Abstract
In the model plant Arabidopsis thaliana, a core eudicot, the floral homeotic C-class gene AGAMOUS (AG) has a dual role specifying reproductive organ identity and floral meristem determinacy. We conduct a functional analysis of the putative AG ortholog ThtAG1 from the ranunculid Thalictrum thalictroides, a representative of the sister lineage to all other eudicots. Down-regulation of ThtAG1 by virus-induced gene silencing resulted in homeotic conversion of stamens and carpels into sepaloid organs and loss of flower determinacy. Moreover, flowers exhibiting strong silencing of ThtAG1 phenocopied the double-flower ornamental cultivar T. thalictroides ‘Double White.’ Molecular analysis of ‘Double White’ ThtAG1 alleles revealed the insertion of a retrotransposon causing either nonsense-mediated decay of transcripts or alternative splicing that results in mutant proteins with K-domain deletions. Biochemical analysis demonstrated that the mutation abolishes protein–protein interactions with the putative E-class protein ThtSEP3. C- and E-class protein heterodimerization is predicted by the floral quartet model, but evidence for the functional importance of this interaction is scarce outside the core eudicots. Our findings therefore corroborate the importance and conservation of the interactions between C- and E-class proteins. This study provides a functional description of a full C-class mutant in a noncore (“basal”) eudicot, an ornamental double flower, affecting both organ identity and meristem determinacy. Using complementary forward and reverse genetic approaches, this study demonstrates deep conservation of the dual C-class gene function and of the interactions between C- and E-class proteins predicted by the floral quartet model.
Keywords: floral organ identity genes, MADS-box genes, solo long terminal repeats, RNA silencing
Current understanding of floral patterning has emerged primarily from studies in the core eudicot model plants Arabidopsis thaliana and Antirrhinum majus. In these species, the genetic ABCE model predicts how combinatorial expression of four classes of transcription factors specifies organ identity in the floral meristem (1–4). According to the latest Arabidopsis model, which incorporates the role of the E-class proteins, once flowering has initiated, A- and E-class proteins specify sepals; A-, B-, and E-class proteins specify petals; B-, C-, and E-class proteins specify stamens; and C- and E-class proteins specify carpels and terminate floral meristem development (2, 5, 6). The underlying biochemical mechanism for specifying organ identity has been described by the floral quartet model, which predicts that correct transcription of organ-specific genetic programs requires the formation of hetero-multimeric complexes between these four interacting classes of transcription factors (5, 7–9). Mutations affecting the class-A, -B, -C, and -E functions are homeotic, resulting in the replacement of one organ type by another. Loss of expression of the Arabidopsis C-class gene AGAMOUS (AG) results in conversion of stamens and carpels to petals and sepals and indeterminacy of the floral meristem (10), leading to showy “double flowers” with excess petals.
Because all but one of the genes in the ABCE model belong to the same gene family, it has been hypothesized that the evolutionary success of the angiosperm clade depends, to a great extent, on the proliferation and coevolution of this MIKC-type MADS-box family of transcription factors (11, 12). However, insufficient taxonomic breadth of functional studies involving organ-identity genes currently hinders the generation of new hypotheses on how genetic-level changes may be responsible for angiosperm floral diversity.
There have been multiple duplication events within the C-class lineage (13) followed by partial redundancy (14–16), potential or proven subfunctionalization (17–24), or switching of functional equivalence among duplicates in different species (25). Even when a single gene is present, alternative transcripts are a source of genetic variation (14). Nonetheless, and presumably related to its crucial role in reproductive organ development, mounting evidence points to high overall functional conservation of C-class gene function across angiosperms (14, 26, 27), including basal eudicots (14, 15) and monocots (16), and possibly more broadly across seed plants (28–30).
In the Ranunculaceae, a duplication event preceding the diversification of the family gave rise to two C-class genes with distinct expression patterns; this duplication is independent of the core eudicot duplication that gave rise to Arabidopsis AG and its Antirrhinum functional equivalent, PLENA (PLE) (13). In Thalictrum, ThtAG1 is expressed in stamens and carpels throughout development, whereas ThtAG2 has ovule-specific expression (20). The fact that recruitment to ovule function has occurred independently multiple times throughout the angiosperm phylogeny (26), combined with the divergent expression data (20), led us to suspect that ThtAG1 is the C-class floral homeotic gene. Here, we focus on the functional evolution of this gene and its interaction with the putative E-class gene ThtSEP3.
Flowers with aberrant phenotypes, also known as “teratomorphs,” have been described for at least the last 2,000 y, double flowers being the first documented (ref. 31 and references therein). Double-flowered cultivars have become popular garden plants because of the attractiveness imparted by the extra petals (e.g., roses, peonies, carnations, and camellias). Moreover, scientists have put similar natural deviations from normal development to good use to help elucidate the genetic basis of normal flower development (32, 33). Although morphological, developmental, and/or genetic aspects of double-flower cultivars have been investigated (22, 34–44), no functional evidence for the underlying molecular mechanism of this familiar phenotype is available to date in a noncore eudicot.
The species Thalictrum thalictroides includes cultivars that exhibit homeotic floral phenotypes suggestive of defects in the canonical organ-identity genes of the ABCE model. Among these cultivars, we identified the Thalictrum thalictroides cultivar ‘Double White’ (also known as ‘Snowball’) as a candidate for loss of C-class function, based on its double-flower phenotype. This cultivar is a sterile homeotic mutant with flowers consisting entirely of multiple white petaloid sepals (the genus Thalictrum is apetalous). It presumably occurred spontaneously in natural populations, where it was collected and clonally propagated by plant enthusiasts because of the attractive nature of its double flowers.
In this study, we set out to characterize functionally ThtAG1, a putative AGAMOUS ortholog from the ranunculid Thalictrum thalictroides, and to test whether a mutation at this locus is responsible for a double-flower variety. In a forward genetic approach, we gathered strong evidence for the ‘Double White’ cultivar being affected in the AGAMOUS-like gene ThtAG1. In the complementary reverse genetics approach, targeted silencing of ThtAG1 caused a double-flowered phenocopy of the cultivar. Importantly, this constitutes a description of a full C-class mutant, affecting both organ identity and determinacy, in a noncore (“basal”) eudicot.
Our results provide strong evidence for high conservation of C-class gene function and of the interaction of C- and E-class proteins between core eudicot model plants and a ranunculid, including comparable roles in reproductive organ identity and flower meristem determinacy. In addition, we identify the genetic and biochemical basis of an ornamental double-flower variety, suggesting that mutations in C-class genes likely underlie other widespread double-flower varieties.
Results
‘Double White’ Flower Morphology and Development Are Consistent with a Loss-of-Function Mutation in a C-Class Gene.
To characterize T. thalictroides ‘Double White,’ we compared its morphology and development with wild type. T. thalictroides is apetalous, with typically 5–12 white or pink petaloid sepals enclosing 45–76 filamentous stamens and 3–11 free, uniovulate carpels (n = 21), the last two spirally arranged (Fig. 1 A, E, and I). T. thalictroides ‘Double White’ flowers are sterile, with 56–105 petaloid sepals (n = 12)—no stamens or carpels are formed—resulting in a double-flower phenotype (Fig. 1 B, F, and J). The outer sepals of ‘Double White’ are comparable to wild-type sepals, whereas internal sepals have a slender base of variable length (Fig. 1 E and F).
Fig. 1.
Silencing of the T. thalictroides AGAMOUS ortholog ThtAG1 results in homeotic conversions of floral organs that phenocopy the ‘Double White’ cultivar. Morphology and development of T. thalictroides wild-type, ‘Double White’ and TRV2-ThtAG1 flowers. (A) Wild-type flower with petaloid sepals (se), stamens (st), and carpels (ca). (B) ‘Double White’ flower with petaloid sepals throughout. (C) Fully silenced TRV2-ThtAG1 flower showing complete homeotic conversion of reproductive organs to sterile organs. (D) Partially silenced TRV2-TtAG1 flower showing incomplete homeotic conversion of reproductive organs. (E–H) Dissected organs, outer to inner (Left to Right). (E) Wild-type sepal, stamen, and carpel. (F) ‘Double White’ outer, middle, and inner sepaloid organs. (G) Fully silenced TRV2-ThtAG1 outer, middle, and inner sepaloid organs. (H) Partially silenced TRV2-ThtAG1 outer sepal, middle sepaloid stamens, and inner sepaloid carpel. (I–L) SEM of developing flowers. (I) Wild-type bud with sepal, stamen, and carpel primordia. (J) ‘Double White’ bud with sepal primordia. (K) TRV2-ThtAG1 bud with sepal primordia. (L) TRV2-ThtAG1 open flower with homeotic organs. (M–Y) Adaxial epidermis cellular types by SEM. (M–O) Wild-type sepal (M), stamen filament (N), and carpel (O). (P–S) ‘Double White’ outer sepal (P), blade of middle homeotic organ (Q), stalk of middle homeotic organ (R), and ventral organ (S). (T–W) TRV2-ThtAG1 outer sepal (T), blade of middle homeotic organ (U), stalk of middle homeotic organ (V), and central homeotic organ (W). (X) Mixed cell types in partially homeotic middle organ depicted in H. (Y) Mixed cell types in stalk of middle organ in H. (Z) Central partially homeotic organ. (Scale bars: 1 cm in A–D; 1 mm in E–H; 100 μm in I–L and Z; and 10 μm in M–Y.)
