Abstract
The anaphase-promoting complex/cyclosome (APC/C) is a large multiprotein E3 ubiquitin ligase involved in ubiquitin-dependent proteolysis of key cell cycle regulatory proteins, including the destruction of mitotic cyclins at the metaphase-to-anaphase transition. Despite its importance, the role of the APC/C in plant cells and the regulation of its activity during cell division remain poorly understood. Here, we describe the identification of a plant-specific negative regulator of the APC/C complex, designated SAMBA. In Arabidopsis thaliana, SAMBA is expressed during embryogenesis and early plant development and plays a key role in organ size control. Samba mutants produced larger seeds, leaves, and roots, which resulted from enlarged root and shoot apical meristems, and, additionally, they had a reduced fertility attributable to a hampered male gametogenesis. Inactivation of SAMBA stabilized A2-type cyclins during early development. Our data suggest that SAMBA regulates cell proliferation during early development by targeting CYCLIN A2 for APC/C-mediated proteolysis.
Plant organ size is determined by the total cell number and final cell size, resulting from cell division and cell expansion, respectively. In most, but not all, cases, the final organ size correlates with cell number, rendering cell division the main driver that controls growth (1).
In all eukaryotes, unidirectional cell cycle progression requires the coordinated destruction of essential cell cycle regulatory proteins by ubiquitin-dependent proteolysis pathways (2–5). Specific E3 ubiquitin ligases mediate the recognition of target proteins (6–8) that are subsequently polyubiquitinated and subjected to proteolysis by the 26S proteasome. One of the most complex ubiquitin ligases involved in cell cycle control is the anaphase-promoting complex/cyclosome (APC/C), of which the composition can vary from 11 to 13 subunits depending on the organism. The APC/C complex plays essential roles in mitosis, meiosis, and postmitotically differentiated cells (3, 9–12).
The APC/C triggers the metaphase-to-anaphase transition and the exit from mitosis by mediating the degradation of proteins such as securin and mitotic cyclins (13). In plants, the A- and B-type cyclins are subjected to proteolysis by APC/C through recognition of specific amino acid motifs, the destruction (D) and KEN boxes (6, 14, 15). Plant A-type cyclins are produced and degraded earlier in the cell cycle than B-type cyclins and have distinct and nonredundant functions in the progression of cell division (16). Based on their primary structures, the plant A-type cyclins are classified into A1, A2, and A3 groups (17). The transcriptional regulation of the CYCLIN A2 (CYCA2) group coordinates cell proliferation during plant development (18, 19) and CYCA2;3 overexpression enhances cell division (20).
The APC/C is regulated partly by two activating proteins CELL DIVISION CYCLE 20 (CDC20) and CDC20 HOMOLOGY1/CELL CYCLE SWITCH 52 (CDH1/CCS52), that also determine substrate specificity (21). Recently, ULTRAVIOLET-B-INSENSITIVE4 (UVI4) and UVI4-like/OMISSION OF SECOND DIVISION1/GIGAS CELL1 (UVI4/OSD1/GIG1) were identified as plant-specific inhibitors of the APC/C that are required for proper mitotic progression in Arabidopsis (22, 23).
In Arabidopsis, the genes encoding the subunits APC2, APC3a, APC3b, APC4, APC6, APC8, and APC10 have been investigated functionally. In all cases, except for APC8, the analysis of the mutants revealed female gametophytic defects, probably as a consequence of the inability to degrade mitotic cyclins (24–28). APC8 was shown to be involved in male gametogenesis (29). Moreover, reduced expression levels of APC6 (10) or APC10 (10, 28) exhibited several developmental abnormalities, including defects in vascular development, whereas an APC4 loss-of-function mutant was defective in embryogenesis (26). APC3b (CDC27b) has a role in the maintenance of cell division in meristems during postembryonic development (30), and overexpression of APC10 enhances cell division and accelerates the degradation of the mitotic CYCB1;1, leading to increased leaf sizes (28).
Here, we functionally analyzed an APC/C regulator of Arabidopsis, designated SAMBA. The samba mutants have an enlarged meristem size and show growth-related phenotypes, including the formation of large seeds, leaves, and roots; additionally, their fertility is reduced because of a defect in male gametogenesis. A biochemical analysis revealed that loss of function of SAMBA stabilizes CYCA2;3. We conclude that SAMBA is a plant-specific negative regulator of APC/C involved in the degradation of A-type cyclins.
Results
SAMBA Is a Plant-Specific APC/C Regulator.
The SAMBA protein, encoded by AT1G32310, had been identified by tandem-affinity purification (TAP) associated with the core APC/C and, more specifically, with the subunits APC3b, APC7, and APC10 in protein complexes purified from Arabidopsis cell suspension cultures (31). In a TAP experiment on Arabidopsis cell cultures with SAMBA as bait, 12 interacting proteins were identified, including the subunits APC1, APC2, APC3b, APC4, APC5, APC6, APC7, APC8, and APC10, except APC11, and the regulators CCS52A2, UVI4, and UVI4-like/OSD1/GIG1 (31).
To confirm that SAMBA forms a complex with APC/C in planta, TAP was carried out with 6-d-old SAMBA-expressing seedlings C-terminally fused to the GS tag (see SI Materials and Methods) and proteins were identified by mass spectrometry (MS). In agreement with previous results, SAMBA interacted with all APC/C subunits in planta, except APC11, which is difficult to identify with the selected analytical approach because of its small size (Table S1). Also, the interaction with the activator CCS52A2 was found, but not with CCS52B, UVI4, and UVI4-like/OSD1/GIG1.
