Abstract
Viroids are a unique class of noncoding RNAs: composed of only a circular, single-stranded molecule of 246–401 nt, they manage to replicate, move, circumvent host defenses, and frequently induce disease in higher plants. Viroids replicate through an RNA-to-RNA rolling-circle mechanism consisting of transcription of oligomeric viroid RNA intermediates, cleavage to unit-length strands, and circularization. Though the host RNA polymerase II (redirected to accept RNA templates) mediates RNA synthesis and a type-III RNase presumably cleavage of Potato spindle tuber viroid (PSTVd) and closely related members of the family Pospiviroidae, the host enzyme catalyzing the final circularization step, has remained elusive. In this study we propose that PSTVd subverts host DNA ligase 1, converting it to an RNA ligase, for the final step. To support this hypothesis, we show that the tomato (Solanum lycopersicum L.) DNA ligase 1 specifically and efficiently catalyzes circularization of the genuine PSTVd monomeric linear replication intermediate opened at position G95–G96 and containing 5′-phosphomonoester and 3′-hydroxyl terminal groups. Moreover, we also show a decreased PSTVd accumulation and a reduced ratio of monomeric circular to total monomeric PSTVd forms in Nicotiana benthamiana Domin plants in which the endogenous DNA ligase 1 was silenced. Thus, in a remarkable example of parasitic strategy, viroids reprogram for their replication the template and substrate specificity of a DNA-dependent RNA polymerase and a DNA ligase to act as RNA-dependent RNA polymerase and RNA ligase, respectively.
Keywords: RNA ligation, RNA replication, RNA processing
Viroids are infectious agents constituted by a circular single-stranded RNA (246-401 nt in currently known species) that despite not coding for any protein can replicate in plants and often induce disease. The unique properties of viroids were uncovered four decades ago when the agents of two plant diseases, potato spindle tuber (1, 2) and citrus exocortis (3), were characterized. Since then, more than 30 viroid species have been described (4, 5). Most, like the 359-nt Potato spindle tuber viroid (PSTVd), have a central conserved region (CCR) in their predicted rod-like secondary structure and are grouped in the family Pospiviroidae. Four other species, including Avocado sunblotch viroid, which lack a CCR and display autocatalytic cleavage, are gathered in the family Avsunviroidae (6). Viroids replicate through a rolling-circle mechanism with only RNA intermediates, consisting of three steps: synthesis of repetitive, oligomeric RNAs of the viroid, cleavage of the oligomers to their monomeric units and subsequent circularization (7). In the Pospiviroidae, which replicate in the nuclei of infected cells and follow an asymmetric pathway of this mechanism (Fig. 1A), the incoming most-abundant circular RNA, arbitrarily assigned (+) polarity, is reiteratively transcribed into oligomeric RNAs of complementary (−) polarity that do not undergo processing and serve directly as templates for transcription of (+) oligomers. These oligomers of (+) polarity are finally processed to circular monomers (8). In the Avsunviroidae, which replicate in the chloroplasts, the oligomeric (−) RNAs self-cleave through hammerhead ribozymes, and the resulting monomers, once circularized, serve as templates for the synthesis of the (+) oligomers in a second rolling-circle symmetric to the first (9).
Fig. 1.
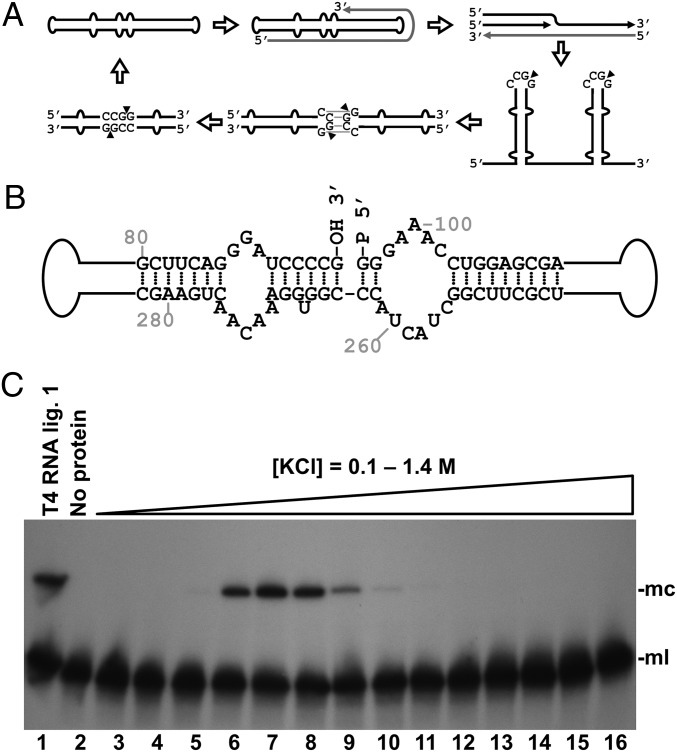
Identification of a tomato RNA ligase activity circularizing the PSTVd linear replication intermediate. (A) Scheme of the asymmetric rolling-circle replication mechanism of PSTVd, including the proposed processing model of viroid oligomeric RNA (14, 15). Black and gray lines represent (+) and (−) PSTVd RNAs, respectively. Arrowheads indicate cleavage sites. (B) Structure of part of the central domain of the PSTVd monomeric linear intermediate, opened between positions G95 and G96 with 5′-phosphomonoester and 3′-hydroxyl termini, which is the physiological substrate circularized by a tomato activity. (C) Tomato RNA ligase activity circularizing the linear PSTVd replication intermediate. Monomeric linear PSTVd opened between positions G95 and G96 and containing 5′-phosphomonoester and 3′-hydroxyl termini was subjected to circularization by chromatographic protein fractions from tomato. Reaction products were separated by denaturing PAGE and PSTVd forms visualized by Northern blot hybridization. Lane 1, control with T4 RNA ligase 1; lane 2, control with no protein added; lanes 3–16, tomato chromatographic fractions eluted with a KCl gradient. The positions of the monomeric circular (mc) and monomeric linear (ml) PSTVd RNAs are shown on the right side of the image.