Scanning electron microscopy (SEM) of young floral buds shows marked differences between wild-type and ‘Double White’ early in development. Wild-type floral meristems develop flattened sepal primordia on the outside, followed by multiple, cylindrical, spirally arranged stamen primordia, then carpel primordia (distinguished by a central depression) (Fig. 1I). In ‘Double White’ all organ primordia are flattened and sepal-like (Fig. 1J).
The development of perianth organs in lieu of stamens and carpels is reminiscent of Arabidopsis C-class ag mutants (10), leading us to hypothesize that ‘Double White’ is mutated in a C-class gene. To test this hypothesis, we (i) silenced the AG ortholog ThtAG1 in wild-type T. thalictroides plants in an attempt to phenocopy the cultivar; (ii) sequenced the ‘Double White’ ThtAG1 genomic locus and transcripts in search of mutations; and (iii) tested the biochemical interactions of mutant proteins as compared with interactions in wild type.
Virus-Induced Gene Silencing of ThtAG1 Phenocopies ‘Double White’ Flowers.
T. thalictroides wild-type plants were subjected to virus-induced gene silencing (VIGS) to test whether the ‘Double White’ floral phenotype results from loss of function of the putative AG ortholog ThtAG1 and, more generally, to test the degree of functional conservation of C-class genes in a ranunculid.
Survival after agroinfiltration with TRV1 and TRV2 silencing vectors was 92% for plants treated with TRV2-ThtAG1, 90% for TRV2-empty controls, and 100% for untreated control plants. Flowering started 6 d after infiltration of the bare root plants. Phenotypes observed in TRV2-empty plants included asymmetric reduction in sepal size and occasional brown necrotic spots, both previously reported as viral effects for this species (45). Initially, 43% of TRV2-ThtAG1–treated plants showed overall stunted growth habit and lack of stamen filament elongation as compared with untreated controls. The early stunted growth phenotype also was observed in 86% of TRV2-empty–treated plants. This early phenotype did not affect subsequent growth; plants initially may have been responding to the vacuum infiltration treatment. The lack of stamen elongation, which was not observed in TRV2-empty plants, was interpreted as a late-stage function of AG consistent with that reported in Arabidopsis (46). This phenotype occurred shortly after infiltration when, in preformed buds, ThtAG1 presumably would have been silenced late in stamen development.
Homeotic floral phenotypes indicative of gene silencing were seen in 53% of treated plants. Flowers showing complete homeotic conversion of stamens and carpels to sterile organs (Fig. 1 C and G) were first observed 4 wk after infiltration and continued for at least 12 wk postinfiltration, resulting in 35 documented silenced flowers across 16 TRV2-ThtAG1–treated plants. In these flowers, outer organs resembled wild-type sepals, middle organs (in the position of stamens) had slender bases as in ‘Double White,’ and central organs (in the position of carpels) resembled small sepals (Fig. 1G). Early development in strongly silenced flowers shows that all primordia initiate with the flattened shape characteristic of perianth organs rather than the cylindrical shape characteristic of reproductive organs (Fig. 1K). Prior to the appearance of the fully homeotic phenotypes described above, approximately 2 wk after infiltration, we began to observe partial homeotic phenotypes consisting of loss of carpel and stamen identity in a chimeric pattern within flowers (Fig. S1). Carpels remained open and lacked their characteristic single apical ovule, no differentiated stigmas with papillae were visible, and nearby carpels were closed and ended in the normal stigmas with white papillae (Fig. S1 A and B). Stamen filaments became flattened and lanceolate, resembling sepals, and anthers were missing or highly reduced (Fig. S1 C and D). This chimeric phenotype was observed in approximately half of TRV2-ThtAG1–treated plants. After 3 wk, 13% of experimental plants had flowers showing intermediate phenotypes, consisting of partial conversion of reproductive organs to sterile organs throughout the whole flower (Fig. 1 D and H). Sepaloid organs replacing stamens were white to light pink, with a long narrow stalk and a distal oval blade (Fig. 1H, two middle organs). Sepaloid organs in the center of the flower, where carpels normally are found, were comparable in shape to outermost sepals, except that they were much smaller, green, and curved inward (Fig. 1H, right-most organ). SEM detail of the center of a strongly silenced mature flower reveals perianth-like organs through the center and a floral meristem still differentiating perianth primordia (Fig. 1L). None of the phenotypes described above were observed on either TRV2-empty or untreated controls.
Close examination of T. thalictroides tubers by dissecting the bud located at the “crown” (at the stage used for agroinfiltration) revealed several floral primordia already present at various stages of development before treatment (Fig. S2). This observation presented a likely explanation for phenotypes becoming stronger over time after infiltration, because flowers emerging after treatment would be affected by the lack of ThtAG1 expression from inception, whereas those present before infiltration would be affected at a later developmental stage.
Wild-type adaxial sepal epidermal cell morphology of T. thalictroides was variable, consisting of regular to interlocking cells (Fig. 1M, Upper) that, in some cases, became asymmetrically papillate toward the base (Fig. 1M, Lower) (20, 47). Stamen filaments had slender, elongated cells (Fig. 1N), whereas carpels had more rounded cells and stomata (Fig. 1O). ‘Double White’ sepals had an epidermal phenotype comparable to that of the more irregular types found in wild type (Fig. 1P). Sepal-like interlocking cells also were found in the blade of organs occupying the position of stamens (Fig. 1Q), whereas slender cells resembling those of stamen filaments were found in their stalks (Fig. 1R). Sepal-like cells also were found in the organs occupying the position of carpels (Fig. 1S). Epidermal morphology of outer sepals in silenced flowers (Fig. 1T) was comparable to the more regular morphology found in wild-type sepals (Fig. 1M, Lower). Homeotic organs that replaced stamens consisted of sepal-like cells in the blade (Fig. 1U) and more elongated cells at the stalk (Fig. 1V), resembling those found in comparable organs of ‘Double White’ (Fig. 1R) and in wild-type stamen filaments (Fig. 1N); organs replacing carpels had regular cells similar to those found in the sepaloid organs (Fig. 1W). Flowers with weak phenotype (Fig. 1D) had partially homeotic organs (Fig. 1H, third from left) exhibiting patchy epidermis with mixed stamen (elongate cells) and sepal cell types (Fig. 1X) and mixed types of elongated cells in their stalks (Fig. 1Y). Partially fused central organs with intermediate carpel–sepal identity also were observed (Fig. 1Z). In summary, the SEM results show that, in both ‘Double White’ and full double flowers resulting from ThtAG1 silencing, cell morphology of homeotic organs matched that of the outer sepals (compare Fig. 1 P, Q, and S with Fig. 1 T, U, and W), except for the stalked bases of organs in place of stamens, which more closely resembled cells in stamen filaments of wild-type plants (compare Fig. 1 N, R, and V).
Most ‘Double White’ flowers had homeotic perianth-like organs through the center (Fig. 2A), and approximately one third of the flowers dissected (n = 14) had a secondary pedicel within the center of the floral receptacle, giving rise to a nested bud that reiterates the pattern of homeotic organs (Fig. 2B) and eventually develops into a mature ‘Double White’ flower (Fig. 2C). Similarly, although most strongly silenced flowers seemed to continue to produce homeotic organs toward the center of the flower (Fig. 2D), beginning at ∼6 wk postinfiltration we observed a floral bud growing on a pedicel from the center of the flower meristem (Fig. 2E). The organs of the secondary flowers also were entirely homeotic, with a whorl of larger outer petaloid sepals surrounding smaller inner perianth-like organs (Fig. 2F). This phenotype was observed in 42% of flowers (n = 19) and 67% of plants surveyed (n = 9). In wild-type flowers, on the other hand, the floral meristem is consumed in the production of carpels, which are surrounded by stamens and sepals, never forming a nested flower (Fig. 2G). Consistent with the indeterminacy that results in a secondary bud within a flower, developmental SEM of VIGS-treated flowers shows an extreme case of overproliferation of the floral meristem, which likely would have led to more than one nested flower (Fig. 2H). Organ counts (nested-flower organs excluded) followed by one-way ANOVA found no significant difference in the total number of organs (P = 0.69, F = 0.36) in wild-type, silenced, and ‘Double White’ flowers (Fig. 2I), consistent with a one-to-one homeotic conversion of reproductive organs into perianth-like organs.
