Abstract
We have selected designed ankyrin repeat proteins (DARPins) from a synthetic library by using ribosome display that selectively bind to the mitogen-activated protein kinase ERK2 (extracellular signal-regulated kinase 2) in either its nonphosphorylated (inactive) or doubly phosphorylated (active) form. They do not bind to other kinases tested. Crystal structures of complexes with two DARPins, each specific for one of the kinase forms, were obtained. The two DARPins bind to essentially the same region of the kinase, but recognize the conformational change within the activation loop and an adjacent area, which is the key structural difference that occurs upon activation. Whereas the rigid phosphorylated activation loop remains in the same form when bound by the DARPin, the more mobile unphosphorylated loop is pushed to a new position. The DARPins can be used to selectively precipitate the cognate form of the kinases from cell lysates. They can also specifically recognize the modification status of the kinase inside the cell. By fusing the kinase with Renilla luciferase and the DARPin to GFP, an energy transfer from luciferase to GFP can be observed in COS-7 cells upon intracellular complex formation. Phosphorylated ERK2 is seen to increase by incubation of the COS-7 cells with FBS and to decrease upon adding the ERK pathway inhibitor PD98509. Furthermore, the anti-ERK2 DARPin is seen to inhibit ERK phosphorylation as it blocks the target inside the cell. This strategy of creating activation-state–specific sensors and kinase-specific inhibitors may add to the repertoire to investigate intracellular signaling in real time.
Keywords: intrabodies, X-ray crystallography
Understanding the integration of a myriad of extracellular and intracellular signals is an important challenge for biological and medical research in the years ahead. Various posttranslational modifications (PTMs) of proteins play a pivotal role in signal propagation and regulate the function of numerous proteins in signaling networks. Protein modifications including phosphorylation, acetylation, ubiquitination, methylation, and glycosylation finely tune protein activity, localization, and interaction in a time-dependent and often spatially controlled manner (1). During the past decade, MS-based proteomic studies have enabled proteome-wide analysis of PTM substrates and modified sites on the proteins (1–3). Large-scale studies can deduce the flow through a network and thus uncover a variety of novel cellular interpendencies, but ultimately cannot assign a function to individual signaling nodes (1). Also, the detection of modifications by MS is still far from comprehensive. Thus, the creation of collections of specific binding molecules targeting a great number of individual proteins, including variants and modifications, will become equally important (4). Such specific affinity reagents would not only be beneficial for traditional biochemical approaches detecting protein modifications and interactions, but could also serve as a starting point for high-throughput assay systems and new protein detection technologies (5–7).
Most PTMs are detected today with conventional monoclonal antibodies that have been raised in mice. For most targets, no specific reagents exist, and the performance of available antibodies, known to be of highly variable quality (8), will have to be tested in every single case. Recombinant technologies with antibody libraries (9, 10) now allow us to specifically select for PTMs on individual targets, but the resulting antibody fragments often lack sufficient conformational stability under harsh conditions, limiting the types of analyses possible. In addition, their framework is stabilized by intradomain disulfide bonds, in general precluding their use in the reducing intracellular environment as “intrabodies” in functional cellular assays (11). To overcome these bottlenecks, new protein scaffolds for molecular recognition have been developed (11). A promising new class of binding proteins is based on naturally occurring ankyrin repeat proteins (12, 13). Exploiting sequence homologies within the structural repeat motif and consensus design, combinatorial libraries of designed ankyrin repeat proteins (DARPins) were generated (14). DARPins consist of modules of 33 aa with fixed framework and randomized potential interaction residues that form elongated protein domains. Members of these libraries show very favorable biophysical properties, are expressed in soluble form with high yields in the cytoplasm of Escherichia coli, and do not contain disulfide bonds, allowing for intracellular applications (12, 15–18). Furthermore, using complex DARPin libraries, specific binders with high affinity and selectivity for their target antigen could be isolated (12, 19, 20).
Extracellular signal-regulated kinase 2 (ERK2), a member of the mitogen-activated protein kinase (MAPK) family, was chosen as a model system for specific intracellular targeting of a protein as a function of its posttranslational modification. For these experiments it was essential that the native, folded protein is recognized. MAPKs are one of the most widely studied classes of signaling proteins (21), and their own activity is controlled by a specific phosphorylation event. They represent a group of evolutionarily conserved serine/threonine kinases that are activated in response to a variety of extracellular stimuli and mediate signal transduction from the cell surface to the nucleus. MAPKs regulate several physiological and pathological phenomena, including inflammation, apoptotic cell death, oncogenic transformation, tumor cell invasion, and metastasis (22). So far, four major types of MAPK cascades have been reported in mammalian cells, designated the ERK1/2, ERK5, p38, and JNK cascades, according to their MAPK component (21).
Protein kinases not only phosphorylate other proteins, but also are themselves targets of phosphorylation, converting them into an active form. In combination with a drastic change of the physicochemical properties of the affected residues, phosphorylation-based modifications are closely linked to reconfiguration of protein interactions and thus regulation of signal processes (23). Activation of the MAPK ERK2 is mediated by phosphorylation of a threonine and tyrosine within a flexible surface loop (activation loop) that undergoes small but significant conformational rearrangements upon modification (Fig. S1), which cause full activation of the kinase by properly aligning active site residues (24).
We report here the results of in vitro selections of target-specific DARPins that can reliably differentiate between the two states of a protein posttranslationally modified by phosphorylation. The crystal structures of two DARPins specifically targeting either the unphosphorylated or the phosphorylated form were determined in a complex with the kinase and revealed insights into the mode of interaction, the reason for selectivity, and potential perturbation of protein function. In addition, selected binders were tested successfully for their anticipated functionality in the cytoplasm of eukaryotic cells.
Results
In Vitro Selections of Phosphorylation-Specific ERK-Binding DARPins.
We used ribosome display to select DARPins against either the unphosphorylated or the phosphorylated form of the MAPK ERK2 (ERK2 or pERK2), using the respective other form as a competitor. Ribosome display (RD), a complete in vitro selection and evolution technology, is based on a ternary complex of ribosome, mRNA without a stop codon, and the newly synthesized polypeptide, emerged from and folded on the ribosome, all formed and staying together in an in vitro translation reaction (7). Selections were carried out with libraries consisting of two or three randomized ankyrin repeat modules between an N-terminal and a C-terminal capping repeat, termed N2C and N3C, respectively. To be able to select for phosphorylation-specific DARPins, we increased the selection pressure after round 1 by introducing a prepanning step, using the nondesired ERK2 form. We observed a significant enrichment of DARPin libraries targeting either ERK2 or pERK2 after four rounds of RD selection, which verified our selection strategy (Fig. S2).
Single clones of enriched selection pools were tested for their specific binding to the target antigen. Crude E. coli extracts of small-scale DARPin expression cultures were analyzed by ELISA screens to identify the most promising binders. We identified binding molecules that can reliably differentiate between ERK2 and pERK2 and those that bind both forms equally well. After round 4 of selections using ERK2 as target antigen, 40 of 96 assayed clones (42%) in the N2C pool and 25 of 96 (26%) in the N3C pool showed specific binding to the desired ERK2 form. For pERK2, 40 of 288 assayed clones (14%) of the N2C pool bound specifically to pERK2, whereas none of the assayed clones of the N3C pool could meet our specificity criteria. In addition, 25 assayed clones (9%) showed binding to both ERK2 and pERK2. None of the analyzed DARPins showed significant binding to microtiter plates coated with neutravidin and BSA. DARPins with the most prominent crude extract ELISA signals were sequenced (Fig. S3). No obvious similarities of randomized positions could be identified for the different binder pools. For both selection outcomes, a few binders contained mutations at nonrandomized positions that were most likely introduced during the numerous PCR amplification cycles and are a hallmark of the evolutionary component of RD.
Characterization of Selected Phosphorylation-Specific ERK-Binding DARPins.
DARPin binders without insertions or deletions were expressed in E. coli XL1-Blue and purified by immobilized metal ion affinity chromatography. For a first-affinity ranking, binders were analyzed in a competition ELISA experiment. Binding of purified DARPins to immobilized ERK2 or pERK2 was assayed in the absence and presence of the corresponding ERK2 form. DARPins with promising competition behavior (significant inhibition by 150 nM ERK2 or pERK2) were analyzed by gel filtration experiments to rule out that detected binding events derive from an oligomeric species. The great majority of the investigated DARPins were monomers, as expected from previous studies (12, 17). Four ERK2-binding (E8, E38, E39, and E40), four pERK2-binding (pE55, pE57, pE59, and pE63), and two ERK2/pERK2-binding (EpE82 and EpE89) DARPins were chosen for subsequent experiments.
