Abstract
Ganoderma lucidum is one of the most important medicinal mushrooms; however, molecular genetics research on this species has been limited due to a lack of reliable reverse genetic tools. In this study, the endogenous orotidine 5′-monophosphate decarboxylase gene (URA3) was cloned as a silencing reporter, and four gene-silencing methods using hairpin, sense, antisense, and dual promoter constructs, were introduced into G. lucidum through a simple electroporation procedure. A comparison and evaluation of silencing efficiency demonstrated that all of the four methods differentially suppressed the expression of URA3. Our data unequivocally indicate that the dual promoter silencing vector yields the highest rate of URA3 silencing compared with other vectors (up to 81.9%). To highlight the advantages of the dual promoter system, we constructed a co-silencing system based on the dual promoter method and succeeded in co-silencing URA3 and laccase in G. lucidum. The reduction of the mRNA levels of the two genes were correlated. Thus, the screening efficiency for RNAi knockdown of multiple genes may be improved by the co-silencing of an endogenous reporter gene. The molecular tools developed in this study should facilitate the isolation of genes and the characterization of the functions of multiple genes in this pharmaceutically important species, and these tools should be highly useful for the study of other basidiomycetes.
Introduction
Ganoderma lucidum is one of the most widely cultivated pharmaceutical mushrooms in East Asia because it contains many pharmacologically active compounds, such as ganoderic acid and polysaccharide [1]–[3]. In recent years, many studies have described the pharmacological activity and chemical composition of G. lucidum [4]–[6]. These pharmacological compounds, which inhibit tumor cell growth [7], modulate the immune system and lower blood sugar levels [8], are thought to be potential candidates for drug discovery, and the pharmacological value of G. lucidum has drawn widespread interest [9].
The number of studies investigating the molecular genetics of G. lucidum has increased and includes genome analysis [10], [11], gene cloning and the characterization of biosynthetic pathways [12]–[17], cloning of other functional genes [18]–[20], and EST analysis [16], [21]. However, further functional analysis of genes of interest has been limited due to the lack of effective genetic tools. G. lucidum has been considered a genetically intractable organism, and tools for investigating the functional genomics of G. lucidum are still at an early stage, although some gene transfer procedures have been established [22]–[24]. The roles of the genes within the biosynthesis network are not yet clear. Improving the genetic tools for this organism will provide significant insight into the mechanism of synthesis of pharmacologically active compounds, open new possibilities for the exploitation of G. lucidum for enhancing pharmacologically active compounds by genetic engineering, and be critical for studying the biology, chemical composition, and evolution of this organism.
Reverse genetic methods, such as gene disruption or RNA interference, can be useful for studying functional genomics [25], [26]. RNA interference (RNAi) is a convenient reverse genetic method that has been widely used in several types of organisms [27], [28] because it is simple to down-regulate the target gene using short DNA sequences, and the use of RNAi circumvents the low homologous recombination rate that occurs with gene disruption. RNAi uses sequence-specific double-stranded RNA (dsRNA) to trigger the degradation of homologous mRNA, thereby reducing gene expression.
Among filamentous fungi, RNAi was first reported in Neurospora crassa [29], and this method has been harnessed to elucidate gene function in an expanding number of organisms [30]–[32]. However, the basidiomycetes group lacks these RNAi methods [33], except for some model species, such as Schizophyllum commune [34], Coprinopsis cinerea [35], Laccaria bicolor [36], and several cultured mushrooms, such as Agaricus bisporus [37], Lentinula edodes [24], and Clitopilus passeckerianus [22]. Although these studies reported wide variation in the frequency of silenced transformants, no clear information exists about the optimal method for silencing a gene in basidiomycetes because the previous studies used different target genes and vector configurations. In G. lucidum, it was unknown which method could induce silencing and which could yield the highest efficiency of silencing. Therefore, the discovery and development of a convenient and efficient RNAi system are important areas for further study in G. lucidum and other basidiomycetes.
In this study, the endogenous orotidine 5′-monophosphate decarboxylase gene (URA3) was cloned as a reporter for monitoring RNAi in G. lucidum, and a convenient RNAi system was established through the development of a simple electroporation procedure. The effectiveness of four different RNAi methods was analyzed, and the dual promoter silencing method was determined to yield the highest fraction of URA3-silenced transformants. Finally, we succeeded in using this dual promoter silencing system to co-silence two endogenous genes in G. lucidum and demonstrated the applicability of this RNAi system.
Results
Isolation of genomic DNA and cDNA of URA3 in G. lucidum
A 400-bp PCR product was amplified using the degenerate primers URA3-1 and URA3-2 (table 1), which were designed based on the conserved regions of the URA3 gene sequence. The 5′ self-formed adaptor PCR (SEFA) and 3′ rapid-amplification of cDNA ends (RACE) methods were utilized to obtain the full-length sequence of the gene. The full-length URA3 genomic sequence was 947 bp in length, including two introns. The full-length URA3 cDNA contained an 840 bp ORF. A BLASTX (http://www.ncbi.nlm.nih.gov/) search confirmed that the sequence was highly homologous to the URA3 genes of Schizophyllum commune (73% identity, 83% homology), which indicated that the amplified sequence was indeed from the G. lucidum URA3 gene. The nucleotide sequences of the URA3 gene and cDNA have been deposited in the GenBank database under accession numbers JQ406674 and JQ406675.
Table 1. Oligonucleotide primers used.
| Primer | Sequence (5′ to 3′)* | Description |
| URA3-1 | AARTTYGCNGAYATHGGNAA | Degenerate primers amplify a specific fragment of URA3 |
| URA3-2 | GGNGTVCGRTAYTGYTGNCC | |
| URA3RACE1 | TAAGATGTAGTCCCTTTGCG | Get the 3′-end of URA3-cDNA |
| URA3RACE2 | CGCCGAGATGAGCACCAAG | |
| URA3-5Sp1 | GCAGTATCCGACGGCATCAGCATTCTATCTC | Amplify the 5′-end of URA3-DNA |
| URA3-5Sp2 | TACAACCTGCGTATCCTTATGCTCCACCCAC | |
| URA3-5Sp3 | GCCTTATCAGACTCAANNNNNNNNNATGTAG | |
| URA3-F | ATGGTGGCCGTGGCCAAGC | Amplify the whole length |
| URA3-R | CTAATCCGAGATCCCAACCC | |
| hp-Anti 1 | GCGCGGTACCTCTCCGCCTGTGCGCGGACT | A antisense fragment of URA3 for the pAN7-ura3-hp plasmid |
| hp-Anti 2 | TCTCCTCGAGACGATCGAGCGCAAGCGCAC | |
| hp-Sense 1 | ATCGCTCGAGGGTGGCACTTCAATATTCTG | A sense fragment of URA3 for the pAN7-ura3-hp plasmid |
| hp-Sense 2 | GCGCAAGCTTTCTCCGCCTGTGCGCGGACT | |
| hp-gpd 1 | TCGAGGATCCGGTATATCGTTACATATATC | The G. lucidum gpd promoter for the pAN7-ura3-hp plasmid |
| hp-gpd 2 | CGATGGTACCAGGGGGATGAAGAGTGAGTA | |
| S-Sense 1 | GCGCGGTACCGGTGGCACTTCAATATTCTG | The sense fragment of URA3 for the pAN7-ura3-s plasmid |
| S-Sense 2 | GCGCAAGCTTTCTCCGCCTGTGCGCGGACT | |
| AS-Anti 1 | GCGCGGTACCTCTCCGCCTGTGCGCGGACT | The antisense fragment of URA3 for the pAN7-ura3-as plasmid |
| AS-Anti 2 | GCGCAAGCTTGGTGGCACTTCAATATTCTG | |
| Dual-35S1 | TCATCTAGAAGAGATAGATTTGTAGAGAG | The 35s promoter for the pAN7-ura3-dual plasmid |
| Dual-35s2 | GATAAGCTTCGTAATCATGGTCATAGCTG | |
| URA3-real1 | TGCCGACATTGGAAACACG | Detects the URA3 expression |
| URA3-real2 | TTGCGAGGCTGCCCTTGGT | |
| Lcc-1 | AGTCCGCGCACAGGCGGAGAATGGTGAAATTCCAATCGTT | Overlap PCR amplify the laccase and URA3 fragment |
| Lcc-2 | TCTAGACACCTTGTTCCCTGTGATGA | |
| URA-LCC | AACGATTGGAATTTCACCATTCTCCGCCTGTGCGCGGACT | |
| Lcc-real 1 | CACACAATGCTGAAGACCAC | Detects the Laccase expression |
| Lcc-real 2 | CGGAACCTGGAAATCGTAGA | |
| Hph 4 | TCGTTATGTTTATCGGCACTTT | Detects the transformants' selectable marker hph (hygromycin phosphotransferase) gene |
| Hph 5 | GATGTTGGCGACCTCGTATT |
R = A, G; Y = T, C; N = A, T, C or G; H = A, T or C; V = A, C or G
Transformation of G. lucidum
In an attempt to detect whether pGL-GPE, which contained the hygromycin phosphotransferase (hph) and enhanced green fluorescence protein (egfp) genes, could be transferred into G. lucidum, the transformants were selected on CYM medium containing 100 µg/mL hygromycin B. Colonies appeared after 7 days, and the transformation efficiency was approximately 30 transformants per µg of plasmid pGL-GPE DNA [31] (data not shown). In addition, the use of higher resistance and field strength may have increased the transformation efficiency; however, the chance of cuvette failure would also have increased.