Fig. 2.
Floral meristem reinitiation and one-to-one homeotic conversion of organs in flowers subjected to VIGS of ThtAG1 compared with that of ’Double White’ flowers. Longitudinal sections of ‘Double White’ (A–C) and silenced (D–F) flowers of T. thalictroides reveal floral meristem determination defects as compared with wild type (G). (A) Multiple sepaloid organs through the flower center in ‘Double White.’ (B) Reinitiation of the floral meristem. (C) Fully developed nested ‘Double White’ flower. (D) Silenced flower with sterile homeotic organs in lieu of stamens and carpels. (E) Reinitiation of the floral meristem in silenced flower leading to another floral bud that reiterates the pattern of homeotic organs. (F) Fully developed silenced nested flower. (G) Wild-type flower with sepals, stamens, and central carpels. (H) SEM detail of extreme overproliferation of the floral meristem in a TRV2-ThtAG1–treated flower. (I) Total number of floral organs (average ± SE) in wild-type (n = 21), ‘Double White’ (n = 21), and silenced pTRV2-ThtAG1 (n = 14) flowers were not significantly different in a one-way ANOVA (P = 0.69, F = 0.36). (Scale bars: 1 mm in A–G; 100 μm in H.)
ThtAG1 Is Down-Regulated in Double Flowers Resulting from VIGS.
To test whether the floral phenotypes described above were, in fact, caused by VIGS, we proceeded first to detect TRV transcripts and then to test for down-regulation of ThtAG1. TRV1 and TRV2 viral transcripts were detected in a subset of treated silenced plants (n = 18) and mock-treated plants (empty TRV2, n = 4) but were absent from untreated controls (n = 4) (Fig. S3).
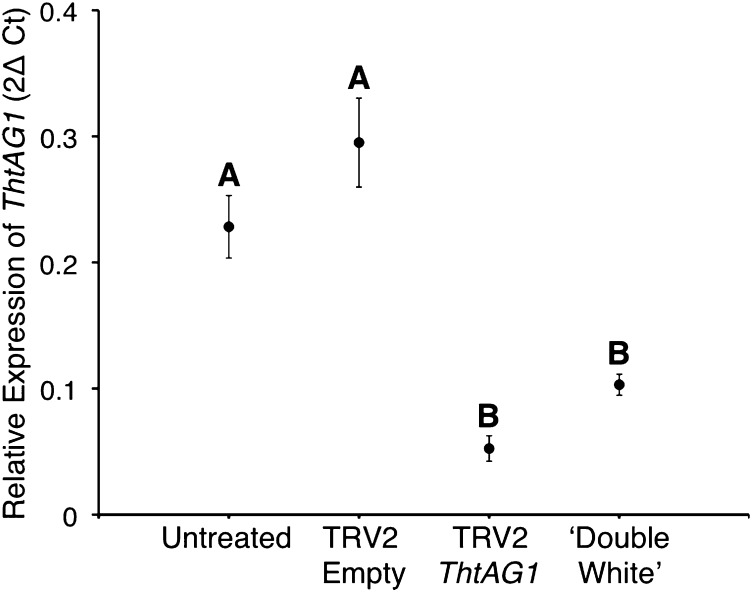
To test whether flowers exhibiting homeotic phenotypes expressed lower levels of ThtAG1 than controls and levels similar to ‘Double White,’ we used quantitative PCR (qPCR) to quantify the expression of ThtAG1 in the different treatments, untreated and empty TRV controls, and the cultivar, relative to housekeeping genes (Fig. 3).
Fig. 3.
Molecular validation of VIGS shows down-regulation of ThtAG1 in homeotic flowers of T. thalictroides compared with controls. Relative levels of ThtAG1 mRNA by qPCR in untreated (n = 12), TRV2-empty (n = 9), TRV2-ThtAG silenced (n = 35), and ‘Double White’ flowers (n = 4), normalized with the housekeeping genes ThtACTIN and ThtEEF1. Mean and SE are shown; different letters indicate a significant difference (P < 0.0001, F = 36.55) in a one-way ANOVA with Tukey contrasts (α = 0.05). The expression in TRV2-ThtAG1 silenced and ‘Double White’ samples (B) is significantly lower than the expression in untreated and TRV2-Empty samples (A). Silenced and ‘Double White’ expression levels were not significantly different from each other, nor were the two controls (untreated and TRV2-empty) (P > 0.05).
As expected from a VIGS experiment, a range of phenotypes (described above) was observed. Flowers with very weak or patchy phenotypes (Fig. S1) were not included in the expression analyses. The remaining silenced flowers collected (n = 35) were divided into the following categories by phenotype level: category 1, partial homeotic conversions of stamens and carpels (e.g., Fig. 1D); category 2, complete homeotic conversion of stamens and carpels to sepaloid organs (e.g., Fig. 1C); and category 3, complete homeotic conversions and containing a secondary nested flower within (e.g., Fig. 2 E and F). To test whether the increased intensity of the phenotypes resulted from increased silencing (lower levels of target gene expression), we compared the relative expression of ThtAG1 in phenotype category levels 1–3 by one-way ANOVA. The result was not significant (P = 0.78, F = 0.25; n = 35), suggesting the phenotype likely is affected by timing rather than by intensity of silencing (Fig. S4).
One-way ANOVA among the four experimental groups—untreated (n = 12), mock-treated (n = 9), TRV2-ThtAG1–treated (n = 35), and ‘Double White’ cultivar (n = 4)—yielded a highly significant effect of treatment (P < 0.0001, F = 36.55). Tukey contrasts resulted in a significant difference between silenced TRV2-ThtAG1–treated plants and controls (α = 0.05). The ThtAG1 expression level in the ‘Double White’ cultivar did not differ significantly from that of silenced flowers and also was significantly lower than controls under Tukey contrasts (α = 0.05) (Fig. 3).
In conclusion, the relative expression level of ThtAG1 was decreased by almost fivefold in double-flower phenotypes resulting from VIGS as compared with the average in controls. These results confirm that the observed phenotypes are caused by the down-regulation of the endogenous copy of ThtAG1 as a result of VIGS (Fig. 3). ‘Double White’ expressed 2.5-fold less ThtAG1 than controls, providing additional evidence supporting the hypothesis that its double-flower phenotype is caused by reduced or lacking C-class function. Although ‘Double White’ appeared to express more ThtAG1 than TRV2-ThtAG1–treated double flowers, the difference was not statistically significant.
Retrotransposon Insertion Underlies the ‘Double White’ Mutation.
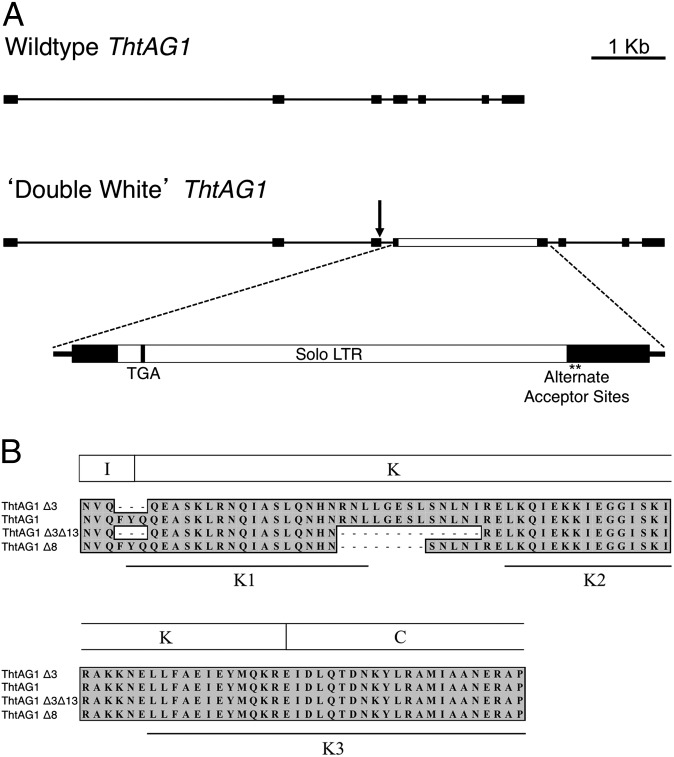
To investigate the molecular basis of the ‘Double White’ mutant, we compared sequences of wild-type and ‘Double White’ ThtAG1 genomic loci. The wild-type gene structure consists of seven exons and six introns, with a total length of 7,394 bp. The ‘Double White’ gene is 9,608 bp, the extra length resulting primarily from an insertion in exon 4 (Fig. 4A). Closer inspection of the 2,209-bp insert revealed that it is the long terminal repeat (LTR) of a retrotransposon, a “solo” LTR, flanked by a 5-bp target site duplication (CTCTC) and exhibiting the signature TG…CA terminal dinucleotides (48). The insertion provides an early stop codon 42 bp into exon 4, resulting in a nonsense mutation that likely causes nonsense-mediated decay of the resulting transcript, since we did not recover it from cDNA (49). Two cryptic splice-acceptor sites (asterisks in Fig. 4A) are present 7 and 22 bp after the LTR insertion.