Soon after viroid discovery, studies with the inhibitor α-amanitin showed that the enzyme catalyzing synthesis of the strands of PSTVd, and of two other members of the Pospiviroidae, is RNA polymerase II (10–12), induced to transcribe an RNA template instead of its natural DNA template. Additional support for this view was obtained with Citrus exocortis viroid, closely related to PSTVd, by immunoprecipitation studies with an antibody against the largest subunit of RNA polymerase II (13). In addition, site-directed mutagenesis analyses suggest that a palindromic quasi–double-stranded structure—formed by the interaction of the upper CCR strand of two consecutive viroid units in the oligomeric intermediates—is cleaved at two symmetric sites by a host type-III RNase, whereas both CCR strands promote ligation by forming a rod-like structure containing a loop E (14, 15) (Fig. 1A). Surprisingly, understanding of the third step of viroid replication, the circularization generating the final product, is very limited.
In this study, we searched for the host enzyme that catalyzes circularization during PSTVd replication.
Results
Detection of a Viroid-Circularizing Activity from Tomato (Solanum lycopersicum L.).
Total soluble leaf proteins were extracted from tomato, the typical experimental host of PSTVd, fractionated by liquid chromatography with a heparin column, and assayed for viroid RNA circularizing activity. Importantly, in this assay we used as substrate an in vitro synthesized monomeric linear PSTVd RNA opened between positions G95 and G96 and containing 5′-phosphomonoester and 3′-hydroxyl termini (Fig. 1B); our previous results indicated that this is the physiological precursor of the circular forms (14, 15). Reaction products were separated by denaturing PAGE and visualized by Northern blot hybridization with a complementary radioactive RNA probe. Detection of a PSTVd form with the expected low mobility of the circular RNA showed that the tomato preparation contained a viroid-circularizing activity eluting from the column at ∼0.5 M KCl (Fig. 1C).
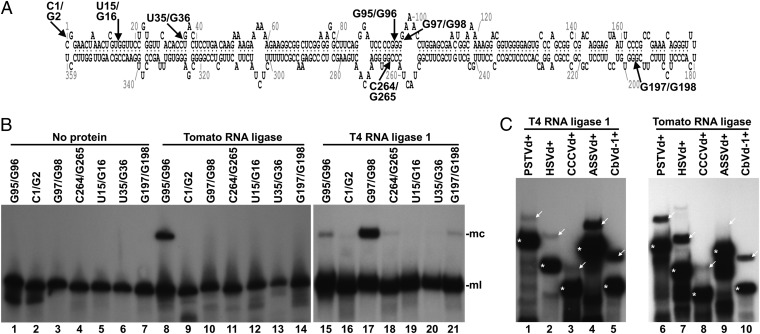
Using the tomato protein fraction with the highest viroid-circularizing activity, we then investigated the biochemical properties of the reaction. First, we observed similar loss of enzymatic activity in both the bacteriophage T4 RNA ligase and the tomato fraction upon digestion with proteinase K and thermal inactivation (Fig. S1A), thus confirming that the tomato activity resided in a protein. Next, we examined the reactivity of the same substrate with different terminal groups. The fraction did not ligate monomeric linear PSTVd RNA substrates opened between positions G95 and G96 when the terminal groups were either 5′-hydroxyl and 2′,5′-cyclic phosphodiester or 5′-hydroxyl and 3′-hydroxyl (Fig. S1B). Ligation in the presence of different nucleoside triphosphates or pairwise combinations thereof indicated a requirement for ATP (Fig. S1C). UTP or GTP could be used less efficiently in ligation, but not CTP, which behaved as a competitive inhibitor. Interestingly, some ligation activity was also detected without externally added NTPs. However, a time-course analysis of the reaction, including a comparison with T4 RNA ligase 1, an enzyme with the same specificity for the terminal groups of the substrate as the tomato activity, indicated that the latter definitively required ATP, but that it was purified, at least in part, as a preadenylated form able to catalyze a single cycle reaction in the absence of added ATP (Fig. S1 D and E). We further studied the substrate specificity of the tomato fraction against different monomeric linear PSTVd RNAs with 5′-phosphomonoester and 3′-hydroxyl termini opened at different positions in the viroid molecule. We randomly chose six different positions mapping at single-stranded loops and double-stranded helices in the predicted minimal free-energy conformation PSTVd, which also included different combinations of terminal nucleotides (Fig. 2A). Intriguingly, the tomato fraction efficiently catalyzed ligation only of the viroid substrate opened between positions G95 and G96 (Fig. 2B), indicating a high preference for the genuine precursor of the circular forms. Moreover, the corresponding (experimentally determined or predicted) physiological monomeric linear precursors of five additional viroid species, representative of the five genera forming the family Pospiviroidae, were all circularized by the tomato fraction, although with variable efficiency (Fig. 2C).
