Abstract
Transgenesis promises a powerful means for assessing gene function during amphibian limb regeneration. This approach is complicated, however, by the need for embryonic appendage development to proceed unimpeded despite the genetic alterations one wishes to test later in the context of regeneration. Achieving conditional gene regulation in this amphibian has not proved to be as straightforward as in many other systems. In this report we describe a unique method for obtaining temporal control over exogenous gene expression in the axolotl. Based on technology derived from the Escherichia coli Lac operon, uninduced transgenes are kept in a repressed state by the binding of constitutively expressed Lac repressor protein (LacI) to operator sequences within the expression construct. Addition of a lactose analog, IPTG, to the swimming water of the axolotl is sufficient for the sugar to be taken up by cells, where it binds the LacI protein, thereby inducing expression of the repressed gene. We use this system to demonstrate an in vivo role for thrombospondin-4 in limb regeneration. This inducible system will allow for systematic analysis of phenotypes at defined developmental or regenerative time points. The tight regulation and robustness of gene induction combined with the simplicity of this strategy will prove invaluable for studying many aspects of axolotl biology.
The urodeles (salamanders, newts, and their close relatives) are a unique clade of vertebrates that stand to broadly advance research in numerous fields including genome evolution, cancer, aging, stem cells, and regeneration. Salamanders are perhaps most well known for their extensive regenerative abilities; they are one of the only groups of higher vertebrates that are capable of regrowing limbs, parts of the eye, portions of the heart, and other structures [reviewed in (1)]. Understanding how salamanders regenerate limbs should provide critical insight into efforts to stimulate these processes in vertebrates that do not regenerate limbs, yet the list of modern molecular genetic tools that can be applied to salamanders is currently very short. Other model systems with a much more sophisticated experimental toolkit, such as zebrafish, have provided valuable clues to the molecular underpinnings of vertebrate appendage regeneration (2–11). However, fin regeneration in teleost fish (such as zebrafish) is not completely analogous to limb development or regeneration. For instance, fish fin bony rays do not form by endochondral ossification (as tetrapod limbs do), and amputations proximal enough to include bones analogous to bones in the tetrapod limb are not followed by regeneration (12). Therefore, although some molecular commonalities between fin and limb regeneration have already been shown to exist (13), key molecular differences are to be expected. Limb regeneration in anurans (frogs) is likely the most similar feat to salamander limb regeneration. Molecular understanding of frog limb regeneration has been boosted by sequenced Xenopus genomes and a handful of genetic tools [reviewed in (14)], but frogs can only regenerate perfect limbs as tadpoles, before they undergo metamorphosis and while the limbs are still developing, and many salamanders can regenerate limbs throughout their entire adult lives.
The axolotl (Ambystoma mexicanum) is a neotenic salamander that can be easily manipulated in the laboratory setting and for which protocols in surgical manipulation, grafting, and qualitative molecular analysis are well established. Decades of research have focused on elucidating the mechanisms by which salamanders can regenerate body parts [reviewed in (15)], and more recent experimentation has brought the study of the phenomenon into the molecular age. For example, using insight into limb development as a starting point, several signaling pathways have been shown to perform similar roles in limb regeneration. When sonic hedgehog (SHH) activity is inhibited by cyclopamine treatment, regenerating axolotls regenerate limbs missing digits (16), a defect similar to genetic loss of shh in developing mouse limbs (17). Conversely, ectopic expression of SHH in regenerating axolotl limbs leads to ectopic digits (18), similar to treatment of developing chick limbs with ectopic SHH (19). Wnt signaling has also been implicated in axolotl regeneration as expression of either of two different inhibitors of Wnt signaling (Axin and Dkk1) following amputation can impair limb regeneration (13). Using an a priori approach to understanding limb regeneration in newts, the cell-surface protein Prod1 was discovered to play a key role in mediating the proximal/distal identities of cultured blastemas (20); nAG, the ligand for Prod1, has also been shown to be sufficient to rescue most aspects of regeneration when a construct encoding it is electroporated into denervated and amputated newt limbs, which are otherwise impaired in regeneration (21). An important recent advance in newt lens regeneration has been the inhibition of expression of particular genes using morpholino injection (22), and this technology might also be applied to limb regeneration in newts and axolotls as well. The methods used in all of these studies relied on tools such as bead implantations, injection of chemical inhibitors, electroporation, or infection with viruses driving constitutive expression of cDNAs—all valuable but imprecise tools that do not allow for fine temporal or spatial manipulation and which are inherently all invasive in some manner.
The development of a protocol for generating transgenic axolotls (and newts) potentially opens the door to more precise genetic dissection of this fascinating process (23, 24). However, it is critical for limb development to proceed normally despite any transgenic manipulations if the impact of a gene is to be assessed in a regenerative setting. Achieving this requires a system for conditional alteration of gene expression, such that genetic pathways essential for the embryonic development of the axolotl or for the initial limb development can be specifically investigated in limb regeneration without disrupting their earlier functions. Additionally, a conditional approach is needed if one wants to manipulate gene activity during the limb-regenerative process, such that distinct temporal requirements for particular genes can be uncovered. For example, a signaling pathway may be necessary after wound healing to create a blastema, but dispensable once the blastema is generated. Since the publication of the first transgenic axolotl, a line of animals constitutively expressing EGFP in all cell types, in 2006, only one new instance of axolotl transgenesis has been described in the literature (25), and no additional F1 animals completely transgenic for a new transgene have been reported. In our experience, this lack of new transgenics is likely due to the labor-intensive process of rearing transgenic F0 animals to adulthood, a process which requires approximately 1 y and considerable space and husbandry efforts, particularly if there is an entire cohort of these animals (which is likely the case if a researcher is waiting a year or more to find out if a founder line has been created).