Fig. 4.
Comparative genomic structure of ThtAG1 reveals a retrotransposon insertion in the ‘Double White’ mutant that disrupts mRNA accumulation and protein function. (A) Genomic structure of the ThtAG1 locus in wild-type (Upper) and ‘Double White’ (Lower). Lines represent introns, black boxes represent exons, arrow represents donor site, and the white box represents an insertion. The K domain is roughly encoded by exons 3–5 shown here. A detail of the insertion shows a solo LTR of a retrotransposon coding for an early stop codon (TGA) and two cryptic splice-acceptor sites (asterisks). (B) Translated amino acid alignment of the K-domain region of T. thalictroides wild-type and ‘Double White’ ThtAG1 recovered from cDNA. Note that two ‘Double White’ transcripts that result from the alternate acceptor sites in (A) translate into shorter proteins, lacking either eight or 13 amino acids. The three K subdomains are indicated (50), showing that the missing amino acids in ‘Double White’ affect the area near the end of the K1 subdomain and the region between K1 and K2 subdomains.
Other differences between wild-type and mutant genomic loci include intronic indels: three in intron 1 (15-bp, 59-bp, and 7-bp long), one in intron 2 (17-bp long), and one in intron 5 (53-bp long). An additional 91 small indels (≤3 bp) and SNPs occur throughout the introns. When all genetic differences were considered, the major insertion in exon 4 was suspected to be the most likely cause of the double-flower phenotype.
mRNA Analysis Reveals Putative Mutant ThtAG1 Proteins in ‘Double White.’
Because the ‘Double White’ cultivar does express a low level of ThtAG1 (Fig. 3), we wanted to investigate whether the transcripts resulted from the use of the two cryptic splice-acceptor sites identified at the genomic level (Fig. 4A). Sequence comparison of cDNA of wild-type T. thalictroides, two accessions of ‘Double White,’ and one of ‘Snowball’ revealed two types of shortened ThtAG1 transcripts that translate into putative proteins lacking either 8 or 13 amino acids within the K domain (ThtAG1∆8 and ThtAG1∆3∆13) (Fig. 4B). These transcripts are consistent with the use of the two cryptic splice-acceptor sites (asterisks in Fig. 4A). The resulting deletion at the protein level encompasses the end of the K1 subdomain and the intervening region between K1 and K2 subdomains (Fig. 4B) (50). In addition, an upstream FYQ sequence at the boundary of the intervening region (the I domain) and the K domain also found in AG from other plants such as Aquilegia, Papaver, Eschscholzia, and Vitis (AY464110, GU123602, DQ088996, and HM192806, respectively), is missing in one of the wild-type alleles (ThtAG1∆3), and occurs in combination with the missing 13-amino acids in one of the ‘Double White’ accessions (ThtAG1∆3∆13) (Fig. 4B). Five additional single-nucleotide changes in the coding region, representing synonymous substitutions, occur between wild type and ‘Double White’/‘Snowball.’ Because the K domain has been implicated in protein–protein interactions (50, 51), the eight- and 13-amino acid deletion alleles in the K domain were considered likely candidates for the loss of AG function in ‘Double White’ (and ‘Snowball’) and were tested further by yeast two-hybrid analysis.
‘Double White’ Mutations Disable Interactions Between C and E Proteins That Are Necessary for Normal Function.
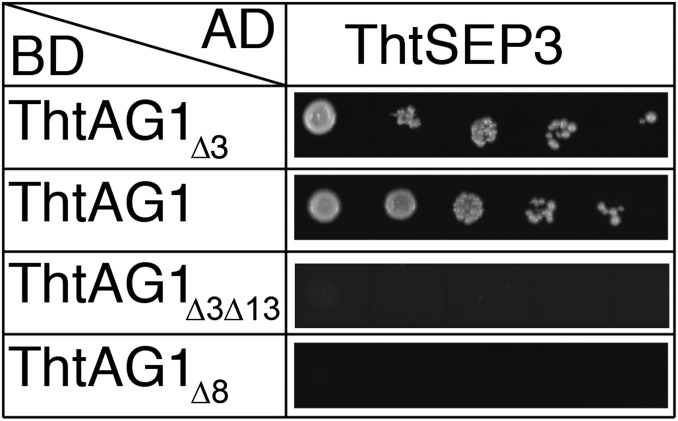
AGAMOUS proteins form homodimers and interact with SEPALLATA proteins to function in floral organ specification (5, 7, 52). To establish whether the lesion in the K domain of ThtAG1 proteins in the double-flowered mutants has a consequence at the level of protein–protein interactions, a GAL4-based yeast two-hybrid system was used. Interactions among the two wild-type alleles ThtAG1∆3 and ThtAG1, the mutant alleles ThtAG1∆3∆13 and ThtAG1∆8, and the SEPALLATA protein ThtSEP3 were assessed (Fig. 5).
Fig. 5.
Mutant C-class proteins of T. thalictroides are unable to interact with their putative E-class partner proteins, whereas wild-type proteins interact normally. Interactions between different ThtAG1 proteins and ThtSEP3 were determined with the yeast two-hybrid system. Colony growth on selective Leu/Trp/His-free + 3 mM 3-AT medium is shown. Yeast cells were spotted in 10-fold serial dilution (from left to right) for each interaction tested. ThtSEP3 was expressed as fusion with the GAL4 activation domain (AD); ThtAG1 proteins were expressed as fusions with the GAL4 DNA-binding domain (BD). For the complete set of interactions tested, see Fig. S5.
ThtSEP3 exhibited autoactivation when fused to the DNA-binding domain of GAL4. Thus, ThtSEP3∆C, a C-terminal–deleted version in which autoactivation was abolished, also was included in the analyses (Fig. S5). The wild-type proteins ThtAG1∆3 and ThtAG1 were equally capable of interacting with ThtSEP3 and with ThtSEP3∆C, albeit only in one direction (i.e., when ThtSEP3∆C was fused to the activation domain of GAL4) (Fig. S5), indicating that the loss of FYQ does not have a detrimental effect on this interaction. In contrast, ThtSEP3 or ThtSEP3∆C did not interact or interacted only very weakly with either of the ‘Double White’ proteins with a mutated K domain (Fig. 5 and Fig. S5). This result suggests that the missing eight or 13 amino acids in the K domain of ‘Double White’ ThtAG1 are essential for mediating interaction with ThtSEP3. Importantly, it also provides much needed biochemical evidence in a noncore eudicot for the C–E interaction predicted by the floral quartet model, in line with findings of high conservation of protein–protein interactions in other basal eudicots (53).
Discussion
We present evidence for strong conservation of C-class function across distantly related angiosperms, i.e., the model core eudicots Arabidopsis thaliana and Antirrhinum majus and the ranunculid Thalictrum thalictroides. Moreover, we present a molecular dissection of a double-flower cultivar, the oldest class of homeotic floral phenotype on record (31).
In a reverse genetics approach, we used VIGS to reduce endogenous levels of ThtAG1 mRNA significantly in wild-type T. thalictroides (Fig. 3). Flowers experiencing silencing of ThtAG1 phenocopied the ‘Double White’ cultivar, with one-to-one conversion of reproductive to sepal-like organs and reinitiation of the floral meristem (Figs. 1 and 2). In the complementary forward genetic approach, we uncovered a solo LTR retrotransposon insertion underlying the double-flower cultivar. A parsimonious explanation of the mutant phenotype comprises a reduction of the amount of functional mRNA caused by nonsense-mediated decay resulting from the insertion. However, even if considerable amounts of a protein were made (i.e., through the use of the cryptic splice-acceptor sites), the results of our yeast two-hybrid assays indicate that the mutant protein would be compromised in its interaction capacities and hence its function (Fig. 5). Therefore, C-class function in the double-flower variety is twice handicapped.