Fig. 2.
Substrate specificity of the viroid-circularizing activity from tomato. (A) Scheme of the minimum free-energy conformation predicted for PSTVd in which arrows indicate the positions wherein the different linear substrates are opened. (B) Monomeric linear PSTVd forms were subjected to circularization by the tomato protein fractions (lanes 8–14) or T4 RNA ligase 1 (lanes 15–21). Lanes 1–7, controls with no protein added. Reaction products were separated by denaturing PAGE, and PSTVd forms visualized by Northern blot hybridization. Positions of PSTVd monomeric circular (mc) and monomeric linear (ml) forms are indicated on the right side of the image. (C) 5′-32P–labeled monomeric linear forms of PSTVd (lanes 1 and 6), Hop stunt viroid (HSVd, lanes 2 and 7), Coconut cadang-cadang viroid (CCCVd, lanes 3 and 8), Apple scar skin viroid (ASSVd, lanes 4 and 9), and Coleus blumei viroid 1 (CbVd-1, lanes 5 and 10) were incubated with T4 RNA ligase 1 (lanes 1–5) or the most active tomato protein fraction eluted from the heparin column (lanes 6–10). Reaction products were separated by denaturing PAGE, and the gel autoradiographed. Positions of the linear and circular forms of the different viroids are indicated with asterisks and arrows, respectively.
Identification of the Tomato Enzyme Circularizing the Linear PSTVd RNA Replication Intermediate.
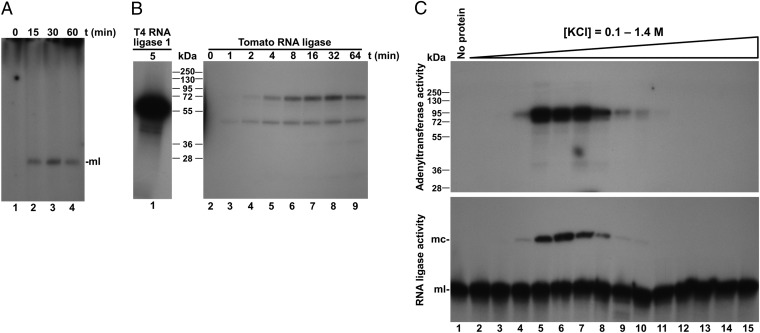
To identify the tomato protein catalyzing PSTVd circularization, we reasoned that an RNA ligase activity requiring ATP may likely follow the conventional nucleotidyltransferase mechanism characteristic of most RNA and DNA ligases as well as of 5′-capping enzymes, in which a lysine in the enzyme active center reacts with ATP to form an enzyme-AMP covalent adduct that subsequently transfers the AMP moiety to the nucleic acid substrate (16). By including in the reaction mix [α-32P]ATP and analyzing the radioactively labeled RNAs and proteins, we detected an adenylated monomeric linear PSTVd RNA (Fig. 3A), as well as two adenylated protein species of about 90 and 50 kDa (Fig. 3B). Consequently, the adenylated protein species—particularly the 90-kDa species that was consistently detected in replicates of this experiment—were considered candidates for the enzyme mediating viroid circularization. Supporting the possibility that the enzymatic activity was associated with the adenylated protein, parallel assays of viroid circularization and protein adenylation revealed a perfect match between the product profiles of both reactions catalyzed by the different chromatographic fractions from tomato leaves (Fig. 3C). To obtain sufficient protein for further characterization, a new chromatographic fractionation was carried out starting from 75 g of tomato leaves. Fractions were assayed for the viroid circularizing activity, and an aliquot from the fraction with the highest activity was separated by preparative SDS/PAGE and stained with Coomassie Blue. The piece of the gel corresponding to a protein size interval that should include the 90-kDa protein-adenylate adduct was excised (Fig. S2), and the proteins were analyzed by tandem mass spectrometry. A search of plant sequence databases using the MASCOT algorithm led to identification of 112 different protein sequences (Table S1). None of these proteins were annotated with an RNA ligase activity; however, the list included Arabidopsis thaliana L. DNA ligase 1. The size of this protein (90 kDa) matched that of the larger tomato protein-adenylate adduct. Also, eukaryotic DNA ligase 1 follows the reaction mechanism described above, including a ligase-AMP intermediate. Moreover, A. thaliana DNA ligase 1 is purified, at least partially, as a preadenylated form, and in vitro assays using oligonucleotide substrates have shown that this enzyme can ligate acceptor RNA to donor DNA when both are hybridized to DNA (17). These observations suggested that tomato DNA ligase 1 may be the host enzyme mediating circularization of PSTVd RNA; and, importantly, they raised the hypothesis that if viroids can subvert host DNA-dependent RNA polymerases to transcribe RNA templates, they could also subvert DNA ligase 1 to ligate (circularize) RNA substrates.
Fig. 3.
