Abstract
Many cyanobacteria have been shown to harbor multiple chromosome copies per cell, yet little is known about the organization, replication, and segregation of these chromosomes. Here, we visualize individual chromosomes in the cyanobacterium Synechococcus elongatus via time-lapse fluorescence microscopy. We find that chromosomes are equally spaced along the long axis of the cell and are interspersed with another regularly spaced subcellular compartment, the carboxysome. This remarkable organization of the cytoplasm along with accurate midcell septum placement allows for near-optimal segregation of chromosomes to daughter cells. Disruption of either chromosome ordering or midcell septum placement significantly increases the chromosome partitioning error. We find that chromosome replication is both asynchronous and independent of the position of the chromosome in the cell and that spatial organization is preserved after replication. Our findings on chromosome organization, replication, and segregation in S. elongatus provide a basis for understanding chromosome dynamics in bacteria with multiple chromosomes.
Several cyanobacterial species contain multiple complete copies of their single chromosome (1–6). This feature is in contrast to more traditionally studied bacteria, such as E. coli, that typically contain only one or two complete chromosome copies (7). The existence of multiple chromosome copies raises intriguing questions concerning chromosome positioning, replication, and segregation. For example, how are multiple copies of a chromosome organized within the bacterial cell? How are chromosomes segregated to daughter cells? How is replication regulated and coordinated between chromosome copies? Here, we investigate such questions by visually tracking chromosomes through multiple cell divisions in the cyanobacterium Synechococcus elongatus PCC 7942 (hereafter, S. elongatus).
S. elongatus is a rod-shaped bacterium with multiple and varying numbers of copies of a ∼2.7-Mb chromosome. The existence of ∼1–6 chromosomes in S. elongatus has been documented by distributions of DNA staining fluorescence (2–4) and PCR-based methods (5, 6). Peaks in DNA staining fluorescence not restricted to 2n chromosomes (2, 3) and whole-genome sequencing of bromodeoxyuridine (BrdU)-labeled DNA (6) have suggested that chromosome replication is asynchronous—not all chromosomes replicate simultaneously. Individual replication loci have been visualized in single cells by comparing isolated BrdU-positive regions to more widespread 4',6-diamidino-2-phenylindole (DAPI) staining (6). Although indirect and static methods have been used to investigate chromosome number and replication in S. elongatus, direct visualization of replication events in live cells has not been reported and may aid in our understanding of DNA replication in cyanobacteria.
DNA staining of another cyanobacterium with multiple chromosome copies, Synechocystis PCC 6803, has shown significant variance in DNA content between daughter cells (8), suggesting unequal partitioning of chromosomes upon cell division. Nucleoid separation in Synechocystis takes place very late in the cell cycle—immediately before completion of cell septum formation—consistent with the idea that the act of constriction partitions chromosomes to daughter cells. These observations led to the proposal that chromosome segregation is random and passive—that there is no specific machinery dedicated to partitioning chromosomes at cell division (8). This random partitioning mechanism may be tolerated in cells harboring many copies of their chromosome because daughters are likely to get at least one chromosome by chance, and subsequent chromosome replication may compensate for variance created during cell division.
Bacteria are now known to have intricate subcellular architectures that play an important role in physiology (9). DAPI staining in S. elongatus has shown DNA to exist throughout the cell volume (6, 10), but little is known about how each individual chromosome copy is organized within the cell and its relation to other cytoplasmic components. The organization of chromosomes within the cytoplasm may play a pivotal role in partitioning during cell division or in the localization of other biomolecules and cytoplasmic components. By observing individual chromosomes within the cell, we investigate the organization of the cyanobacterial cytoplasm and its role in chromosome segregation.
Results
Spatial Organization of Chromosomes.
To investigate chromosome location, replication and segregation in S. elongatus, we visualized individual genomic loci in live cells by using the fluorescent repressor-operator system (11–13). Individual chromosomal loci are visualized by binding of fluorescently tagged repressor proteins to a tandem array of operator sites. Specifically, we inserted lactose (lac) operator arrays and tetracycline (tet) operator arrays into different chromosomal sites, ∼200° apart along the circular chromosome (Fig. 1A). The tet operator arrays are positioned 11° from the putative replication origin (oriC) (6, 14) and the lac operators 33° from the putative replication terminus (terC) (6). Tandem arrays with 120 operators were interspersed with heterogeneous sequences to reduce instability (15). Expression of tagged repressor proteins, EYFP-LacI and TetR-ECFP, allowed for the simultaneous tracking of genomic loci near the origin and terminus in live cells (Fig. 1B). Fluorescent loci depend on binding of the repressor proteins, because tet or lac loci disappear upon addition of saturating anhydrotetracycline (aTC) or isopropyl β-d-thiogalactoside (IPTG), respectively.
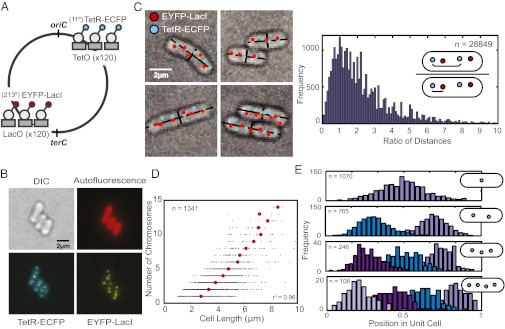
Fig. 1.