Here we describe efforts to develop an inducible genetic system for axolotls that would enable precise temporal control of transgene expression, thus facilitating the analysis of specific genetic pathways during distinct phases of regeneration. We first considered and tested various components of a series of inducible gene expression systems previously developed for use in other vertebrates, including the tamoxifen-inducible Cre-lox, doxycycline inducible tet-on, and IPTG-inducible LacI repressor systems. In axolotls we found the CreERT2/lox system to be leaky and doxycycline was toxic, which rendered neither system useful to build upon. However, IPTG proved to be innocuous to axolotls, thus making the classical LacI-repressor/IPTG-inducer system the most promising of those we tested. Therefore, the fact that an induction system has not yet been developed for axolotl is underpinned by both technical considerations, some of which are described in this manuscript, as well as facility and personnel considerations.
In bacteria, the genes of the Lac operon are repressed in the absence of lactose by the binding of the Lac repressor protein (LacI) to the Lac operator (26). These genes are induced in the presence of lactose, which directly binds LacI, thus altering its conformation and preventing operator binding. As adapted for use in higher organisms, this system uses a synthetic analog of lactose, IPTG that is not metabolized by cells. Recent work in both transgenic mice and in mammalian cell culture has demonstrated that IPTG treatment can robustly induce the expression of genes positioned downstream of Lac operator sequences, whose expression would otherwise be inhibited by coexpression of the repressor molecule (27, 28). Here we demonstrate that the system can be successfully used in axolotls as well. No other inducible systems have been reported for regulating transgene expression in axolotls, and of all the mainstream inducible systems applicable to higher vertebrate genetic study, it may be the only one unambiguously functional in this species. This inducible system opens many doors for axolotl research, including studies of regeneration, as we demonstrate in this report.
Results
Testing the Ability of Various Inducible Genetic Systems to Regulate Gene Expression in the Axolotl.
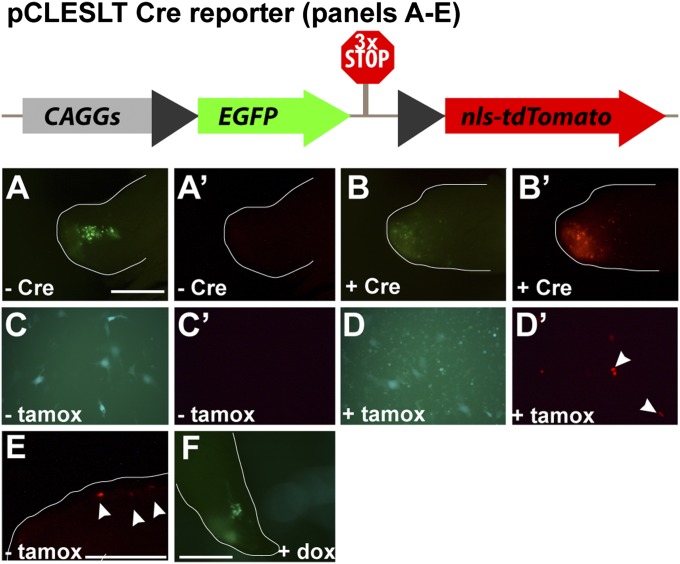
To establish a conditional genetic system that could be used for the study of regeneration, we first considered induction strategies commonly used in other vertebrates, and attempted to apply them to axolotls. For example, creERT2/loxP-based recombination is a powerful means by which to control gene expression at precise times via administration of tamoxifen in mice. In this system, a fusion protein between Cre recombinase and the estrogen receptor is rendered inert by being sequestered in the cytoplasm in the absence of an estrogen analog, tamoxifen (29). Tamoxifen was not toxic in axolotls when administered by i.p. injection at doses reported to induce recombination in mammalian systems (it did not kill or outwardly sicken them). We next constructed a Cre-responsive plasmid, pCLESLT, that is fully operational in axolotl limb blastemas (Fig. 1 A–B′) and in axolotl AL1 cells when cotransformed with pCAG-Cre, a plasmid encoding a constitutively expressed Cre (Fig. 1 C–D′, see also SI Data and Fig. S1). We never observed leakiness of the pCLESLT plasmid without cotransfection of a Cre-encoding plasmid; transfected cells were always green and never red. To be useful as a means of temporally controlling gene expression, a means of activating Cre protein or transcript expression is necessary. The most common method for achieving temporal control is to express Cre as a fusion protein to a portion of the estrogen receptor, ERT2, which results in targeting of the recombinase to the cytosol. Because the recombinase needs to be inside the nucleus to catalyze recombination at LoxP sites, in theory it should be inert until an estrogen analog such as tamoxifen binds the ERT2-Cre fusion protein, instigating its translocation to the nucleus. To be most useful, promiscuous entry of the Cre fusion protein into the nucleus must be eliminated or minimized as much as possible. Hence, we chose to test the pCLESLT reporter in conjunction with a tighter version of the fusion protein, one with two ERT2 elements, encoded by pCAG-ERT2CreERT2 (30). We found the pCLESLT reporter is inducible by tamoxifen when cotransfected with pCAG-ERT2CreERT2; however, a few mammalian 293T cells transfected with both plasmids produced both GFP and tdTomato in the absence of tamoxifen, similar to what we have seen with other Cre reporters in 293T cells. In regenerating axolotl limb blastemas, this effect was exacerbated and we often observed expression of the recombined pCLESLT reporter even without tamoxifen administration (Fig. 1E). Although it might be possible to tighten the control by tinkering with this system, for this and other reasons mentioned in Discussion, we instead chose to explore other strategies for achieving conditional gene expression.
Fig. 1.