Although TAP allowed the detection of the entire APC/C complex, it did not provide insight into the direct interactions between core subunits and associated proteins. Therefore, we performed yeast two-hybrid (Y2H) assays with the SAMBA protein against all APC/C subunits identified in Arabidopsis and its two coactivators, CCS52A2 and CCS52B. The results revealed a direct interaction of SAMBA with APC3b (Fig. S1A) but not with APC1, APC2, APC3a, APC4, APC5, APC6, APC7, APC8, APC10, APC11, or the activators CCS52A2 and CCS52B. Taken together, the protein interaction data indicate that SAMBA is a component of the APC/C complex in plants, by binding APC3b.
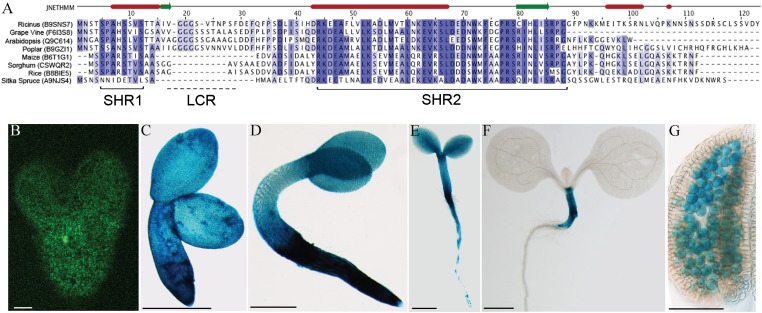
SAMBA is 100 aa in length (10.8 kDa) and resembles APC16, a recently described APC/C subunit conserved throughout metazoans (32–34) that is 110–120 residues in size and characterized by four regions of sequence similarity, referred to as AH1 to AH4 (33). Based on these regions of sequence similarity, several candidate APC16 subunits can be found in plants, including in Arabidopsis (Fig. S1B), but except for some remote similarity at the AH2 and AH3 regions, SAMBA lacks sequence homology with APC16, particularly in the extended homology region AH4 located toward its C terminus. Inversely, homology searches identified single SAMBA-like proteins in various other plant genomes. Sequence alignments of SAMBA homologs revealed two regions of sequence conservation [SAMBA homology region (SHR)1 and SHR2], spaced by a 10- to 20-residue-long low-complexity region (Fig. 1A). Because no homologous sequences were found in metazoans or protista, SAMBA constitutes an APC/C regulator unique to plants.
Fig. 1.
SAMBA orthologs and in vivo expression profile. (A) ClustalW2 multiple sequence alignment of putative SAMBA orthologs identified in selected plant genomes. A low-complexity region (LCR) that separates two regions of increased sequence similarity is marked as SHR1 and SHR2. (B) Expression of the pSAMBA-GUS-GFP reporter gene at different developmental stages. Embryo at heart stage visualized by confocal microscopy. (C) GUS activity in mature embryo. (D) Seedling at 3 DAS. (E) Seedling at 5 DAS. (F) Seedling at 8 DAS. (G) Mature pollen grains. [Scale bars: 20 μm (B); 500 μm (C and D); 1 mm (E and F); 100 μm (G).]
SAMBA Expression.
Analysis of published microarray datasets and use of the BioArray (http://www.bar.utoronto.ca) and Genevestigator (https://www.genevestigator.com) tools revealed that the SAMBA expression was very weak in all tissues, except during seed development. To study the expression pattern of SAMBA, a 1.8-kb fragment upstream of the ATG codon of the SAMBA gene was fused to a β-glucuronidase (GUS)-GFP tandem reporter cassette and introduced into Arabidopsis plants. SAMBA expression was high during embryogenesis (Fig. 1 B and C) but decreased gradually when seedlings germinated (Fig. 1 D and F). At 3 d after stratification (DAS) (Fig. 1D), GUS staining was still well visible in all tissues; at 5 DAS, it diminished and became patchy in root tissues (Fig. 1E); finally, at 8 DAS, it was present only in the hypocotyls (Fig. 1F); and, at later stages of development, the expression was only observed in pollen grains (Fig. 1G).
samba Mutants Develop Enlarged Organs.
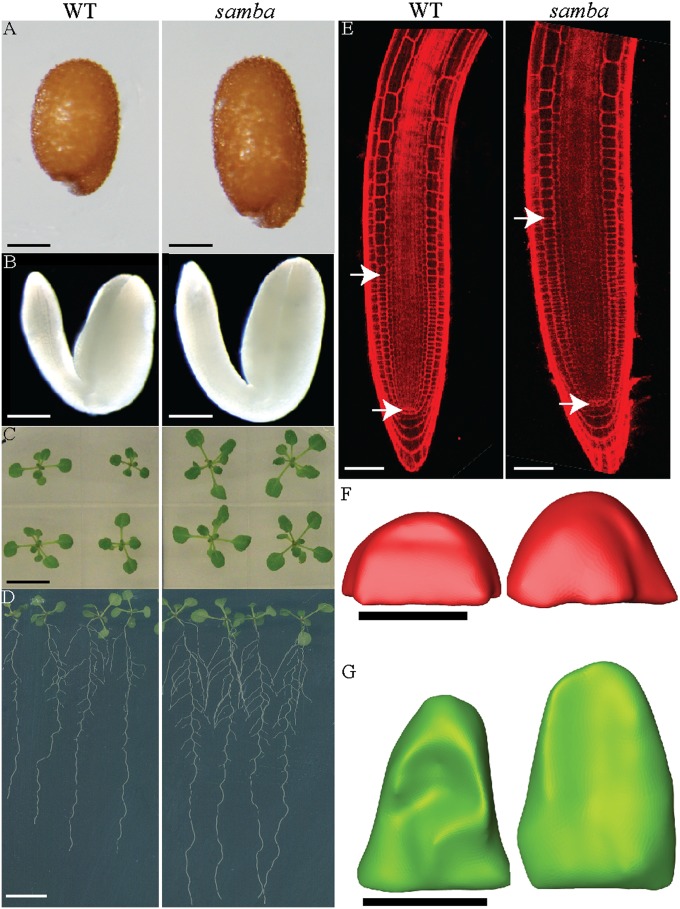
To assess the role of the SAMBA gene in plant growth and development, we analyzed its loss-of-function phenotype in two independent T-DNA insertion lines. Both lines were inserted into the first intron (Fig. S1C), completely abolishing gene expression. Because both mutants had the same phenotype, they will be further referred to as samba, unless specified. Interestingly, the analysis of samba showed an increased seed size [up to 143% of the wild type (WT)] (Fig. 2A and Fig. S2A) and embryo size (Fig. 2B). Additionally, samba rosettes were visibly larger (Fig. 2C) and roots were longer (Fig. 2D and Fig. S2B) than those of WT plants. As a consequence of the increased root length, the number of lateral roots in samba mutants was higher than that in the WT (Fig. S2C). The increased growth of samba mutants also led to a significant increase in fresh and dry weight of shoots and roots (Fig. S2 D–G).
