities (34). Second, in cellular and animal models of polyglutamine expansion diseases, the number of aggregates detectable by light microscopy can be reduced by several means, all of which lead to increased cell death, consistent with an accumulated intermediate being toxic yet undetectable (31, 32). Third, the brain of a transgenic mouse model of familial prion disease contains a form of the prion protein at birth that is abnormally protease-resistant, albeit not nearly as resistant as the disease-associated form (PrPSc) (35). As the animal ages, the protease-resistance of brain PrP progressively increases and symptoms become apparent before PrP-containing fibrils can be detected. Fourth, a study of the cortex of a patient with diffuse Lewy body disease (2) showed that apoptotic markers were more frequently observed in neurons not containing Lewy bodies, consistent with the idea that fibrillar inclusions are neuroprotective (36). Finally, detailed behavioral and electrophysiological studies of transgenic animals that overexpress the amyloid precursor protein and develop amyloid plaques show that certain changes precede the detection of amyloid (37). In conclusion, this “toxic intermediate” scenario clearly points to a series of diagnostic experiments that will test aspects of the proposal and will motivate future drug discovery efforts. It also is consistent with both extreme models that have dominated debate in the AD field (10) and, recently, the Huntington’s disease field (31, 32): first, that fibrils are an epiphenomenon linked to disease, and second, that fibril formation causes disease. These are not, in fact, mutually exclusive. The simple proposal depicted in Fig. 2 argues that both of these statements may be correct, since fibrils are an epiphenomenon of protofibril formation and their formation is linked to that of the pathogenic protofibril.
Acknowledgments
I am grateful for illuminating conversations with my research group and with Michael Hecht, Eugene Shakhnovich, Michael Morrisey, Dominic Walsh, and Dean Hartley. I thank Dominic Walsh and David Teplow for sharing information before its publication.
Footnotes
The companion to this Commentary is published on page 3590.
References
- 1.Kelly J W, Lansbury P T., Jr Amyloid Int J Exp Clin Invest. 1994;1:186–205. [Google Scholar]
- 2.Lansbury P T., Jr Neuron. 1997;19:1151–1154. doi: 10.1016/s0896-6273(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 3.Blake C, Serpell L. Structure (London) 1996;4:989–998. doi: 10.1016/s0969-2126(96)00104-9. [DOI] [PubMed] [Google Scholar]
- 4.Harper J D, Wong S S, Lieber C M, Lansbury P T., Jr Chem Biol. 1997;4:119–125. doi: 10.1016/s1074-5521(97)90255-6. [DOI] [PubMed] [Google Scholar]
- 5.Harper J D, Lieber C M, Lansbury P T., Jr Chem Biol. 1997;4:951–959. doi: 10.1016/s1074-5521(97)90303-3. [DOI] [PubMed] [Google Scholar]
- 6.Conway K, Harper J, Lansbury P T., Jr Nat Med. 1998;4:1318–1320. doi: 10.1038/3311. [DOI] [PubMed] [Google Scholar]
- 7.Chiti F, Webster P, Taddei N, Clark A, Stefani M, Ramponi G, Dobson C M. Proc Natl Acad Sci USA. 1999;96:3590–3594. doi: 10.1073/pnas.96.7.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guijarro J I, Sunde M, Jones J A, Campbell I D, Dobson C M. Proc Natl Acad Sci USA. 1998;95:4224–4228. doi: 10.1073/pnas.95.8.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolynes P G, Onuchic J N, Thirumalai D. Science. 1995;267:1619–1620. doi: 10.1126/science.7886447. [DOI] [PubMed] [Google Scholar]
- 10.Selkoe D. Science. 1997;275:630–631. doi: 10.1126/science.275.5300.630. [DOI] [PubMed] [Google Scholar]