LTR retrotransposons have been implicated in inducing alternative splicing when inserted in an intron, disrupting the original splice-acceptor site (54). Although in ‘Double White’ the insertion is in an exon, and the original splice acceptor site is maintained (Fig. 4A), the recovery of almost full-length transcripts from cDNA of ‘Double White’ (Fig. 4B) implies that two cryptic splice-acceptor sites downstream of the inserted transposon are active. Although LTR retrotransposons are abundant in plant genomes (48), they rarely have been shown to affect phenotype significantly, a notable exception being the loss of red skin color during the evolution of white grape varieties (55). Therefore, our demonstration that an LTR retrotransposon is the cause of a flower homeotic mutation provides further evidence supporting the potential role of these elements in plant domestication and evolution.
Protein–protein interactions are essential for the function of MADS-domain transcription factors (5, 8, 56, 57). Importantly, the K domain is involved in conferring protein–protein interactions among MADS-domain proteins (12, 50, 58). Evidence for the functional importance of the K domain in AG comes from Arabidopsis thaliana, where the partial loss-of-function allele ag-4 possesses a lesion in the K domain (59). Moreover, the presence of a single additional amino acid in the K domain of the C-class–related protein FARINELLI (FAR) from Antirrhinum, also resulting from a change in splicing, alters its interaction affinities and influences its ability to specify floral organ identity (60). Thus, a deletion in the K domain of ThtAG1 may well explain the mutant phenotype observed in ‘Double White’ cultivars. This possibility is supported by the notion that the interaction between AG and SEP orthologs, considered essential for determination of reproductive organ identity (5, 8, 60), is impaired in ThtAG1-mutant proteins that lack part of the K domain. In addition, AG expression levels were significantly lower in ‘Double White’ than in wild-type plants (Fig. 3). In Arabidopsis, AG forms protein complexes with other transcription factors to act in a positive autoregulatory loop that maintains and amplifies AG expression (61). If the K-domain mutation affects the ability of the AG protein to form such complexes by inhibiting protein–protein interactions, this effect also would explain the low expression rates in ‘Double White’ flowers. However, an alternative explanation (which is not mutually exclusive with the former one) for the reduced ThtAG1 levels in ‘Double White’ flowers is that the nonsense mutation generated by the insertion of a solo LTR (Fig. 4A) renders the truncated ThtAG1 transcripts unstable so that they are targeted for degradation.
Homeotic mutants have been invaluable in increasing the understanding of the role of floral organ-identity genes, including the elucidation of the central ABC model of flower development (1, 3). Partial C-class mutants have been described previously in the monocot grasses rice (24) and maize (21), and recently, the rice double-mutant mads3 mads58 was shown to have the full C-class loss-of-function phenotype, including loss of organ identity and loss of floral determinacy (16). Such mutations typically have been ignored as potential mechanisms of evolution because of the assumption that such a radical transformation of floral organs invariably would cause a decrease in fitness (62). For this reason, homeotic mutants occurring in the wild typically have been considered evolutionary dead ends, and such a fate would seem particularly likely for any C-class mutant lacking reproductive organs that might arise spontaneously within a given population. ‘Double White’ originally was isolated by growers from the wild as a spontaneous mutant; the clonal growth habit of the species makes it possible for the mutant to persist in a population despite being sterile. The existence of similar mutants has been documented in natural populations of other plant species; in one notable instance, a double-flowered Vinca mutant has persisted for more than 160 y (22).
Functional studies in the Papaveraceae have found that lack of C function in the fourth whorl results in petals (14, 15), as in the snapdragon ple far double mutant (23), rather than in sepals, as in Arabidopsis (10). In Thalictrum the perianth is unipartite; only one type of organ is present, historically described as sepals, based on evolutionary and developmental evidence (63). The sepals of T. thalictroides, as is typical of Ranunculaceae, are the attractive part of the flower: They are large, white or pink, and sometimes have conical-papillate cells (47). In our VIGS experiments, stamens and carpels were replaced by organs that resembled additional sepals, except that those in the position of stamens were stalked, which appears to be a staminoid characteristic based on epidermal morphology (compare Fig. 1 N and U). Because petals have been lost before the diversification of Thalictrum (64), the gene and protein networks required for petal identity presumably would no longer be functional. Two other double flowers of Ranunculaceae have been described in which carpels were interpreted as suppressed and stamens as replaced by petals (44). In ‘Double White,’ we find no evidence of carpel suppression or petals being formed; instead, the meristem produces sepaloid organs in lieu of stamens and carpels.
The preformation of floral meristems in the tubers of our perennial-plant study system (Fig. S2) provides an opportunity to dissect the timing of C-class gene functions in Thalictrum. Two distinct roles have been attributed to AGAMOUS in core eudicots: reproductive-organ identity and flower determinacy (10, 23, 65–67). Evidence from Arabidopsis indicates that AG has different timing and threshold levels for its identity vs. determinacy roles, with determinacy occurring earlier (stage 3) (68) and organ identity later (stage 6) (46). Although our experiments did not test directly for AG thresholds, they do provide insight into the timing of these two fundamental functions. The fact that flowers displaying phenotypes of different severity had comparable levels of ThtAG1 mRNA (Fig. S4) does suggest a timing effect, as opposed to a delay in the triggering of silencing. In Thalictrum, loss of reproductive-organ differentiation was the first evident phenotype (Fig. S1), indicating a developmentally late function of AG, because the first flowers to emerge presumably were preformed at treatment and therefore were affected by silencing late in their development. In fact, lack of filament elongation was among the earliest phenotypes observed, consistent with the late role of AG in Arabidopsis stamen maturation (46). Floral meristem reinitiation, on the other hand, occurred later after the day of infiltration; these flowers would have emerged after silencing had initiated and therefore show the early effects of ThtAG1. This timing for the determinacy and identity functions therefore is similar to that reported for Arabidopsis and further confirms the conservation not only of overall function but also of the timing C-class roles, despite the distinct floral traits present in these early- and late-diverging lineages of eudicot angiosperms.
Variable numbers of spirally arranged floral organs and free uniovulate carpels with ascidiate development are ancestral floral traits present in Thalictrum and are distinct from the fixed number of whorled organs and the syncarpous gynoecia with multiple ovules of core eudicots (69). These marked differences in flower morphology and development in the two lineages make the similarity in underlying genetic mechanisms of reproductive-organ identity and flower meristem determinacy demonstrated in this study especially remarkable.
None of the myriad of double-flowered horticultural varieties has been dissected at a molecular level comparable to agamous in Arabidopsis (3) or plena in Antirrhinum (67). Here we provide a likely explanation for a case of double flower formation in the ranunculid Thalictrum thalictroides. More broadly, functional analysis of a C-class gene showed strong conservation of the reproductive-organ identity and floral-determinacy roles described for core eudicots (10, 23, 67) and provided vital experimental evidence for the importance of the interaction between C- and E-class proteins in a noncore eudicot.
Materials and Methods
Plant Materials.
T. thalictroides wild-type, ‘Double White,’ and ‘Snowball’ plants were purchased from nurseries and grown in the University of Washington greenhouses. Voucher specimens (WTU 376535, WTU 376544, and WTU-387674) were deposited in the University of Washington Herbaria.
Cloning and Sequencing of the ThtAG1 Genomic Locus.
The complete wild-type and ‘Double White’ ThtAG1 locus was PCR amplified and sequenced from genomic DNA. Intron 1 was amplified using primers ThtAG1int-cloneF7 and ThtAG1int-cloneR7, cloned, and sequenced using M13 and internal primers. The remainder of the wild-type gene was amplified using primers ThtAG1int-cloneF6 and ThtAG1int-cloneR4, cloned, and sequenced using M13F and internal primers. The remainder of the ‘Double White’ gene was PCR amplified and direct sequenced using the primer combinations ThtAG1 Exon 2 forward and ThtAG1 Exon 3 reverse; ThtDWAG1 Intron 2 forward and ThtDWAG1 Intron 4 reverse; and ThtAG1 Exon 4 forward and ThtAG1 coding stop reverse (Table S1). Sequence fragments were assembled into a contig in Sequencher (v. 4.9), and consensus sequences by plurality were deposited in GenBank (accession nos. JQ002519 and JQ002520).
Cloning of ‘Double White’ ThtAG1 Transcripts from cDNA.