Fig. 2.
Representative pictures of the phenotypical analysis of samba. (A) Seed size of WT and samba. (B) Mature embryos of WT and samba. (C) Seedlings of WT and samba at 15 DAS. (D) Root phenotype of WT and samba at 10 DAS. (E) Propidium iodide–stained root meristems of WT and samba at 5 DAS. Arrowheads mark the QC position and the meristem end, defined by the position where cells start elongating. (F) Side view of a reconstructed samba and WT SAM at 12 DAS. (G) Adaxial view of reconstructed primordia of leaves 1 and 2 of WT and samba at 4 DAS. [Scale bars: 100 μm (A and B); 1 mm (C and D); 20 μm (E); 50 μm (F and G).] The number of samples analyzed and the statistical analysis are provided in Fig. S2.
To understand the samba growth phenotype, we analyzed both root and shoot meristem sizes. The root apical meristem (RAM), measured from the quiescent center (QC) toward the point where cortical cells start to elongate, was on average 26% larger in samba than in WT (Fig. 2E and Fig. S2H). The shoot apical meristem (SAM) of seedlings at 12 DAS was analyzed by means of a 3D reconstruction made from serial sections. Volumetric measurements of seedlings at 12 DAS revealed that the mean SAM volume of samba was ∼40% higher (139 × 103 μm3; n = 19) than that of the WT (99 × 103 μm3; n = 16) (Fig. 2F and Fig. S2I). Also when 3D reconstructions of the first leaf initials were compared at 4 DAS, leaf primordia of samba plants were on average 80% larger than those of the WT (Fig. 2G and Fig. S2J). In conclusion, SAMBA appears to negatively regulate early seedling growth.
To demonstrate that the observed growth phenotype is caused by the samba-1 and samba-2 mutations, we engineered a rescue construct (pSAMBA:SAMBA) consisting of 1,817 bp of the promoter sequence fused to the SAMBA full-length genomic sequence. Transgenic lines expressing this construction complemented the samba growth phenotype in both mutant lines analyzed (Fig. S3A).
Cellular Analysis of the Samba Leaf Phenotype.
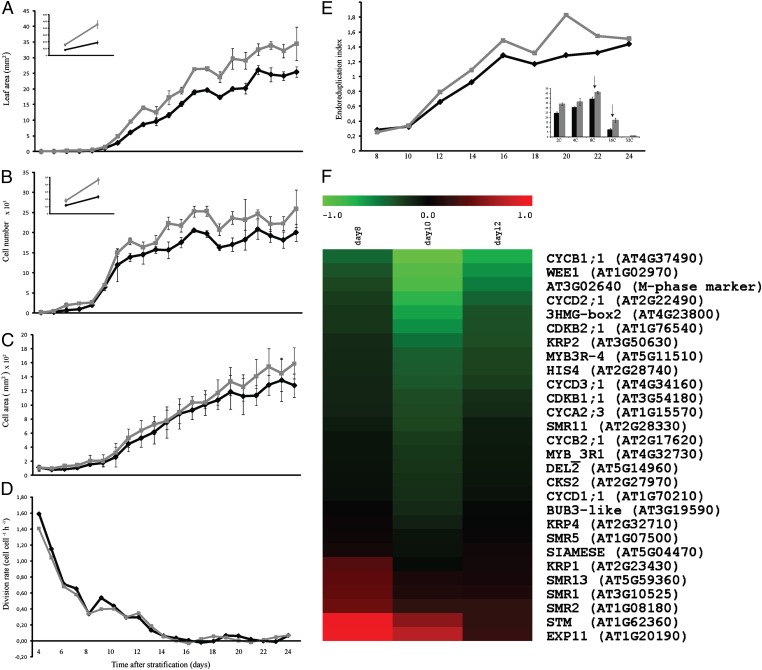
To investigate the effect of SAMBA inactivation at later stages of vegetative development, we analyzed leaf growth kinematically. From 4 until 24 DAS, the first leaf pair of samba and WT plants, grown side by side, was harvested and leaf blade area, cell number, cell area, and stomatal index were measured. In agreement with the measurements of the shoot apex by 3D reconstructions, the leaf size of the first leaf pair of samba plants was already larger during the initial stages of development (Fig. 3A, Inset). At 4 DAS, the size of the samba leaf initials was on average 94% larger than that of the WT (Table S2), but, during further development, it became relatively less pronounced and at maturity (24 DAS) had increased ∼36% (Fig. 3A). Cellular measurements demonstrated that cell number (Fig. 3B) and, to a lesser extent, cell size (Fig. 3C) contributed to the increased leaf area. At 4 DAS, samba leaf initials already contained ∼56% more cells than the WT (Fig. 3B Inset and Table S2). However, calculated on the basis of the increase in cell number over time, the average cell division rate for the entire leaf in samba and control plants was similar (Fig. 3D), meaning that the difference observed in samba mutants happened already before analysis of the first time point (4 DAS).