RNA and protein adenylation during PSTVd circularization. (A and B) Monomeric linear PSTVd was incubated in the presence of [α-32P]ATP, with the most active tomato fraction eluted from the heparin column. (A) RNAs were separated by denaturing PAGE, and the gel autoradiographed. Lanes 1–4, aliquots of the reaction taken at 0, 15, 30, and 60 min. (B) Proteins were separated by SDS/PAGE, and the gel autoradiographed. Lane 1, control reaction with T4 RNA ligase 1. Lanes 2–9, aliquots of the reaction taken at 0, 1, 2, 4, 8, 16, 32, and 64 min. (C) The tomato chromatographic fractions were simultaneously assayed for protein adenylation and PSTVd circularization. In the adenylation assay (Upper), reaction products were separated by SDS/PAGE, and the gel autoradiographed. In the circularization assay (Lower), reaction products were separated by denaturing PAGE, and PSTVd detected by Northern blot hybridization. Lane 1, control with no protein added. Lanes 2–15, tomato chromatographic fractions eluted with increasing concentrations of KCl. The positions of some protein markers with their sizes in kilodaltons, and the positions of monomeric circular (mc) and monomeric linear (ml) PSTVd forms are indicated.
In Vitro and in Vivo Analysis of the Viroid RNA Circularizing Activity of Tomato DNA Ligase 1.
To test the hypothesis that viroids subvert host DNA ligase 1 to mediate their circularization, we first retrieved the tomato sequence from GenBank (accession no. BT014510) that is orthologous to A. thaliana DNA ligase 1. Next, using a tomato RNA preparation, we obtained by RT-PCR amplification the full-length cDNA corresponding to the DNA ligase 1 ORF and cloned it in a plasmid to express in Escherichia coli a recombinant version of the protein with a carboxyl-terminal His6 tag. Subsequent purification by affinity chromatography with a nickel column (Fig. S3A) and in vitro assays showed that the purified recombinant protein very efficiently catalyzed circularization of the genuine linear PSTVd RNA precursor (Fig. S3B). A protein preparation obtained through the same procedure from E. coli transformed with empty vector failed to catalyze this reaction (Fig. S3C). The recombinant protein was also adenylated during the circularization reaction in the presence of [α-32P]ATP (Fig. S3D); interestingly, it displayed the same exquisite substrate specificity as the protein extracted from tomato leaves, being unable to circularize the other monomeric linear PSTVd RNAs opened at positions other than G95–G96 (Fig. S3E).
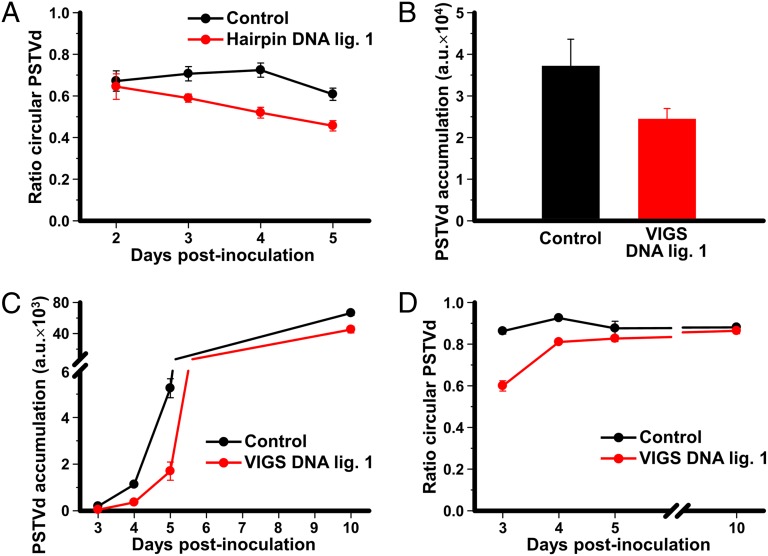
To complement these data with in vivo approaches, we next coexpressed an infectious dimeric head-to-tail PSTVd RNA together with a hairpin construct designed to silence the gene encoding DNA ligase 1 in Nicotiana benthamiana Domin (another experimental host for PSTVd in the family Solanaceae) by Agrobacterium tumefaciens-mediated transient transformation. The nucleotide sequences of N. benthamiana and tomato DNA ligase 1 are highly identical in that part of the sequence used to produce the hairpin construct. RNA was purified from infiltrated areas at different time intervals and analyzed by denaturing PAGE and Northern blot hybridization. When coexpressed with the empty vector, the dimeric PSTVd RNA was processed properly in N. benthamiana, and the monomeric linear and circular PSTVd forms were detected. However, when coexpressed with the DNA ligase 1 hairpin construct, the ratios of the monomeric circular form to total monomeric PSTVd RNAs (circular plus linear forms) were significantly lower (Fig. 4A and Fig. S4). RT-quantitative PCR (qPCR) analysis of N. benthamiana DNA ligase 1 mRNA indicated a 2.4-fold reduction at 4 d postinfiltration in tissues infiltrated with the hairpin construct respect to those infiltrated with the empty vector.
Fig. 4.