Chromosomes form domains and are ordered along the length of the cyanobacterial cell. (A) Genomic loci are labeled by using fluorescently tagged repressor proteins and corresponding operator arrays. The tet operator array is located at 11° and the lac operator array at 213° on the chromosome relative to the putative replication origin (oriC). The lac operator array is 33° from the putative replication terminus (terC) (6, 14). (B) Distinct genomic loci can be visualized when tagged repressors are bound to operator arrays (TetR-ECFP bound to tet operators and EYFP-LacI bound to lac operators). A single z-section is shown. (C Left) Computational identification of cells and operator arrays (cyan and red dots), and calculation of cell width and length (black lines). Multiple z-sections are used to identify chromosomes within the cell (Materials and Methods). (C Right) The distances between a given tet operator (cyan dot) and (i) the nearest tet operator (cyan dot) and (ii) the nearest lac operator (red dot) were calculated. The ratio of these distances (see schematic) is plotted as a histogram. More than 75% of ratios are greater than one, which implies that identical genomic loci are generally further apart than different genomic loci. A total of 28,849 pairs were analyzed. (D) The number of chromosome copies in a cell as a function of cell length. Red dots represent the average cell length for cells with a given number of chromosomes. The correlation between the average cell length and chromosome number is r2 = 0.96. A total of 1,341 cells were analyzed. (E) The position of chromosomes along the major axis of the cell (normalized for cell length) for cells containing one, two, three, or four chromosomes.
DAPI staining has suggested that S. elongatus chromosomes occupy a large volume (6, 10). Imaging of strains with both lac and tet operators revealed that identical genomic loci are generally further apart than different genomic loci (Fig. 1C), suggesting that interchromosomal distances are greater than intrachromosomal ones. The tet and lac genomic loci appear to exist in pairs within the cell (Fig. 1C), implying that individual chromosomes occupy separate territories within a cell, rather than being randomly dispersed or being clustered by genomic position. The existence of pairs of tet and lac genomic loci also suggests that most chromosomes in S. elongatus are fully or close-to-fully replicated because the tet operators are positioned close to the putative terminus. Occasional unpaired tet loci represent chromosomes undergoing replication (see Fig. 3). On average there are 3.3 ± 2.5 chromosomes in exponentially growing cells (Fig. S1). There is a linear relationship between chromosome number and mean cell length (r2 = 0.96) (Fig. 1D).
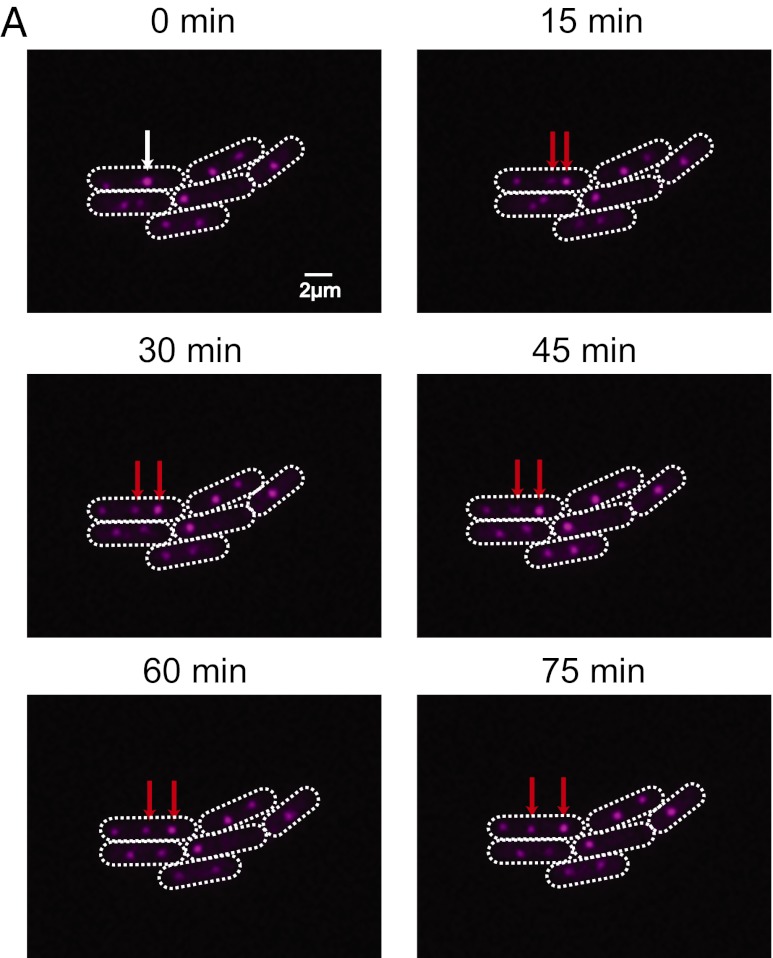
Fig. 3.