Fig. 3.
Kinematic analysis of leaf growth in samba and WT plants. (A) Leaf blade area of the first leaf pair of samba (gray) and WT (black) plants grown in vitro from 4 to 24 DAS. (Inset) Measurement at 4 and 5 DAS. (B) Number of cells on the abaxial side of leaves. (Inset) Cell numbers at 4 and 5 DAS. (C) Cell area. (D) Cell division rate. (E) Ploidy distribution of the first leaf pair of samba and WT plants. (Inset) Ploidy distribution measured by flow cytometry of the first leaves from 20-d-old samba and WT plants. (F) Transcript analysis of selected cell cycle genes in samba mutants measured by nCounter analysis. All values were normalized against the expression level of the housekeeping genes and expression was compared with the expression data in the WT. Data are means ± SE (n = 3).
An additional parameter involved in leaf development is the onset of endoreduplication that marks the transition between cell division and cell differentiation. Cells from leaves 1 and 2 divide up to 10 DAS, after which they gradually exit the cell cycle and start to endoreduplicate (35). At 8 DAS, leaves 1 and 2 of samba and control lines had an equally low endoreduplication index (EI), representing the mean number of endoreduplication cycles per nucleus. The EI of samba leaves was significantly higher between 12 and 20 DAS than that of equivalent control leaves, particularly because of an increase in the 8C and 16C cell population (Fig. 3E, Inset).
SAMBA Loss of Function Affects a Diversity of Transcripts.
To understand the molecular mechanisms associated with the vegetative samba mutant phenotype, we extracted RNA from leaves 1 and 2 at 10 DAS for microarray transcript profiling. At 10 DAS, leaves 1 and 2 of samba mutants were clearly larger than the WT leaves. A total of 298 genes were differentially expressed (fold change 1.5; false discovery rate < 0.05): 287 were up-regulated, and 11 down-regulated. PageMan (36) analysis indicated that among the 287 up-regulated genes, the most overrepresented classes were ethylene signal transduction, stress response, transcription, and calcium signaling (Dataset S1).
Because the samba mutants were affected in cell number, as well as endoreduplication, we analyzed the expression profile of a set of 28 cell cycle marker and growth-related genes (Table S3). WT and samba plants were grown in vitro for 8, 10, and 12 DAS, and RNA levels in dissected leaves 1 and 2 were measured with the nCounter system that allows a direct multiplexed analysis of selected transcripts (37). Most of the G2/M-phase–related genes analyzed, such as CYCB1;1, cyclin-dependent kinase CDKB2;1, and 3xHMG-box2 (38–40), were significantly down-regulated at all three developmental time points in samba mutants, whereas genes encoding cell cycle inhibitors, such as KIP-RELATED PROTEIN 1, SIAMESE RELATED 1 (SMR1), SMR2, and SMR3, were up-regulated, in particular at 8 DAS. A similar expression profile was found for the SHOOT MERISTEMLESS and EXPANSIN11 (EXP11) genes (Fig. 3F). In conclusion, the expression analysis is consistent with the hypothesis that loss of SAMBA advances early development as seen by the down-regulation of mitotic cell cycle genes and the up-regulation of genes encoding cell cycle inhibitors, expansin, and ethylene-responsive genes.
Loss of SAMBA Stabilizes CYCA2;3.
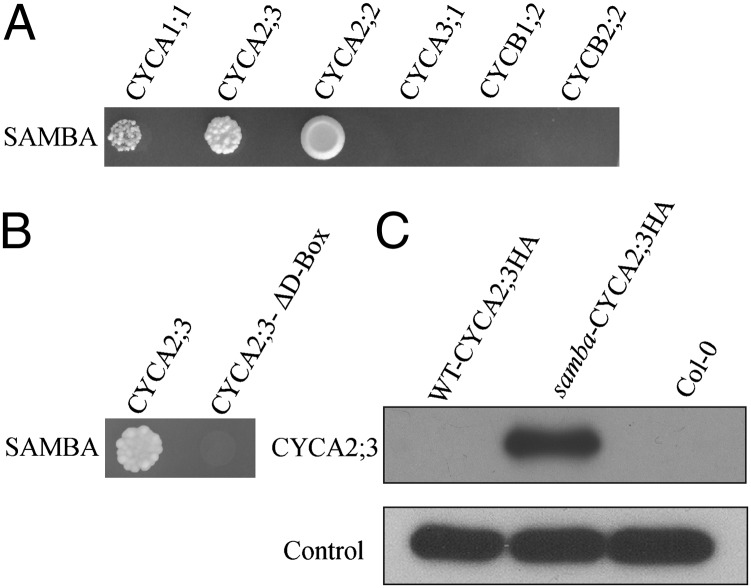
Recently, APC10 had been shown to regulate the CYCB1;1 abundance (28), and the two APC/C inhibitors UVI4 and UVI-like/OSD1/GIG1 stabilize CYCA2;3 and CYCB1;2, respectively (22, 23). Therefore, we tested whether SAMBA interacts with mitotic cyclins and affect their stability. Y2H assays between SAMBA and six different mitotic cyclins (CYCA1;1, CYCA2;2, CYCA2;3, CYCA3;1, CYCB1;2, and CYCB2;2) revealed that SAMBA interacted with CYCA2;2 and CYCA2;3, weakly with CYCA1;1, and not with CYCA3;1, CYCB1;2, or CYCB2;2 (Fig. 4A). When the D-box in CYCA2;3 was deleted, no interaction with SAMBA was observed (Fig. 4B).