PCR analysis was performed to detect the presence of the fusion fragment containing the hph gene in genomic DNA isolated from the hygromycin B-resistant transformants. The hph gene was detected in 23 of 34 transformants (data not shown). To determine whether the putative transformants could stably maintain hygromycin B resistance, 10 randomly selected transformants were grown on CYM plates without hygromycin B and re-plated for 3 months. We recovered 8 mitotically stable transformants. These results indicate that the transformants can maintain the desired phenotypes long enough for further study.
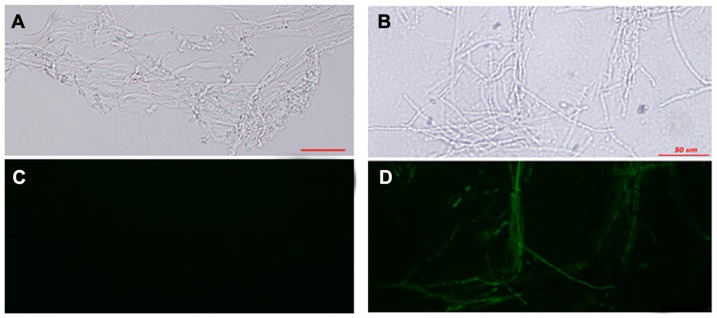
To determine whether exogenous genes were expressed in the pGL-GP transformants, the expression of GFP in transformants cultured on CYM plates without hygromycin B and re-plated for 10 days was detected by fluorescence imaging (Fig. 1). While no GFP fluorescence was detected in wild-type mycelia, the transformants exhibited a positive GFP signal. These results indicate that the method is reliable for the transformation of G. lucidum.
Figure 1. Expression of GFP in G. lucidum.
A GFP-expressing G. lucidum transformant was cultivated on CYM agar medium at 28°C and examined microscopically. Fluorescence of wild-type G. lucidum (A and C) and the GFP transformant (B and D) was examined. Mycelia were removed from actively growing colonies, suspended in sterile water, and viewed microscopically. Samples shown in panels A and B were examined using bright-field microscopy, and samples in panels C and D were viewed using blue-field microscopy. Bars = 50 µm.
Presence of a Dicer-1 homologue in G. lucidum
Genome sequence alignment, which was supplied by Shilin Chen and Haibin Xu (The Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences), was used to identify a Dicer-1 homolog in the G. lucidum genome. Fig. S1 shows the conserved domains detected in this protein fragment using the NCBI Conserved Domain Database. Sequence analysis identified 4 domains characteristic of DCL proteins: a DEAD domain, a helicase C domain, a dsRNA-binding domain and two RNase 3 domains. This sequence is of a 4635 bp fragment that encodes 1544 amino acids, corresponding to a central internal fragment of a Dicer-1 protein homolog (Fig. S2). The Panther Classification System identified this protein as Dicer-1 with an E value of 6.5 e−73.
Fig. S3 shows the amino acid sequence alignment of the GLDCL-1 fragment to other fungal DCL homologues. This alignment demonstrates that these proteins are conserved among fungi, specifically in the regions of the previously mentioned domains.
Construction of RNAi vectors
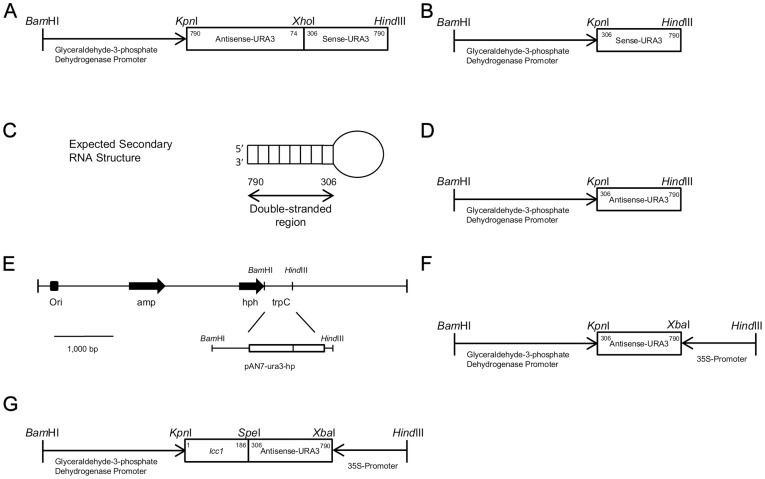
To examine the variables associated with silencing, four vectors were generated that expressed a 484 bp fragment of the URA3 gene in hairpin, sense, antisense, and dual promoter constructs, named pAN7-ura3-hp, pAN7-ura3-s, pAN7-ura3-as, and pAN7-ura3-dual, respectively (Fig. 2).
Figure 2. Construction of the URA3 RNAi expression cassette plasmids used to silence URA3.
In all plasmids except for the dual promoter plasmid, URA3 transcription is driven by the glyceraldehyde-3-phosphate dehydrogenase promoter. pAN7-ura3-hp contains the antisense fragment of URA3 from 790-74 bp and the sense fragment from 306-790 bp (A). pAN7-ura3-s contains 484 bp of URA3 in the sense orientation (B). Complementary portions of the antisense and sense regions are paired in the 306-790 nucleotide region of the URA3 gene, forming a hairpin loop with a 484 bp complementary region (C). pAN7-ura3-as contains URA3 in the antisense orientation (D). Maps of the pAN7-1 vector and the inserted RNAi cassettes (E). The dual promoter plasmid contains 484 bp of URA3 between the gpd promoter and 35S promoter (F). The ura3-lcc co-silencing vector used the dual promoter method (G). Bar = 1000 bp
The pAN7-ura3-hp plasmid was expected to encode a hairpin RNA comprised of two 484 bp complementary regions separated by a 232 bp spacer fragment (Fig. 2A). The pAN7-ura3-hp RNA transcript structure is typical of many functional RNAi products, and the 484 bp complementary stem region is sufficiently long to silence the target gene [32]. The remainder of the antisense fragment forms a loop (Fig. 2C), which contributes to the stability of the inverted repeat sequence [38]. pAN7-ura3-s and pAN7-ura3-as contain the 484 bp fragment of URA3 from position 306-790 (Fig. 2B; Fig. 2D). The pAN7-ura3-dual plasmid was constructed with the 484 bp fragment of URA3 and two promoters: Pgpd (gpd promoter) and P35s (35s promoter) (Fig. 2F). Maps of the pAN7-1 vector and the inserted RNAi cassettes are shown in Fig. 2E.