Common induction systems do not easily transfer to axolotl use. The pCLESLT reporter is diagrammed. In the absence of Cre activity, CAG promoter drives expression of EGFP. In the presence of Cre activity, recombination at LoxP sites results in removal of EGFP and the stop cassette, allowing for TdTomato expression in the nucleus. pCLESLT reporter shows expression of EGFP in blastemas (A), but no expression of TdTomato (A′). When Cre is coexpressed from a constitutive promoter, recombination results in loss of EGFP in blastemas (B), and a gain of TdTomato expression (B′). Expression of TdTomato from the pCLESLT reporter can be induced in axolotl AL1cells only in the presence of tamoxifen; when cotransfected with pCAG-ERT2-Cre-ERT2, cells express only EGFP (C) and not TdTomato (C′); however, if tamoxifen is added (1 mM), although some EGFP persists (D), many cells can be seen expressing nuclear tdTomato (D′) due to Cre-mediated recombination at the LoxP sites. (E) Coelectroporation of pCLESLT and pCAG-ERT2-Cre-ERT2 in blastemas showed some leaky expression of the TdTomato (arrowheads). (F) Although a tet-responsive EGFP reporter electroporated into blastemas could be activated by doxycycline, induction was only achieved at doses of doxycycline lethal to the animals. Scale bar is 1 mm.
We next tested the doxycycline-inducible tet-on system using the constructs pCAG-rtTA and pTRE2-EGFP. In this system, the transactivator is constitutively produced but gene expression is only driven from a tetracycline response element-containing promoter in the presence of a tetracycline analog (31). In this case, we found the reciprocal problem: induction in vivo was only possible at concentrations of doxycycline that were lethal to the axolotls (e.g., 500 μg/animal by i.p. injection; Fig. 1F). Lower amounts of doxycycline, for example, 50 μg/animal by i.p. injection, resulted in no induction yet still left the animals dead or dying within 40 h postinjection.
Having encountered problems with two of the most common inducible systems used in higher vertebrates, we turned our attention to a less widely used, but equally powerful strategy: controlling gene expression by coopting the bacterial Lac operator/Lac repressor system. This strategy has been successfully used in mice to precisely control the expression of a tyrosinase transgene that enables otherwise albino mice grow to wild-type agouti coat hair (27). Furthermore, in mice and in mammalian cell culture, gene induction by IPTG is completely reversible (27, 28). We found that IPTG is innocuous to axolotls and can be administered in their water at concentrations up to at least 2 mg/mL (10 mM) with no overtly deleterious effects to the animals’ health. We examined limbs regenerated at 1 mM IPTG (the concentration at which induction plateaus in the mammalian cell culture system) and found no overt defects.
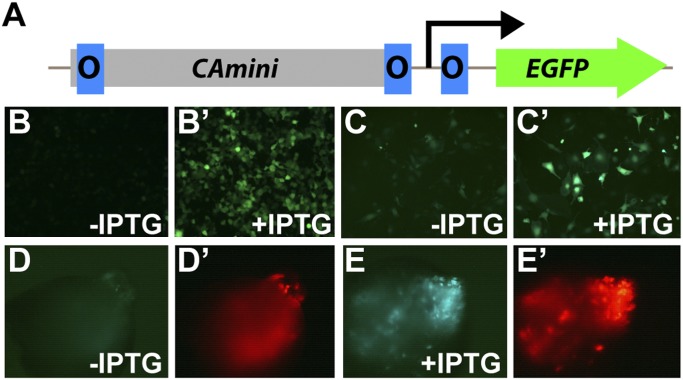
We tested the basic components of a Lac operator/IPTG system in axolotls and found them to be operational. In this system, one or more Lac operator elements are placed upstream of the transcriptional start site of a gene. When bound to the Lac repressor, the Lac operator prevents gene expression. However, in the presence of the inducer IPTG, the repressor can no longer bind its DNA target and transcription is derepressed. In axolotl blastemas expressing GFP from ubiquitous promoters separated by single Lac operator elements (e.g., CAmini-O-EGFP), expression was indeed diminished when pCAG-LacI was coelectroporated, and could be induced following administration of IPTG. However, in this initial test of the system, although repression was evident it was not complete. Therefore, we created and tested numerous alternate configurations of the reporter construct containing either additional numbers of operator sequences and/or varied spacing of these sequences (Figs. S2–S4 and SI Data). Ultimately, we achieved the best combination of near-complete repression without IPTG and robust induction with IPTG using a configuration similar to that used in producing the IPTG-inducible tyrosinase mouse (27) (Fig. 2A), CAmini-O1O2O3-GFP (termed O-GFP). This reporter worked well in both mammalian cells (Fig. 2 B and B′) and AL1 axolotl cells (Fig. 2 C and C′) in tissue culture, with robust expression only evident in the presence of IPTG.
Fig. 2.
The Lac operator/Lac repressor system is functional in axolotl. (A) The pCAmini-O1O2O3-EGFP LacI-repressible construct. (B–C′) Expression of EGFP from the pCAmini-O1O2O3-EGFP reporter when cotransfected with pCAG-LacI in the absence and presence of 1 mM IPTG in mammalian 293T cells (B and B′) and axolotl AL1 cells (C and C′). (D) Regenerating axolotl limb blastemas electroporated with CAmini-O1O2O3-EGFP reporter and pCAG-LacI express very little EGFP in the absence of IPTG. (D′) Coelectroporation of a pCAG-tdTomato control for blastema shown in D. (E) When housed in water containing 1 mM IPTG, blastemas electroporated with the pCAmini-O1O2O3-EGFP reporter and pCAGG-LacI show robust expression of EGFP. (E′) tdTomato coelectroporation control for E.
To test whether the Lac operator/IPTG system would function in vivo in the setting of axolotl limb regeneration, we coelectroporated the reporter into blastema cells with a construct-driving constitutive expression of the LacI repressor. As found in vitro, we saw negligible expression of the GFP reporter in the absence of IPTG, but when 1 mM IPTG was added to the water housing the animals, the reporter was strongly expressed (Fig. 2 D and E′).
LacI-Inducible System Can Be Used to Test Gene Function During Axolotl Limb Regeneration.