Chromosome replication is asynchronous within individual cells. (A) A time course of wild-type cells. A single genomic locus proximal to the origin is labeled by using tet operator arrays (pink dots). White arrows point to a replicating chromosome and red arrows point to the resulting, replicated chromosomes. A single z-section is shown.
Because chromosomes form distinct territories within cells, subsequent experiments were performed with cells containing only tet operators and TetR-EYFP, and these foci were used as a proxy of chromosome location. We measured the position of chromosomes within cells and found them to be equally spaced along the long axis of the cell (Fig. 1E).
Chromosome Ordering and Midcell Septum Placement Minimize Chromosome Partitioning Error.
We followed chromosomes through multiple generations of cell division (Movies S1 and S2). Cells were grown on an agarose pad under continuous light, resulting in a doubling time of ∼12 h. In wild-type cells, the cell septum forms precisely at the midcell position (Fig. 2A). The combination of spatial ordering of chromosomes and accurate midcell placement of the septum may allow for nearly equal partitioning of chromosomes upon cell division. Indeed, quantification of chromosomes partitioned to each daughter cell shows nearly equal partitioning (Fig. 2B). In addition, we find that cells have an average of 5.2 ± 1.5 chromosomes before cell division (Fig. 2B), which suggests that cells do not commit to cell division immediately after reaching a particular number of chromosomes.
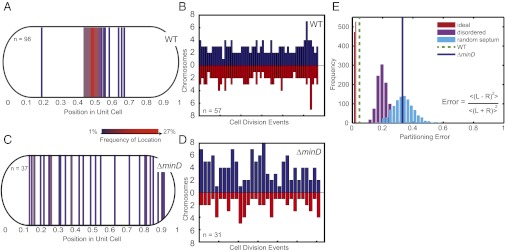
Fig. 2.
Chromosome ordering and midcell septum formation enhance the accuracy of chromosome partitioning. (A) The site of septum formation in wild-type cells during cell division. Each line corresponds to a single cell division site, and the color represents the frequency of the septum site. (B) The number of chromosomes partitioned to pairs of daughter cells upon cell division. Each pair of bars represents a single cell division event, with the height corresponding to the number of chromosomes received by a daughter cell. (C) The site of septum formation in the ΔminD strain. (D) The number of chromosomes partitioned to pairs of daughter cells upon cell division in the ΔminD strain. (E) The partitioning error for chromosome segregation in wild-type and the ΔminD strain. Simulations of partitioning error for: (i) ideal chromosome segregation (perfect ordering and perfect midcell septa) (red histogram); (ii) disordered chromosome segregation (disordered chromosomes and wild-type distribution of septa) (purple histogram); and (iii) random septa (perfect ordering and random septa) (cyan histogram). Inset gives equation for partitioning error with R and L representing the number of chromosomes segregated to each individual right and left daughter cell pair. Brackets represent averages of all cell division events of the population.
To investigate the role of septum placement in chromosome segregation, we followed segregation in a strain lacking minD (Synpcc7942_0896) (Movies S3 and S4), which encodes a protein that plays a role in regulating cell septum position in bacteria (16, 17). We find that S. elongatus ΔminD cells inaccurately position the cell septum along the cell length (Fig. 2C). Despite inaccurate septum placement, chromosomes still remain ordered along the length of the cell in the ΔminD strain (Fig. S2), suggesting that chromosome ordering and cell septum placement are independent events. Misplacement of the cell septum in ΔminD cells results in daughter cells receiving unequal numbers of chromosomes (Fig. 2D), with the number of chromosomes partitioned to each daughter cell determined by the position of the septum (Fig. S3 A and B). Thus, no active mechanism exists to ensure that equal numbers of chromosomes are partitioned to daughter cells—if the septum is misplaced, chromosomes are simply partitioned based on their initial location in the cell and the position of the septum.
To quantify the partitioning error in cells, we defined the error as the statistical difference in chromosome number between daughters averaged across cell division events (18) (Fig. 2E). An error of zero corresponds to perfectly equal segregation to daughter cells and higher error values correspond to greater disparity between the number of chromosomes segregated to daughter cells. To compare the segregation error of wild-type cells to cases of disordered chromosomes or random septum placement, we performed the following simulations: (i) perfectly ordered chromosomes and perfect midcell septum position; (ii) disordered chromosomes and midcell septum drawn from the wild-type distribution; and (iii) perfectly ordered chromosomes and random cell septum placement (Fig. 2E and Materials and Methods). For each case, the result of 1,000 independent simulations of cell populations is plotted as a histogram. The ideal partitioning error is not zero because cells may have an odd number of chromosomes at the time of cell division. The observed chromosome partitioning error in wild-type cells is slightly greater than the ideal scenario, possibly due to slight errors in ordering (Fig. 1E), cell septum placement (Fig. 2A), or miscounting of insufficiently separated chromosomal loci. However, the experimental partitioning error for wild-type cells is significantly lower than for simulations of disordered chromosomes or misplaced septa. The partitioning error for simulations of cells with ordered chromosomes and random cell septum placement is in agreement with experimental observations of ΔminD cells. Together, these results imply that chromosome ordering combined with accurate midcell septum placement permits near-ideal accuracy of chromosome partitioning to daughter cells. Disruption of either process results in partitioning errors that may adversely affect the fitness of a cell population.