Selection of RNAi transformants and phenotypic analysis
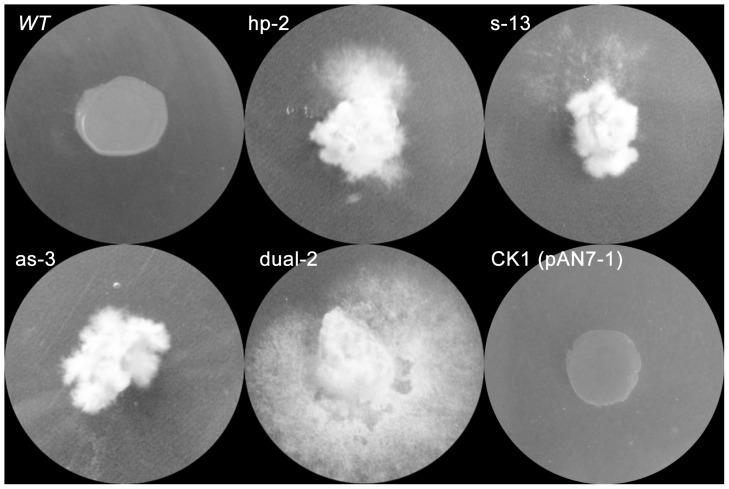
The four silencing plasmids (pAN7-ura3-hp, pAN7-ura3-s, pAN7-ura3-as, and pAN7-ura3-dual) were introduced by electroporation into G. lucidum. To reduce the false positive rate, all of the hygromycin B-resistant transformants were subcultured on CYM medium without selection for 6 generations (approximately 3 months) and then re-cultured on CYM with 600 µg/mL 5-FOA (5-fluoroorotic acid) to select for the silenced transformants. Fig. 3 shows that different transformants and the wt strain exhibited different levels of 5-FOA tolerance. The ck1 strain, which contains the empty vector (pAN7-1) as a control, exhibited the same phenotype as the wt strain. The wt strain and the ck1 strain did not grow or form colonies on CYM containing 600 µg/mL 5-FOA, whereas the URA3-silenced transformants did grow after approximately two weeks of culturing. These results demonstrate that the RNAi remained stable across mitotic cell divisions for at least 3 months. However, not all of the transformants obtained stable silencing or could grow on CYM containing 600 µg/mL 5-FOA (data not shown).
Figure 3. Phenotypic assays of the RNAi transformants on 5-FOA plates.
URA3-silenced transformants, wild-type (wt) and control transformants (CK1, transformed with plasmid pAN7-1) were cultured on CYM medium containing 600 µg/mL 5-FOA for ten days. Hp-2 was one of the pAN7-ura3-hp transformants, s-13 was one of the pAN7-ura3-s transformants, as-3 was one of the pAN7-ura3-as transformants, and dual-2 was one of the pAN7-ura3-dual transformants.
Transformants were categorized as silenced and non-silenced based on their tolerance to 5-FOA: the representative transformants could be cultured on CYM containing 600 µg/mL 5-FOA, whereas the mycelia of non-silenced transformants could not germinate on this selective medium (table 2; Fig. 3). The effectiveness of the silencing was determined in 202 transformants. Both silenced and non-silenced transformants were generated with all four of the silencing plasmids and an empty vector (pAN7-1). The dual promoter silencing method had the highest silencing efficiency, with a mean silencing value of 81.9%; in addition, the dual promoter method allows single-step non-oriented cloning for further vector construction (Table 2). All 40 of the ck1 transformants exhibited the non-silenced phenotype (Table 2). This indicates that all four of the RNAi methods can produce the silenced phenotype and that the dual promoter silencing method has the highest silencing efficiency in G. lucidum. The control results demonstrate that the silenced phenotype was not caused by the transfer procedures.
Table 2. Number of transformants with silenced and no-silenced.
| Plasmid Configuration | Construction steps for further study | Replicate | silenced | non-silenced |
| Hairpin (pAN7-ura3-hp) | 2–3 | I | 7 | 4 |
| II | 8 | 7 | ||
| III | 9 | 5 | ||
| Mean(%) | 60.4 | 39.6 | ||
| Sense (pAN7-ura3-s) | 1 | I | 8 | 5 |
| II | 6 | 11 | ||
| III | 8 | 3 | ||
| Mean(%) | 56.5 | 43.5 | ||
| Antisense (pAN7-ura3-as) | 1 | I | 11 | 7 |
| II | 8 | 6 | ||
| III | 4 | 7 | ||
| Mean(%) | 51.5 | 48.5 | ||
| Dual-promoter (pAN7-ura3-dual) | 1 | I | 10 | 0 |
| II | 19 | 5 | ||
| III | 16 | 8 | ||
| Mean(%) | 81.9 | 18.1 | ||
| CK1 (pAN7-1) | 0 | I | 0 | 11 |
| II | 0 | 15 | ||
| III | 0 | 14 | ||
| Mean(%) | 0 | 100 |
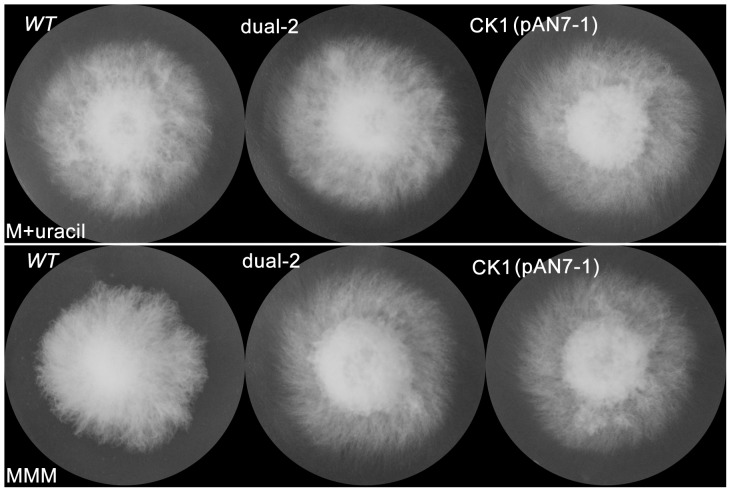
If URA3 were fully silenced, the silenced transformants would be expected to behave as orotidine 5′-monophosphate decarboxylase (OMPdecase) auxotrophs and be unable to grow on mushroom minimal medium (MMM, which is commonly used for detecting auxotrophic strains of mushrooms). However, the growth rate of the RNAi transformants did not differ from that of the wt strain or the ck1 transformant on MMM with or without uracil (100 mg/L) (Fig. 4). This result is the same as was obtained with Agaricus bisporus [37].
Figure 4. Phenotypic assays of the RNAi transformants on MMM plates.
URA3-silenced transformants (dual-2), wild-type (wt) and control transformants (CK1, transformed with the pAN7-1 plasmid) were cultured on MMM medium with or without uracil (100 mg/L) for 7 days at 28°C. The upper row (M+uracil) is MMM medium with uracil, and the lower row (MMM) is MMM medium.
Quantification of URA3 expression in RNAi transformants using the four RNAi methods
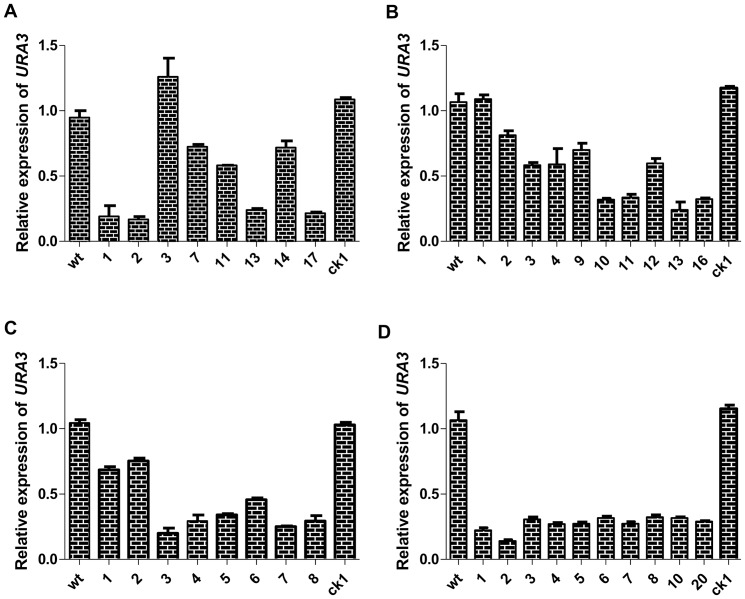
To examine the RNAi transformants on the molecular level, transcription in the URA3-silenced transformants was examined by real-time PCR. The URA3 transcription levels were normalized to those of the endogenous 18S rRNA gene. Transformants were randomly chosen from each RNAi method to quantify the reduction in mRNA transcript levels at this locus. All four methods downregulated the expression of URA3, albeit to varying degrees (Fig. 5).