Previously, we had found that two genes encoding axolotl thrombospondins, thrombospondin-1 (tsp-1) and thrombospondin-4 (tsp-4), displayed dynamic expression patterns in discrete cell types during the course of limb regeneration (32). Although thrombospondins have been implicated in numerous aspects of mammalian biology (33), including some that may be pertinent to regeneration such as wound healing and angiogenesis, nothing is known about their potential role in appendage regeneration.
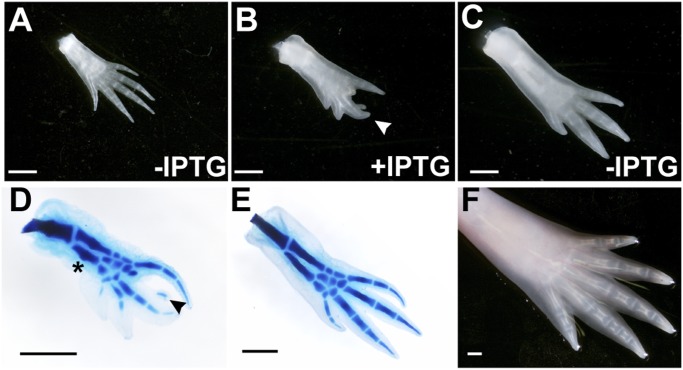
Expression of the tsp-4 mRNA appears in the regenerating axolotl blastema in the mesenchymal cells accumulating beneath in the wound epidermis 1 wk postamputation. This expression intensifies as the blastema cells proliferate over the next 2 wk. However, once differentiation commences within the regenerate, tsp-4 expression becomes restricted to the perichondrium around the developing cartilage, consistent with, for instance, a role in the branching of the skeletal elements (32). We cloned the axolotl tsp-4 ORF downstream of our CAmini-O1O2O3 promoter/operator sequence (termed pO-Tsp4) and coinjected newly fertilized axolotl eggs with pO-Tsp4 and pCAG-LacI to generate mosaic F0 cotransgenic axolotls. As expected, all of the F0 animals developed limbs with normal morphologies in the absence of IPTG (Fig. 3A). However, when we amputated the limbs and allowed them to regenerate in the presence of IPTG, we found ∼10% of the limbs to have deformities including forked and ectopic digits (n = 44, Fig. 3B). The variability and low penetrance of the phenotypes was expected, given the mosaic expression of transgenes in F0 animals. However, the phenotypes are indeed consistent with what one would expect for a molecule playing a key role in patterning the skeletal structures.
Fig. 3.
Administration of IPTG can induce regeneration defects in an animal with a LacI-dependent transgene. Scale bar in all images is 1 mm in length. (A) Right hind limb from O-Tsp4; CAG-LacI F0 transgenic animal reared in the absence of IPTG shows a normal morphology. (B) Following amputation and subsequent housing in IPTG, the same limb regenerated with several defects including truncated posterior digits, missing digit 2, and a bifurcation on another digit (arrowhead). (C) When placed immediately into water without IPTG following the amputation of the limb shown in B, an almost entirely normal limb regenerated. (D) Skeletal preparation of the limb shown in B revealed the bifurcated minidigit contained a cartilaginous condensation (arrowhead); additionally, long bones in the stylopod and zeugopod regrew with a nodule near the center (asterisk). (E) Skeletal preparation of the limb shown in C demonstrated that even a malformed, IPTG-induced limb can regenerate an almost perfect limb in the absence of IPTG (normal skeletal morphology, missing one posterior digit). (F) The same limb was amputated again and allowed to regenerate in the absence of IPTG, producing a morphologically perfect limb.
To demonstrate that the phenotypes were indeed dependent upon Tsp4 activity, limbs with extreme branching phenotypes were reamputated and allowed to regenerate a second time in the absence of IPTG. Nearly normal skeletal patterns were obtained following regeneration of these limbs (Fig. 3C). Skeletal preparations of a limb with an extreme phenotype regenerated in the presence of IPTG revealed that the ectopic digital growth contained a cartilaginous condensation (Fig. 3D, arrowhead), and that the long bones had a nodular aspect where ossification would be expected to later commence (Fig. 3D, asterisk); this nodular appearance was not observed when the limb was regenerated in the absence of IPTG (Fig. 3E), nor were ectopic digits. Completely normal regeneration was observed following another amputation and regrowth in the absence of IPTG (Fig. 3F, shown on the living animal). Although a detailed analysis of the Tsp4 phenotype is beyond the scope of the current study, requiring germline transgenics carrying these constructs, these preliminary experiments do provide strong evidence for the in vivo utility of the LacI/IPTG system for regulating transgenes during axolotl limb regeneration.