Asynchronous and Position-Independent Chromosome Replication.
The repressor-operator system allows for visualization of replicating chromosomes. Because our tet operator arrays are positioned only 11° from the putative replication origin (6, 14), we can visualize chromosomes undergoing replication. We visualized replicating chromosomes by imaging cells every 15 min under constant light conditions and observe that nascent (newly replicated or replicating) chromosomes become spatially ordered within a 45-min window, and typically only one chromosome replicates at a given time (Fig. 3 and Fig. S4). The 45 min it takes to order a nascent chromosome is much shorter than the average cell generation time of 12 h, suggesting that ordering is a dynamic process, not necessarily coupled to cell growth. Upon following different cells, each containing three chromosomes and undergoing a single replication event, we found the first, second, and third chromosomes replicated 17, 18, and 12 times, respectively. Thus, chromosome replication is independent of the position of the chromosome in the cell.
Spatial Organization of Chromosomes and Carboxysomes.
Cyanobacteria have carbon fixation organelles known as carboxysomes, which were recently shown to be ordered along the long axis of the S. elongatus cell (19). Because we found chromosomes to display a similar pattern of ordering, we investigated the spatial relationship between these two components. To visualize carboxysomes, we expressed an ECFP fusion of the ribulose-1,5-bisphosphate carboxylase oxygenase (RuBisCO) large chain protein—RbcL (19), which localizes to the carboxysomes. When RbcL-ECFP is expressed in the repressor-operator strain, chromosomes and carboxysomes show a striking, alternating pattern (Fig. 4A) with carboxysomes positioned closer to chromosomes than to other carboxysomes (Fig. 4B).
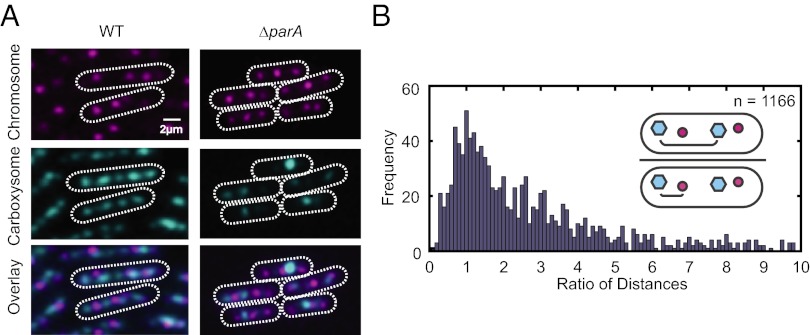
Fig. 4.
Chromosomes and carboxysomes are spatially mutually exclusive. (A) Chromosome and carboxysome positioning in wild-type compared with parA mutants. A single z-section is shown. (B) The distances between a given carboxysome (cyan dot) and (i) the nearest carboxysome (cyan dot) and (ii) the nearest chromosome (red dot) were calculated. The ratio of these distances (see schematic) is plotted as a histogram. More than 75% of ratios are greater than one, which implies that carboxysomes are generally further apart than a carboxysome and the nearest chromosome. A total of 1,166 pairs were analyzed.
To further investigate the relationship between chromosomes and carboxysomes, we deleted the parA-like gene Synpcc7942_1833 (hereafter parA), encoding a member of a family of proteins known to play a role in chromosome and plasmid segregation in many bacteria (20). Although the deletion of parA in S. elongatus affects carboxysome alignment (19), it does not affect chromosome positioning (Fig. 4A). Interestingly, the position of chromosomes and carboxysomes still remains mutually exclusive in the ΔparA strain. Thus, either independent mechanisms regulate the spacing of carboxysomes and chromosomes or carboxysomes are organized by chromosomes and this organization is mediated via ParA (21–23).
To identify a possible mechanism for spatial chromosome organization, we attempted to disrupt three additional cytoskeletal genes that may have involvement in chromosome organization [ftsZ (Synpcc7942_2378), mreB (Synpcc7942_0300), and a parA-like gene (Synpcc7942_0220); ref. 24]. Complete deletions of mreB and ftsZ could not be obtained, suggesting that these genes may be essential for viability in S. elongatus. Deletion of the parA-like gene had no effect on chromosome alignment and did not result in any detectable phenotypes. The parA-like genes (Synpcc7942_1833 and Synpcc7942_0220) in S. elongatus differ from those with a role in plasmid or chromosome segregation in other bacteria (20) in that they do not have an associated parB and are located more than 30° from the origin. A parAB gene (Synpcc7942_B2637 and Synpcc7942_B2626) exists on a 46-kb endogenous plasmid in S. elongatus, but its function was not investigated.