Figure 5. Measurement of URA3 gene expression as determined by real-time PCR.
Real-time PCR analysis was performed on pAN7-ura3-hp (A), pAN7-ura3-s (B), pAN7-ura3-as (C) and pAN7-ura3-dual (D) transformants. The relative mRNA levels of URA3 were calculated as the ratio of G. lucidum URA3 mRNA to endogenous 18S rRNA. The upper running title of each panel is the ratio of URA3 mRNA and 18S rRNA mRNA compared with wt. The lower running title of each panel is the silenced transformant, wt and ck1 strains. Wild type was considered to have a value of 1.0 for all experiments trials because the mRNA expression in all other isolates was relative to wt. Values are the means ± SE (n = 3).
Among the selected pAN7-ura3-hp transformants, 4 of 8 exhibited significant suppression of URA3, with relative expression reduced 70% compared with the wt and ck1 strains (Fig. 5A). The pAN7-ura3-s and pAN7-ura3-as transformants exhibited results similar to the pAN7-ura3-hp transformants, with a reduction in the relative expression of URA3 ranging from 30% to 75% (Fig. 5B; Fig. 5C). The mRNA transcripts of the ten randomly selected pAN7-ura3-dual silenced transformants were downregulated, and our results show that all ten clones appeared to have achieved a fairly high efficiency of URA3 knockdown compared with the wt or ck1 strains (Fig. 5D). These results also indicate that the dual promoter silencing vector yields the highest rate of URA3 silencing.
Multiple genes can be co-silenced using the dual promoter silencing vector
Given that the silencing of URA3 could be selected with 5-FOA, we constructed a co-silencing vector (pAN7-lcc1-ura3-dual) using URA3 as a selectable marker to facilitate the silencing of other genes (Fig. 2G). The co-silencing transformants would exhibit both the URA3- and lcc1- silenced phenotypes when the two genes were both silenced. To determine the silencing efficiency of the transformants, 30 of 36 hygromycin B-resistant transformants were selected on CYM medium containing 600 µg/mL 5-FOA. Then, 10 of the phenotypically 5-FOA-positive transformants were randomly chosen for further study.
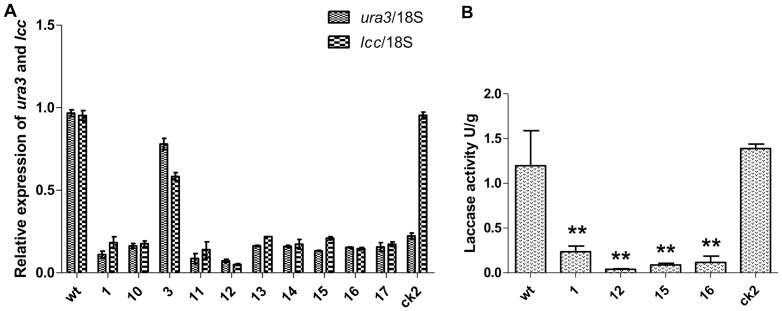
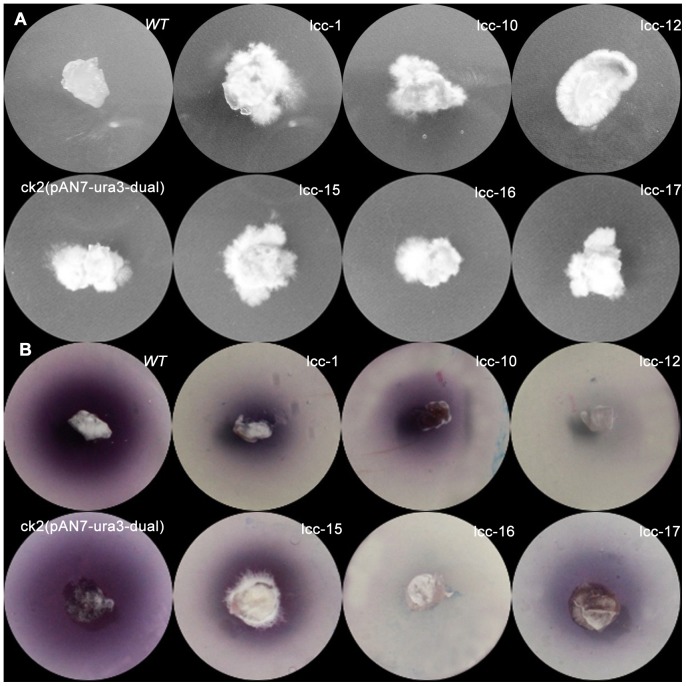
Real-time PCR analysis of the URA3 and lcc1 transcripts demonstrated that the URA3 and lcc1 transcripts were downregulated in most of the transformants compared with the wt strain. The ck2 strains containing the pAN7-ura3-dual plasmid exhibited no difference in lcc1 expression compared with wt, whereas lcc1 expression in co-silenced transformants was reduced. Furthermore, the reduction in levels of the URA3 and lcc1 mRNAs was correlated (Fig. 6A). The silencing of laccase was examined using ABTS-Petri dishes (2,2′-azino-di-3-ethylbenzothiazoline-6-sulfonate, Sigma), in which the chromogen ABTS is a very sensitive substrate that allows the rapid screening of fungal strains that produce extracellular laccases by means of a chromogenic reaction. All of the randomly selected co-silenced transformants that were 5-FOA-tolerant (Fig. 7A) had significantly fewer colored circles than the wt and ck2 strains (Fig. 7B). In two of the URA3-LCC co-silenced transformants (lcc-12, lcc-16), the color change was barely detectable. Upon further analysis, the laccase assay, which was measured by monitoring the increase in absorbance at 420 nm using ABTS as a substrate, demonstrated that the enzymatic activity of RNAi transformants was greatly reduced when compared with wt or ck2 (Fig. 6B). Laccase activity was consistent with the phenotypes (Fig. 7B). These results indicate that it is possible to use URA3 as a reporter for co-silencing another gene, and the control results with ck2 demonstrate that co-silencing was not caused by the transfer procedures.
Figure 6. Measurement of co-silenced gene expression as determined by real-time PCR and laccase enzyme assays.
(A) Real-time PCR analysis was performed in dual promoter co-silenced transformants. The relative mRNA levels of URA3 and lcc1 were calculated as the ratio of URA3 and lcc1 mRNA to endogenous 18S rRNA. The upper running title of each panel is the ratio of target genes and 18S rRNA mRNA compared with wt. The lower running title of each panel is the silenced transformant, wt and ck2 strains. The dotted column is ura3/18s, and the black rectangle column is lcc1/18s. (B) Laccase activity was monitored in the filtrate cultures using an ABTS oxidization test. Wt represents the wild-type strain, ck2 strains contain the pAN7-ura3-dual vector, and strains 1, 12, 15, and 16 represent ura3-lcc co-silenced transformants. Values are means ± SE (n = 3). Asterisks represent significant differences among the strains according to a one-way ANOVA test (P<0.01).
Figure 7. Phenotypic assays of co-silenced transformants.