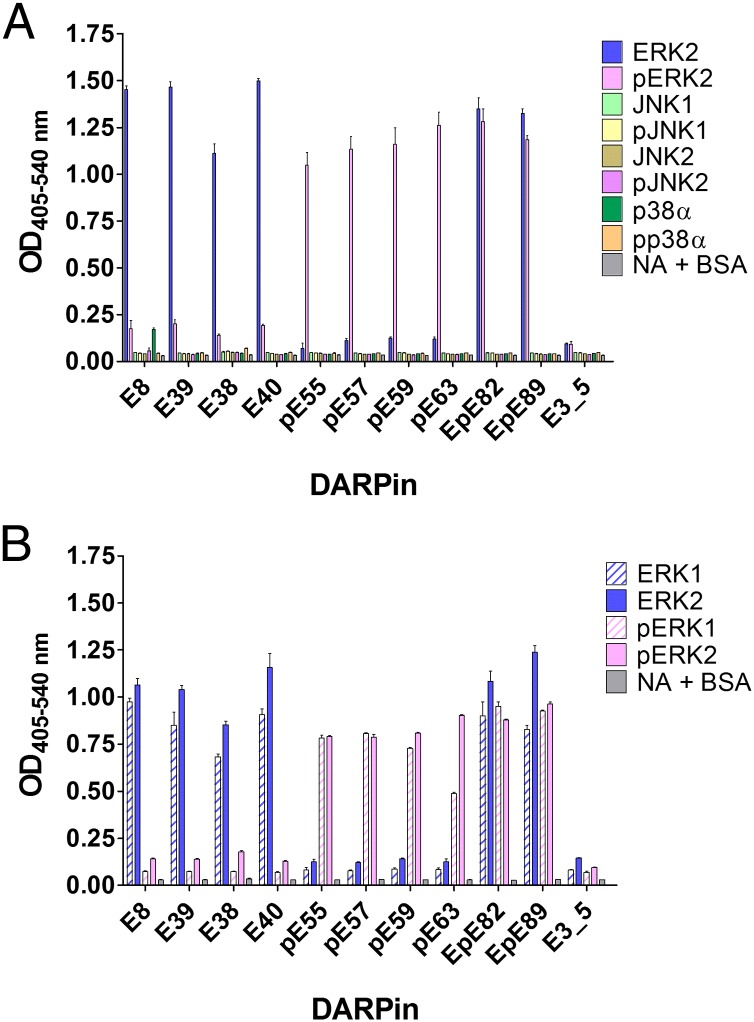
To investigate the binding specificity of the selected DARPins in more detail, ELISA experiments were carried out with purified inactive and active forms of the MAPK ERK2, JNK1α1, JNK2α1, and p38α. The different MAPK family members have marked sequence homologies, with an overall sequence identity of >40% over the highly conserved catalytic core. All tested DARPins were highly specific for their cognate antigens ERK2 and pERK2 and did not interact with the inactive or active form of any other MAPK (Fig. 1A). In a second ELISA experiment, binding of the selected DARPins to ERK2 was compared with that to ERK1, two closely related kinases with 85% sequence identity on the amino acid level. Tested DARPins did not discriminate between ERK1 and ERK2, but retained the phosphorylation status specificity observed for ERK2 and also for ERK1 (Fig. 1B). Our result is not surprising, because the selections focused on epitopes that are specific for the unphosphorylated and phosphorylated states of ERK2, where both kinases are almost identical, but not on epitopes that distinguish ERK1 and ERK2 from each other. Sequence alignment analysis of ERK1 and ERK2 together with previously determined 3D structures of both the inactive and active state of ERK2 (24, 25) showed that potential binding epitopes, where changes occur upon phosphorylation, are highly similar for both ERK kinases, and indeed these structural elements constitute the binding epitope as determined by X-ray crystallography for DARPins E40 and pE59 (see below).
Fig. 1.
Binding specificity analysis of selected DARPins by ELISA. (A) DARPins derived from RD selection with ERK2 (names starting with E) and phosphorylated ERK2 (pERK2) (names starting with pE) as targets were tested for interaction with ERK2 and pERK2. In addition, binding to the MAPK JNK1α1, JNK2α1, and p38α in the unphosphorylated and the phosphorylated state as well as to wells containing only neutravidin and BSA (NA+BSA) was assessed. As a further control, the unselected DARPin library member E3_5 was included. (B) Binding of selected DARPins to ERK2 and pERK2 was compared with the closely related MAPK ERK1 in the unphosphorylated and the phosphorylated state. The unselected DARPin E3_5 and the interaction with NA+BSA served as negative controls.
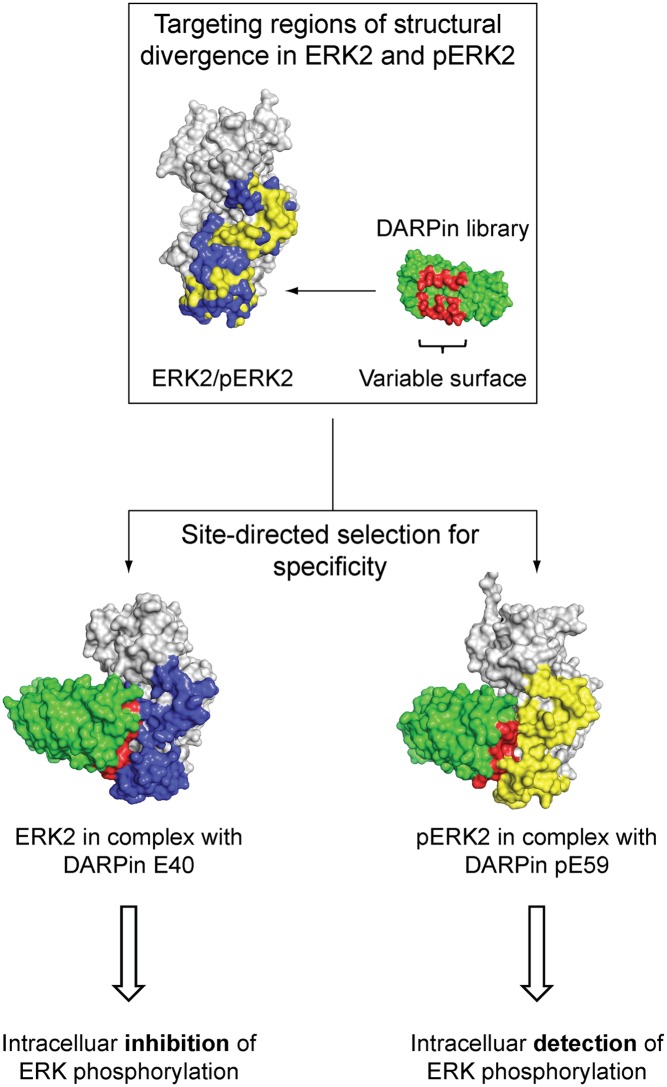
Furthermore, the affinities of ERK- (E8, E38, E39, and E40), pERK- (pE55, pE57, pE59, and pE63) and ERK/pERK (EpE82 and EpE89)-binding DARPins were compared qualitatively by a competition ELISA experiment using ERK2 and pERK2 as antigens (Fig. S4). ERK2- and pERK2-specific DARPins showed very similar competition behavior when incubated with low nanomolar concentrations of the respective ERK2 forms. ERK2/pERK2-binding DARPin EpE89 bound both ERK2 forms with an affinity comparable to that of phosphorylation-specific ERK2 DARPins, whereas EpE82 showed preferred binding to pERK2. We aimed to further characterize the epitope of each group of selected DARPins (ERK-binding, pERK-binding and ERK/pERK-binding DARPins). Therefore, we set up a sandwich ELISA experiment to elucidate whether binders within one type of specificity bind to a similar epitope and whether binders from the three groups share an overlapping binding epitope. DARPin binders E40 and pE59, for which structural information of the binding mode to ERK2/pERK2 is available (see below), were biotinylated and immobilized on microtiter plates via neutravidin (Fig. S5A). After letting ERK2 or pERK2 bind to the immobilized DARPins, ERK- or pERK-DARPins were added to test whether any of them bind to a different epitope than do E40 and pE59. As a control for epitope blocking E40 and pE59 themselves were used. The unselected library member E3_5 served as a control for unspecific binding. In both setups, with E40 or pE59 immobilized in the first step, none of the tested DARPins showed binding to captured ERK2 or pERK2 (Fig. S5 C and D). The two DARPins EpE82 and EpE89, which recognize both ERK and pERK, also do not show any binding in both setups and thus compete with E40 and pE59 (Fig. S5 C and D). We therefore conclude that DARPins from all three groups target a similar or at least an overlapping region on both kinase forms. Nonetheless, the epitope must undergo conformational changes to allow activation-specific recognition and contain parts that have enough conformational freedom to allow the selection of binders that recognize both forms. Superposition of the 3D structures of ERK2 and pERK2 revealed that major changes can be located to three regions: the activation loop, the MAPK insertion, and the L16 segment from the N-terminal domain (Fig. S1) (24). Upon phosphorylation, the activation loop takes on a defined structure, loses contacts present in the unphosphorylated state, and forms new contacts with both the N- and the C-terminal domain. Furthermore, conformational changes in the neighboring MAPK insertion and the formation of a 3/10 helix in L16 occur, making it likely that the epitope that accounts for the observed binding specificity of selected binders will be in these areas. Indeed, the structures of the ERK/DARPin complexes (see below) confirm these expectations.
E40/ERK2 and pE59/pERK2 Complex Structures.
To validate our selection strategy and to explain the binding specificity on the atomic level, we determined the crystal structures of two complexes of ERK2 with DARPins, which recognize the kinase ERK2 as a function of phosphorylation status: E40 in complex with ERK2 and pE59 in complex with pERK2. DARPins E40 and pE59 were chosen for detailed characterization, because both DARPins readily yielded high-quality crystals in complex with the corresponding form of ERK2. The structures of the complexes were solved by molecular replacement and refined to 1.9 Å for E40/ERK2 [Protein Data Bank (PDB) ID 3zu7] and 2.7 Å for pE59/pERK2 (PDB ID 3zuv). A summary of the data collection statistics and the refinement results are listed in Table S1.
The asymmetric unit in the E40/ERK2 complex contains one heterodimeric complex with nine hydrogen bonds (Table S2). Two pE59/pERK2 heterodimers, named AB and CD, are present in the pE59/pERK2 complex structure. Chains A and C represent pERK2, whereas B and D represent the N2C DARPin pE59. The A/B and C/D interfaces display seven defined hydrogen bonds each, but differ in two of them (Table S3). The information of both interfaces was used for further structural analysis.
The global fold of the kinase domains is conserved in both structures, consisting of an N-terminal (composed mainly of β-strands) and a C-terminal domain (formed primarily by α-helices) (Fig. 2). The two DARPins E40 and pE59 in the complex have the expected ankyrin repeat domain fold (Fig. 2 and Fig. 3A). The interaction of E40 with ERK2 and pE59 with pERK2 involves residues from all randomized repeat modules and the C-capping repeat (Tables S2 and S3). In both complex structures, the C-capping repeat, which does not contain any randomized residues, is involved significantly in contact formation with the kinase (Fig. 3A).