The coding sequence for the ThtAG1 locus (GenBank accession no. AY867878) was completed on the 5′ end by PCR on two accessions of wild-type cDNA using a forward primer on the 5′UTR of the related species Thalictrum dioicum: TdAG-1–5′UTR-F2: 5′-TGATCATTCCCCCCAAAGA-3′ and the reverse primer TdAG-1-R-Whole Gene: 5′TAACAAAGTCCAGTTTGAAGGCA-3′. The same primers were used on two accessions of ’Double White’ to obtain complete coding regions. PCR products were cloned into pCRII using the TA cloning kit (Invitrogen), following the manufacturer’s instructions. Three to six clones from each of the two accessions were sequenced at the University of Washington High-throughput Genomics Unit (Seattle). Sequences were aligned in Sequencher (v. 4.9), and consensus sequences by plurality were deposited in GenBank (accession nos. JN887118– JN887121).
The expressed allelic forms of ThtAG1 were amplified from cDNA of two wild-type and three ‘Double White’ individuals using primers specific to each of two wild-type and two mutant transcripts (Table S2).
VIGS of ThtAG1.
A TRV2-ThtAG1 construct was prepared using a 421-bp fragment of the cDNA (AY867878) (20), half of which comprised the end of the coding region and the other half most of the 3′UTR. The fragment was amplified by PCR from a representative clone using primers with added XbaI and BamHI restriction sites, was ligated to TRV2, and was transformed into Agrobacterium tumefaciens strain GV3101.
VIGS was performed on 60 T. thalictroides tubers (bare-root plants) that had been kept at 4 °C for 8 wk, as described previously (45). Tubers were wounded lightly near the bud using a clean razor blade, were placed in infiltration medium so that all parts were submerged, and were infiltrated under full vacuum (−100 kPa) for 10 min. Five untreated plants were potted and grown alongside treated plants. Ten mock-treated control plants were infiltrated identically, with empty TRV2 vector. Plants were placed in a Conviron growth chamber at 21 °C and 16 h light and were monitored daily for the following 2 mo. Homeotic floral phenotypes were first observed 2.5 wk after infiltration. When flowers appeared to show an abnormal phenotype (shortly after anthesis), they were photographed, collected, and frozen for molecular validation or were fixed in formaldehyde/acetic acid/alcohol (FAA) for SEM.
Morphological Characterization of Floral Phenotypes.
Flowers of T. thalictroides ‘Double White,’ silenced plants, and controls were photographed using a Nikon SMZ800 dissecting microscope equipped with a QImaging MicroPublisher 3.3 RTV digital camera.
In preparation for SEM, floral tissue was fixed overnight or longer in FAA, dehydrated for 30 min through an alcohol series, critical-point dried, mounted, and sputter coated. Tissue preparation and observations were conducted in a JEOL JSM-840A scanning electron microscope at the University of Washington microscopy facility. Images were processed minimally in Adobe Photoshop CS5 v.11.0 and assembled using Adobe Illustrator CS5 v14.0.0.
Flower Organ Counts.
Floral organs were counted once flowers had reached maturity, defined as having fully developed anthers before dehiscence in wild-type flowers or expanded outer sepals in fully silenced flowers that lacked stamens. Organs were identified as sepals, stamens, carpels, or homeotic organs. A flower was considered to possess a secondary (nested) flower if a pedicel was observed separating the innermost whorls; organs within these secondary flowers were not included in counts. One-way ANOVA was performed to test the statistical significance of differences in the total number of flower organs among silenced TRV2-ThtAG1 (n = 14), wild-type (n = 21), and ‘Double White’ (n = 21) flowers using JMP v. 7 statistical software (SAS Institute, Inc).
Molecular Validation of VIGS.
Total RNA was prepared from 50–100 mg of frozen floral tissue using TRIzol (Invitrogen), following the manufacturer’s instructions. One microgram of the resulting total RNA was treated with amplification-grade DNase I (Invitrogen) to eliminate potential genomic contamination and was reverse-transcribed to cDNA using the SuperScript III first-strand synthesis kit (Invitrogen) with Oligo(dT)20 or specific primers to pTRV1, OYL 198 (5′-GTAAAATCATTGATAACAACACAGACAAAC-3′) (70), or pTRV2, PYL156R (5′-GGACCGTAGTTTAATGTCTTCGGG-3′) (71).
To test for the presence of the TRV1 and TRV2 viral RNA in treated plants, RT-PCR was carried out on 1 μL of cDNA using the TRV1-specific primers OYL195 (5′-CTTGAAGAAGAAGACTTTCGAAGTCTC-3′) (70) and OYL198 and the TRV2-specific primers pYL156F (5′-TTACTCAAGGAAGCACGATGAGC-3′) (71) and pYL156R for 30 cycles at 51 °C annealing temperature.
Quantification of ThtAG-1 expression in the different treatments was performed using real-time PCR, as previously described (47). Briefly, each 30-μL reaction contained 15 μL of SYBR Green PCR Master Mix (Bio-Rad), 0.9 μL (10 μM) of the gene-specific primers, 1 μL of template cDNA, and 12.2 μL of water. Locus-specific primers used for qPCR were ThtAG forward 5′-AGTCTCTCAGCAATCTCAATATCAGG-3′ and ThtAG1 reverse 5′-GCCCTGAGATACTTGTTATCAGTCTGC-3′.
Samples were amplified by 40 cycles in triplicate, including a no-template control, under the following conditions: 94 °C for 10 min, followed by 45 cycles of 94 °C for 30 s, 54 °C for 30 s, and 72 °C for 30 s on the MJ Research Chromo4 PCR at the University of Washington Comparative Genome Center. Melting curve analysis of the primers (50–95 °C in 0.5-°C increments of 1 s each) yielded a single peak. Reactions were normalized to two housekeeping genes, ACTIN and ELONGATION FACTOR 1 (EEF1), using the ΔΔCT relative quantification method (72). Average values and SEs were graphed and compared statistically by one-way ANOVA with Tukey contrasts using JMP v. 7.
Plasmid Construction for Yeast Two-Hybrid Assays.
cDNA sequences encoding T. thalictroides MADS-domain proteins were cloned into plasmids pGADT7 and pGBKT7 for yeast two-hybrid analyses using EcoRI and BamHI recognition sites. Exceptions are ThtAG1, ThtAG1∆3∆13, and ThtSEP3, which were cloned into pGBKT7 using NcoI and SalI recognition sites. ThtSEP3 was cloned into pGADT7 using EcoRI and SacI recognition sites. Primers used for the PCR cloning procedure are listed in Table S3. Except for ThtSEP3∆C, which lacked the last 195 bp of the coding sequence of ThtSEP3, all sequences cloned spanned the region from the MADS domain to at least the stop codon of the respective cDNAs. The region encoding the N-terminal extension present in some AG-like genes was not included.
Yeast Two-Hybrid Assays.
Yeast two-hybrid assays were carried out as described previously (73) with the following modifications: Cells were dissolved in 100 μL water and were serially diluted 10-fold to up to 1:10,000 in water before 2 μL of the undiluted suspension and of each dilution was plated on Leu/Trp/His-free medium supplemented with 3mM 3-Amino-1,2,4-triazole (3-AT) to test for an interaction and on Leu/Trp-free plates to control for yeast growth. Interactions were scored after 6–9 d of incubation at 28 °C.
Supplementary Material
Acknowledgments
We thank Dr. Dinesh Kumar for allowing the use of his TRV1-TRV2 vector system for virus-induced gene silencing; Kacie Mc Carty for her contribution to scoring phenotypes; Horacio de la Iglesia for assistance with statistical analysis; Rachana Kumar for preliminary sequencing of the ‘Double White’ transcripts; Yaowu Yuan for identification of the LTR retrotransposon; Doug Ewing and the staff of the University of Washington Botany greenhouses for assistance with plant care; Pang Chan (University of Washington microscopy facility) for assistance with scanning electron microscopy; David Baum, Dick Olmstead, and Valerie Soza for advice on the use of phylogenetic terminology; and Christian Gafert for help with the yeast two-hybrid analyses. This work was supported by National Science Foundation Grant IOS-1121669 (to V.S.D.). T.R.T was supported by a Research Experience for Undergraduates-linked-to-National Science Foundation Grant IOS-RIG 0818836 (to V.S.D.). R.M. received a postdoctoral fellowship from the Carl Zeiss Stiftung.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. JQ002519, JQ002520, and JN887118–JN887121).
See Author Summary on page 13478 (volume 109, number 34).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1203686109/-/DCSupplemental.