(A) URA3-lcc1 co-silenced transformants, wild-type and ck2 transformants were cultured on CYM medium containing 600 µg/mL 5-FOA for ten days. Wt represents the wild-type strain, ck2 strains contain the pAN7-ura3-dual vector, and lcc-1, 10, 12, 15, 16, and 17 represent ura3-lcc co-silenced transformants. (B) An ABTS-Petri dish was used to determine whether laccase was silenced in the co-silencing transformants. Lcc-1, lcc-10 lcc-12, lcc-15, lcc-16, and lcc-17 had significantly smaller colored circles compared with the wt and control strains.
Discussion
In this study, we have shown that the transformation procedure based on the electroporation of mycelial fragments is a convenient method for introducing transgenes into G. lucidum because it obviates the complicated production of protoplasts in previously described methods [39] and results in hygromycin B resistance and GFP expression in the transformants. Using this method, we succeeded in silencing the endogenous URA3 gene and co-silencing lcc1 and URA3 by reducing their mRNA and enzyme levels. Because of the uncertainty regarding the presence of a gene-silencing mechanism in some fungi, such as Saccharomyces cerevisiae and Ustilago maydis [40], we identified the presence of a Dicer-1 homolog in the G. lucidum genome (Fig. S1, Fig. S2, Fig. S3). The Dicer enzymes are important components of the mechanism that processes double-stranded RNA precursors into small RNAs during gene silencing [41], [42], and the identification of this homolog implies that G. lucidum can use RNAi for gene silencing, which is also supported by our results.
Although most RNAi studies performed in filamentous fungi report the use of sense, antisense, dual promoter, or hairpin constructs, previous publications did not directly compare the efficiency of these silencing constructs in filamentous fungi. To systematically examine vectors associated with silencing in G. lucidum, four RNAi plasmids carrying the silencing fragment of the same gene were transferred into this species. Our data unequivocally indicate that a dual promoter silencing vector yields the highest rate of URA3 silencing (Table 2). The independent transcription of a target gene from each promoter may produce a pool of sense and antisense RNAs in the cell that combine to form long dsRNAs, which can then be processed into small interfering RNAs (siRNAs) by Dicer [43]. These dual promoter transformants appear to produce much more uniform results than other vectors (Fig. 5D, Fig. 6A) and make it possible to select silenced transformants by fewer tests. Our data suggest that the dual promoter silencing system may be a powerful tool for loss-of-function analysis in G. lucidum. In addition to the high silencing efficiency, the greatest advantage of the dual promoter system is that it allows single-step non-oriented cloning for vector construction (table 2). This advantage allows not only fewer cloning steps but also the construction of multiple silencing vectors in a single transformation experiment. To highlight this advantage, we constructed a co-silencing system using mixed PCR (Fig. 2G) and were able to co-silence two genes simultaneously. When cDNA inserts are made from a normalized mRNA pool, a silencing library consisting of thousands of genes may be constructed in a single transformation [12].
However, the silencing induced by dual promoter as well as other RNAi systems was not always absolute. Some of the transformants did not grow on 5-FOA medium (data not shown), or the silencing levels were different between transformants. These results were consistent with those of our predecessors [34], [37]. The phenotypic variability among RNAi transformants may be related to the condition of the RNAi cassettes; for example, the site of transgene integration, which influences the expression level of the silencing RNA, affects the silencing level [25]. In addition, the copy number of the RNAi cassettes would also affect the silencing level of independent transformants [27]. Furthermore, the variable activity of the RNA-dependent RNA polymerase (RdRP) in different transformants would also affect the silencing level between independent transformants because the as RdRP can extend dsRNA synthesis from the antisense primer into the regions upstream of the target gene [27]. The incomplete silencing of the URA3 locus observed in our study is not an unusual result; indeed, many species of fungi have shown incomplete gene suppression by RNAi [24], [36], [44], although most gene functions can be studied only with preselected strains [45]. To overcome these difficulties, several previous publications have used vector systems in which a reporter gene is downregulated simultaneously with an endogenous target gene. Such RNAi systems use either strain-specific marker genes that can be identified phenotypically [46] or, alternatively, a generally applicable exogenous reporter gene, such as GFP or red fluorescent protein (DsRed) [33], [45]. However, these RNAi systems are strain-specific or require the construction of an exogenous gene expression strain with multiple resistance markers. In this report, endogenous URA3, which encodes OMPdecase and catalyzes the conversion of orotidylic acid to ruidylic acid during uracil biosynthesis in fungi, was cloned and silenced The OMPdecase of fungi can convert 5-FOA into the toxic compound 5-fluorouracil (a suicide inhibitor) causing death (negative selection) [22]. Our results show that the URA3-silenced transformants could be selected on 5-FOA medium (Table 2; Fig. 3) and this phenotype was easy to detect, so we used this characteristic to construct a co-silencing RNAi system containing this gene as a reporter gene. To monitor the efficiency of the co-silencing system, URA3 was co-silenced with Lcc1, which encodes one of the laccase genes in G. lucidum and can be easily detected using ABTS [47]. All of the randomly selected ura3-lcc1 co-silenced transformants, which were selected after the 5-FOA assay, exhibited a significant reduction in mRNA levels and enzymatic activity of laccase, and the reduction in levels of the mRNA of lcc1 correlated with those of URA3 (Fig. 6; Fig. 7). Therefore, co-silencing URA3 could improve the monitoring efficiency of RNAi for another gene linked with URA3, and constructing other co-silencing victors by replacing lcc1 would be straightforward. The lcc1-silenced transformants will help us to better understand its function in G. lucidum in the future.
In addition to the advantages of RNAi, these technique also has limitations, which have been mentioned in many reports [32], [48], and some of these limitations were also observed in this study. The disadvantages primarily include inconsistency due to incomplete and/or reversible silencing [49] and variations in the silencing specificity of different genes. The latter shortcoming can actually be harnessed as an additional advantage, especially when silencing of multiple genes is required [48]. Due to incomplete or reversible silencing, RNAi is not preferred for studying non-essential gene functions compared with gene deletion or knockout.
URA3 mutant strains cannot grow on minimal medium without uracil [22]. However, in our study, there was no difference between the growth rate of RNAi transformants and the wt strain or the ck1 transformant on MMM medium with and without uracil. This result was the same as that observed in Agaricus bisporus [37]. Similarly, the use of antisense constructs to suppress another uracil biosynthesis gene, PYR3 (dihydroorotase), in the heterobasidiomycete Ustilago maydis failed to yield mutant phenotypes [50]. Collectively, these results suggest that although substantial downregulation of mRNAs can be achieved through RNAi transformations, very low-level transcription of key biosynthetic genes may be sufficient to permit the growth of the organism on selective media. However, URA3 can be used as a selectable marker using 5-FOA medium.
The molecular tools that we have described in this paper may aid in identifying and manipulating the genes of this pharmaceutically important species. More importantly, this co-silencing system will make it possible for us to silence genes in a high-throughput manner, and these tools should also facilitate studies aimed at gene isolation or characterization in other mushrooms.
Materials and Methods
Strains and culture conditions
The DH5α strain of Escherichia coli was used for plasmid amplification and was grown in Luria–Bertani (LB) medium containing 100 µg/mL ampicillin or 50 µg/mL kanamycin as required. G. lucidum strain HG was grown at 28°C in CYM medium (1% maltose, 2% glucose, 0.2% yeast extract, 0.2% tryptone, 0.05% MgSO4·7H2O, 0.46% KH2PO4) or MMM medium (2% glucose, 0.2% DL-asparagine, 0.05% MgSO4·7H2O, 0.046% KH2PO4, 0.1%K2HPO4, 120 µg/L thiamin-HCl) and was used as the recipient host strain for transformation and gene silencing.