Discussion
Chromosome organization and segregation have not been previously characterized in single S. elongatus cells. The principles of chromosome organization, segregation, and replication in S. elognatus may also be applicable to other bacteria with multiple chromosome copies. By tracking genomic loci through cell divisions, we find that chromosomes form separate domains that are linearly ordered along the cell length. The spatial organization of chromosomes combined with accurate midcell septum placement aids in near-optimal partitioning of chromosomes to daughter cells. In addition, we find that chromosome replication is asynchronous and position-independent and that the bacterial cytoplasm is arranged with chromosomes and carboxysomes alternating along the long axis of the cell.
One potential benefit of chromosome ordering is that the concentration of molecules that localize near chromosomes may be uniform across the cell length (25). In addition, the observed correlation between chromosome number and cell length suggests a possible mechanism for gene dosage compensation. As the cell grows, a chromosome is replicated, maintaining a relatively constant ratio of genetic material to cell volume. The dilution of a molecule during cell growth may trigger chromosome replication. After a chromosome is replicated, the concentration of the molecule may be restored by the additional genetic material. In this manner, a coupling of chromosome replication to cell growth may provide the basis for gene dosage compensation in bacteria with multiple chromosome copies. Our observation of asynchronous chromosome replication is unique to S. elongatus. Well-studied bacteria such as Escherichia coli undergo synchronous DNA replication through the combined actions of SeqA and DnaA (26). The mechanism of asynchronous chromosome replication remains to be determined in S. elongatus.
When combined with accurate midcell septum placement, chromosome ordering also plays an important role in segregation of chromosomes to daughter cells. Previous studies in the spherical cyanobacterium Synechocystis showed random segregation of chromosomes to daughters (8). The difference in segregation patterns between Synechocystis and S. elongatus may be related to their respective chromosome copy numbers—Synechocystis has 10 times as many chromosome copies (5) as S. elongatus and, therefore, may be able to safely rely on random chromosome partitioning. Spatial ordering of chromosomes and accurate midcell septum placement may be a prominent feature of bacteria with an intermediate number of chromosomes where random segregation can lead to large variation between daughters and occasional anucleate cells. Inaccurate septum placement may be a tolerated feature when chromosomes are ordered because the number of chromosomes per unit of cell volume remains the same. However, inaccurate septum placement increases the likelihood of anucleate daughter cells, as observed in the ΔminD strain (Fig. S3C), thereby decreasing the fitness of the population.
The natural question that arises is as follows: What mechanism is responsible for chromosome ordering? In one model, chromosomes can be represented as self-avoiding polymers. When placed under spatial constraints, conformational entropy may drive chromosome segregation into ordered domains (27). Alternatively, there may be a molecular explanation for chromosome repulsion. In several bacterial species, the filamentous protein ParA drives chromosome and/or plasmid segregation (20, 28). Although deletion of parA and other candidate cytoskeletal genes did not perturb chromosome ordering in S. elongatus, we cannot eliminate the possibility of such a system.
We find that chromosomes and carboxysomes are ordered in the cytoplasm in a manner that is spatially mutually exclusive, suggesting that spatial constraints may influence ordering in the cell. Each S. elongatus cell is roughly 700 nm in width, but the majority of this space is occupied by thylakoid membranes (29). The ∼100-nm diameter carboxysomes appear to occupy a significant fraction of the cytoplasm in transverse section (19, 29), possibly occluding the chromosome. As a result, the simultaneous ordering of the two components may increase the accuracy of each individual component’s ordering. The observation that parA deletion perturbs carboxysome ordering, but not chromosome ordering suggests two possible models. Either ParA is responsible for ordering carboxysomes using chromosome ordering as a template (21, 22), or chromosomes and carboxysomes are ordered independently, but spatial constraints influence their ultimate positioning.
In this study, we characterize chromosome ordering, replication, and segregation in S. elongatus. We observe an intricate organization of the cyanobacterial cytoplasm and demonstrate its role in chromosome segregation. However, the molecular mechanisms underlying the organization of chromosomes in the cytoplasm remain unknown. Further investigation of the mechanisms responsible for chromosome ordering in S. elongatus will improve our understanding of chromosome segregation and spatial organization in bacteria.
Materials and Methods
Strain Construction and Cloning.
Relevant plasmids, strains, and primers are presented in Tables S1–S3, respectively. Wild-type S. elongatus PCC 7942 was acquired from ATCC catalog no. 33912. Unless otherwise noted, S. elongatus were grown in modified BG-11 media (BG-11M) (30) at 30 °C with cool-white fluorescent illumination of 4,000–6,000 lx and all appropriate antibiotics. Antibiotic concentrations were 2.5 μg/mL each spectinomycin/streptomycin, 5 μg/mL chloramphenicol, 5 μg/mL kanamycin, and 2 μg/mL gentamycin. Transformations were performed by following standard conditions (30). If additional selection was necessary to reduce false-positive colonies, transformations were plated onto a sterile nitrocellulose membrane placed on top of a BG-11M agar plate and kept in low light (1,500 lx) for 2 d before transfer to normal light conditions. On the third and fifth days, the nitrocellulose membrane was moved to a fresh BG-11M agar plate to ensure continuous selection. After 10 d, individual colonies were isolated and patched.