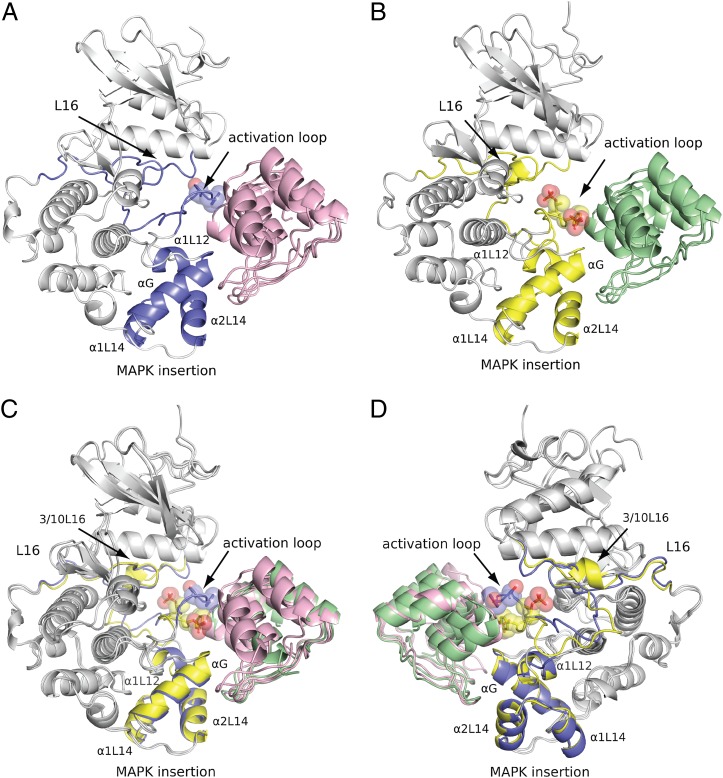
Fig. 2.
Crystal structures of phosphorylation status-specific DARPins E40 and pE59 in complex with ERK2 and phosphorylated ERK2. Segments of structural divergence taking part in interaction with DARPins are labeled and highlighted in ERK2 (blue) and pERK2 (yellow). The side chains of Thr-183, Tyr-185, pThr-183, and pTyr-185 are shown. (A) Ribbon diagram of E40 (magenta) in complex with ERK2 (gray and blue). (B) Ribbon diagram of pE59 (green) in complex with pERK2 (gray and yellow). (C and D) Two views (180° rotated) of the superimposed complexes E40/ERK2 and pE59/pERK2 are shown.
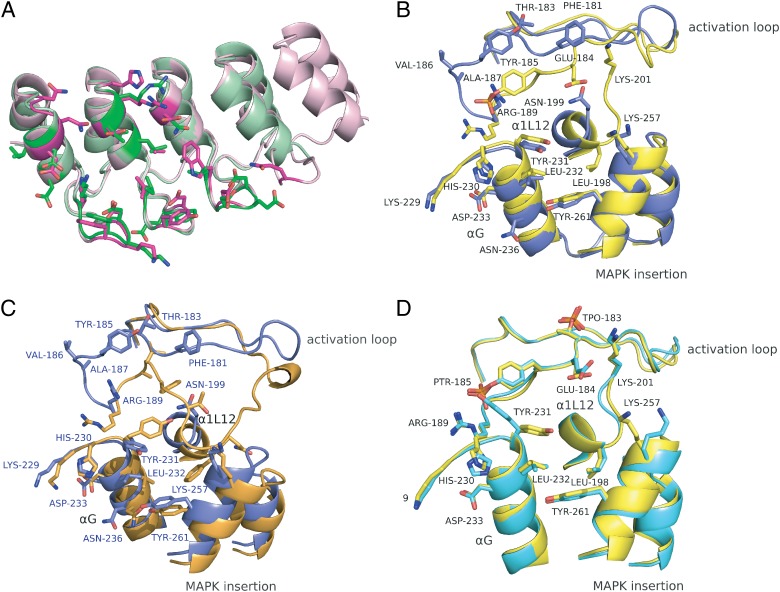
Fig. 3.
Conformational differences of the structures of complexes E40/ERK2 and pE59/pERK2. Different superpositions are shown with the interaction residues highlighted in stick mode. The color scheme is the same as in Fig. 2. Structural elements and interaction residues are labeled. (A) Superposition of ERK2-specific DARPin E40 (magenta, an N3C molecule) and pERK2-specific DARPin pE59 (green, an N2C DARPin), each in the kinase-bound state. (B) Contact regions of DARPin-bound ERK2 (blue) and pERK2 (yellow) are superposed. (C) The contact region of DARPin-bound ERK2 (blue) is superposed with unbound ERK2 (PDB ID 1ERK, orange). Interaction residues of the E40/ERK2 complex are labeled (blue). (D) Superposition of DARPin-bound pERK2 (yellow) with unbound pERK2 (PDB ID 2ERK, cyan). pThr-183 (TPO-183) is highlighted, but it is not involved in pE59/pERK2 interaction.
The main differences of the kinase in the E40/ERK2 and pE59/pERK2 structures compared with ERK2 and pERK2 alone are located in the E40- and pE59-binding region (Figs. 2 and 3). Both DARPins target the same region on the kinase as suggested by the epitope characterization with ELISA (Fig. S5). The binding region of DARPins includes the activation loop, the MAPK insertion, α-helix G, and partially α-helix 1L12 (Figs. 2 and 3). Superposition of E40/ERK2 and pE59/pERK2 showed that the binding specificity of E40 and pE59 is mainly due to different conformations of the activation loop (Figs. 2 C and D and 3B). Additional subtle changes were observed for helix 1L12. The remaining two contact regions, helix G and the MAPK insertion, are highly similar in both complex structures. Consistent with the previously determined structures of ERK2 and pERK2, the DARPin-binding region includes the most profound changes between the inactive and the active state of ERK2, indicating that the selection has been successfully steered toward this region (Fig. 2 and Fig. S1) (24, 25).
Analysis of the Interaction of E40 with ERK2.
Upon binding of DARPin E40, the activation loop of ERK2 adopts a conformation different from the arrangement in ERK2 alone (Fig. 3C). In unphosphorylated ERK2, the loop is folded down over the C-terminal domain to block the P+1 site (25). In the complex E40/ERK2, DARPin E40 makes contact with Phe-181, Thr-183, Tyr-185, Val-186, Ala-187, and Arg-189 in the activation loop of ERK2, causing the displacement of interactions of the loop as it is present in free ERK2. E40 binding to Arg-189 prevents its interaction with residues of the preceding α-helix G, causing a 90° rotation of this side chain and the formation of a new contact with Tyr-185. In addition, the refolding of the loop interrupts contacts with Tyr-231 that stabilize the folding down over the C-terminal domain and the buried location of Tyr-185. Binding of E40 to Phe-181, Thr-183, Tyr-185, Val-186, and Ala-187 stabilizes the rotated conformation of the loop in the complex. Further conformational changes were observed for the extension of the activation loop (Asp-175 to Thr-179, Fig. 2D), a region for which the observed electron density was weak. Changes in the other contact regions of the E40/ERK2 complex are more subtle. The DARPin E40 interacts with several residues in α-helix G and α-helix 2L14 of the MAPK insertion (Fig. 3B and Table S2). In free ERK2 these regions stabilize the down-folded conformation of the loop. Binding of E40 to Tyr-231 of helix G and Lys-257 of helix 2L14 directly interrupts stabilizing contacts of the loop arrangement in ERK2. In addition, the helices G, 1L12, and 1L14/2L14 of the MAPK insertion are rearranged upon DARPin binding (Fig. 3C). Most significant changes were observed in helix 1L12 and the MAPK insertion. In summary, DARPin E40 makes contacts to both the activation loop and residues in the C-terminal domain, thus interrupting their interaction and enforcing a new conformation of the activation loop, different from free ERK2.
Analysis of the Interaction of pE59 with pERK2.
The interaction of pE59 with pERK2 relies on contact formation with identical regions and residues as identified for the E40/ERK2 complex (Fig. 3B and Table S3). Thus, specificity of DARPins E40 and pE59 results from binding to discriminating structural elements, which have changed in spatial conformation, but not from interaction with different amino acid residues on the target. In contrast to the E40/ERK2 complex, where the activation loop has moved with respect to free ERK2, no significant conformational changes could be observed in pERK2 upon binding of DARPin pE59 (Fig. 3D). This observation is consistent with the fact that, although the inactive state of the loop in different kinases can assume many conformations, the active form of the loop is rather similar (26).
The activation loop, helices G and 1L12, and the MAPK insertion region retain their conformation from free pERK2 in the complex with DARPin pE59. DARPin pE59 makes contact with Arg-189, pTyr-185, and Glu-184 in the activation loop but does not prevent conserved interactions of this loop with both kinase domains. The most pronounced changes from free pERK2 are movements of the Tyr-231 and Lys-257 side chains that are in direct contact with the DARPin. The DARPin does not make direct contacts with pThr-183, as it is pointing away from the DARPin paratope.
Affinity and Selectivity of DARPins E40 and pE59.