Amplification of the full-length sequence of URA3
Based on the highly conserved amino acid regions of known orotidine 5′-monophosphate decarboxylase genes (URA3) from other fungi, two degenerate primers, URA3-1 and URA3-2 (Table 1), were synthesized for the amplification of a specific 400 bp fragment of the URA3 gene. A BLASTX search confirmed that the specific fragment of the G. lucidum URA3 gene was highly homologous with other fungal URA3 genes. Subsequently, this sequence was used to design and synthesize gene-specific primers to clone the full-length sequences. Rapid amplification of cDNA ends (RACE-PCR) was performed to obtain the sequence of the 3′ end of the cDNA using the specific primers URA3-RACE1 and URA3-RACE2 (Table 1). Self-formed adaptor PCR (SEFA-PCR) [51] was performed to amplify the 5′ end of the DNA and promoter region using three gene-specific primers (URA3-5Sp1, URA3-5Sp2, URA3-5Sp3) (Table 1). By aligning and assembling the sequences of the 3′ RACE and 5′ SEFA products and the specific fragment, the full-length sequence of URA3 was deduced and subsequently amplified by PCR using the primers URA3-F and URA3-R (Table 1). The amplified product was purified, cloned into the pMD18-T vector (TARAKA, Dalian, China), and sequenced.
Construction of RNA interference cassettes for URA3 and lcc1
RNAi cassettes were designed according to the URA3 gene of G. lucidum (GenBank accession number JQ406675). Expression cassettes were designed to downregulate the expression of the URA3 gene of G. lucidum strain HG. Constructs incorporated the 484nt URA3 coding region in the expression plasmid pAN7-1 [52] (Fig. 2). The hairpin plasmid, which was defined as pAN7-ura3-hp, was constructed by amplifying a 716 bp antisense and a 484 bp sense fragment of the URA3 gene using the primers shown in table 1. Expression of the linked antisense/sense URA3 gene fragments was driven by the glyceraldehyde-3-phosphate dehydrogenase (gpd) promoter (GenBank: DQ404345.1) [10] (Fig. 2A). The gpd promoter, antisense URA3 and sense URA3 fragments were sequentially ligated into the pAN7-1 vector by replacing the TrpC terminator. The sense plasmid, called pAN7-ura3-s, was produced by amplifying the 484 bp sense fragment flanked with the restriction sites KpnI and BamHI and inserting it into the plasmid pAN7-ura3-hp after KpnI/HindIII restriction digestion (Fig. 2B). The antisense vector pAN7-ura3-as was constructed using the same method (Fig. 2D). The dual promoter plasmid pAN7-ura3-dual was generated by the amplification of three DNA fragments: the gpd promoter (Pgpd), the antisense fragment and the 35S promoter (P35S) (table 1). The 35S promoter was amplified using the plasmid pCAMBIA-1300 (CAMBIA, Canberra) as the template (table 1) and was subsequently cloned into pAN7-1 (Fig. 2F). The URA3 and lcc1 co-silencing cassettes (GenBank: FJ656307.1) were produced by mixed PCR, cloned into pAN7-ura3-dual and renamed pAN7-lcc1-ura3-dual, and this vector contained the RNAi cassettes of URA3 linked with lcc1 (Fig. 2G). The constructs were verified by nucleotide sequence analysis to confirm the presence of all of the required elements of a functional cassette prior to their transformation.
Transformation procedure
All plasmids were transferred into G. lucidum by electroporation [39] with some modifications. The procedure and electrical pulse delivery test conditions included several settings [53], and the appropriate modifications were as follows:
First, four-day-old liquid cultures of G. lucidum mycelia were incubated at 28°C in 250 mL conical beakers containing 100 mL of CYM medium in static culture that was shaken three times a day, for 30 s. Mycelial fragments were collected by centrifugation at 3000 g for 10 min, washed twice with 10.93% mannitol and then resuspended in 1 mL 2% lysing enzymes (Guang Dong Institute of Microbiology, China) containing 10.93% mannitol. After incubation for 1.5 h, the mycelia pellets were washed twice with electroporation buffer (10.93% mannitol, 10 mM sodium phosphate [pH 7]) to remove the enzymes, pelleted by centrifugation at 3000 g for 10 min and resuspended in 1 mL of electroporation buffer per 0.3 g of wet weight of mycelia.
Next, ever 200 µL of mycelia was mixed with 10 µL of plasmid DNA (0.5 µg/µL PEG-purified plasmid pGL-GPE, which contains the EGFP and hph genes [31], in sterilized water). The mycelia fragments were incubated on ice for 10 min before electroporation. Exponential decay high-voltage electric pulses were delivered by a BTX ECM 630 using 2 mm cuvettes (BTX, San Diego, CA). The electroporation settings used for transformation were 10 kV/cm field strength, 25 μF capacitance, and 400 Ω resistance.
After pulse delivery, the mycelia fragments were incubated on ice for 10 min and mixed with 5 mL of CYM selective medium (37°C) containing 10.93% mannitol, 1% LMP (low melting point) agarose, and 100 µg/mL hygromycin B. The mixture was poured onto a CYM plate containing 10.93% mannitol, 1% agarose, and 100 µg/mL hygromycin B. The plate was incubated at 28°C for 10 d. To estimate viability, small samples of mycelia fragments were cultured on hygromycin B-free CYM plates before and after electroporation.
RNAi vectors were also transformed into G. lucidum using these electroporation conditions.
Detection and stability of the introduced sequence in transformants
The transformants were first selected on a CYM plate containing 100 µg/mL hygromycin B and then subcultured on media without selection for 6 generations (approximately 3 months), or the RNAi transformants were cultured on CYM with 600 µg/mL 5-FOA (SIGMA). Genomic DNA isolated from putative hygromycin B-resistant or 5-FOA-selected transformants was analyzed by PCR. The hph gene was amplified using the primers HPH-4 and HPH-5, which were previously used for plasmid construction (table 1).
Phenotypic analysis of transformants and laccase enzyme assay
Colonies on CYM selective medium were transferred to nonselective medium and incubated for 10 days before they were analyzed by fluorescence microscopy. The green fluorescence emission from EGFP was detected using a Nikon Eclipse Ti–S microscope. Images were recorded and processed using the NIS-Elements F package.
The URA3-RNAi phenotype was tested by growing transformants on MMM with and without uracil (100 mg/L) and also by growing transformants on CYM containing 600 µg/mL 5-FOA for 2 weeks.
Laccase activity on solid medium was measured as described [54] with little modification. The hyphal tip plug was inoculated onto CYM medium supplemented with 10.97% 2,2′-azino-di-3-ethylbenzothiazoline-6-sulfonate (ABTS, Sigma) for 2 days. The laccase enzyme assay was performed as previously described [47]. One unit of laccase activity was defined as a one-point increase in absorbance at 420 nm at 28°C in 1.0 min.
Real-time PCR analysis of gene expression
The levels of URA3 and lcc1-specific messenger RNA (mRNA) expressed by the wild-type strain (wt) and the isolated RNAi transformants were assessed using quantitative real-time PCR.
Aliquots of 0.2 g of mycelia were collected by filtration from the culture medium and frozen in liquid nitrogen. Total RNA was extracted using an RNA isolation kit (TaKaRa, China), treated with RNase-free DNaseI (TaKaRa) and then reverse-transcribed to cDNA using an oligo (dT)17 primer. Subsequently, 18S [6] and URA3 transcript levels were determined by quantitative real-time PCR using SYBR Green I on the Eppendorf Mastercycler ep realplex (Eppendorf, Germany). The primer sequences are presented in Table 1.
PCR reactions were performed using the SYBR Green Real Time PCR Master Mix (TOYOBO, Japan) according to the manufacturer's protocol. After an initial denaturation step at 95°C for 5 min, amplification was performed in three steps: 30 s of denaturation at 95°C, 60 s of annealing at 60°C, and 30 s of extension at 72°C for a total of 40 cycles. Identical PCR conditions were used for all targets. Transcript levels were calculated using the standard curve method and normalized against the G. lucidum 18S rRNA (18S) gene as an internal control. Wt mycelia served as the reference sample against which all other transformants were compared; expression of the reference sample was defined as 1.0 and the expression of the URA3 or laccase genes in all other transformants is reported as the fold increase over the reference sample. Post-qRT-PCR calculations of relative gene expression levels were performed according to the 2-ΔΔCT method described by Livak and Schmittgen [55].