Effect of DNA ligase 1 silencing on the in vivo accumulation and circularization of PSTVd. (A) Interference with circularization of PSTVd RNA caused by the expression of a DNA ligase 1 hairpin. A dimeric PSTVd construct was coagroinoculated in N. benthamiana with either a tomato DNA ligase 1 hairpin construct to silence the endogenous gene (red), or with the empty vector (black). RNAs were purified from the agroinfiltrated areas of triplicate plants and separated by denaturing PAGE. Monomeric circular and linear PSTVd forms were visualized by Northern blot hybridization and quantified by phosphorimetry. The plots represent the ratio of monomeric circular to total monomeric PSTVd forms (circular plus linear) at several days postinoculation (dpi). (B–D) Interference with PSTVd accumulation and circularization caused by the expression of a VIGS construct specific for the endogenous DNA ligase 1. (B) A dimeric PSTVd construct was coagroinfiltrated in N. benthamiana 16c plants constitutively expressing GFP with either a Tobacco rattle virus construct to induce VIGS of DNA ligase 1 and GFP (red histogram) or a control construct to induce VIGS of GFP alone (black histogram). At 1 mo postinfiltration, RNA was extracted from quadruplicate samples of upper noninoculated leaves, separated by denaturing PAGE, and the monomeric circular and linear PSTVd forms detected by Northern blot hybridization and quantified as indicated above. Histograms represent PSTVd accumulation in arbitrary units (a.u.). (C and D) N. benthamiana 16c plants constitutively expressing GFP were preinoculated with the VIGS vectors to induce silencing of either the endogenous DNA ligase 1 and GFP (red) or GFP alone (control, black). At 3–4 wk later, GFP silenced plants were agroinfiltrated with a construct to express a dimeric PSTVd RNA. RNA was extracted at different time intervals from the agroinoculated areas of triplicate plants, and PSTVd forms were visualized by Northern blot hybridization and quantified. (C) Plot of total PSTVd RNA accumulation (in a.u.) vs. dpi. (D) Plot of the ratio of monomeric circular to total PSTVd monomeric forms vs. dpi. All plots include bars indicating the SD of the replicate measures.
We also applied virus-induced gene silencing (VIGS), using a vector based on Tobacco rattle virus (18). The construct included two cDNA fragments to silence the genes coding for DNA ligase 1 and Aequorea victoria GFP, the latter serving as a silencing reporter. N. benthamiana transgenic 16c plants constitutively expressing GFP (19) were either coinoculated with the VIGS vector and the PSTVd dimeric construct, or they were preinoculated with the VIGS vector, and 3–4 wk later selected leaves were infiltrated with the infectious PSTVd dimeric construct. In all cases, VIGS effectiveness was screened by monitoring GFP fluorescence decay. In the coinoculation format, quantitative determinations using PAGE and Northern blot hybridization (Fig. S5) showed that the upper noninoculated leaves accumulated significantly less PSTVd in plants coinoculated with the VIGS vector containing the DNA ligase 1 cDNA fragment than in those coinoculated with the control VIGS vector without the DNA ligase 1 fragment (Fig. 4B). In the preinoculation format (Fig. S6), a similar analysis showed that total PSTVd RNA, as well as the ratio of the monomeric circular form to total monomeric RNAs, were significantly lower in the infiltrated leaf areas of plants preinoculated with the VIGS vector expressing the DNA ligase 1 cDNA fragment than in those from plants preinoculated with the VIGS vector containing only the GFP cDNA fragment (Fig. 4 C and D). In this case, quantification of N. benthamiana DNA ligase 1 mRNA indicated an approximately sevenfold reduction at 4 wk postinfiltration in plants infiltrated to silence GFP and DNA ligase 1 with respect to those infiltrated to silence only GFP.
Finally, we mechanically inoculated tomato plants with the circular RNA generated in vitro by the recombinant DNA ligase 1 and purified by PAGE. One month later, the plants exhibited typical PSTVd symptoms (stunting and epinasty; Fig. S7), confirming that the product of the recombinant DNA ligase 1 was a bona fide infectious RNA.
Discussion
The identity of the host activity mediating the circularization step during replication of viroids belonging to the family Pospiviroidae has been a long-standing mystery. The situation became even more complicated when the monomeric linear precursor was shown to have 5′-phosphomonoester and 3′-hydroxyl ends (14, 15), because the only RNA ligase described so far in plants, the tRNA ligase, specifically requires 5′-hydroxyl and 2′,3′-cyclic phosphodiester termini (20). This observation led to the prediction (15) that higher plants must contain an RNA ligase activity similar to T4 bacteriophage RNA ligases, with the ability to catalyze joining of 5′-phosphomonoester and 3′-hydroxyl termini (21, 22). The results reported here using a combination of in vitro and in vivo approaches show that the host DNA ligase 1 is the long-sought enzyme involved in circularizing the monomeric PSTVd RNA during replication. Moreover, because the cleavage mechanism of the oligomeric RNA intermediates appears conserved in the family Pospiviroidae (14), the ligation mechanism that we propose for PSTVd most likely also operates for the rest of the viroid species in this family (Fig. 2). In contrast, we have recently reported that the Avsunviroidae recruit the host chloroplastic tRNA ligase, an isoform that includes a transit peptide that targets it to the chloroplast, for the ligation of the (+) and (−) monomeric linear replication intermediates (23). Our in vivo approaches did not completely abolish PSTVd circularization (Fig. 4), suggesting incomplete silencing of N. benthamiana DNA ligase 1, a key enzyme essential for cell viability (24). Nonetheless, the possibility of another host DNA ligase taking the role of the silenced DNA ligase 1 could not be excluded.