In ELISA experiments, the selected DARPin binders E40 and pE59 interacted only weakly with the noncognate ERK form, i.e., active ERK in the case of E40 and inactive ERK2 in the case of pE59 (Fig. 1). Residual binding to the nontargeted ERK form may occur via interaction with helix G and the MAPK insertion region, which are both highly similar in the relevant contact regions of the E40/ERK2 and pE59/pERK2 complex (Fig. 3B). To quantify the difference in binding, the equilibrium dissociation constant KD was determined for E40 and pE59 binding to the antigens ERK2 and pERK2 by kinetic surface plasmon resonance (SPR) measurements on a Biacore instrument. All data were evaluated by fitting the equilibrium responses (Fig. S6), except the DARPin E40/ERK2 interaction, which was analyzed with a modified heterogeneous ligand model, accounting for the previously proposed less rigid conformation of the activation loop in nonphosphorylated ERK2 (26). Indeed, we could observe two binding reactions with different affinity values (Fig. S6A). The high-affinity binding with a low nanomolar KD most likely reflects the binding observed in the E40/ERK2 complex in accordance with results obtained in ELISA experiments (Fig. S4) and was thus used for quantifying E40 selectivity. The observed low-affinity binding might derive from a different loop conformation in unbound ERK2, which possibly diminishes the E40 binding epitope. Overall, it can be seen that the discrimination between ERK2 and pERK2 is based on a 182-fold difference in KD value for E40 and a >74-fold difference in KD value for pE59 (Table 1). The apparent selectivity for pE59 may be even higher, because SPR signals for the noncognate ERK2 form were very low and thus led to an imprecise estimation of KD (Fig. S6D).
Table 1.
Affinity and selectivity of DARPins E40 and pE59
| DARPin | KD, M: ERK2 | KD, M: pERK2 | Selectivity |
| E40 | 6.6 × 10−9 | 1.2 × 10−6 | 182 |
| pE59 | >8.7 × 10−6 | 117 × 10−9 | >74 |
Affinity Precipitation with Selected ERK Phosphorylation-Specific DARPins.
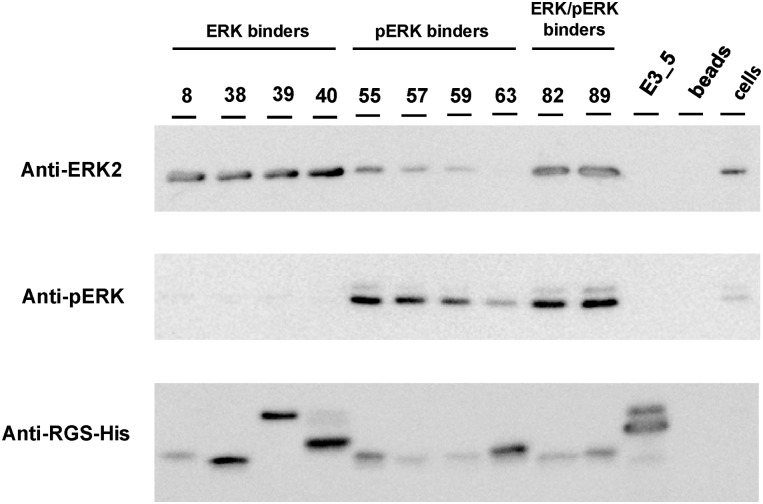
To test whether the selected DARPins retain their binding and specificity in eukaryotic cells, DARPins were tested in affinity precipitation experiments. Cell lysates from HEK293T cells were incubated with His-tagged DARPins, which were subsequently captured by Ni-NTA beads. Affinity-precipitated ERK and pERK were detected on Western blots with ERK2- and pERK1/2-specific antibodies. The final orthogonal detection is limited by the ERK2 antibody, which detects both inactive and active ERK2, as tested with purified ERK2 and phosphorylated ERK2. Analyzed DARPins selectively precipitated ERK2, pERK, or both forms from cell lysates (Fig. 4), in accordance with their binding specificity observed in ELISA experiments (Fig. 1). No precipitation of ERK/pERK was observed for the controls, the nonbinding DARPin E3_5, and beads without DARPin added.
Fig. 4.
Affinity precipitation. ERK and pERK were immunoprecipitated from human embryo kidney (HEK293T) cells by the selected ERK, pERK, and ERK/pERK DARPins indicated. Precipitated ERK2 and pERK were detected by Western blot, using an ERK2- and pERK-specific antibody. Tested DARPins precipitate unphosphorylated, phosphorylated, or both ERK forms from cell lysates, giving rise to the corresponding bands on a Western blot, whereas the controls, the nonbinding DARPin E3_5 and beads with no DARPin, did not. The presence of DARPins was confirmed by detection of the N-terminal RGS-His tag. Note that E39, E40, and E3_5 are N3C molecules, which because of their stability do not completely denature in SDS/PAGE.
ERK2-binding DARPins interacted specifically with unphosphorylated ERK2, resulting in the corresponding band in the anti-ERK2 but not in the anti-pERK1/2 Western blot (Fig. 4). In contrast, DARPins selected for specific binding to pERK gave rise to bands in both ERK blots, but the intensity of the ERK band is significantly lower compared with those of ERK- and ERK/pERK-binding DARPins (Fig. 4). The band detected in the ERK2 blot thus most likely reflects pERK2 precipitated by the anti-pERK DARPin, which is detected by cross-reactivity for pERK2 by the anti-ERK2 antibody. Because the ERK signaling pathway was not activated by exogenous stimuli, the level of pERK1/2 was rather low in the given experimental setup, as seen when detecting the level of ERK and pERK in lysates of HEK293T cells without immunoprecipitation (Fig. 4, lane cells). The two ERK/pERK-binding DARPins EpE82 and EpE89 precipitated both inactive and active kinase from cell lysates (Fig. 4). In the ERK2 blot, both DARPins gave rise to the strongest band, caused by the simultaneous pull-down of ERK and pERK. The band intensity is only slightly increased, compared with that of ERK-binding DARPins, because of the low level of pERK present in the cell lysate. The presence of DARPins in the samples was confirmed by detection of the N-terminal RGS-His-tag (Fig. 4). The different amounts of captured DARPins did not influence the amount of precipitated kinase (e.g., comparison of E8 with E40, pE59 with pE63, or EpE82 with pE55). Therefore, the differences in band intensities seen with ERK2 and pERK2 antibodies did not correlate with the amounts of captured DARPins, but may result from different binding affinities of the individual DARPins under the given experimental conditions.
Selectivity and Function of Selected DARPins in Cells.
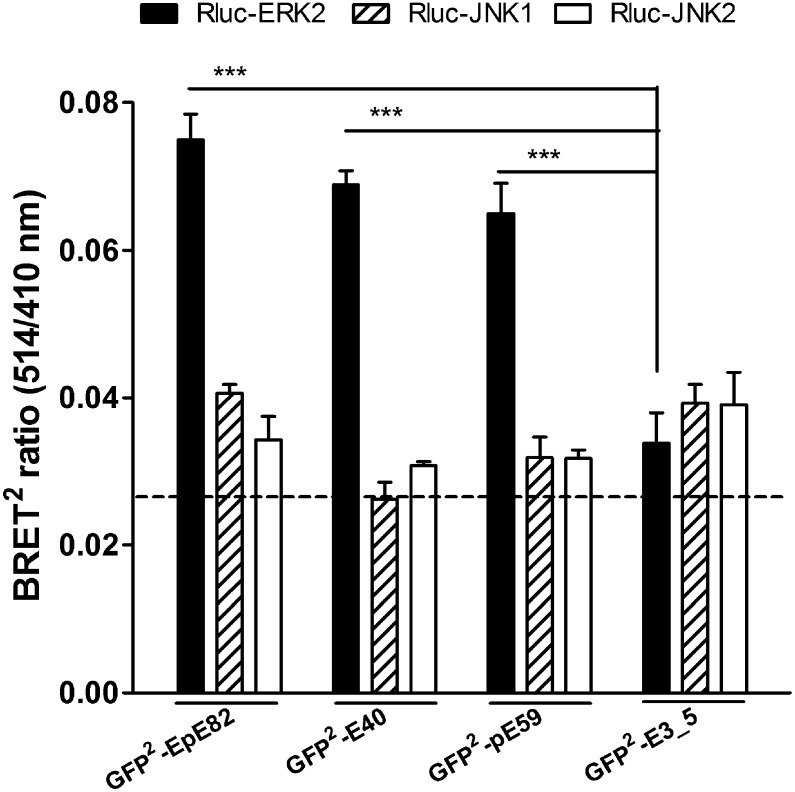
To further complement in vitro studies of DARPin/ERK interaction, we designed bioluminescence resonance energy transfer (BRET) sensors to verify specific binding of selected DARPins to ERK and pERK directly within COS-7 cells. Several chimeras of selected DARPins E40, pE59, EpE82, the unselected library member E3_5, and ERK2 fused to the N and C termini of either Renilla luciferase (Rluc) (27) or a variant of green fluorescent protein (termed GFP2) (27) were created. Successful expression of the combination of fusion proteins in COS-7 cells, to be used for BRET2 assays (the superscript denoting the use of GFP2; see Materials and Methods), was confirmed by Western blot analysis and immunofluorescence microscopy. The combination of Rluc-ERK2 and GFP2-DARPins resulted in the best BRET2 signals in intact cells, which were about two to three times stronger than the background signal (Fig. 5). This preferred orientation is consistent with the crystal structure of the complexes, as in these orientations, Rluc and GFP2 would be closest. Furthermore, GFP2-DARPin fusions of E40, pE59, and EpE82 were specific for ERK2 but did not interact with JNK1 or JNK2 (Fig. 5), thus retaining their ERK2 specificity in living cells. The nonbinding DARPin control GFP2-E3_5 did not show any significant increase of BRET2 signals when coexpressed with Rluc-MAPK fusions.
Fig. 5.
Specificity comparison of selected DARPins in living cells by BRET2 assays. The indicated fusion proteins were coexpressed for 24 h by transfecting a 1:1 DNA ratio in COS-7 cells. Control transfections using vectors encoding solely the fusion protein Rluc were used to determine the background signal (dashed line). Data are means with SEM of at least three independent experiments with n = 6. ***P < 0.001.