Bioinformatic Sequence Analysis
The theoretical molecular weights of the proteins were calculated using the on-line ExPASy tool http://expasy.org/tools/pi_tool.html. On-line NCBI Conserved Domains Database http://www.ncbi.nlm.nih.gov/cdd [56], and Pfam http://pfam.sanger.ac.uk/ [57] searches were used to identify potential motifs present in GLDCL-1. Multiple sequence alignments were built using M-COFFEE http://www.tcoffee.org
Supporting Information
Protein domains analysis of G. lucidum DCL-1 homologue. This figure shows the 4 domains that characterize the Dicer-2 proteins that were present in the G. lucidum DCL-1 homologue fragment. The domains were identified using the NCBI Conserved Domain Database. The domains in the 1544 amino acid fragment were: DEXDc (DEAD Like), HELIC_c (helicase domain), dsRNA binding, and the 2 RIBOc domains.
(PDF)
DNA and Amino acid sequence GLDCL-1. The partial DNA and derived amino acid sequence of the gldcl-1 gene. coding regions and amino acids are given in upper case letters. The DEAD domain is shadowed in blue-green, the helicase domain is shadowed in purple, the dsRNA binding domain is shadowed in green and the RNAse 3 domain is shadowed in red and gray.
(PDF)
The predicted amino acid sequence of G. lucidum DCL-1 and DCL-1 homologues from other fungi. In the alignment, blue shading to red shading indicates BAD identity to GOOD identity. Important domains are highlighted in colored boxes. The DEAD domain, the helicase domain, dsRNA binding domain and the two RNAse III domains are highlighted in blue-green, purple, green, red and gray boxes, respectively. blue-green, purple, green, red and gray.
(PDF)
Acknowledgments
We thank the Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences for DNA sequence data.
Funding Statement
This work was supported by the National Natural Science Foundation of China (Project No. 30970042), the Fundamental Research Funds for the Central Universities (Project No. KYZ201121), the Science and Technology Pillar Program of Jiangsu Province, China (Project No. BE2011312) and the Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Eo SK, Kim YS, Lee CK, Han SS (1999) Antiviral activities of various water and methanol soluble substances isolated from Ganoderma lucidum . Journal of Ethnopharmacology 68: 129–136. [DOI] [PubMed] [Google Scholar]
- 2. Mizushina Y, Hanashima L, Yamaguchi T, Takemura M, Sugawara F, et al. (1998) A mushroom fruiting body-inducing substance inhibits activities of replicative DNA polymerases. Biochem Biophys Res Commun 249: 17–22. [DOI] [PubMed] [Google Scholar]
- 3. Wang H, Ng TB (2006) Ganodermin, an antifungal protein from fruiting bodies of the medicinal mushroom Ganoderma lucidum . Peptides 27: 27–30. [DOI] [PubMed] [Google Scholar]
- 4. Adamec J, Jannasch A, Dudhgaonkar S, Jedinak A, Sedlak M, et al. (2009) Development of a new method for improved identification and relative quantification of unknown metabolites in complex samples: determination of a triterpenoid metabolic fingerprint for the in situ characterization of Ganoderma bioactive compounds. J Sep Sci 32: 4052–4058. [DOI] [PubMed] [Google Scholar]
- 5. Lin KI, Kao YY, Kuo HK, Yang WB, Chou A, et al. (2006) Reishi polysaccharides induce immunoglobulin production through the TLR4/TLR2-mediated induction of transcription factor Blimp-1. J Biol Chem 281: 24111–24123. [DOI] [PubMed] [Google Scholar]
- 6. Xu JW, Xu YN, Zhong JJ (2010) Production of individual ganoderic acids and expression of biosynthetic genes in liquid static and shaking cultures of Ganoderma lucidum . Appl Microbiol Biotechnol 85: 941–948. [DOI] [PubMed] [Google Scholar]
- 7. Lin CC, Yu YL, Shih CC, Liu KJ, Ou KL, et al. (2011) A novel adjuvant Ling Zhi-8 enhances the efficacy of DNA cancer vaccine by activating dendritic cells. Cancer Immunol Immunother 60: 1019–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tsai CC, Yang FL, Huang ZY, Chen CS, Yang YL, et al. (2012) Oligosaccharide and peptidoglycan of Ganoderma lucidum activate the immune response in human mononuclear cells. J Agric Food Chem 60: 2830–2837. [DOI] [PubMed] [Google Scholar]
- 9. Shi L, Ren A, Mu DS, Zhao MW (2010) Current progress in the study on biosynthesis and regulation of ganoderic acids. Appl Microbiol Biotechnol 88: 1243–1251. [DOI] [PubMed] [Google Scholar]
- 10. Hseu RS, Wang HH, Wang HF, Moncalvo JM (1996) Differentiation and grouping of isolates of the Ganoderma lucidum complex by random amplified polymorphic DNA-PCR compared with grouping on the basis of internal transcribed spacer sequences. Appl Environ Microbiol 62: 1354–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sun SJ, Gao W, Lin SQ, Zhu J, Xie BG, et al. (2006) Analysis of genetic diversity in Ganoderma population with a novel molecular marker SRAP. Appl Microbiol Biotechnol 72: 537–543. [DOI] [PubMed] [Google Scholar]
- 12. Ding YX, Ou-Yang X, Shang CH, Ren A, Shi L, et al. (2008) Molecular cloning, characterization, and differential expression of a farnesyl-diphosphate synthase gene from the basidiomycetous fungus Ganoderma lucidum . Bioscience Biotechnology and Biochemistry 72: 1571–1579. [DOI] [PubMed] [Google Scholar]
- 13. Fei X, Zhao MW, Li YX (2006) Cloning and sequence analysis of a glyceraldehyde-3-phosphate dehydrogenase gene from Ganoderma lucidum . J Microbiol 44: 515–522. [PubMed] [Google Scholar]
- 14. Shang CH, Shi L, Ren A, Qin L, Zhao MW (2010) Molecular cloning, characterization, and differential expression of a lanosterol synthase gene from Ganoderma lucidum . Biosci Biotechnol Biochem 74: 974–978. [DOI] [PubMed] [Google Scholar]
- 15. Shang CH, Zhu F, Li N, Ou-Yang X, Shi L, et al. (2008) Cloning and characterization of a gene encoding HMG-CoA reductase from Ganoderma lucidum and its functional identification in yeast. Biosci Biotechnol Biochem 72: 1333–1339. [DOI] [PubMed] [Google Scholar]
- 16. Xu JW, Zhao W, Xu YN, Zhong JJ (2012) Isolation and analysis of differentially expressed genes during asexual sporulation in liquid static culture of Ganoderma lucidum by suppression subtractive hybridization. Mol Biol Rep 39: 3603–3610. [DOI] [PubMed] [Google Scholar]
- 17. Zhao MW, Liang WQ, Zhang DB, Wang N, Wang CG, et al. (2007) Cloning and characterization of squalene synthase (SQS) gene from Ganoderma lucidum . J Microbiol Biotechnol 17: 1106–1112. [PubMed] [Google Scholar]
- 18. Joo SS, Ryu IW, Park JK, Yoo YM, Lee DH, et al. (2008) Molecular cloning and expression of a laccase from Ganoderma lucidum, and its antioxidative properties. Mol Cells 25: 112–118. [PubMed] [Google Scholar]
- 19. Murasugi A, Tanaka S, Komiyama N, Iwata N, Kino K, et al. (1991) Molecular cloning of a cDNA and a gene encoding an immunomodulatory protein, Ling Zhi-8, from a fungus, Ganoderma lucidum . J Biol Chem 266: 2486–2493. [PubMed] [Google Scholar]