Our finding raises several considerations. During replication, PSTVd (and presumably the other Pospiviroidae) parasitizes two crucial components of the nuclear replication and transcription machinery of the host—namely, the DNA-dependent RNA polymerase II and the DNA ligase 1—and it redirects these enzymes to function on RNA templates and substrates, respectively. Targeting such key host proteins suggests a robust parasitic strategy, leaving little opportunity for the host to evade viroid infection; it also suggests an intricate evolutionary adaptation of the parasite to the host that may also be ancient, considering the very old origin proposed for viroids (25). Viroids must have evolved for finely adapting their processing intermediates to be specifically recognized by DNA ligase 1. Our observation about the specificity of the DNA ligase 1 for just one of the several circular permutations of the viroid sequence tested (Fig. 2) supports the notion that it is not enough for the DNA ligase to have access to the two termini of the same RNA molecule; rather, a specific sequence and presumably a specific conformation is needed for the DNA ligase to accept an RNA substrate. Note that, upon binding DNA, human DNA ligase 1 induces a localized RNA-like A-form conformation around the DNA break (26, 27), and it has been proposed that DNA ligases, RNA ligases, and RNA-capping enzymes evolved by fusion of ancillary effector domains to an ancestral catalytic module involved in RNA repair (16). Thus, viroids would profit the retention of a primordial function of DNA ligase 1. Finally, DNA ligase 1-mediated joining of viroid RNAs may represent the tip of the iceberg of host RNA metabolism, with this enzyme playing roles as RNA ligase unknown so far. Pertinent to this context is Hepatitis delta virus (HDV), an important human pathogen resembling plant viroids in many aspects (28). HDV replicates in the nucleus of infected cells, and elongation of one or both of its RNA strands is also catalyzed by the host RNA polymerase II. And although HDV ribozymes produce 5′-hydroxyl and 2′,3′-cyclic phosphodiester instead of 5′-phosphomonoester and 3′-hydroxyl termini, our finding opens the possibility that DNA ligase 1—after RNA termini modification—or another human DNA ligase might be involved in HDV replication.
Materials and Methods
Tomato Protein Extraction and Fractionation.
Young leaves (15 g) from 1-mo-old tomato plants [cultivar (cv.) Rutgers] were harvested, ground in a mortar in the presence of liquid N2, and homogenized with 3 vol of cold extraction buffer [100 mM Tris⋅HCl (pH 8.0), 100 mM KCl, 10 mM MgCl2, 10 mM DTT, 1% Nonidet P-40] plus a mixture of protease inhibitors (Complete; Roche)]. The crude extract was clarified twice by centrifugation, first at 8,000 × g for 15 min and then at 100,000 × g for 30 min. The supernatant was fractionated by chromatography using a 1-mL heparin column (HiTrap Heparin HP; GE Healthcare) with an ÄKTA Prime Plus (GE Healthcare) liquid chromatography system operated at 4 °C at a flow rate of 1 mL/min. After equilibration with 10 mL of extraction buffer, the column was loaded with the clarified extract and then washed with 20 mL of extraction buffer. Proteins were eluted with 10 mL of a linear gradient from 0.1 to 2 M KCl in extraction buffer, and 20 fractions of 0.5 mL were collected. For some experiments, 50-μL aliquots of specific fractions were dialyzed (10,000 kDa; Pierce) 3× for 1 h against 1 L of dialysis buffer [50 mM Tris⋅HCl (pH 7.5), 25 mM KCl, 1 mM DTT, 0.1% Nonidet P-40] at 4 °C.
Viroid RNA Circularization Assay.
Reaction mixtures (20 μL) in 50 mM Tris⋅HCl (pH 8.0), 75 mM KCl, 5 mM MgCl2, 5 mM DTT, and 1 mM ATP included 25 ng RNA substrate and 4 μL protein fraction, and were incubated for 30 min at 30 °C. See SI Materials and Methods related to viroid RNA transcripts, in vitro transcription of viroid RNAs, and terminal group modification of in vitro transcribed viroid RNAs. Control reactions with T4 RNA ligase 1 (Fermentas) were carried out similarly in 50 mM Tris⋅HCl (pH 7.5), 10 mM MgCl2, 1 mM ATP, and 10 U of the enzyme. Reactions were stopped by adding 20 μg (1 μL) of proteinase K (New England Biolabs) and 2 μL of 100 mM Tris⋅HCl (pH 8.0), 50 mM EDTA, 10 mM 2-mercaptoethanol, 0.5% SDS, and incubating successively for 15 min at 42 °C and 15 min at 55 °C. Reaction products were mixed with 1 vol of loading buffer [98% (vol/vol) formamide, 10 mM Tris⋅HCl (pH 8.0), 1 mM EDTA, 0.0025% xylene cyanol, 0.0025% bromophenol blue] and separated by denaturing PAGE in 5% (wt/vol) gels including 8 M urea in TBE buffer. RNAs were electroblotted to positively charged nylon membranes (Nytran SPC; Whatman), cross-linked by irradiation with 1.2 J/cm2 UV light (Vilber Lourmat), and analyzed by hybridization using a complementary 32P-labeled RNA probe as previously described (29). In some experiments, 1 mM ATP was excluded from the RNA circularization assay or replaced by 1 mM CTP, 1 mM GTP, 1 mM UTP, or all of the possible combinations of each two NTPs at 1 mM each. Proteinase K digestion and thermal inactivation of the tomato RNA ligase activity is described in SI Materials and Methods.