Fig. 4.
CYCA2;3 stabilization. (A and B) Y2H interactions between SAMBA and different cyclins and CYCA2;3∆D-box (D-box–mutated), respectively. (C) Detection of CYCA2;3-HA protein levels in etiolated seedlings of WT CYCA2;3-HA (control line) and samba CYCA2;3-HA at 5 DAS.
To substantiate the Y2H results, WT and samba mutant plants were stably transformed with a hemagglutinin (HA)-tagged CYCA2;3 under control of the CaMV35S promoter. Subsequently, the CYCA2;3 abundance was analyzed with a commercially available anti-HA antibody. Because SAMBA is strongly expressed during early developmental stages, protein extracts of etiolated plants at 5 DAS, were used (Fig. S4A), avoiding interference with the large subunit of Rubisco. After fractionation by SDS/PAGE, Western blotting revealed a 51-kDa band specific to CYCA2;3-HA only in the samba mutant (Fig. 4C), indicating the stabilization of CYCA2;3 in samba plants. Quantitative (q)RT-PCR analysis confirmed the overexpression of CYCA2;3 and the expression levels of SAMBA in the lines analyzed (Fig. S4 B and C). This result, together with the Y2H experiments, implies that SAMBA targets CYCA2;3 for degradation.
To assess the specificity of the CYCA2;3 stabilization in samba mutant plants, we crossed transgenic plants carrying the CYCB1;1 promoter fused to the CYCB1;1 D-box-GUS/GFP construct with samba plants. Expression of the pCYCB1;1:CYCB1;1 D-box-GUS/GFP construct allowed us to estimate the rate of CYCB1;1 degradation in proliferating cells (28, 41). The CYCB1;1 degradation rate did not differ in the leaf primordia or in the RAM of WT and samba mutant plants (Fig. S4 D and E), suggesting that SAMBA specifically targets A2-type and not B-type cyclins for degradation.
SAMBA Is Required for Male Gametogenesis.
Although homozygous samba mutants could be obtained, immature siliques after self-fertilization of the samba-1 and samba-2 mutants contained ∼46–47% (403 of 875 and 249 of 531, respectively) of aborted ovules, which were small, whitish, and shrunken (Fig. S5A). This phenotype was probably attributable to malfunctioning of the male gametogenesis, because samba mutants produced considerably less viable pollen than WT plants. Indeed, in anthers of samba mutants, only 29% of pollen grains colored red after Alexander’s staining, indicating viability (n = 1,606; Fig. 5 A and B).
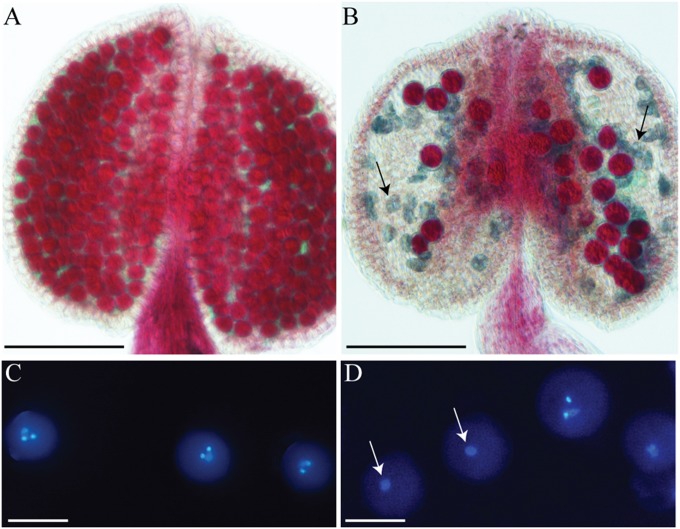
Fig. 5.
Pollen phenotype of samba mutants and WT. The viability of pollen grains was tested by coloration with Alexander’s stain in WT (A) and samba (B) plants. The red staining indicates that the pollen is viable. (C and D) WT and samba pollen stained with DAPI and observed under UV fluorescence, respectively. Two densely stained sperm nuclei and one large diffuse vegetative nucleus are visible in the WT but frequently only one single vegetative nucleus in samba (arrows). [Scale bars: 500 μm (A and B); 10 μm (C and D).]
Reciprocal crosses between samba-2/+ and WT plants were carried out to determine the T-DNA transmission. The transmission rate was severely reduced only when heterozygous samba mutants were used as paternal donor, confirming the dysfunction of the male gametogenesis (Fig. S5B). Furthermore, the construct pSAMBA:SAMBA fully complemented the ovule abortion and the pollen viability phenotypes, showing that they were caused by the SAMBA inactivation (Fig. S3 B and C). To understand which stage of pollen development is affected in the samba mutants, we analyzed the progression through male meiosis. In both samba-1 and samba-2 homozygous mutants, meiosis was normal and formed balanced spore-containing tetrads (Fig. S5C). Also, analysis of chromosome spreads revealed no abnormalities (Fig. S5D). We conclude that SAMBA is not required for meiosis, and, therefore, the defective male gametogenesis must be postmeiotic. Indeed, analysis of the pollen grains at the mature stage revealed that WT pollen contained one vegetative nucleus and two sperm nuclei (Fig. 5C), whereas ∼21% (n = 682) of the samba pollen contained only one vegetative nucleus (Fig. 5D). These results indicate that the lack of SAMBA expression interferes with mitosis I of pollen microspore development.