References
- 1.Coen ES, Meyerowitz EM. The war of the whorls: Genetic interactions controlling flower development. Nature. 1991;353:31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- 2.Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature. 2000;405:200–203. doi: 10.1038/35012103. [DOI] [PubMed] [Google Scholar]
- 3.Bowman JL, Smyth DR, Meyerowitz EM. Genetic interactions among floral homeotic genes of Arabidopsis. Development. 1991;112(1):1–20. doi: 10.1242/dev.112.1.1. [DOI] [PubMed] [Google Scholar]
- 4.Schwarz-Sommer Z, Huijser P, Nacken W, Saedler H, Sommer H. Genetic control of flower development by homeotic genes in Antirrhinum majus. Science. 1990;250:931–936. doi: 10.1126/science.250.4983.931. [DOI] [PubMed] [Google Scholar]
- 5.Honma T, Goto K. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature. 2001;409:525–529. doi: 10.1038/35054083. [DOI] [PubMed] [Google Scholar]
- 6.Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky MF. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr Biol. 2004;14:1935–1940. doi: 10.1016/j.cub.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 7.Theissen G. Development of floral organ identity: Stories from the MADS house. Curr Opin Plant Biol. 2001;4(1):75–85. doi: 10.1016/s1369-5266(00)00139-4. [DOI] [PubMed] [Google Scholar]
- 8.Theissen G, Saedler H. Plant biology. Floral quartets. Nature. 2001;409:469–471. doi: 10.1038/35054172. [DOI] [PubMed] [Google Scholar]
- 9.Melzer R, Theissen G. Reconstitution of ‘floral quartets’ in vitro involving class B and class E floral homeotic proteins. Nucleic Acids Res. 2009;37:2723–2736. doi: 10.1093/nar/gkp129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowman JL, Smyth DR, Meyerowitz EM. Genes directing flower development in Arabidopsis. Plant Cell. 1989;1:37–52. doi: 10.1105/tpc.1.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kramer EM, Hall JC. Evolutionary dynamics of genes controlling floral development. Curr Opin Plant Biol. 2005;8:13–18. doi: 10.1016/j.pbi.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 12.Kaufmann K, Melzer R, Theissen G. MIKC-type MADS-domain proteins: Structural modularity, protein interactions and network evolution in land plants. Gene. 2005;347:183–198. doi: 10.1016/j.gene.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 13.Kramer EM, Jaramillo MA, Di Stilio VS. Patterns of gene duplication and functional evolution during the diversification of the AGAMOUS subfamily of MADS box genes in angiosperms. Genetics. 2004;166:1011–1023. doi: 10.1534/genetics.166.2.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hands P, Vosnakis N, Betts D, Irish VF, Drea S. Alternate transcripts of a floral developmental regulator have both distinct and redundant functions in opium poppy. Ann Bot (Lond) 2011;107:1557–1566. doi: 10.1093/aob/mcr045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yellina AL, et al. Floral homeotic C function genes repress specific B function genes in the carpel whorl of the basal eudicot California poppy (Eschscholzia californica) Evodevo. 2010;1:13. doi: 10.1186/2041-9139-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dreni L, et al. 2011. Functional analysis of all AGAMOUS subfamily members in rice reveals their roles in reproductive organ identity determination and meristem determinacy. The Plant Cell 23(8):2850–2863.
- 17.Lü S, Du X, Lu W, Chong K, Meng Z. Two AGAMOUS-like MADS-box genes from Taihangia rupestris (Rosaceae) reveal independent trajectories in the evolution of class C and class D floral homeotic functions. Evol Dev. 2007;9:92–104. doi: 10.1111/j.1525-142X.2006.00140.x. [DOI] [PubMed] [Google Scholar]
- 18.Pan IL, McQuinn R, Giovannoni JJ, Irish VF. Functional diversification of AGAMOUS lineage genes in regulating tomato flower and fruit development. J Exp Bot. 2010;61:1795–1806. doi: 10.1093/jxb/erq046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang SY, et al. Duplicated C-class MADS-box genes reveal distinct roles in gynostemium development in Cymbidium ensifolium (Orchidaceae) Plant Cell Physiol. 2011;52:563–577. doi: 10.1093/pcp/pcr015. [DOI] [PubMed] [Google Scholar]
- 20.Di Stilio VS, Kramer EM, Baum DA. Floral MADS box genes and homeotic gender dimorphism in Thalictrum dioicum (Ranunculaceae) - a new model for the study of dioecy. Plant J. 2005;41:755–766. doi: 10.1111/j.1365-313X.2005.02336.x. [DOI] [PubMed] [Google Scholar]
- 21.Mena M, et al. Diversification of C-function activity in maize flower development. Science. 1996;274:1537–1540. doi: 10.1126/science.274.5292.1537. [DOI] [PubMed] [Google Scholar]
- 22.Wang YQ, Melzer R, Theissen G. A double-flowered variety of lesser periwinkle (Vinca minor fl. pl.) that has persisted in the wild for more than 160 years. Ann Bot (Lond) 2011;107:1445–1452. doi: 10.1093/aob/mcr090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davies B, et al. PLENA and FARINELLI: Redundancy and regulatory interactions between two Antirrhinum MADS-box factors controlling flower development. EMBO J. 1999;18:4023–4034. doi: 10.1093/emboj/18.14.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamaguchi T, et al. Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa. Plant Cell. 2006;18:15–28. doi: 10.1105/tpc.105.037200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Causier B, et al. Evolution in action: Following function in duplicated floral homeotic genes. Curr Biol. 2005;15:1508–1512. doi: 10.1016/j.cub.2005.07.063. [DOI] [PubMed] [Google Scholar]
- 26.Zahn LM, et al. Conservation and divergence in the AGAMOUS subfamily of MADS-box genes: Evidence of independent sub- and neofunctionalization events. Evol Dev. 2006;8:30–45. doi: 10.1111/j.1525-142X.2006.05073.x. [DOI] [PubMed] [Google Scholar]
- 27.Ferrario S, Immink RG, Angenent GC. Conservation and diversity in flower land. Curr Opin Plant Biol. 2004;7:84–91. doi: 10.1016/j.pbi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 28.Zhang PY, Tan HTW, Pwee KH, Kumar PP. Conservation of class C function of floral organ development during 300 million years of evolution from gymnosperms to angiosperms. Plant J. 2004;37:566–577. doi: 10.1046/j.1365-313x.2003.01983.x. [DOI] [PubMed] [Google Scholar]
- 29.Tandre K, Svenson M, Svensson ME, Engström P. Conservation of gene structure and activity in the regulation of reproductive organ development of conifers and angiosperms. Plant J. 1998;15:615–623. doi: 10.1046/j.1365-313x.1998.00236.x. [DOI] [PubMed] [Google Scholar]
- 30.Rutledge R, et al. Characterization of an AGAMOUS homologue from the conifer black spruce (Picea mariana) that produces floral homeotic conversions when expressed in Arabidopsis. Plant J. 1998;15:625–634. doi: 10.1046/j.1365-313x.1998.00250.x. [DOI] [PubMed] [Google Scholar]
- 31.Meyerowitz EM, Smyth DR, Bowman JL. Abnormal flowers and pattern-formation in floral development. Development. 1989;106(2):209–217. [Google Scholar]
- 32.Hintz M, et al. Catching a ‘hopeful monster’: Shepherd’s purse (Capsella bursa-pastoris) as a model system to study the evolution of flower development. J Exp Bot. 2006;57:3531–3542. doi: 10.1093/jxb/erl158. [DOI] [PubMed] [Google Scholar]
- 33.Cubas P, Vincent C, Coen E. An epigenetic mutation responsible for natural variation in floral symmetry. Nature. 1999;401:157–161. doi: 10.1038/43657. [DOI] [PubMed] [Google Scholar]
- 34.Akita Y, Horikawa Y, Kanno A. Comparative analysis of floral MADS-box genes between wild-type and a putative homeotic mutant in lily. J Hortic Sci Biotechnol. 2008;83(4):453–461. [Google Scholar]
- 35.Innes RL, Remphrey WR, Lenz LM. An analysis of the development of single and double flowers in Potentilla fruticosa. Can J Bot. 1989;67(4):1071–1079. [Google Scholar]
- 36.MacIntyre JP, Lacroix CR. Comparative development of perianth and androecial primordia of the single flower and the homeotic double-flowered mutant in Hibiscus rosa-sinensis (Malvaceae) Can J Bot. 1996;74(12):1871–1882. [Google Scholar]