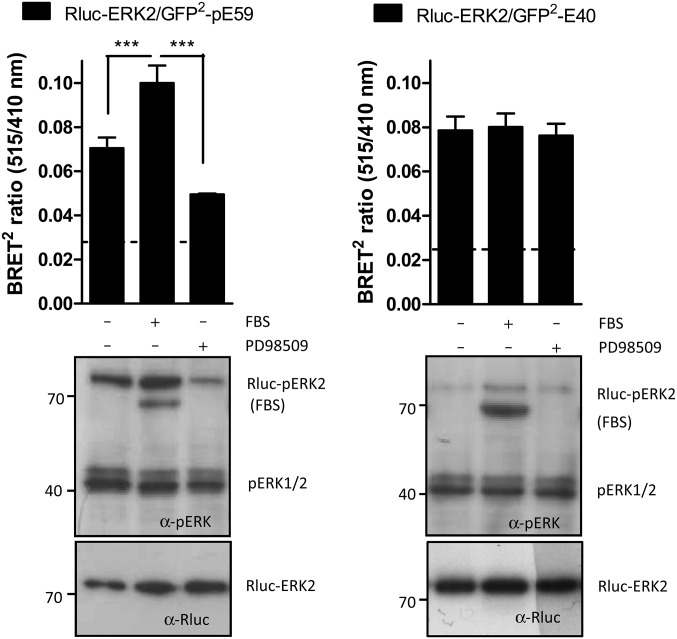
To determine whether the BRET2 DARPin reporters GFP2-E40 and -pE59 retain their specificity for ERK or pERK, ERK phosphorylation in COS-7 cells was either stimulated by addition of FBS or diminished by the ERK pathway inhibitor PD98509, which inhibits the upstream kinase MEK (28). Observed BRET2 signals were compared with Rluc-ERK2 and Rluc-pERK levels obtained from Western blot analysis, performed with aliquots taken from corresponding BRET2 experiments (Fig. 6). For both DARPins, the BRET2 signals correlate with the Western blot results. In BRET2 assays with pERK-binding DARPin pE59, both Western blot and BRET2 revealed an intermediate level of basal Rluc-ERK phosphorylation for COS-7 cells in the given experimental setup (Fig. 6, Left). The addition of FBS to starved cells caused a robust phosphorylation of Rluc-ERK as detected by the BRET2 DARPin reporter and by probing with an anti-pERK2 primary antibody (Fig. 6, Left). Consistently, in the presence of the ERK pathway inhibitor P98509, cells showed a lower level of Rluc-ERK2 phosphorylation in comparison with control cells (Fig. 6, Left). Because total expression of Rluc-ERK2 remained mostly unchanged, whereas Rluc-pERK2 varied due to exogenous stimuli, we conclude that DARPin pE59 indeed selectively binds activated ERK2 in living cells. In assays with ERK-binding DARPin E40 neither a decrease of BRET2 signals after stimulation nor an increase after inhibition of ERK activation was observed in comparison with control cells (Fig. 6, Right). The BRET2 results were supported by the corresponding immunoblot data. The level of phosphorylated Rluc-ERK was generally very low for the assays with DARPin E40 compared with pE59, suggesting an inhibitory effect of E40 on Rluc-ERK2 phosphorylation.
Fig. 6.
Functionality of selected DARPins in living cells. (Upper) BRET2 assays were performed in COS-7 cells 24 h after transfection of the indicated fusion constructs at a 1:1 DNA ratio. Before measurements, cells were starved for 6 h in serum-free media and treated for 10 min with PBS (−), 100% FBS, or 10 μM of the MEK1 inhibitor PD98509. The background BRET2 signal deriving from control transfections with vector coding for unfused Rluc is indicated by dashed lines. Data are means with SEM of at least three independent experiment with n = 6. ***P < 0.001. (Lower) Levels of phospho-Rluc-ERK2 (Rluc-pERK2) and Rluc-ERK2 were monitored for the different treatments by Western blot, using anti–phospho-p44/42 (α-pERK) and anti-Rluc (α-Rluc) primary antibodies.
Discussion
In this study, we demonstrate the selection of DARPins that can reliably differentiate between the two states of a posttranslationally modified protein, the MAPK ERK. Protein modifications finely tune protein functions, by causing changes in protein activity, cellular localization, and interactions with other proteins (29). Thus, binding proteins that target sites of PTMs or specifically recognize changes in tertiary and quaternary structures of modified proteins would be valuable tools both to detect different protein states and to interfere with their function in living cells (4). In contrast to antibodies and their derivatives, DARPins are superior for such intracellular applications, because their conformational and functional integrity does not rely on disulfide bond formation and they fold well when expressed in the cytoplasm.
The choice of ERK2 as a target for creating PTM-specific DARPins was guided by several considerations. We have previously demonstrated the selection of MAPK isoform-specific binders from DARPin libraries, proving that DARPins are able to discriminate subtle differences in highly similar proteins (30). In addition, expression and purification of homogenous species of inactive and active ERK2, an essential prerequisite for a successful selection, have been described in the literature (31, 32). Most importantly, 3D structures of both ERK2 states (24, 25) revealed small but substantial conformational changes upon phosphorylation, which could serve as binding epitopes to allow selection of phosphorylation-specific DARPins. Major structural rearrangements upon ERK2 phosphorylation are located in three regions, the activation loop, the MAPK insertion and the L16 segment from the N-terminal domain (24). In unphosphorylated ERK2, the activation loop extends from the active site over the C-terminal domain, interacting with residues of the N terminus of α-helix G and the MAPK insertion. Upon dual phosphorylation of ERK2 on residues Thr-183 and Tyr-185 within the activation loop, the phosphorylation lip rearranges and gains new contacts within both kinase domains.
If selection of DARPins can be directed to such defined epitopes, specificity should result. The results obtained in the present study validate this approach. The activation loop differs significantly in sequence and structure between MAPKs and protein kinases in general (25, 33, 34) and is thus a handle of specificity. Furthermore, the 50-residue α-helical MAPK insertion and the extended C terminus (L16 segment) are unique to MAPKs and distinguish them from other members of the protein kinase family (24).
Our selection strategy was validated by the structures of two phosphorylation-specific DARPin binders in complex with their cognate targets (Fig. 2). Binding specificity to ERK2 or pERK2 is mainly mediated by contact formation with the activation loop, the TEY-phosphorylation motif (constituted by residues Thr-183, Glu-184, and Tyr-185), and MAPK insertion. Upon binding of DARPin E40 to ERK2, the activation loop is stabilized in a conformation remarkably different from that of unbound ERK2 and from that of the pE59/pERK2 complex. More subtle changes occurred in the MAPK insertion region. In contrast, no significant changes were observed for active ERK2 with or without DARPin pE59, pointing to a more rigid loop conformation, as has been suggested previously (26). DARPin pE59 thus recognizes a predefined loop conformation, whereas a loop rearrangement appears to occur in nonphosphorylated ERK2 when interacting with the rigid DARPin. Interactions with structurally conserved contact regions in both complexes, helix G and part of the MAPK insertion, and contact formation via the conserved DARPin C-capping repeat may contribute to weak cross-binding to the nontargeted ERK form (Fig. S6). Nonetheless, the observed difference in KD (Table 1) is sufficient to achieve remarkable specificity in different assay formats (ELISA, IP, and BRET2) (Figs. 1, 4, and 6). Other selected DARPins behave similarly and target the same structural differences in ERK2 and pERK2 (Fig. S5) as the ones whose structures were determined, underlining that DARPin libraries in combination with ribosome display offer multiple solutions for a certain binding epitope. Interestingly, DARPins targeting two other protein kinases with structural homology to MAPK (35, 36) bind to an epitope located near the observed structural changes in ERK2 upon phosphorylation. Thus, this region appears to be amenable for the comparatively rigid-body interaction surface of DARPins and provides specificity through structural changes in the activation loop.
Importantly, selected DARPins retain their binding and specificity in eukaryotic cell extracts, as shown by affinity precipitation (Fig. 4). As the DARPins have been selected to bind to the native state of the target proteins, and in combination with their superior stability (37), DARPins specifically targeting PTM states of proteins may represent a promising tool for use in proteomic applications, such as affinity reagent microarrays (5, 38). Because of their small molecular size, defined attachment of DARPins to a support surface is expected to be fully compatible with current designs of dense protein arrays.
Apart from being useful as capturing molecules in protein arrays or affinity precipitations, DARPins are especially suited for functional studies as intracellular protein-specific reagents (intrabodies), because they do not require stabilizing disulfide bonds (12, 18, 39) and because they can thus fold in the cytoplasm, where they neither aggregate nor are degraded. Intrabodies targeting PTM sites are of particular importance for detection of target protein level and the present status of modification. In addition, if the DARPin binds to regulatory structural elements, such as the activation loop of a kinase, it will probably interfere with activation in a predictable manner. In this particular case, it will inhibit the activation reaction.
We proved these two aspects by designing BRET2 sensors of ERK phosphorylation-specific DARPins E40 and pE59. Selected DARPins interacted specifically with the cognate ERK2 form in living cells (Fig. 5). Upon stimulation of ERK signaling, an increased level of Rluc-pERK2 was observed in both BRET2 and Western blot experiments with the pERK-binding DARPin pE59 (Fig. 6, Left). The level of Rluc-pERK2 diminished after addition of an ERK pathway inhibitor, thus allowing us to sensitively monitor changes of Rluc-pERK2 in both directions in living cells. In assays with the ERK-binding DARPin E40, no Rluc-ERK2 phosphorylation was expected. E40 directly interacts with the TEY motif in the activation loop and thus blocks putative contact regions of the MAP/ERK kinase (MEK) 1 (40). In all experiments with DARPin E40 (Fig. 6, Right), the level of Rluc-pERK2 was consistently and significantly lower, suggesting an inhibitory effect, by blocking the substrate Rluc-ERK from phosphorylation by upstream kinases. Future experiments with nontagged versions of DARPins targeting endogenous ERK are needed to confirm the observed inhibitory effect by applying an experimental approach recently developed for DARPin-based inhibitors of JNK phosphorylation (30). In summary, we have shown in our approach that both sensors and intracellular inhibitors can be obtained by selecting DARPins to the native targets in their modified and their unmodified form.