Discussion
Although the APC/C plays an important regulatory role in the eukaryotic cell cycle and has been subject to numerous studies (12), still much has to be learned on its biochemical structure and regulation. For example, only recently, APC16 had been identified as a functional APC/C component in animals (33, 34) and, similarly, UVI4 and UVI4-like/OSD1/GIG1 as plant-specific inhibitors of APC/C (22, 23). Here, we identified SAMBA as a negative regulator of APC/C also unique to plants. TAP of protein complexes with SAMBA as bait allowed in planta isolation of the entire APC/C complex.
The SAMBA protein associates with the APC/C through direct interaction with APC3b that also binds to the CCS52 and CDC20 proteins throughout the common C-box and IR motifs (25). Although SAMBA does not have these motifs, it might interact with APC3b via the highly conserved SHR region found in all putative SAMBA orthologs. We speculate that SAMBA competes through interaction with APC3b with CDC20/CCS52 in targeting substrates for APC/C-mediated proteolysis.
The SAMBA gene is highly expressed in developing seeds and during early plant development, indicating a specific regulatory role at these early developmental stages. In concert, loss of function of SAMBA increases seed, embryo, and seedling sizes, suggesting that SAMBA acts as a negative regulator of growth. Later in development, the size differences between samba mutants and WT plants decrease. No size difference can be observed during flower development. Other APC/C regulators might act at different developmental stages. For example, APC10 is involved in CYCB1;1 degradation and has a role during vascular development (10, 28), whereas the APC/C inhibitor UVI4 is important for trichome branching and UVI4-like/OSD1/GIG1 is required for the proper development of guard cells (22, 23).
A detailed kinematics analysis revealed that the increased growth in samba plants results from enhanced cell proliferation during embryogenesis and very early, postembryogenic development, noticeable by the difference in leaf size and cell number at the first time point analyzed (4 DAS). The size of the SAM and early leaf primordia in the samba plants was on average 40% and 94% larger than that of the WT plants, respectively. Moreover, neither cell division rates nor timing were affected in the samba mutants. We interpret that the loss of function of SAMBA stimulates cell proliferation in developing seeds and early seedlings, and this initial size difference is maintained throughout leaf development until maturity.
Whereas differences in cell number already occurred at early developmental stages, differences in cell size became significant only at later stages of leaf development. Moreover, the endoreduplication rate of samba is higher, resulting from increased fractions of 8C, 16C, and 32C nuclei, suggesting that despite the increased cell number cells exit the division cycle faster. This hypothesis is supported by the expression analysis of cell cycle marker genes that revealed a more rapid down-regulation of mitotic genes and an up-regulation of the cell cycle inhibitory genes in the samba mutants. The EXP11 gene expression was also up-regulated, in agreement with data showing that the ectopic expression of EXPANSIN genes stimulate plant growth and suppression by gene silencing reduces it (42, 43). Furthermore, microarray analysis showed that in samba knockout plants, the ethylene signal transduction was clearly overrepresented, which plays a prominent role in cell division regulation during leaf development by arresting the cell cycle (44).
SAMBA expression does not appear to be cell cycle–regulated in cell cultures (45) or in root tips synchronized with hydroxyurea (HU) treatment. SAMBA interacts with A-type cyclins, and, at least for CYCA2;3, this interaction depends on the presence of the D-box, suggesting that SAMBA might target A-type cyclins for the APC/C-mediated proteolysis. In support of this hypothesis, samba mutants highly accumulate the A2-type cyclins. This CYCA2 accumulation during early seed and seedling development is likely responsible for the stimulation of cell division, as corroborated by the enhanced cell proliferation in plants with increased CYCA2;3 levels, attributable to a mutation in the D-box (20). Also, other APC/C components were shown to control cyclin stabilization. UVI4 negatively regulates the endoreduplication onset in Arabidopsis by restraining the activity of the CCS52A1 activator subunit. As a result, uvi4 mutants fail to accumulate CYCA2;3 during the S phase (22). Conversely, UVI4-like/OSD1/GIG1 may prevent the occurrence of endoreduplication by selective inhibition of CCS52A2 activity during mitosis, thereby promoting the accumulation of CYCB1;2 (23).
Mutations in most APC/C subunits affect female and/or male gametogenesis (24–29). Here, we show that samba is defective in mitosis I of pollen development, resulting in pollen without sperm nuclei. Mutants of APC8 and APC13 (29, 46) show phenotypes similar to those described here for the samba plants. Both mutants are affected in pollen development, leading to an increased proportion of uninucleated mature pollen, indicating that the APC/C is required during the first mitosis of the male gametophytic development. Also, in agreement with the samba male phenotype, SAMBA was expressed strongly in pollen grains but not in female organs.
Materials and Methods
A full discussion of materials and methods can be found in SI Materials and Methods.
Plant Material and Production of Transgenic Plants.
Samba mutant plants (seed code SALK_018488 and SALK_048833) were obtained from the SALK collection (http://signal.salk.edu).
Kinematic Analysis.
The complete kinematics was analyzed on leaves 1 and 2 of 8–10 samba and WT plants grown in vitro and harvested daily from 4 to 24 DAS.
Phenotypical Analysis of samba.
For measurements of different parameters of the samba mutant phenotype, see SI Materials and Methods.
Immunoprecipitation and Protein Gel Blotting.
Whole etiolated plant tissue (5 DAS) was ground with 4-mm metal beads. Proteins were extracted with homogenization buffer. After centrifugation at 20,000 × g, 300 μg of total protein extract was incubated overnight with Anti-HA Affinity Matrix according to the instructions of the manufacturer (Roche).