S. elongatus cells were always transformed in the following order: (i) lac and/or tet operator arrays, (ii) deletion vector (if necessary), (iii) carboxysome marker (if necessary), and (iv) TetR and/or LacI fusion proteins. TetR and/or LacI fusion proteins were always transformed last, and aTC and/or IPTG were required to mitigate growth defects during transformation and propagation of strains with both DNA-binding protein and operator array. TetR/TetO strains required aTC at 5–7.5 μg/mL, and LacI/LacO strains required IPTG at 1 mM. Patched colonies were generally prepared for microscopy 3–5 d after patching, because cells stored for longer periods of time often lost operator arrays.
Two new S. elongatus integration sites were developed to insert lac operators into the genome. These sites, A and B, were strategically chosen not to affect transcription. They are situated in a region of negligible transcription between convergent transcripts as verified by RNA sequencing (31). Multiple-cloning-site and chloramphenicol cassette for integration vectors were obtained from pAM1573 (30) and cloned between 1 and 1.5 kb of upstream and downstream homologous sequence. Individual PCR products were first assembled by using fusion PCR and subsequently integrated into pBR322 by using GeneArt Seamless Cloning and Assembly Kit (Invitrogen). All PCR primers are provided in Table S3.
One hundred twenty tet and 120 lac operator repeats with interspersed heterogeneous sequences were obtained from eBB110 (32) and pLAU43 (15), respectively. Integration plasmids with operators were designed as shown in Tables S1 and S3. Fig. 1 shows cells with lac operators in site A and tet operators in neutral site 2.1 (NS 2.1) (30). All other figures and analysis was performed on cells with only tet operators in NS 2.1.
C-terminal TetR and N-terminal LacI fluorescent protein fusions were used to visualize chromosomes. The tetramerization domain (last 12 amino acids) of LacI was deleted. Expression of fusion proteins was driven by the kaiBC promoter. Levels of fusion proteins did not appear to have appreciable circadian oscillations (33). Sequences and annotation for TetR and LacI fusion protein constructs are provided in Table S4.
To visualize carboxysomes, RbcL-ECFP was expressed from neutral site 2.2 (NS 2.2) (30) whereas the native copy of RbcL remained intact (19). The fusion protein was expressed by using the apcA promoter. ECFP was obtained from pJRC23 (33). Individual PCR products were assembled by using fusion PCR and subsequently ligated between SmaI and XhoI of EB2065 (NS 2.2). All PCR primers are provided in Table S3.
Gene knockouts were generated by deletion of the gene with a gentamycin resistance cassette obtained from pAM2055 (30). Constructs were created by flanking the gentamycin cassette with at least 700 bases of upstream and downstream homologous sequence by fusion PCR. The PCR product was subsequently ligated into pUC18. All PCR primers are provided in Table S3. Knockouts were created by transforming S. elongatus with the appropriate plasmid, patching single colonies, and verifying segregation by PCR.
Microscopy.
Cells growing on plates were scraped from patched colonies within 3–5 d of patching and washed three times in 50 μL of BG-11M and resuspended in 5 μL of BG-11M. One microliter of cells were placed in a Lab-Tek II Chamber Coverglass Chamber (Electron Microscopy Sciences; catalog no. 70377–11) and overlaid with a 1.5 cm × 1.5 cm × 1 cm 2% (wt/vol) ultrapure agarose (Invitrogen) pad with antibiotics. In general, to minimize stress, only the antibiotics selecting for operator arrays and repressor proteins were used during imaging. Pads for TetR/TetO strains were supplemented with 0.005–0.01 μg/mL aTC to allow for growth. LacI/LacO strains did not require supplementation with IPTG for growth. Still images were taken on pads without aTC or IPTG. To prevent drying, sponges were soaked in water and placed along the inside of the chamber. Cells were allowed to equilibrate in chamber under 4,000 lx illumination for at least 8 h before imaging.
Samples were imaged on a Zeiss Inverted scope with a 100× 1.4 N.A. objective and equipped with a Photometrics Evolve 512 EMCCD camera. The microscope was controlled by using Axiovision software (Zeiss). Photosynthetic lighting at ∼4,000 lx was constantly provided by a white LED ring light (Advanced Illumination; part no. RL1360-WHI-C2) and turned off only before exposures by Axiovision software. Cells were maintained in a 30 °C, 0.5–1% CO2 chamber throughout the experiment. Multiple positions (1–10) were imaged at regular time intervals between 15 and 120 min. At each time-point, RFP (autofluorescence), CFP, YFP, and differential interference contrast (DIC) were measured at five different z-stacks (300-nm intervals). For longer time courses, CFP exposure was limited to the middle z-stack to limit phototoxicity. Definite focus (Zeiss) was used to maintain focus over the duration of the experiment. Division times varied between 10 and 14 h for wild-type cells with the operator-repressor system.
Image Analysis.
Image analysis was performed by using custom software written in Matlab (MathWorks). Images were corrected for x-y drift and segmented. DIC and CFP images were used to distinguish cell boundaries, and the autofluorescence at RFP emission wavelength was used to eliminate background noise. Identified objects were filtered by size and defined as cells. Inaccurately segmented cells were manually corrected. Custom software tracked the mother-daughter lineage by determining the maximum overlap in coordinates of cells through different time-points. In this manner, lineage was tracked through multiple generations.