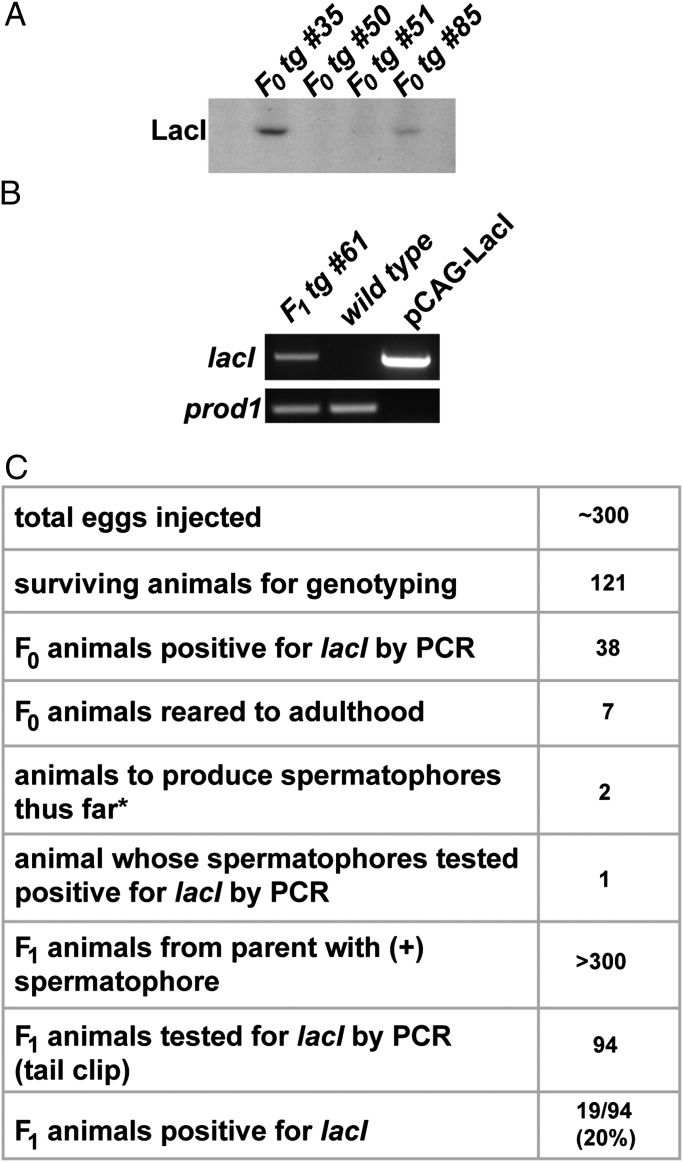
To facilitate future studies using the LacI repressor in axolotls, a genetic line of axolotls harboring a CAG-LacI transgene was created. The pCAG-LacI plasmid was injected into newly fertilized axolotl eggs, and 121 F0 juveniles were screened by PCR and by Western blot for production of LacI protein (Fig. 4A) for transgene integration. A CAG-LacI F0 animal was bred to a white mutant (otherwise wild type and referred to as “wild type” in the figure) female, and progeny were screened by genomic DNA isolation and PCR genotyping to identify animals positive for the LacI transgene (Fig. 4B), hence demonstrating germline transmission of this transgene. We have since identified several other transgenic F1 siblings, and we are planning to mate them once mature to homozygose the line. The transgenic efficiency (∼20% of the progeny from an F0 animal prescreened by PCR and Western blot) is comparable with levels previously reported in the axolotl system (23). Although we did not have a fluorescent reporter to gauge which F0 mosaic animals had the most transgenic cells—and therefore perhaps most likely to have transgenic gametes—we were able to use a variety of molecular tests to identify the animal most likely to transmit the transgene, and it did indeed produce transgenic F1 offspring.
Fig. 4.
A founding genetic line of LacI-expressing transgenic axolotls. (A) Protein purified from the tails of two F0 CAG-LacI mosaic transgenics (35 and 85) shows detectable levels of LacI protein on a Western blot. (B) PCR analysis of genomic DNA from an F1 juvenile sired by CAG-LacI F0 #35 (Left) and a wild-type juvenile (Middle), as well as a positive control (pCAG-LacI plasmid, 50 ng). The prod1 promoter sequence served as a positive control for genomic DNA preps (40). (C) Table summarizing process of creating CAG-LacI founder line. Numbers of animals for each category are listed at right. *Only 2/7 animals are males; male gametes can be easily genotyped because sperm is laid in a visible gelatinous packet outside the body. Fertilization is internal and occurs after the female retrieves the spermatophore. Hence, female gametes have not been genotyped.
Discussion
To tease apart the intricacies of a gene’s function during a complex process such as limb regeneration, the ability to precisely control timing of gene expression is absolutely necessary. Here we describe a system for achieving that goal in the axolotl, a salamander that regenerates entire limbs as robustly as any known organism and whose limbs are exceedingly similar to those of humans in both form and cellular constitution. Although the axolotl’s regenerative abilities are impressive, genetic manipulation of particular elements at particular times during regeneration has been hitherto impossible; indeed, many of the tools available to researchers working in other model organisms may not be easily transferrable to the axolotl (29, 31). Mitotic recombination systems now commonplace in other systems, such as Cre/Lox in mice, are operational in axolotls, but coupling these to temporal controls, such as CreERT2/tamoxifen, has proven to not be a straightforward endeavor as these temporal control elements behave differently in a salamander than they do in a mouse. We speculate that perhaps the axolotls have an endogenous hormone that can functionally mimic estrogen and drive the Cre into the nucleus in the absence of tamoxifen. Further modifications to the CreERT2 constructs (or a different strategy for inducing Cre) may indeed more thoroughly restrict the recombinase from the nucleus in the absence of tamoxifen. The constructs used to test the CreERT2/tamoxifen system might behave differently in a transgenic setting; we only tested constructs by transient transfection in cell culture and by electroporation into blastemas, and in both scenarios many copies of the plasmids are expected to be propagating inside these rapidly dividing cells. However, it is important to note that the tamoxifen-induction strategy, at heart, is not reversible: once the intervening genetic element is recombined out it is permanently excised. Hence, should a tighter version be developed, it might still be most useful as a lineage-tracing device rather than a means to transiently misexpress genetic elements. Finally, the technical consideration of ease of use for the inducing compound is also starkly different between the LacO (IPTG) and CreERT2/Lox (tamoxifen) systems; IPTG is a nonhazardous sugar that can simply be added to the axolotl’s water, and tamoxifen is not water-soluble, requiring animals to be injected or gavaged, and although a water-soluble analog is available, it is prohibitively expensive considering the volumes necessary. Tamoxifen is excreted by the animals, rendering housing water hazardous, and used water would need to be disposed as hazardous waste.