In the experiment in which five different linear viroid RNAs representative of the five different genera in the family Pospiviroidae were subjected to the RNA circularization assays (Fig. 2C), viroid RNAs were initially produced with 5′- and 3′-hydroxyl termini, and then 5′-phosphorylated with T4 polynucleotide kinase (Fermentas) in the presence of 5 μCi [γ-32P]ATP (3,000 Ci/mmol). The polyacrylamide gel used to separate the reaction products was fixed for 30 min in 10% (vol/vol) acetic acid, 20% (vol/vol) methanol, dried under vacuum, and imaged by autoradiography.
Viroid RNA and Protein Adenylation Assays.
The viroid RNA and protein adenylation assays were performed exactly like the viroid RNA circularization assay, but the 1 mM ATP was replaced by 5 μCi of [α-32P]ATP (800 Ci/mmol). In the RNA adenylation assay, the substrate consisted of 250 ng of linear PSTVd RNA from position G96 to G95 with 5′-phosphomonoester and 3′-hydroxyl termini. The reaction was stopped and the products analyzed as described for the viroid RNA circularization assay, but polyacrylamide gels were fixed, dried, and imaged by autoradiography. In the protein adenylation assay, reactions were stopped by adding Laemmli loading buffer and heating at 95 °C for 2 min. The reactions products were separated by SDS/PAGE in 12.5% (wt/vol) polyacrylamide gels that were fixed, dried, and imaged by autoradiography.
Identification of Tomato DNA Ligase 1, Cloning, and Expression of a Recombinant Version of the Protein.
The viroid-circularizing activity was identified by mass spectrometry analysis of the proteins contained in a piece of a preparative polyacrylamide gel. The tomato DNA ligase 1 cDNA was amplified by RT-PCR from a tomato RNA preparation and cloned in a plasmid for expressing a recombinant version of the protein in E. coli. The recombinant protein fused to a carboxyl-terminal His6 tag was purified by chromatography using a 1-mL Ni-Sepharose column (HisTrap HP; GE Healthcare). Details are provided in SI Materials and Methods.
RNA Silencing Assays.
The effect of DNA ligase 1 silencing on the in vivo accumulation and circularization of PSTVd was tested by transient expression of PSTVd and DNA ligase 1 constructs in N. benthamiana 16c and by VIGS of N. benthamiana DNA ligase 1. The constructs used in these experiments are provided in SI Materials and Methods.
Circular PSTVd RNA Purification and Tomato Infectivity Assay.
PSTVd RNAs circularized in vitro by the recombinant tomato DNA ligase 1 and bacteriophage T4 RNA ligase 1 were separated by denaturing PAGE in a 5% (wt/vol) polyacrylamide gel. The gel was stained with ethidium bromide, and the bands corresponding to the circular PSTVd RNA excised. RNAs in those bands were eluted by diffusion and used to mechanically inoculate tomato plants. Inoculation media consisted of a suspension of 10% (wt/vol) carborundum in 50 mM K2HPO4. Plants were grown in a greenhouse at 25 °C with a photoperiod of 16 h day and 8 h night.
Supplementary Material
Acknowledgments
We thank Verónica Aragonés and Teresa Cordero for excellent technical assistance, and Colleen J. Doherty and C. Douglas Grubb for critical review of the manuscript. Proteomic analyses were performed in the proteomics laboratory of Centro de Investigación Príncipe Felipe de Valencia, a member of Spanish ProteoRed. This research was funded by the Spanish Ministerio de Economía y Competitividad (formerly Ministerio de Ciencia e Innovación) Grants BIO2008-01986 and BIO2011-26741 and also partially by Grant BFU2008-03154. M.-Á.N. was the recipient of a predoctoral fellowship from the Spanish Ministerio de Educación y Ciencia.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1206187109/-/DCSupplemental.