The approach presented here is not restricted at all to phosphorylation, but could be easily extended to other proteins of interest with different PTMs. Many PTMs have regulatory roles and in turn mediate the interaction with other molecules. In combination with recent progress in cell-type–specific DNA delivery strategies (41), intrabodies perturbing specifically PTMs may become a valuable tool in signaling research or human and animal health. Furthermore, the uniformity and stability of the DARPin scaffold may greatly simplify the construction and optimization of fluorescent biosensors for living cells based on resonance energy transfer or attachment of environmentally sensitive dyes (42).
Materials and Methods
Molecular Biology.
Unless stated otherwise, all experiments were done according to standard protocols (43). Enzymes and buffers were purchased from New England Biolabs (NEB) or Fermentas. Oligonucleotides were obtained from Microsynth and MWG Operon. Sequences of oligonucleotides used for cloning are listed in Table S4.
Cloning of Recombinant MAPK and DARPins.
pAT222 (GeneBank accession no. AY327137) was used as a backbone for the construction of expression vectors to produce unphosphorylated and phosphorylated human ERK1, rat ERK2, human JNK1α1, human JNK2α1, and mouse p38α. Vectors used for encoding MAPKs and MEKs, used as PCR templates, were kind gifts from Melanie H. Cobb (University of Texas Southwestern, Dallas) (32). The ERK1 and ERK2 genes were amplified from the plasmids NpT7-5ERK1 and NpT75-ERK2 with oligonucleotides ERK1f, ERK1r, ERK2f, and ERK2r. The JNK1α1, JNK2α1, and p38α genes were amplified from the plasmids pAT222_huJNK1α1, pAT222_huJNK2α1, and pAT222_p38 (12, 30) with oligonucleotides JNK1α1f, JNK1α1r, JNK2α1f, JNK2α1r, p38αf, and p38αr. PCR products were cut with NcoI/HindIII and ligated into pAT222, yielding pLK1_ERK1, pLK1_ERK2, pLK1_JNK1α1, pLK1_JNK2α1, and pLK1_p38α.
The expression from these vectors results in fusion proteins with an N-terminal avi tag for in vivo biotinylation followed by the respective MAPK and a C-terminal His tag for purification (avi-MAPK-His6). Production of activated MAPKs requires the coexpression of upstream kinases (32). A constitutively active form of MEK1, MEK1R4F, was used for activation of ERK1 and ERK2. Human MEK1R4F was amplified from pETHis6/MEK1R4F+ERK2 (32), using oligonucleotides MEK1R4Ff and MEK1R4Fr. The PCR product was cut with BlpI and ligated into pLK1_ERK1 and pLK1_ERK2, yielding pLK1_ERK1+MEK1R4F and pLK1_ERK2+MEK1R4F. For activation of JNK1α1, JNK2α1, and p38α, human MEK4 was amplified from NpT7-5/α-SAPK+MEK4, using oligonucleotides MEK4f and MEK4r. The PCR product was digested with BlpI and ligated into pAT222, yielding pAT222_MEK4. PCR products of JNK1α1, JNK2α1, and p38α digested with NcoI/HindIII were ligated into pAT222_MEK4, yielding pLK1_JNK1α1+MEK4, pLK1_JNK2α1+MEK4, and pLK1_p38α+MEK4. pBB131/MEKK-C [kind gift from Melanie H. Cobb (University of Texas Southwestern, Dallas)] (32) was used for coexpression to activate MEK4. pBirAcm (Avidity) was used for in vivo biotinylation of MAPKs.
For BRET2 experiments, DARPins were amplified from plasmids pDST67 (E40, pE59, EpE82) and pQIss (E3_5) (30) and each cloned into vectors pCMV-Rluc-N3, pCMV-Rluc-C2, pCMV-GFP2-N3, and pCMV-GFP2-C1 (PerkinElmer Life Sciences). To obtain N-terminal DARPin fusion expression constructs, we used the primer combination darpin2f-BglII and darpin2r-HindIII. C-terminally fused DARPins were amplified with darpin1f-XhoI and darpin1rstop-HindIII and cloned via XhoI and HindIII. ERK2 was amplified from pLK1_ERK2, using the primers erk2f-XhoI and erk2rstop-HindIII, cut with XhoI and HindIII to allow ligation into the vectors pCMV-GFP2-C2 and pCMV-Rluc-C3. JNK1α1 was amplified using the primer pair jnk1f-XhoI and jnk1rstop-HindIII and pAT222_huJNK1α1 as the template. The PCR fragments were digested with XhoI and HindIII and subcloned into pCMV-GFP-C1 and pCMV-Rluc-C2. JNK2α1 was amplified using the primer pair jnk2f-XhoI and jnk2rstop-HindIII and pAT222_huJNK2α1 as the template. The PCR fragments were again digested with XhoI and HindIII and subcloned into pCMV-GFP-C1 and pCMV-Rluc-C2.
Protein Production and Purification.
Biotinylated ERK1, pERK1, ERK2, pERK2, JNK1α1, JNK2α1, and p38α (plasmids pLK1_ERK1, pLK1_ERK1+MEK1R4F, pLK1_ERK2, pLK1_ERK2+MEK1R4F, pLK1_JNK1α1, pLK1_JNK2α1, and pLK1_p38α) were produced by in vivo biotinylation according to the protocols of Avidity and as described previously (32) in E. coli BL21DE3 with plasmid pBirAcm. Using the same conditions as for production of pERK1/2 (32), biotinylated pJNK1α1, pJNK2α1, and pp38α were expressed in E. coli AVB100 (Avidity), which was cotransformed with pBB131/MEKK-C and pLK1_JNK1α1+MEK4, pLK1_JNK2α1+MEK4, or pLK1_p38α+MEK4. Purification of His6-tagged biotinylated MAPKs was performed as described in ref. 32. Biotinylation of MAPKs was confirmed by ELISA, Western blot analysis with an anti-biotin antibody (Cell Signaling), and mass spectrometry analysis. Successful phosphorylation of MAPKs was verified by Western blot analysis with phosphospecific ERK1/2, JNK and p38 antibodies (Cell Signaling), mass spectrometry, and a kinase activity assay (44).
Nonbiotinylated ERK2 and pERK2, used for competition ELISA experiments and crystallization, were produced in BL21(DE3) and purified as described in refs. 24, 25, 31, and 32, using the plasmids NpT75-ERK2 and pETHis6/MEK1R4F+ERK2. Purification and quality tests were performed in the same way as for biotinylated ERK2 and pERK2.
For the expression of DARPins, selected DARPin genes were cloned into pDST67 (20). To generate biotinylated DARPins E40 and pE59, coding sequences were cloned into pBD001 (45). DARPins were expressed and purified as described in refs. 12 and 45.
Ribosome Display Selections.
DARPin N2C and N3C libraries (12) were transcribed and selections were performed using RD as described in refs. 7, 12, and 46. For the selection of phosphorylation-specific DARPins, biotinylated ERK2 or pERK2 was used as the target molecule. Four rounds of RD were carried out under reducing conditions for each library with the corresponding two targets. After the first round an additional prepanning step on the nondesired ERK2 form (pERK2 for ERK2 selections and ERK2 for pERK2 selections) was included to enrich for binders targeting conformational differences. After each RD round, the RT-PCR products were purified on an agarose gel to prevent enrichment of molecules with recombined modules that might occur during PCR due to the repetitive nature of DNA sequences in DARPin genes.
Analysis of Selected DARPin Binders.
From the enriched DARPin DNA pools, N2C and N3C DARPin ORFs were amplified by PCR as described in ref. 12 and cloned into pDST67 (20). DARPin pools were screened by a crude extract ELISA according to previous studies (12, 47). Protein sequences of DARPins subjected to detailed analysis in this study are listed in Fig. S3. Selected DARPins were expressed on a liter scale and purified as described in ref. 37 for quantitative ELISA, analytical gel filtration, and crystallization. Analytical gel filtration was performed as described in ref. 37, using 15 μM protein in PBS (pH 7.4).
ELISA.
Biotinylated antigens were immobilized on neutravidin-coated ELISA plates blocked with BSA. Target immobilization and all subsequent ELISA steps were performed at 4 °C in PBS (pH 7.4) with 1 mM DTT and 0.05% Tween-20. For screening of single clones from enriched DARPin selection pools, 100 μL of crude extract was applied to wells with or without ERK2 or pERK2 (added as 50 nM each in biotinylated form to neutravidin-coated wells) for 1 h at 4 °C. DARPin binding was detected by an anti–RGS-His antibody (Qiagen) followed by a secondary antibody/alkaline phosphatase conjugate (Thermo Scientific), both in PBS (pH 7.4) with 0.05% Tween-20. Incubation with the substrate p-nitrophenylphosphate (Sigma) was performed at 37 °C. ELISAs for phosphorylation status and ERK specificity of selected DARPins were carried out in the same manner, except that purified DARPins (100 nM) were used. In competition ELISA, selected purified DARPins (10 nM) were incubated with varying amounts of nonbiotinylated ERK2 or pERK2 before (4 °C, 120 min) and during the binding reaction on immobilized ERK2 or pERK2 (10 nM each, 4 °C, 10 min).
For sandwich epitope mapping ELISAs, biotinylated DARPins E40 and pE59 (1 μM each) were immobilized on microtiter plates via neutravidin. They carry a C-terminal His6-tag, which is not detected by the anti–RGS-His antibody. To verify their correct immobilization, they were detected by an anti-His4 antibody (Qiagen) followed by a secondary antibody/alkaline phosphatase conjugate (Pierce). Residual neutravidin sites were blocked with free biotin. Biotinylated kinases ERK2 or pERK2 (50 nM each) were added for capturing by immobilized DARPins. The efficiency of capturing ERK2 and pERK2 could be verified with an anti-biotin HRP-linked antibody (Cell Signaling), because the biotin of DARPins E40 and pE59 is buried within neutravidin. Finally, DARPins to be tested (100 nM) were added after the kinase capture step. DARPin binding to captured kinase was detected by an anti–RGS-His antibody (Qiagen).