- 11.Spillantini M G, Murrell J R, Goedert M, Farlow M R, Klug A, Ghetti B. Proc Natl Acad Sci USA. 1998;95:7737–7741. doi: 10.1073/pnas.95.13.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polymeropoulos M H, Lavedan C, Leroy E, Ide S E, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, et al. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 13.Kruger R, Kuhn W, Muller T, Woitalla D, Graeber M, Kosel S, Przuntek H, Epplen J T, Schols L, Riess O. Nat Genet. 1998;18:106–108. doi: 10.1038/ng0298-106. [DOI] [PubMed] [Google Scholar]
- 14.Jarrett J T, Berger E P, Lansbury P T., Jr Biochemistry. 1993;32:4693–4697. doi: 10.1021/bi00069a001. [DOI] [PubMed] [Google Scholar]
- 15.Scherzinger E, Lurz R, Turmaine M, Mangiarini L, Hollenbach B, Hasenbank R, Bates G P, Davies S W, Lehrach H, Wanker E E. Cell. 1997;90:549–558. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- 16.Lashuel H A, Lai Z, Kelley J W. Biochemistry. 1998;37:17851–17864. doi: 10.1021/bi981876+. [DOI] [PubMed] [Google Scholar]
- 17.Booth D R, Sunde M, Bellotti V, Robinson C V, Hutchinson W L, Fraser P E, Hawkins P N, Dobson C M, Radford S E, Blake C E, Pepys M B. Nature (London) 1997;385:787–793. doi: 10.1038/385787a0. [DOI] [PubMed] [Google Scholar]
- 18.Harper J D, Lansbury P T., Jr Annu Rev Biochem. 1997;66:385–407. doi: 10.1146/annurev.biochem.66.1.385. [DOI] [PubMed] [Google Scholar]
- 19.Jarrett J T, Lansbury P T., Jr Cell. 1993;73:1055–1058. doi: 10.1016/0092-8674(93)90635-4. [DOI] [PubMed] [Google Scholar]
- 20.Herzfeld J. Acc Chem Res. 1996;29:31–37. doi: 10.1021/ar9500224. [DOI] [PubMed] [Google Scholar]
- 21.Friedhoff P, Schneider A, Mandelkow E M, Mandelkow E. Biochemistry. 1998;37:10223–10230. doi: 10.1021/bi980537d. [DOI] [PubMed] [Google Scholar]
- 22.Weinreb P H, Zhen W, Poon A W, Conway K A, Lansbury P T., Jr Biochemistry. 1996;35:13709–13715. doi: 10.1021/bi961799n. [DOI] [PubMed] [Google Scholar]
- 23.Chothia C. J Mol Biol. 1973;75:295–302. doi: 10.1016/0022-2836(73)90022-3. [DOI] [PubMed] [Google Scholar]
- 24.Salemme F R. Prog Biophys Mol Biol. 1983;42:95–133. doi: 10.1016/0079-6107(83)90005-6. [DOI] [PubMed] [Google Scholar]
- 25.Minor D L, Jr, Kim P S. Nature (London) 1994;371:264–267. doi: 10.1038/371264a0. [DOI] [PubMed] [Google Scholar]
- 26.Wetzel R. Cell. 1996;86:699–702. doi: 10.1016/s0092-8674(00)80143-9. [DOI] [PubMed] [Google Scholar]
- 27.Asakura S. J Mol Biol. 1968;35:101–135. doi: 10.1016/s0022-2836(68)80051-8. [DOI] [PubMed] [Google Scholar]
- 28.Eaton W A, Hofrichter J. Science. 1995;268:1142–1143. doi: 10.1126/science.7539154. [DOI] [PubMed] [Google Scholar]
- 29.Walsh D M, Lomakin A, Benedek G B, Condron M M, Teplow D B. J Biol Chem. 1997;272:22364–22372. doi: 10.1074/jbc.272.35.22364. [DOI] [PubMed] [Google Scholar]
- 30.Goldsbury C, Kistler J, Aebi U, Arvinte T, Cooper G J S. J Mol Biol. 1999;285:33–39. doi: 10.1006/jmbi.1998.2299. [DOI] [PubMed] [Google Scholar]
- 31.Klement I A, Skinner P J, Kaytor M D, Yi H, Hersch S M, Clark H B, Zoghbi H Y, Orr H T. Cell. 1998;95:41–53. doi: 10.1016/s0092-8674(00)81781-x. [DOI] [PubMed] [Google Scholar]
- 32.Saudou F, Finkbeiner S, Devys D, Greenberg M E. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 33.Crawford F, Soto C, Suo Z, Fang C, Parker T, Sawar A, Frangione B, Mullan M. FEBS Lett. 1998;436:445–448. doi: 10.1016/s0014-5793(98)01170-3. [DOI] [PubMed] [Google Scholar]
- 34.Lambert M P, Barlow A K, Chromy B A, Edwards C, Freed R, Liosatos M, Morgan T E, Rozovsky I, Trommer B, Viola K L, et al. Proc Natl Acad Sci USA. 1998;95:6448–6453. doi: 10.1073/pnas.95.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiesa R, Piccardo P, Ghetti B, Harris D A. Neuron. 1998;21:1339–1351. doi: 10.1016/s0896-6273(00)80653-4. [DOI] [PubMed] [Google Scholar]
- 36.Tompkins M M, Hill W D. Brain Res. 1997;775:24–29. doi: 10.1016/s0006-8993(97)00874-3. [DOI] [PubMed] [Google Scholar]
- 37.Moechars D, Dewachter I, Lorent K, Reverse D, Baekelandt V, Naidu A, Tesseur I, Spittels K, Van Den Haute C, Checler F, Godaux E, Cordell B, Van Leuven F. J Biol Chem. 1999;274:6483–6492. doi: 10.1074/jbc.274.10.6483. [DOI] [PubMed] [Google Scholar]