Chromosomes and carboxysomes were assigned coordinates by using custom Matlab software. Briefly, the YFP or CFP image was convolved with a Gaussian filter. The Laplace operator was applied to the filtered image (using an approximation kernel) to sharpen maxima and minima, defining boundaries between dots and the surrounding regions. Subsequently, a threshold was applied to create a binary image. Appropriate threshold values were determined by finding the number of foci identified in an image for a range of threshold values. The inflection point of this distribution was used as the final threshold value. Finally, the Matlab function regionprops was used to determine properties of closed regions (foci). To ensure that chromosomal foci on different focal planes were being identified, five 300-nm z-stacks were acquired at each time-point. Centroids were projected from 3D to 2D space. Foci that were within 320 nm of each other along the x-y plane were collapsed into the same foci so that chromosomes moving during z-stack acquisition were not identified as multiple chromosomes. After cell segmentation, regionprops was used to determine cell length and superimpose a rectangular coordinate system on each cell. The location of dots were determined in polar coordinates and transformed to a unit cell for analysis of relative spacing of dots within a cell.
Partitioning Error Simulations.
The partitioning error was calculated to represent the difference in chromosomes partitioned to daughter cells, normalized for the total number of chromosomes in the mother cell (Fig. 2E, Inset) (18). The partitioning error was calculated for wild-type and ΔminD cells using experimental data from Fig. 2 B and D, respectively. Chromosome partitioning error was also simulated for the following conditions: (i) ordered chromosomes and perfect midcell septum placement, (ii) disordered chromosomes and a wild-type distribution of midcell septum placement and (iii) ordered chromosomes with random septum placement. For each scenario, 1,000 independent experiments were simulated, with each simulated experiment consisting of the same number of cell divisions and chromosome numbers as the actual experiments [wild-type conditions for (i) and (ii) and ΔminD conditions for (iii)]. Disordered chromosome partitioning was modeled by using a binomial distribution with each chromosome representing an independent trial and the septum location determining the probability of segregation to a daughter cell (for example, perfect midcell septum corresponds to probability of 0.5). Random cell septum placement was modeled by using a uniform distribution from 0.05 to 0.95 cell lengths, representing the observed range of septum placements in ΔminD cells.
Supplementary Material
Acknowledgments
We thank members of the E.K.O. laboratory for discussions, Shankar Mukherji for help with image processing, and Harvard Center for Biological Imaging and Bernhard Goetze for microscope access and assistance. This work was funded by The Howard Hughes Medical Institute, National Defense Science and Engineering (to V.V.), and National Science Foundation Graduate Research Fellowships (to V.V.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211144109/-/DCSupplemental.
References
- 1.Mann N, Carr NG. Control of macromolecular composition and cell division in the blue-green algae Anacystis nidulans. J Gen Microbiol. 1974;83:399–405. doi: 10.1099/00221287-83-2-399. [DOI] [PubMed] [Google Scholar]
- 2.Binder BJ, Chisholm SW. Relationship between DNA cycle and growth rate in Synechococcus sp. strain PCC 6301. J Bacteriol. 1990;172:2313–2319. doi: 10.1128/jb.172.5.2313-2319.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binder BJ, Chisholm SW. Cell cycle regulation in marine Synechococcus sp. strains. Appl Environ Microbiol. 1995;61:708–717. doi: 10.1128/aem.61.2.708-717.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mori T, Binder B, Johnson CH. Circadian gating of cell division in cyanobacteria growing with average doubling times of less than 24 hours. Proc Natl Acad Sci USA. 1996;93:10183–10188. doi: 10.1073/pnas.93.19.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Griese M, Lange C, Soppa J. Ploidy in cyanobacteria. FEMS Microbiol Lett. 2011;323:124–131. doi: 10.1111/j.1574-6968.2011.02368.x. [DOI] [PubMed] [Google Scholar]
- 6.Watanabe S, et al. Light-dependent and asynchronous replication of cyanobacterial multi-copy chromosomes. Mol Microbiol. 2012;83:856–865. doi: 10.1111/j.1365-2958.2012.07971.x. [DOI] [PubMed] [Google Scholar]
- 7.Sherratt DJ. Bacterial chromosome dynamics. Science. 2003;301:780–785. doi: 10.1126/science.1084780. [DOI] [PubMed] [Google Scholar]