SPR.
SPR was measured using a Biacore 2000 instrument (GE Healthcare) for DARPin pE59 and a Biacore 3000 instrument (GE Healthcare) for DARPin E40 interactions. The running buffer was 50 mM Tris (pH 7.4), 150 mM NaCl, 0.05 mM EDTA, and 0.005% Tween-20 for pE59 and PBS (pH 7.4), 1 mM DTT, 0.05 mM EDTA, and 0.005% Tween-20 for E40 measurements. Biotinylated ERK2 or pERK2 was immobilized on a streptavidin SA chip (GE Healthcare) to ∼300 response units (RU). The interactions were determined at a flow rate of 50 μL/min for E40 and 30 μL/min for pE59, with 3–5 min injection of DARPin in varying concentrations and an off-rate measurement of 45–60 min. The signal of an uncoated reference cell was subtracted from the sensorgrams. Zero-concentration samples (blanks) were included for “double referencing.” The kinetic data of the interaction were evaluated with Scrubber2 (BioLogic Software) and BIAevaluation 4.1 (GE Healthcare).
Complex Purification and Crystallization.
Nonbiotinylated ERK2, pERK2, and DARPins E40 and pE59 (with N-terminal MRGSH6-tag) were produced as described above. Cell pellets of bacterial cultures of MAPKs and DARPins were lysed by sonification, using a Sonifier 250 (Branson). Proteins were purified using immobilized metal-ion affinity chromatography (IMAC) as described in refs. 25, 32, and 37. IMAC-purified ERK2 and pERK2 were subjected to ion exchange chromatography as described in ref. 32. Peak fractions were pooled and dialyzed against 20 mM Tris⋅HCl (pH 7.5 at 4 °C), 150 mM NaCl, and 5 mM DTT for ERK2 and 10 mM Hepes (pH 7.5 at 4 °C), 150 mM NaCl, 0.1 mM EDTA, and 5 mM DTT for pERK2. DARPins E40 and pE59 were purified by IMAC as described in ref. 12. DARPin E40 was dialyzed against the ERK2 buffer and DARPin pE59 against the pERK2 buffer. After dialysis, E40 and ERK2, or pE59 and pERK2, were mixed in a 2:1 molar ratio (DARPin:MAPK) and incubated for 4 h at 4 °C. The mixtures were further purified by preparative Superdex-200 (GE Healthcare) size-exclusion chromatography in the respective buffers used for dialysis. For both protein mixtures, the peak fraction containing the DARPin/ERK2 complexes was collected and used for crystallization. The retention volume of the peak fraction matched the expected molecular weight of the DARPin/ERK2 complexes and contained both the MAPK and the DARPin in equimolar amounts (determined by SDS/PAGE). The DARPin/MAPK complexes were concentrated to 6 mg/mL (E40/ERK2) and 8 mg/mL (pE59/pERK2). Initial crystallization screening was performed in 96-well sitting-drop crystallization plates (Corning). The initial crystallization conditions were refined using standard techniques. Crystals used for data collection were grown using the sitting-drop–vapor diffusion method at 4 °C in 24-well crystallization plates. Two microliters of protein solution was mixed with 1 μL reservoir solution. Crystals for E40/ERK2 appeared in 100 mM Tris⋅HOAc (pH 8.5), 20% PEG-4000, 5 mM CdCl2. Crystals for pE59/pERK2 grew in 100 mM Tris⋅HOAc (pH 7.5), 2 M sodium formate. Crystals were soaked in the reservoir buffers supplemented with 10% ethylene glycol (vol/vol) for E40/ERK2 or 20% ethylene glycol (vol/vol) for pE59/pERK2 as cryo-protectant and flash frozen in liquid nitrogen.
Structure Determination and Analysis.
X-ray diffraction data were collected using an in-house Microstar Microfocus rotating anode X-ray generator together with a MAR Research MAR-345dtb image plate detector or at the Swiss Light Source beamline PXIII on a CCD detector. Data were processed using the program XDS (48). Crystal structures were solved by molecular replacement, using the program Phaser (49) in combination with the structures of the unselected N3C DARPin E3_5 (PDB ID 1MJ0) (50), ERK2 (PDB ID 1ERK) (25), and pERK2 (PDB ID 2ERK) (24) as search models. The structures were refined using the programs PHENIX (51) and Coot (52). Protein contacts were calculated using the Protein Interfaces, Surfaces, and Assemblies service (PISA) at the European Bioinformatics Institute (http://www.ebi.ac.uk/pdbe/prot_int/pistart.html) (53) and the CCP4 software suite (54). PyMOL was used to prepare figures. The atomic coordinates of the described complexes were deposited in the PDB (PDB IDs 3zuv and 3zu7).
Affinity Precipitation.
Affinity precipitation experiments using selected DARPins were performed as described in ref. 19. Endogenous ERK2, pERK, and recombinant DARPins were detected by Western blotting. Primary antibodies against ERK2 (Cell Signaling), pERK (Cell Signaling), and the DARPin RGS-His tag (Qiagen) were used. Secondary anti-rabbit and anti-mouse antibodies, coupled to horseradish peroxidase, were visualized by enhanced chemiluminescence (Cell Signaling).
Cell Culture and BRET2 Assays.
COS-7 cells (ATCC CRL-1651) were cultivated in DMEM including 5% FBS Gold (PAA) as described before (27) and transfected in white 96-well microplates (Nunc), using 1 μL PEI (1 mg/mL; Polysciences) and 0.2 μg DNA per plasmid and well. About 1 h after transfection, cells were rinsed with PBS and cultivated for another ∼6 h in medium without FBS and subjected to treatments as indicated in Fig. 6. Immediately before BRET2 readout, 3 μM Coelenterazine 400a (Biotrend) (final concentration) was added as luciferase substrate. The light outputs of Rluc and GFP2 were taken simultaneously (read time 1 s) for each well with filters at 410 nm (± 80 nm band pass) for the Rluc and 515 nm (± 30 nm band pass) for the GFP2 emission, using a POLARstar Omega microplate reader (BMG). As a control, wells containing cells expressing Rluc alone were included in every experiment (background BRET2 signal). From the emission values of transfected cells (em) those from nontransfected (nt) cells were subtracted, and BRET signals were calculated as follows: BRET ratio = (em515 nm – nt515 nm)/(em410 nm – nt410 nm). Experiments were repeated at least three times with six wells per experimental condition. Statistical evaluation (one-way ANOVA with Newman–Keuls posttests) was carried out with GraphPad Prism software version 5.01 (GraphPad Software).
Western Blot Analyses.
Western blot analyses were performed as published before (30) with samples prepared from cells grown in microplates and treated as described above for BRET2 experiments. Antibodies against pERK (Cell Signaling) and Rluc (Millipore) were detected with mouse secondary antibodies and visualized by enhanced chemiluminescence (Western Lightning ECL; Perkin-Elmer).
Supplementary Material
Acknowledgments
We thank Céline Stutz-Ducommun and Beat Blattmann for help in protein crystallization and the staff of beamline PXIII at the Swiss Light Source for support during data collection. Prof. Melanie H. Cobb (University of Texas Southwestern) provided plasmids encoding kinases used in this study. L.K. was supported by a doctoral fellowship of the Ernst Schering Foundation. This work was supported by the Swiss National Center of Competence in Research in Structural Biology, Schweizerische Nationalfonds Grant 3100A0_128671, the PhosphoNetX project in SystemsX (all to A.P.), and the European Union Seventh Framework Programme collaborative project AffinityProteome (Contract 222635 to A.P. and F.W.H.) and the Federal Ministry of Education and Research Grant 0315449E (to F.W.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3ZU7 and 3ZUV).
See Author Summary on page 13475 (volume 109, number 34).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205399109/-/DCSupplemental.