- 37.Lehmann NL, Sattler R. Homeosisin floral development of Sanguinaria canadensis and S. canadensis 'Multiplex' (Papaveraceae) Am J Bot. 1993;80(11):1323–1335. [Google Scholar]
- 38.Ma GY, et al. Analysis of the Petunia hybrida double flower transcriptome using suppression subtractive hybridization. Sci Hortic (Amsterdam) 2011;127(3):398–404. [Google Scholar]
- 39.Akita Y, Nakada M, Kanno A. Effect of the expression level of an AGAMOUS-like gene on the petaloidy of stamens in the double-flowered lily, 'Elodie'. Sci Hortic (Amsterdam) 2011;128(1):48–53. [Google Scholar]
- 40.Dubois A, et al. Tinkering with the C-function: A molecular frame for the selection of double flowers in cultivated roses. PLoS ONE. 2010;5:e9288. doi: 10.1371/journal.pone.0009288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lehmann NL, Sattler R. Floral Development and homeosis in Actaea rubra (Ranunculaceae) Int J Plant Sci. 1994;155(6):658–671. [Google Scholar]
- 42.Scutt CP, Oliveira M, Gilmartin PM, Negrutiu I. Morphological and molecular analysis of a double-flowered mutant of the dioecious plant white campion showing both meristic and homeotic effects. Dev Genet. 1999;25:267–279. doi: 10.1002/(SICI)1520-6408(1999)25:3<267::AID-DVG9>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 43.Scovel G, Ben-Meir H, Ovadis M, Itzhaki H, Vainstein A. RAPD and RFLP markers tightly linked to the locus controlling carnation (Dianthus caryophyllus) flower type. Theor Appl Genet. 1998;96(1):117–122. [Google Scholar]
- 44.Smith GH. Vascular Anatomy of Ranalian Flowers. II. Ranunculaceae (Continued), Menispermaceae, Calycanthaceae, Annonaceae. Bot Gaz. 1928;85(2):152–177. [Google Scholar]
- 45.Di Stilio V, et al. Virus-induced gene silencing as a tool for comparative functional studies in Thalictrum. PloS ONE. 2010;5(8):e12064. doi: 10.1371/journal.pone.0012064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ito T, Ng KH, Lim TS, Yu H, Meyerowitz EM. The homeotic protein AGAMOUS controls late stamen development by regulating a jasmonate biosynthetic gene in Arabidopsis. Plant Cell. 2007;19:3516–3529. doi: 10.1105/tpc.107.055467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Di Stilio VS, Martin C, Schulfer AF, Connelly CF. An ortholog of MIXTA-like2 controls epidermal cell shape in flowers of Thalictrum. New Phytol. 2009;183:718–728. doi: 10.1111/j.1469-8137.2009.02945.x. [DOI] [PubMed] [Google Scholar]
- 48.Kumar A, Bennetzen JL. Plant retrotransposons. Annu Rev Genet. 1999;33:479–532. doi: 10.1146/annurev.genet.33.1.479. [DOI] [PubMed] [Google Scholar]
- 49.Chang Y-F, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 50.Yang YZ, Jack T. Defining subdomains of the K domain important for protein-protein interactions of plant MADS proteins. Plant Mol Biol. 2004;55:45–59. doi: 10.1007/s11103-004-0416-7. [DOI] [PubMed] [Google Scholar]
- 51.Fan HY, Hu Y, Tudor M, Ma H. Specific interactions between the K domains of AG and AGLs, members of the MADS domain family of DNA binding proteins. Plant J. 1997;12:999–1010. doi: 10.1046/j.1365-313x.1997.12050999.x. [DOI] [PubMed] [Google Scholar]
- 52.Melzer R, Verelst W, Theissen G. The class E floral homeotic protein SEPALLATA3 is sufficient to loop DNA in ‘floral quartet’-like complexes in vitro. Nucleic Acids Res. 2009;37:144–157. doi: 10.1093/nar/gkn900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu C, et al. Interactions among proteins of floral MADS-box genes in basal eudicots: Implications for evolution of the regulatory network for flower development. Mol Biol Evol. 2010;27:1598–1611. doi: 10.1093/molbev/msq044. [DOI] [PubMed] [Google Scholar]
- 54.Varagona MJ, Purugganan M, Wessler SR. Alternative splicing induced by insertion of retrotransposons into the maize waxy gene. Plant Cell. 1992;4:811–820. doi: 10.1105/tpc.4.7.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kobayashi S, Goto-Yamamoto N, Hirochika H. Retrotransposon-induced mutations in grape skin color. Science. 2004;304:982–982. doi: 10.1126/science.1095011. [DOI] [PubMed] [Google Scholar]
- 56.de Folter S, et al. Comprehensive interaction map of the Arabidopsis MADS Box transcription factors. Plant Cell. 2005;17:1424–1433. doi: 10.1105/tpc.105.031831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Immink RGH, Kaufmann K, Angenent GC. The ‘ABC’ of MADS domain protein behaviour and interactions. Semin Cell Dev Biol. 2010;21:87–93. doi: 10.1016/j.semcdb.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 58.Yang YZ, Fanning L, Jack T. The K domain mediates heterodimerization of the Arabidopsis floral organ identity proteins, APETALA3 and PISTILLATA. Plant J. 2003;33:47–59. doi: 10.1046/j.0960-7412.2003.01473.x. [DOI] [PubMed] [Google Scholar]
- 59.Sieburth LE, Running MP, Meyerowitz EM. Genetic separation of third and fourth whorl functions of AGAMOUS. Plant Cell. 1995;7:1249–1258. doi: 10.1105/tpc.7.8.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Airoldi CA, Bergonzi S, Davies B. Single amino acid change alters the ability to specify male or female organ identity. Proc Natl Acad Sci USA. 2010;107:18898–18902. doi: 10.1073/pnas.1009050107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gómez-Mena C, de Folter S, Costa MMR, Angenent GC, Sablowski R. Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development. 2005;132:429–438. doi: 10.1242/dev.01600. [DOI] [PubMed] [Google Scholar]
- 62.Theissen G. The proper place of hopeful monsters in evolutionary biology. Theory Biosci. 2006;124:349–369. doi: 10.1016/j.thbio.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 63.Tamura M. Morphology, ecology, and phylogeny of the Ranunculaceae IV. Scientific Reports of Osaka University. 1965;14(1):53–71. [Google Scholar]
- 64.Wang W, Chen Z-D. Generic level phylogeny of Thalictroideae (Ranunculaceae) - implications for the taxonomic status of Paropyrum and petal evolution. Taxon. 2007;56(3):811–821. [Google Scholar]
- 65.Mizukami Y, Ma H. Determination of Arabidopsis floral meristem identity by AGAMOUS. Plant Cell. 1997;9:393–408. doi: 10.1105/tpc.9.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mizukami Y, Ma H. Separation of AG function in floral meristem determinacy from that in reproductive organ identity by expressing antisense AG RNA. Plant Mol Biol. 1995;28:767–784. doi: 10.1007/BF00042064. [DOI] [PubMed] [Google Scholar]
- 67.Bradley D, Carpenter R, Sommer H, Hartley N, Coen E. Complementary floral homeotic phenotypes result from opposite orientations of a transposon at the plena locus of Antirrhinum. Cell. 1993;72:85–95. doi: 10.1016/0092-8674(93)90052-r. [DOI] [PubMed] [Google Scholar]
- 68.Sun B, Xu YF, Ng KH, Ito T. A timing mechanism for stem cell maintenance and differentiation in the Arabidopsis floral meristem. Genes Dev. 2009;23:1791–1804. doi: 10.1101/gad.1800409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Endress PK, Doyle JA. Reconstructing the ancestral angiosperm flower and its initial specializations. Am J Bot. 2009;96:22–66. doi: 10.3732/ajb.0800047. [DOI] [PubMed] [Google Scholar]
- 70.Hileman LC, Drea S, Martino G, Litt A, Irish VF. Virus-induced gene silencing is an effective tool for assaying gene function in the basal eudicot species Papaver somniferum (opium poppy) Plant J. 2005;44:334–341. doi: 10.1111/j.1365-313X.2005.02520.x. [DOI] [PubMed] [Google Scholar]
- 71.Gould B, Kramer EM. Virus-induced gene silencing as a tool for functional analyses in the emerging model plant Aquilegia (columbine, Ranunculaceae) Plant Methods. 2007;3:6. doi: 10.1186/1746-4811-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 73.Wang YQ, Melzer R, Theissen G. Molecular interactions of orthologues of floral homeotic proteins from the gymnosperm Gnetum gnemon provide a clue to the evolutionary origin of ‘floral quartets’. Plant J. 2010;64:177–190. doi: 10.1111/j.1365-313X.2010.04325.x. [DOI] [PubMed] [Google Scholar]