Systems such as the tet-on system are also functional in axolotl cells, but the inducer has proven toxic in vivo. Recently, two doxycycline-inducible transgenic lines have been reported in Xenopus (34); for these transgenes, doxycycline was administered via the water, an administration method we did not attempt and which might be less toxic than the injections we used in axolotls. Although this may prove less toxic, we anticipate these conditions would not be sufficient for transgene induction in the axolotl based on the high inducer concentration necessary in our preliminary experiments.
Following our initial observations regarding the aforementioned induction systems, we modified our approach to first consider the action of the inducer on living axolotls, and we found that IPTG, a synthetic analog of lactose, was very well tolerated by the animals. This finding opened the door to the Lac operator system as an elegant and simple means of controlling transgene expression in the axolotl. We have shown that the expression of both a reporter—EGFP—as well as a gene involved in regeneration—thrombospondin-4—can be induced by simply adding IPTG to the axolotl’s water when genetic elements encoding them are downstream of Lac operators. For thrombospondin-4, our studies provide evidence that expression of this gene must be tightly controlled during regeneration for the process to proceed perfectly. Establishing LacI-repressed lines of O-Tsp4 will enable future studies that will elucidate the precise steps during regeneration at which misexpression of Tsp4 results in deformities as the IPTG inducer can be added and removed from the water at different times.
The CAmini-O1O2O3 repressible promoter described here represents an ideal backbone for researchers wishing to examine the activity of specific genetic elements during limb regeneration (or any other process in axolotl). Upon sexual maturity the F1 CAG-LacI animals can be bred for production of unique F0 mosaic transgenics. An especially fruitful approach may be to inject CAmini-O1O2O3 derivatives directly into CAG-LacI eggs and screen for integration events on this repressed background. Future development of tissue-specific means of expressing transgenes can also be coupled with temporal control to uncover exactly where particular genetic pathways function during regeneration. Future possible improvements on a researcher’s ability to predict which animals are most likely to harbor transgenic gametes—and are therefore worth rearing to adulthood—will greatly facilitate the process of generating novel founder lines in the LacI and other genetic backgrounds. With the advent of new technologies such as deep sequencing that are enabling more a priori approaches to uncovering the mysteries of salamander limb regeneration [reviewed in (35)] comes an even more heightened need for new tools that can be used for assessing the function of the genes that might be uncovered. We believe that the LacI/IPTG system will prove broadly useful for future studies, we anticipate the development of additional tools in the field, and we hope that this work might inspire other laboratories to invest in the development of technologies to enable molecular insight into limb regeneration in these remarkable animals.
Materials and Methods
Constructs and Cloning.
The Cre-reporter plasmid (pCAG-loxP-EGFP-3xSTOP-loxP-NLStdTomato) was created by PCR amplification of loxP-EGFP-3xSTOP-NotI-loxP with EcoRI and KpnI in the forward and reverse primer, respectively, from a shuttle vector and ligation into a pCAG-containing plasmid via EcoRI and KpnI. The original SV40 triple stop cassette was amplified from pFREPE (36). tdTomato was amplified with LoxP and NLS in the forward primer and NotI sites in both primers, and this fragment was cloned into pCAG-loxP-EGFP-3xSTOP-NotI-loxP via the NotI site and a downstream NotI site. The pCAG-ERT2CreERT2 plasmid is described in (30).
The tet system was tested with the following plasmids: pCAG-rtTA (to produce the transactivator), pTRE2-EGFP (Clontech), and pCAG-DsRed2 [electroporation control (30)]. The pCAG-rtTA construct was generated by replacing the hCMV enhancer/promoter in pUHrT62-1 (37) with CAG (excised from pCAG-DsRed2) via the SpeI and EcoRI restriction sites.
The original CAmini-O-EGFP plasmid was created by PCR amplification of the CMV enhancer and chicken β-actin promoter regions of the CAGGS promoter (i.e., the CAGGS but without the intron), 607 bp, and insertion into the pStagia3 vector (38) upstream of the minimal TATA promoter sequence and the EGFP reporter via the SalI site. The tail end of this PCR fragment was outfitted with a PacI and XbaI restriction site just upstream of the SalI to facilitate future cloning operations. The operator sequence was cloned into the PacI site via direct ligation of two partially complementary synthetic oligonucleotides: 5′TAAGAATTGTGAGCGCTCACAATTATTTAAATGCGGCCGCCGATCGTTAAT3′ and 5′TAACGATCGGCGGCCGCATTTAAATAATTGTGAGCGCTCACAATCTTAAT 3′. The CAmini-O1O2O3-EGFP plasmid was created by modifying CAmini-O-EGFP. Nucleotide linkers containing Lac operator sequences, as reported elsewhere (27), were ligated upstream of the GFP coding sequence. Using the HindIII and AgeI restriction sites, one operator (5′-gtggaattgtgagcggataacaatttcac-3′) was centered 55 nt upstream of the ATG start codon. The operator sequence of the original construct was removed from within the PacI sites and a different operator sequence was introduced (5′-agatctgtggaattgtgagcggataacaatttcacggatccagatct-3′), centered 145 nt upstream of the ATG, and destroying the 5′ PacI site. A final, identical operator (5′-agatctgtggaattgtgagcggataacaatttcacggatccagatct-3′) was placed within the SpeI sites centered 772 nt upstream of the ATG, and destroying both restriction sites. The bacterial Lac operator sequences endogenous to the plasmid backbone were excised with BfuAI and SfiI, and replaced with a PacI site. To generate the Tsp4 construct, the GFP cDNA was excised with AgeI and NruI replaced with a modified cloning site (AgeI-NotI-PmeI-NsiI-NruI). The Tsp4 cDNA was PCR amplified and ligated into the NotI and NsiI restriction sites.
The pCAG-LacI plasmid was created by replacing the CMV enhancer/promoter from TLD-027 [a CMV-LacI-nls plasmid (28)] with a PCR-amplified CAG enhancer/promoter via the EcoRI and BamHI sites. Operator sequences in the backbone of this plasmid were excised by HindIII and PciI, and replaced with a HindIII-PacI-PciI linker.