References
- 1.Choudhary C, Mann M. Decoding signalling networks by mass spectrometry-based proteomics. Nat Rev Mol Cell Biol. 2010;11:427–439. doi: 10.1038/nrm2900. [DOI] [PubMed] [Google Scholar]
- 2.Jensen ON. Interpreting the protein language using proteomics. Nat Rev Mol Cell Biol. 2006;7:391–403. doi: 10.1038/nrm1939. [DOI] [PubMed] [Google Scholar]
- 3.Witze ES, Old WM, Resing KA, Ahn NG. Mapping protein post-translational modifications with mass spectrometry. Nat Methods. 2007;4:798–806. doi: 10.1038/nmeth1100. [DOI] [PubMed] [Google Scholar]
- 4.Taussig MJ, et al. ProteomeBinders: Planning a European resource of affinity reagents for analysis of the human proteome. Nat Methods. 2007;4:13–17. doi: 10.1038/nmeth0107-13. [DOI] [PubMed] [Google Scholar]
- 5.Borrebaeck CA, Wingren C. Design of high-density antibody microarrays for disease proteomics: Key technological issues. J Proteomics. 2009;72:928–935. doi: 10.1016/j.jprot.2009.01.027. [DOI] [PubMed] [Google Scholar]
- 6.Renard M, et al. Knowledge-based design of reagentless fluorescent biosensors from recombinant antibodies. J Mol Biol. 2002;318:429–442. doi: 10.1016/S0022-2836(02)00023-2. [DOI] [PubMed] [Google Scholar]
- 7.Hanes J, Plückthun A. In vitro selection and evolution of functional proteins by using ribosome display. Proc Natl Acad Sci USA. 1997;94:4937–4942. doi: 10.1073/pnas.94.10.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uhlén M, Hober S. Generation and validation of affinity reagents on a proteome-wide level. J Mol Recognit. 2009;22:57–64. doi: 10.1002/jmr.891. [DOI] [PubMed] [Google Scholar]
- 9.Vaughan TJ, et al. Human antibodies with sub-nanomolar affinities isolated from a large non-immunized phage display library. Nat Biotechnol. 1996;14:309–314. doi: 10.1038/nbt0396-309. [DOI] [PubMed] [Google Scholar]
- 10.Knappik A, et al. Fully synthetic human combinatorial antibody libraries (HuCAL) based on modular consensus frameworks and CDRs randomized with trinucleotides. J Mol Biol. 2000;296:57–86. doi: 10.1006/jmbi.1999.3444. [DOI] [PubMed] [Google Scholar]
- 11.Binz HK, Amstutz P, Plückthun A. Engineering novel binding proteins from nonimmunoglobulin domains. Nat Biotechnol. 2005;23:1257–1268. doi: 10.1038/nbt1127. [DOI] [PubMed] [Google Scholar]
- 12.Binz HK, et al. High-affinity binders selected from designed ankyrin repeat protein libraries. Nat Biotechnol. 2004;22:575–582. doi: 10.1038/nbt962. [DOI] [PubMed] [Google Scholar]
- 13.Boersma YL, Plückthun A. DARPins and other repeat protein scaffolds: Advances in engineering and applications. Curr Opin Biotechnol. 2011;22:849–857. doi: 10.1016/j.copbio.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 14.Binz HK, Stumpp MT, Forrer P, Amstutz P, Plückthun A. Designing repeat proteins: Well-expressed, soluble and stable proteins from combinatorial libraries of consensus ankyrin repeat proteins. J Mol Biol. 2003;332:489–503. doi: 10.1016/s0022-2836(03)00896-9. [DOI] [PubMed] [Google Scholar]
- 15.Wetzel SK, Settanni G, Kenig M, Binz HK, Plückthun A. Folding and unfolding mechanism of highly stable full-consensus ankyrin repeat proteins. J Mol Biol. 2008;376:241–257. doi: 10.1016/j.jmb.2007.11.046. [DOI] [PubMed] [Google Scholar]
- 16.Wyler E, et al. Inhibition of NF-kappaB activation with designed ankyrin-repeat proteins targeting the ubiquitin-binding/oligomerization domain of NEMO. Protein Sci. 2007;16:2013–2022. doi: 10.1110/ps.072924907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawe M, Forrer P, Amstutz P, Plückthun A. Isolation of intracellular proteinase inhibitors derived from designed ankyrin repeat proteins by genetic screening. J Biol Chem. 2006;281:40252–40263. doi: 10.1074/jbc.M602506200. [DOI] [PubMed] [Google Scholar]
- 18.Amstutz P, et al. Intracellular kinase inhibitors selected from combinatorial libraries of designed ankyrin repeat proteins. J Biol Chem. 2005;280:24715–24722. doi: 10.1074/jbc.M501746200. [DOI] [PubMed] [Google Scholar]
- 19.Amstutz P, Koch H, Binz HK, Deuber SA, Plückthun A. Rapid selection of specific MAP kinase-binders from designed ankyrin repeat protein libraries. Protein Eng Des Sel. 2006;19:219–229. doi: 10.1093/protein/gzl004. [DOI] [PubMed] [Google Scholar]
- 20.Steiner D, Forrer P, Plückthun A. Efficient selection of DARPins with sub-nanomolar affinities using SRP phage display. J Mol Biol. 2008;382:1211–1227. doi: 10.1016/j.jmb.2008.07.085. [DOI] [PubMed] [Google Scholar]
- 21.Chen Z, et al. MAP kinases. Chem Rev. 2001;101:2449–2476. doi: 10.1021/cr000241p. [DOI] [PubMed] [Google Scholar]
- 22.Lawrence MC, et al. The roles of MAPKs in disease. Cell Res. 2008;18:436–442. doi: 10.1038/cr.2008.37. [DOI] [PubMed] [Google Scholar]
- 23.Olsen JV, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 24.Canagarajah BJ, Khokhlatchev A, Cobb MH, Goldsmith EJ. Activation mechanism of the MAP kinase ERK2 by dual phosphorylation. Cell. 1997;90:859–869. doi: 10.1016/s0092-8674(00)80351-7. [DOI] [PubMed] [Google Scholar]
- 25.Zhang F, Strand A, Robbins D, Cobb MH, Goldsmith EJ. Atomic structure of the MAP kinase ERK2 at 2.3 Å resolution. Nature. 1994;367:704–711. doi: 10.1038/367704a0. [DOI] [PubMed] [Google Scholar]
- 26.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 27.Prinz A, Diskar M, Erlbruch A, Herberg FW. Novel, isotype-specific sensors for protein kinase A subunit interaction based on bioluminescence resonance energy transfer (BRET) Cell Signal. 2006;18:1616–1625. doi: 10.1016/j.cellsig.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 28.Reiners JJ, Jr, Lee JY, Clift RE, Dudley DT, Myrand SP. PD98059 is an equipotent antagonist of the aryl hydrocarbon receptor and inhibitor of mitogen-activated protein kinase kinase. Mol Pharmacol. 1998;53:438–445. doi: 10.1124/mol.53.3.438. [DOI] [PubMed] [Google Scholar]
- 29.Seo J, Lee KJ. Post-translational modifications and their biological functions: Proteomic analysis and systematic approaches. J Biochem Mol Biol. 2004;37:35–44. doi: 10.5483/bmbrep.2004.37.1.035. [DOI] [PubMed] [Google Scholar]
- 30.Parizek P, et al. Designed Ankyrin Repeat Proteins (DARPins) as novel isoform-specific intracellular inhibitors of c-Jun N-terminal kinases. ACS Chem Biol. 2012 doi: 10.1021/cb3001167. [DOI] [PubMed] [Google Scholar]
- 31.Heise CJ, Cobb MH. Expression and characterization of MAP kinases in bacteria. Methods. 2006;40:209–212. doi: 10.1016/j.ymeth.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 32.Wilsbacher JL, Cobb MH. Bacterial expression of activated mitogen-activated protein kinases. Methods Enzymol. 2001;332:387–400. doi: 10.1016/s0076-6879(01)32217-6. [DOI] [PubMed] [Google Scholar]
- 33.Wilson KP, et al. Crystal structure of p38 mitogen-activated protein kinase. J Biol Chem. 1996;271:27696–27700. doi: 10.1074/jbc.271.44.27696. [DOI] [PubMed] [Google Scholar]
- 34.Xie X, et al. Crystal structure of JNK3: A kinase implicated in neuronal apoptosis. Structure. 1998;6:983–991. doi: 10.1016/s0969-2126(98)00100-2. [DOI] [PubMed] [Google Scholar]
- 35.Kohl A, et al. Allosteric inhibition of aminoglycoside phosphotransferase by a designed ankyrin repeat protein. Structure. 2005;13:1131–1141. doi: 10.1016/j.str.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 36.Bandeiras TM, et al. Structure of wild-type Plk-1 kinase domain in complex with a selective DARPin. Acta Crystallogr D Biol Crystallogr. 2008;64:339–353. doi: 10.1107/S0907444907068217. [DOI] [PubMed] [Google Scholar]
- 37.Zahnd C, et al. A designed ankyrin repeat protein evolved to picomolar affinity to Her2. J Mol Biol. 2007;369:1015–1028. doi: 10.1016/j.jmb.2007.03.028. [DOI] [PubMed] [Google Scholar]
- 38.Haab BB. Applications of antibody array platforms. Curr Opin Biotechnol. 2006;17:415–421. doi: 10.1016/j.copbio.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 39.Schweizer A, et al. Inhibition of caspase-2 by a designed ankyrin repeat protein: Specificity, structure, and inhibition mechanism. Structure. 2007;15:625–636. doi: 10.1016/j.str.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 40.Robinson FL, Whitehurst AW, Raman M, Cobb MH. Identification of novel point mutations in ERK2 that selectively disrupt binding to MEK1. J Biol Chem. 2002;277:14844–14852. doi: 10.1074/jbc.M107776200. [DOI] [PubMed] [Google Scholar]
- 41.Patil SD, Rhodes DG, Burgess DJ. DNA-based therapeutics and DNA delivery systems: A comprehensive review. AAPS J. 2005;7:E61–E77. doi: 10.1208/aapsj070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gulyani A, et al. A biosensor generated via high-throughput screening quantifies cell edge Src dynamics. Nat Chem Biol. 2011;7:437–444. doi: 10.1038/nchembio.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1989. [Google Scholar]
- 44.Forrer P, Tamaskovic R, Jaussi R. Enzyme-linked immunosorbent assay for measurement of JNK, ERK, and p38 kinase activities. Biol Chem. 1998;379:1101–1111. doi: 10.1515/bchm.1998.379.8-9.1101. [DOI] [PubMed] [Google Scholar]
- 45.Theurillat JP, et al. Designed ankyrin repeat proteins: A novel tool for testing epidermal growth factor receptor 2 expression in breast cancer. Mod Pathol. 2010;23:1289–1297. doi: 10.1038/modpathol.2010.103. [DOI] [PubMed] [Google Scholar]
- 46.Dreier B, Plückthun A. Ribosome display: A technology for selecting and evolving proteins from large libraries. Methods Mol Biol. 2011;687:283–306. doi: 10.1007/978-1-60761-944-4_21. [DOI] [PubMed] [Google Scholar]
- 47.Zahnd C, Pecorari F, Straumann N, Wyler E, Plückthun A. Selection and characterization of Her2 binding-designed ankyrin repeat proteins. J Biol Chem. 2006;281:35167–35175. doi: 10.1074/jbc.M602547200. [DOI] [PubMed] [Google Scholar]
- 48.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Cryst. 1993;26:795–800. [Google Scholar]
- 49.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kohl A, et al. Designed to be stable: Crystal structure of a consensus ankyrin repeat protein. Proc Natl Acad Sci USA. 2003;100:1700–1705. doi: 10.1073/pnas.0337680100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 52.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 53.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 54.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]