- 8.Schneider D, Fuhrmann E, Scholz I, Hess WR, Graumann PL. Fluorescence staining of live cyanobacterial cells suggest non-stringent chromosome segregation and absence of a connection between cytoplasmic and thylakoid membranes. BMC Cell Biol. 2007;8:39. doi: 10.1186/1471-2121-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gitai Z. The new bacterial cell biology: Moving parts and subcellular architecture. Cell. 2005;120:577–586. doi: 10.1016/j.cell.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 10.Smith RM, Williams SB. Circadian rhythms in gene transcription imparted by chromosome compaction in the cyanobacterium Synechococcus elongatus. Proc Natl Acad Sci USA. 2006;103:8564–8569. doi: 10.1073/pnas.0508696103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinett CC, et al. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J Cell Biol. 1996;135:1685–1700. doi: 10.1083/jcb.135.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Straight AF, Belmont AS, Robinett CC, Murray AW. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- 13.Gordon GS, et al. Chromosome and low copy plasmid segregation in E. coli: Visual evidence for distinct mechanisms. Cell. 1997;90:1113–1121. doi: 10.1016/s0092-8674(00)80377-3. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Tsinoremas NF. An unusual gene arrangement for the putative chromosome replication origin and circadian expression of dnaN in Synechococcus sp. strain PCC 7942. Gene. 1996;172:105–109. doi: 10.1016/0378-1119(96)00160-6. [DOI] [PubMed] [Google Scholar]
- 15.Lau IF, et al. Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol Microbiol. 2003;49:731–743. doi: 10.1046/j.1365-2958.2003.03640.x. [DOI] [PubMed] [Google Scholar]
- 16.Lutkenhaus J. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu Rev Biochem. 2007;76:539–562. doi: 10.1146/annurev.biochem.75.103004.142652. [DOI] [PubMed] [Google Scholar]
- 17.Miyagishima SY, Wolk CP, Osteryoung KW. Identification of cyanobacterial cell division genes by comparative and mutational analyses. Mol Microbiol. 2005;56:126–143. doi: 10.1111/j.1365-2958.2005.04548.x. [DOI] [PubMed] [Google Scholar]
- 18.Huh D, Paulsson J. Random partitioning of molecules at cell division. Proc Natl Acad Sci USA. 2011;108:15004–15009. doi: 10.1073/pnas.1013171108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Savage DF, Afonso B, Chen AH, Silver PA. Spatially ordered dynamics of the bacterial carbon fixation machinery. Science. 2010;327:1258–1261. doi: 10.1126/science.1186090. [DOI] [PubMed] [Google Scholar]
- 20.Toro E, Shapiro L. Bacterial chromosome organization and segregation. Cold Spring Harb Perspect Biol. 2010;2:a000349. doi: 10.1101/cshperspect.a000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson SR, Wadhams GH, Armitage JP. The positioning of cytoplasmic protein clusters in bacteria. Proc Natl Acad Sci USA. 2006;103:8209–8214. doi: 10.1073/pnas.0600919103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roberts MA, Wadhams GH, Hadfield KA, Tickner S, Armitage JP. ParA-like protein uses nonspecific chromosomal DNA binding to partition protein complexes. Proc Natl Acad Sci USA. 2012;109:6698–6703. doi: 10.1073/pnas.1114000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thanbichler M, Shapiro L. Getting organized—how bacterial cells move proteins and DNA. Nat Rev Microbiol. 2008;6:28–40. doi: 10.1038/nrmicro1795. [DOI] [PubMed] [Google Scholar]
- 24.Nakao M, et al. CyanoBase: The cyanobacteria genome database update 2010. Nucleic Acids Res. 2010;38(Database issue):D379–D381. doi: 10.1093/nar/gkp915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montero Llopis P, et al. Spatial organization of the flow of genetic information in bacteria. Nature. 2010;466:77–81. doi: 10.1038/nature09152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaguni JM. DnaA: Controlling the initiation of bacterial DNA replication and more. Annu Rev Microbiol. 2006;60:351–375. doi: 10.1146/annurev.micro.60.080805.142111. [DOI] [PubMed] [Google Scholar]
- 27.Jun S, Mulder B. Entropy-driven spatial organization of highly confined polymers: Lessons for the bacterial chromosome. Proc Natl Acad Sci USA. 2006;103:12388–12393. doi: 10.1073/pnas.0605305103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ebersbach G, Gerdes K. Plasmid segregation mechanisms. Annu Rev Genet. 2005;39:453–479. doi: 10.1146/annurev.genet.38.072902.091252. [DOI] [PubMed] [Google Scholar]
- 29.Schwarz R, Forchhammer K. Acclimation of unicellular cyanobacteria to macronutrient deficiency: Emergence of a complex network of cellular responses. Microbiology. 2005;151:2503–2514. doi: 10.1099/mic.0.27883-0. [DOI] [PubMed] [Google Scholar]
- 30.Mackey SR, Ditty JL, Clerico EM, Golden SS. Detection of rhythmic bioluminescence from luciferase reporters in cyanobacteria. Methods Mol Biol. 2007;362:115–129. doi: 10.1007/978-1-59745-257-1_8. [DOI] [PubMed] [Google Scholar]
- 31.Vijayan V, Jain IH, O’Shea EK. A high resolution map of a cyanobacterial transcriptome. Genome Biol. 2011;12:R47. doi: 10.1186/gb-2011-12-5-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marquis KA, et al. SpoIIIE strips proteins off the DNA during chromosome translocation. Genes Dev. 2008;22:1786–1795. doi: 10.1101/gad.1684008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chabot JR, Pedraza JM, Luitel P, van Oudenaarden A. Stochastic gene expression out-of-steady-state in the cyanobacterial circadian clock. Nature. 2007;450:1249–1252. doi: 10.1038/nature06395. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.