Animal Experiments.
Limbs were amputated using microdissection scissors under a stereodissecting microscope while the animals were anesthetized in 0.1% tricaine (pH 7, in axolotl water). Animals were housed in 0.5% sulfamerazine (Sigma) for 24 h following amputations. Electroporations were performed by injection of plasmid DNA into blastemas (DNA concentration at 100 ng/μL each, in water, with Fast Green for visualization) using a pulled glass microcapillary needle and a pressurized injector, followed by five pulses at 35 mV each, with 1-s intervals between pulses, using a BTX 830 electroporation device (Harvard Apparatus) and gold-plated forcep-style electrodes submerged in PBS. Tamoxifen was dissolved in DMSO (stock concentration was 10 μg/μL) and 50 μg per gram of body weight was injected intraperiotoneally. IPTG was dissolved in water (for cell culture) or axolotl water (40% Holfretters, for animal experiments). Transgenics were produced according to (23). All animal work was carried out in accordance with the Harvard Medical School Institutional Animal Care and Use Committee regulations.
Genotyping.
Genomic DNA was prepared by incubating tissue sample in 50 mM NaOH at 95 °C for 10 min followed by cooling sample on ice and adding 1/10 volume of 1M Tris (pH 8). Samples were centrifuged at 16,400 × g for 5 min to pellet out the cellular debris. PCR reactions were seeded with 1 μL of genomic DNA. Primers for detecting LacI insertion were 5′CGTTATACGATGTCGCAGAGTATGCC3′ and 5′GATGCTCCACGCCCAGTCGC3′, both of which bind within the coding sequence.
Skeletal Preparations.
Alcian blue staining was performed as follows: limbs incubated with rocking overnight in 95% ethanol, overnight in acetone, and 7 d in alcian blue/alizarin red at 37 °C. Following staining, limbs were cleared by incubation in 1% (wt/vol) KOH, followed by 1% (vol/vol) KOH/25% glycerol, then 1% KOH/50% glycerol, then 1% KOH/75% glycerol, in which they were also imaged. Alcian blue stock was 0.3% alcian blue in 70% ethanol; alizarin red stock was 0.1% alizarin red 95%; the working solution was 5% alcian blue stock/5% alizarin red stock/5% glacial acetic acid/volume in 70% ethanol.
Cell Culture.
Mammalian 293T cells were maintained according to established protocols and transfected with PEI (Polethylenimine; Polysciences Inc., 1 μg/μL diluted 1:1,000 in serum-free DMEM). AL1 cells (gift of Stephane Roy, University of Montreal, Montreal, Canada) were maintained and transfected according to (39). A separate stock of AL1 cells was established in galactose-free media for LacI-dependent experiments [custom gal(-) Leibovitz’s L-15 media, Gibco, supplemented with 4.5 g/L glucose].
Supplementary Material
Acknowledgments
We thank Jourdan White and Melissa Werner for animal husbandry assistance, Johanna Kowalko for advice on the manuscript, Stephane Roy for the AL1 cells, and Elly Tanaka for helpful discussions in the early stages of the work. This work was supported by Grants F32HD054082 (to J.L.W.), F32AR056149 (to J.A.L.), R21AR059884 (to C.J.T.), and R01HD045499 (to C.J.T.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211816109/-/DCSupplemental.
References
- 1.Brockes JP, Kumar A. Appendage regeneration in adult vertebrates and implications for regenerative medicine. Science. 2005;310:1919–1923. doi: 10.1126/science.1115200. [DOI] [PubMed] [Google Scholar]
- 2.Stewart S, Tsun ZY, Izpisua Belmonte JC. A histone demethylase is necessary for regeneration in zebrafish. Proc Natl Acad Sci USA. 2009;106:19889–19894. doi: 10.1073/pnas.0904132106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeLaurier A, et al. Zebrafish sp7:EGFP: A transgenic for studying otic vesicle formation, skeletogenesis, and bone regeneration. Genesis. 2010;48:505–511. doi: 10.1002/dvg.20639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chablais F, Jazwinska A. IGF signaling between blastema and wound epidermis is required for fin regeneration. Development. 2010;137:871–879. doi: 10.1242/dev.043885. [DOI] [PubMed] [Google Scholar]
- 5.Knopf F, et al. Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin. Dev Cell. 2011;20:713–724. doi: 10.1016/j.devcel.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 6.Tu S, Johnson SL. Fate restriction in the growing and regenerating zebrafish fin. Dev Cell. 2011;20:725–732. doi: 10.1016/j.devcel.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sousa S, et al. Differentiated skeletal cells contribute to blastema formation during zebrafish fin regeneration. Development. 2011;138:3897–3905. doi: 10.1242/dev.064717. [DOI] [PubMed] [Google Scholar]
- 8.Blum N, Begemann G. Retinoic acid signaling controls the formation, proliferation and survival of the blastema during adult zebrafish fin regeneration. Development. 2012;139:107–116. doi: 10.1242/dev.065391. [DOI] [PubMed] [Google Scholar]
- 9.Singh SP, Holdway JE, Poss KD. Regeneration of amputated zebrafish fin rays from de novo osteoblasts. Dev Cell. 2012;22:879–886. doi: 10.1016/j.devcel.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rojas-Muñoz A, et al. ErbB2 and ErbB3 regulate amputation-induced proliferation and migration during vertebrate regeneration. Dev Biol. 2009;327:177–190. doi: 10.1016/j.ydbio.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 11.Wills AA, Kidd AR, 3rd, Lepilina A, Poss KD. Fgfs control homeostatic regeneration in adult zebrafish fins. Development. 2008;135:3063–3070. doi: 10.1242/dev.024588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morgan TH. Regeneration in teleosts. Arch Entwicklungsmech Org. 1900;10:120–134. [Google Scholar]