Supplementary Material
Acknowledgments
We thank Moritz Nowack for advice with pollen analysis and Martine De Cock for help in preparing the manuscript. This work was supported by Ghent University (“Bijzonder Onderzoeksfonds Methusalem” Project Grant BOF08/01M00408 and Multidisciplinary Research Partnership “Biotechnology for a Sustainable Economy” Project 01MRB510W), the Belgian Science Policy Office (BELSPO) [Interuniversity Attraction Poles Programme (IUAP VI/33) and a postdoctoral fellowship to N.B.E.], and the European Union Sixth Framework Programme (“AGRON-OMICS” Grant LSHG-CT-2006-037704). S.D. and H.V. were supported by a predoctoral fellowship from the Agency for Innovation by Science and Technology. J.V.L. is a postdoctoral fellow of the Research Foundation-Flanders.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211418109/-/DCSupplemental.
References
- 1.Breuninger H, Lenhard M. Control of tissue and organ growth in plants. Curr Top Dev Biol. 2010;91:185–220. doi: 10.1016/S0070-2153(10)91007-7. [DOI] [PubMed] [Google Scholar]
- 2.Zachariae W, Schwab M, Nasmyth K, Seufert W. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science. 1998;282:1721–1724. doi: 10.1126/science.282.5394.1721. [DOI] [PubMed] [Google Scholar]
- 3.Capron A, Ökrész L, Genschik P. First glance at the plant APC/C, a highly conserved ubiquitin-protein ligase. Trends Plant Sci. 2003;8:83–89. doi: 10.1016/S1360-1385(02)00028-6. [DOI] [PubMed] [Google Scholar]
- 4.Peters J-M. The anaphase promoting complex/cyclosome: A machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- 5.Marrocco K, Bergdoll M, Achard P, Criqui M-C, Genschik P. Selective proteolysis sets the tempo of the cell cycle. Curr Opin Plant Biol. 2010;13:631–639. doi: 10.1016/j.pbi.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 6.Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 7.Hershko A, et al. Components of a system that ligates cyclin to ubiquitin and their regulation by the protein kinase cdc2. J Biol Chem. 1994;269:4940–4946. [PubMed] [Google Scholar]
- 8.Pickart CM, Fushman D. Polyubiquitin chains: Polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 9.Eloy NB, Coppens F, Beemster GTS, Hemerly AS, Ferreira PCG. The Arabidopsis anaphase promoting complex (APC): Regulation through subunit availability in plant tissues. Cell Cycle. 2006;5:1957–1965. doi: 10.4161/cc.5.17.3125. [DOI] [PubMed] [Google Scholar]
- 10.Marrocco K, Thomann A, Parmentier Y, Genschik P, Criqui MC. The APC/C E3 ligase remains active in most post-mitotic Arabidopsis cells and is required for proper vasculature development and organization. Development. 2009;136:1475–1485. doi: 10.1242/dev.035535. [DOI] [PubMed] [Google Scholar]
- 11.Yang Y, Kim AH, Bonni A. The dynamic ubiquitin ligase duo: Cdh1-APC and Cdc20-APC regulate neuronal morphogenesis and connectivity. Curr Opin Neurobiol. 2010;20:92–99. doi: 10.1016/j.conb.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McLean JR, Chaix D, Ohi MD, Gould KL. State of the APC/C: Organization, function, and structure. Crit Rev Biochem Mol Biol. 2011;46:118–136. doi: 10.3109/10409238.2010.541420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harper JW, Burton JL, Solomon MJ. The anaphase-promoting complex: It’s not just for mitosis any more. Genes Dev. 2002;16:2179–2206. doi: 10.1101/gad.1013102. [DOI] [PubMed] [Google Scholar]
- 14.Pfleger CM, Kirschner MW. The KEN box: An APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 2000;14:655–665. [PMC free article] [PubMed] [Google Scholar]
- 15.Criqui MC, et al. Cell cycle-dependent proteolysis and ectopic overexpression of cyclin B1 in tobacco BY2 cells. Plant J. 2000;24:763–773. doi: 10.1111/j.1365-313x.2000.t01-1-.x. [DOI] [PubMed] [Google Scholar]
- 16.De Veylder L, Joubès J, Inzé D. Plant cell cycle transitions. Curr Opin Plant Biol. 2003;6:536–543. doi: 10.1016/j.pbi.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Vandepoele K, et al. Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell. 2002;14:903–916. doi: 10.1105/tpc.010445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vanneste S, et al. Developmental regulation of CYCA2s contributes to tissue-specific proliferation in Arabidopsis. EMBO J. 2011;30:3430–3441. doi: 10.1038/emboj.2011.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshizumi T, et al. INCREASED LEVEL OF POLYPLOIDY1, a conserved repressor of CYCLINA2 transcription, controls endoreduplication in Arabidopsis. Plant Cell. 2006;18:2452–2468. doi: 10.1105/tpc.106.043869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imai KK, et al. The A-type cyclin CYCA2;3 is a key regulator of ploidy levels in Arabidopsis endoreduplication. Plant Cell. 2006;18:382–396. doi: 10.1105/tpc.105.037309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kramer ER, Scheuringer N, Podtelejnikov AV, Mann M, Peters J-M. Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol Biol Cell. 2000;11:1555–1569. doi: 10.1091/mbc.11.5.