References
- 1.Diener TO, Raymer WB. Potato spindle tuber virus: A plant virus with properties of a free nucleic acid. Science. 1967;158:378–381. doi: 10.1126/science.158.3799.378. [DOI] [PubMed] [Google Scholar]
- 2.Gross HJ, et al. Nucleotide sequence and secondary structure of potato spindle tuber viroid. Nature. 1978;273:203–208. doi: 10.1038/273203a0. [DOI] [PubMed] [Google Scholar]
- 3.Semancik JS, Weathers LG. Exocortis virus of citrus: Association of infectivity with nucleic acid preparations. Virology. 1968;36:326–328. doi: 10.1016/0042-6822(68)90156-6. [DOI] [PubMed] [Google Scholar]
- 4.Flores R, Hernández C, Martínez de Alba AE, Daròs JA, Di Serio F. Viroids and viroid-host interactions. Annu Rev Phytopathol. 2005;43:117–139. doi: 10.1146/annurev.phyto.43.040204.140243. [DOI] [PubMed] [Google Scholar]
- 5.Ding B. The biology of viroid-host interactions. Annu Rev Phytopathol. 2009;47:105–131. doi: 10.1146/annurev-phyto-080508-081927. [DOI] [PubMed] [Google Scholar]
- 6.Flores R, Owens RA. Viroids. In: Mahy BWJ, Van Regenmortel MHV, editors. Encyclopedia of Virology. Oxford: Elsevier; 2008. pp. 332–342. [Google Scholar]
- 7.Branch AD, Robertson HD. A replication cycle for viroids and other small infectious RNA’s. Science. 1984;223:450–455. doi: 10.1126/science.6197756. [DOI] [PubMed] [Google Scholar]
- 8.Branch AD, Benenfeld BJ, Robertson HD. Evidence for a single rolling circle in the replication of potato spindle tuber viroid. Proc Natl Acad Sci USA. 1988;85:9128–9132. doi: 10.1073/pnas.85.23.9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daròs JA, Marcos JF, Hernández C, Flores R. Replication of avocado sunblotch viroid: Evidence for a symmetric pathway with two rolling circles and hammerhead ribozyme processing. Proc Natl Acad Sci USA. 1994;91:12813–12817. doi: 10.1073/pnas.91.26.12813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flores R, Semancik JS. Properties of a cell-free system for synthesis of citrus exocortis viroid. Proc Natl Acad Sci USA. 1982;79:6285–6288. doi: 10.1073/pnas.79.20.6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mühlbach HP, Sänger HL. Viroid replication is inhibited by α-amanitin. Nature. 1979;278:185–188. doi: 10.1038/278185a0. [DOI] [PubMed] [Google Scholar]
- 12.Schindler IM, Mühlbach HP. Involvement of nuclear DNA-dependent RNA-polymerases in potato spindle tuber viroid replication: A reevaluation. Plant Sci. 1992;84:221–229. [Google Scholar]
- 13.Warrilow D, Symons RH. Citrus exocortis viroid RNA is associated with the largest subunit of RNA polymerase II in tomato in vivo. Arch Virol. 1999;144:2367–2375. doi: 10.1007/s007050050650. [DOI] [PubMed] [Google Scholar]
- 14.Gas ME, Hernández C, Flores R, Daròs JA. Processing of nuclear viroids in vivo: An interplay between RNA conformations. PLoS Pathog. 2007;3:e182. doi: 10.1371/journal.ppat.0030182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gas ME, Molina-Serrano D, Hernández C, Flores R, Daròs JA. Monomeric linear RNA of citrus exocortis viroid resulting from processing in vivo has 5′-phosphomonoester and 3′-hydroxyl termini: Implications for the RNase and RNA ligase involved in replication. J Virol. 2008;82:10321–10325. doi: 10.1128/JVI.01229-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shuman S, Lima CD. The polynucleotide ligase and RNA capping enzyme superfamily of covalent nucleotidyltransferases. Curr Opin Struct Biol. 2004;14:757–764. doi: 10.1016/j.sbi.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 17.Wu YQ, Hohn B, Ziemienowic A. Characterization of an ATP-dependent type I DNA ligase from Arabidopsis thaliana. Plant Mol Biol. 2001;46:161–170. doi: 10.1023/a:1010679901911. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 2002;30:415–429. doi: 10.1046/j.1365-313x.2002.01297.x. [DOI] [PubMed] [Google Scholar]
- 19.Ruiz MT, Voinnet O, Baulcombe DC. Initiation and maintenance of virus-induced gene silencing. Plant Cell. 1998;10:937–946. doi: 10.1105/tpc.10.6.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Englert M, Beier H. Plant tRNA ligases are multifunctional enzymes that have diverged in sequence and substrate specificity from RNA ligases of other phylogenetic origins. Nucleic Acids Res. 2005;33:388–399. doi: 10.1093/nar/gki174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang LK, Schwer B, Shuman S. Structure-guided mutational analysis of T4 RNA ligase 1. RNA. 2006;12:2126–2134. doi: 10.1261/rna.271706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho CK, Shuman S. Bacteriophage T4 RNA ligase 2 (gp24.1) exemplifies a family of RNA ligases found in all phylogenetic domains. Proc Natl Acad Sci USA. 2002;99:12709–12714. doi: 10.1073/pnas.192184699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nohales MA, Molina-Serrano D, Flores R, Daròs JA. Involvement of the chloroplastic isoform of tRNA ligase in the replication of the viroids belonging to the family Avsunviroidae. J Virol. 2012;86:8269–8276. doi: 10.1128/JVI.00629-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waterworth WM, et al. DNA ligase 1 deficient plants display severe growth defects and delayed repair of both DNA single and double strand breaks. BMC Plant Biol. 2009;9:79. doi: 10.1186/1471-2229-9-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diener TO. Circular RNAs: Relics of precellular evolution? Proc Natl Acad Sci USA. 1989;86:9370–9374. doi: 10.1073/pnas.86.23.9370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pascal JM. DNA and RNA ligases: Structural variations and shared mechanisms. Curr Opin Struct Biol. 2008;18:96–105. doi: 10.1016/j.sbi.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 27.Pascal JM, O’Brien PJ, Tomkinson AE, Ellenberger T. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature. 2004;432:473–478. doi: 10.1038/nature03082. [DOI] [PubMed] [Google Scholar]
- 28.Taylor JM. Replication of the hepatitis delta virus RNA genome. Adv Virus Res. 2009;74:103–121. doi: 10.1016/S0065-3527(09)74003-5. [DOI] [PubMed] [Google Scholar]
- 29.Martínez F, Marqués J, Salvador ML, Daròs JA. Mutational analysis of eggplant latent viroid RNA processing in Chlamydomonas reinhardtii chloroplast. J Gen Virol. 2009;90:3057–3065. doi: 10.1099/vir.0.013425-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.