- 13.Kawakami Y, et al. Wnt/beta-catenin signaling regulates vertebrate limb regeneration. Genes Dev. 2006;20:3232–3237. doi: 10.1101/gad.1475106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tseng AS, Levin M. Tail regeneration in Xenopus laevis as a model for understanding tissue repair. J Dent Res. 2008;87:806–816. doi: 10.1177/154405910808700909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brockes JP. Amphibian limb regeneration: Rebuilding a complex structure. Science. 1997;276:81–87. doi: 10.1126/science.276.5309.81. [DOI] [PubMed] [Google Scholar]
- 16.Roy S, Gardiner DM. Cyclopamine induces digit loss in regenerating axolotl limbs. J Exp Zool. 2002;293:186–190. doi: 10.1002/jez.10110. [DOI] [PubMed] [Google Scholar]
- 17.Chiang C, et al. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- 18.Roy S, Gardiner DM, Bryant SV. Vaccinia as a tool for functional analysis in regenerating limbs: Ectopic expression of Shh. Dev Biol. 2000;218:199–205. doi: 10.1006/dbio.1999.9556. [DOI] [PubMed] [Google Scholar]
- 19.Riddle RD, Johnson RL, Laufer E, Tabin C. Sonic hedgehog mediates the polarizing activity of the ZPA. Cell. 1993;75:1401–1416. doi: 10.1016/0092-8674(93)90626-2. [DOI] [PubMed] [Google Scholar]
- 20.da Silva SM, Gates PB, Brockes JP. The newt ortholog of CD59 is implicated in proximodistal identity during amphibian limb regeneration. Dev Cell. 2002;3:547–555. doi: 10.1016/s1534-5807(02)00288-5. [DOI] [PubMed] [Google Scholar]
- 21.Kumar A, Godwin JW, Gates PB, Garza-Garcia AA, Brockes JP. Molecular basis for the nerve dependence of limb regeneration in an adult vertebrate. Science. 2007;318:772–777. doi: 10.1126/science.1147710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsonis PA, et al. Controlling gene loss of function in newts with emphasis on lens regeneration. Nat Protoc. 2011;6:593–599. doi: 10.1038/nprot.2011.341. [DOI] [PubMed] [Google Scholar]
- 23.Sobkow L, Epperlein HH, Herklotz S, Straube WL, Tanaka EM. A germline GFP transgenic axolotl and its use to track cell fate: Dual origin of the fin mesenchyme during development and the fate of blood cells during regeneration. Dev Biol. 2006;290:386–397. doi: 10.1016/j.ydbio.2005.11.037. [DOI] [PubMed] [Google Scholar]
- 24.Casco-Robles MM, et al. Expressing exogenous genes in newts by transgenesis. Nat Protoc. 2011;6:600–608. doi: 10.1038/nprot.2011.334. [DOI] [PubMed] [Google Scholar]
- 25.Monaghan JR, Maden M. Visualization of retinoic acid signaling in transgenic axolotls during limb development and regeneration. Dev Biol. 2012;368:63–75. doi: 10.1016/j.ydbio.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 27.Cronin CA, Gluba W, Scrable H. The lac operator-repressor system is functional in the mouse. Genes Dev. 2001;15:1506–1517. doi: 10.1101/gad.892001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deans TL, Cantor CR, Collins JJ. A tunable genetic switch based on RNAi and repressor proteins for regulating gene expression in mammalian cells. Cell. 2007;130:363–372. doi: 10.1016/j.cell.2007.05.045. [DOI] [PubMed] [Google Scholar]
- 29.Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: A tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- 30.Matsuda T, Cepko CL. Controlled expression of transgenes introduced by in vivo electroporation. Proc Natl Acad Sci USA. 2007;104:1027–1032. doi: 10.1073/pnas.0610155104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kistner A, et al. Doxycycline-mediated quantitative and tissue-specific control of gene expression in transgenic mice. Proc Natl Acad Sci USA. 1996;93:10933–10938. doi: 10.1073/pnas.93.20.10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whited JL, Lehoczky JA, Austin CA, Tabin CJ. Dynamic expression of two thrombospondins during axolotl limb regeneration. Dev Dyn. 2011;240:1249–1258. doi: 10.1002/dvdy.22548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adams JC, Lawler J. The thrombospondins. Int J Biochem Cell Biol. 2004;36:961–968. doi: 10.1016/j.biocel.2004.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rankin SA, Zorn AM, Buchholz DR. New doxycycline-inducible transgenic lines in Xenopus. Dev Dyn. 2011;240:1467–1474. doi: 10.1002/dvdy.22642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whited JL, Tabin CJ. Regeneration review reprise. J Biol. 2010;9:15. doi: 10.1186/jbiol224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dymecki SM, Ray RS, Kim JC. Mapping cell fate and function using recombinase-based intersectional strategies. Methods Enzymol. 2010;477:183–213. doi: 10.1016/S0076-6879(10)77011-7. [DOI] [PubMed] [Google Scholar]
- 37.Urlinger S, et al. Exploring the sequence space for tetracycline-dependent transcriptional activators: Novel mutations yield expanded range and sensitivity. Proc Natl Acad Sci USA. 2000;97:7963–7968. doi: 10.1073/pnas.130192197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Billings NA, Emerson MM, Cepko CL. Analysis of thyroid response element activity during retinal development. PLoS ONE. 2010;5:e13739. doi: 10.1371/journal.pone.0013739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Villiard E, et al. Urodele p53 tolerates amino acid changes found in p53 variants linked to human cancer. BMC Evol Biol. 2007;7:180. doi: 10.1186/1471-2148-7-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaikh N, Gates PB, Brockes JP. The Meis homeoprotein regulates the axolotl Prod 1 promoter during limb regeneration. Gene. 2011;484:69–74. doi: 10.1016/j.gene.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.