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heyman J, et al. Arabidopsis ULTRAVIOLET-B-INSENSITIVE4 maintains cell division activity by temporal inhibition of the anaphase-promoting complex/cyclosome. Plant Cell. 2011;23:4394–4410. doi: 10.1105/tpc.111.091793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwata E, et al. GIGAS CELL1, a novel negative regulator of the anaphase-promoting complex/cyclosome, is required for proper mitotic progression and cell fate determination in Arabidopsis. Plant Cell. 2011;23:4382–4393. doi: 10.1105/tpc.111.092049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Capron A, et al. The Arabidopsis anaphase-promoting complex or cyclosome: Molecular and genetic characterization of the APC2 subunit. Plant Cell. 2003;15:2370–2382. doi: 10.1105/tpc.013847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pérez-Pérez JM, et al. Specialization of CDC27 function in the Arabidopsis thaliana anaphase-promoting complex (APC/C) Plant J. 2008;53:78–89. doi: 10.1111/j.1365-313X.2007.03312.x. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, et al. The Arabidopsis APC4 subunit of the anaphase-promoting complex/cyclosome (APC/C) is critical for both female gametogenesis and embryogenesis. Plant J. 2012;69:227–240. doi: 10.1111/j.1365-313X.2011.04785.x. [DOI] [PubMed] [Google Scholar]
- 27.Kwee H-S, Sundaresan V. The NOMEGA gene required for female gametophyte development encodes the putative APC6/CDC16 component of the Anaphase Promoting Complex in Arabidopsis. Plant J. 2003;36:853–866. doi: 10.1046/j.1365-313x.2003.01925.x. [DOI] [PubMed] [Google Scholar]
- 28.Eloy NB, et al. The APC/C subunit 10 plays an essential role in cell proliferation during leaf development. Plant J. 2011;68:351–363. doi: 10.1111/j.1365-313X.2011.04691.x. [DOI] [PubMed] [Google Scholar]
- 29.Zheng B, Chen X, McCormick S. The anaphase-promoting complex is a dual integrator that regulates both microRNA-mediated transcriptional regulation of cyclin B1 and degradation of Cyclin B1 during Arabidopsis male gametophyte development. Plant Cell. 2011;23:1033–1046. doi: 10.1105/tpc.111.083980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blilou I, et al. The Arabidopsis HOBBIT gene encodes a CDC27 homolog that links the plant cell cycle to progression of cell differentiation. Genes Dev. 2002;16:2566–2575. doi: 10.1101/gad.237302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Leene J, et al. Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol Syst Biol. 2010;6:397. doi: 10.1038/msb.2010.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hutchins JRA, et al. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science. 2010;328:593–599. doi: 10.1126/science.1181348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kops GJPL, et al. APC16 is a conserved subunit of the anaphase-promoting complex/cyclosome. J Cell Sci. 2010;123:1623–1633. doi: 10.1242/jcs.061549. [Erratum J Cell Sci 123:1875] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shakes DC, Allen AK, Albert KM, Golden A. emb-1 encodes the APC16 subunit of the Caenorhabditis elegans anaphase-promoting complex. Genetics. 2011;189:549–560. doi: 10.1534/genetics.111.131714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beemster GTS, et al. Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol. 2005;138:734–743. doi: 10.1104/pp.104.053884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Usadel B, et al. PageMan: An interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics. 2006;7:535. doi: 10.1186/1471-2105-7-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geiss GK, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 38.Hemerly A, Bergounioux C, Van Montagu M, Inzé D, Ferreira P. Genes regulating the plant cell cycle: Isolation of a mitotic-like cyclin from Arabidopsis thaliana. Proc Natl Acad Sci USA. 1992;89:3295–3299. doi: 10.1073/pnas.89.8.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Segers G, et al. The Arabidopsis cyclin-dependent kinase gene cdc2bAt is preferentially expressed during S and G2 phases of the cell cycle. Plant J. 1996;10:601–612. doi: 10.1046/j.1365-313x.1996.10040601.x. [DOI] [PubMed] [Google Scholar]
- 40.Pedersen DS, et al. The plant-specific family of DNA-binding proteins containing three HMG-box domains interacts with mitotic and meiotic chromosomes. New Phytol. 2011;192:577–589. doi: 10.1111/j.1469-8137.2011.03828.x. [DOI] [PubMed] [Google Scholar]
- 41.Genschik P, Criqui MC, Parmentier Y, Derevier A, Fleck J. Cell cycle -dependent proteolysis in plants. Identification of the destruction box pathway and metaphase arrest produced by the proteasome inhibitor mg132. Plant Cell. 1998;10:2063–2075. doi: 10.1105/tpc.10.12.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pien S, Wyrzykowska J, McQueen-Mason S, Smart C, Fleming A. Local expression of expansin induces the entire process of leaf development and modifies leaf shape. Proc Natl Acad Sci USA. 2001;98:11812–11817. doi: 10.1073/pnas.191380498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi D, Lee Y, Cho HT, Kende H. Regulation of expansin gene expression affects growth and development in transgenic rice plants. Plant Cell. 2003;15:1386–1398. doi: 10.1105/tpc.011965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skirycz A, et al. Pause-and-stop: The effects of osmotic stress on cell proliferation during early leaf development in Arabidopsis and a role for ethylene signaling in cell cycle arrest. Plant Cell. 2011;23:1876–1888. doi: 10.1105/tpc.111.084160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Menges M, Hennig L, Gruissem W, Murray JA. Genome-wide gene expression in an Arabidopsis cell suspension. Plant Mol Biol. 2003;53:423–442. doi: 10.1023/B:PLAN.0000019059.56489.ca. [DOI] [PubMed] [Google Scholar]
- 46.Saze H, Kakutani T. Epigenetic mutation of a transposon-flanked gene. EMBO J. 2007;26:3641–3652. doi: 10.1038/sj.emboj.7601788. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.