- 20.Xu YN, Zhong JJ (2011) Impacts of calcium signal transduction on the fermentation production of antitumor ganoderic acids by medicinal mushroom Ganoderma lucidum Biotechnol Adv. Available: http://dx.doi.org/10.1016/j.biotechadv.2011.10.001. Accessed 17 October 2011. [DOI] [PubMed]
- 21. Chuang HW, Wang IW, Lin SY, Chang YL (2009) Transcriptome analysis of cadmium response in Ganoderma lucidum . FEMS Microbiol Lett 293: 205–213. [DOI] [PubMed] [Google Scholar]
- 22. Kim S, Song J, Choi HT (2004) Genetic transformation and mutant isolation in Ganoderma lucidum by restriction enzyme-mediated integration. FEMS Microbiol Lett 233: 201–204. [DOI] [PubMed] [Google Scholar]
- 23. Shi L, Qin L, Xu YJ, Ren A, Fang X, Mu DS, Tan Q, Zhao MW (2012) Molecular cloning, characterization, and function analysis of a mevalonate pyrophosphate decarboxylase gene from Ganoderma lucidum . Mol Biol Rep 39: 6149–6159. [DOI] [PubMed] [Google Scholar]
- 24. Shi L, Fang X, Li MJ, Mu DS, Ren A, Tan Q, Zhao MW (2012) Development of a simple and efficient transformation system for the basidiomycetous medicinal fungus Ganoderma lucidum. . World J Microbiol Biotechnol 28: 283–291. [DOI] [PubMed] [Google Scholar]
- 25. Kilaru S, Collins CM, Hartley AJ, Bailey AM, Foster GD (2009) Establishing molecular tools for genetic manipulation of the pleuromutilin-producing fungus Clitopilus passeckerianus . Appl Environ Microbiol 75: 7196–7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Staab JF, Sundstrom P (2003) URA3 as a selectable marker for disruption and virulence assessment of Candida albicans genes. Trends Microbiol 11: 69–73. [DOI] [PubMed] [Google Scholar]
- 27. Carneiro JS, de la Bastide PY, Chabot M, Lerch L, Hintz WE (2010) Suppression of polygalacturonase gene expression in the phytopathogenic fungus Ophiostoma novo-ulmi by RNA interference. Fungal Genet Biol 47: 399–405. [DOI] [PubMed] [Google Scholar]
- 28. Nakade K, Watanabe H, Sakamoto Y, Sato T (2011) Gene silencing of the Lentinula edodes lcc1 gene by expression of a homologous inverted repeat sequence. Microbiol Res 166: 484–493. [DOI] [PubMed] [Google Scholar]
- 29. Cogoni C, Macino G (1997) Isolation of quelling-defective (qde) mutants impaired in posttranscriptional transgene-induced gene silencing in Neurospora crassa . Proc Natl Acad Sci U S A 94: 10233–10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cottrell TR, Doering TL (2003) Silence of the strands: RNA interference in eukaryotic pathogens. Trends Microbiol 11: 37–43. [DOI] [PubMed] [Google Scholar]
- 31. Hutvagner G, Zamore PD (2002) RNAi: nature abhors a double-strand. Curr Opin Genet Dev 12: 225–232. [DOI] [PubMed] [Google Scholar]
- 32. Nakayashiki H, Nguyen QB (2008) RNA interference: roles in fungal biology. Curr Opin Microbiol 11: 494–502. [DOI] [PubMed] [Google Scholar]
- 33. Kemppainen MJ, Pardo AG (2010) pHg/pSILBAgamma vector system for efficient gene silencing in homobasidiomycetes: optimization of ihpRNA - triggering in the mycorrhizal fungus Laccaria bicolor . Microb Biotechnol 3: 178–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. de Jong JF, Deelstra HJ, Wosten HA, Lugones LG (2006) RNA-mediated gene silencing in monokaryons and dikaryons of Schizophyllum commune . Appl Environ Microbiol 72: 1267–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Namekawa SH, Iwabata K, Sugawara H, Hamada FN, Koshiyama A, et al. (2005) Knockdown of LIM15/DMC1 in the mushroom Coprinus cinereus by double-stranded RNA-mediated gene silencing. Microbiology 151: 3669–3678. [DOI] [PubMed] [Google Scholar]
- 36. Kemppainen M, Duplessis S, Martin F, Pardo AG (2009) RNA silencing in the model mycorrhizal fungus Laccaria bicolor: gene knock-down of nitrate reductase results in inhibition of symbiosis with Populus. Environ Microbiol 11: 1878–1896. [DOI] [PubMed] [Google Scholar]
- 37. Costa AS, Thomas DJ, Eastwood D, Cutler SB, Bailey AM, et al. (2009) Quantifiable downregulation of endogenous genes in Agaricus bisporus mediated by expression of RNA hairpins. J Microbiol Biotechnol 19: 271–276. [PubMed] [Google Scholar]
- 38. Smith NA, Singh SP, Wang MB, Stoutjesdijk PA, Green AG, et al. (2000) Total silencing by intron-spliced hairpin RNAs. Nature 407: 319–320. [DOI] [PubMed] [Google Scholar]
- 39. Sun L, Cai HQ, Xu WH, Hu YL, Gao Y, et al. (2001) Efficient transformaion of the medicinal Mushroom Ganoderma lucidum . Plant Mol Biol Rep 19: 383a–383j. [Google Scholar]
- 40. Harrison BR, Yazgan O, Krebs JE (2009) Life without RNAi: noncoding RNAs and their functions in Saccharomyces cerevisiae . Biochemistry and Cell Biology-Biochimie Et Biologie Cellulaire 87: 767–779. [DOI] [PubMed] [Google Scholar]
- 41. Jinek M, Doudna JA (2009) A three-dimensional view of the molecular machinery of RNA interference. Nature 457: 405–412. [DOI] [PubMed] [Google Scholar]
- 42. Meister G, Tuschl T (2004) Mechanisms of gene silencing by double-stranded RNA. Nature 431: 343–349. [DOI] [PubMed] [Google Scholar]
- 43. Nguyen QB, Kadotani N, Kasahara S, Tosa Y, Mayama S, et al. (2008) Systematic functional analysis of calcium-signalling proteins in the genome of the rice-blast fungus, Magnaporthe oryzae, using a high-throughput RNA-silencing system. Mol Microbiol 68: 1348–1365. [DOI] [PubMed] [Google Scholar]
- 44. Bose I, Doering TL (2011) Efficient implementation of RNA interference in the pathogenic yeast Cryptococcus neoformans . J Microbiol Methods 86: 156–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Janus D, Hoff B, Hofmann E, Kuck U (2007) An efficient fungal RNA-silencing system using the DsRed reporter gene. Appl Environ Microbiol 73: 962–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Goldoni M, Azzalin G, Macino G, Cogoni C (2004) Efficient gene silencing by expression of double stranded RNA in Neurospora crassa . Fungal Genet Biol 41: 1016–1024. [DOI] [PubMed] [Google Scholar]
- 47. Guo M, Chen Y, Du Y, Dong Y, Guo W, et al. (2011) The bZIP transcription factor MoAP1 mediates the oxidative stress response and is critical for pathogenicity of the rice blast fungus Magnaporthe oryzae . PLoS Pathog 7: e1001302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Salame TM, Ziv C, Hadar Y, Yarden O (2011) RNAi as a potential tool for biotechnological applications in fungi. Appl Microbiol Biotechnol 89: 501–512. [DOI] [PubMed] [Google Scholar]
- 49. Nakayashiki H (2005) RNA silencing in fungi: mechanisms and applications. FEBS Lett 579: 5950–5957. [DOI] [PubMed] [Google Scholar]
- 50. Keon JP, Owen JW, Hargreaves JA (1999) Lack of evidence for antisense suppression in the fungal plant pathogen Ustilago maydis . Antisense Nucleic Acid Drug Dev 9: 101–104. [DOI] [PubMed] [Google Scholar]
- 51. Wang S, He J, Cui Z, Li S (2007) Self-formed adaptor PCR: a simple and efficient method for chromosome walking. Appl Environ Microbiol 73: 5048–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ah-Fong AM, Bormann-Chung CA, Judelson HS (2008) Optimization of transgene-mediated silencing in Phytophthora infestans and its association with small-interfering RNAs. Fungal Genet Biol 45: 1197–1205. [DOI] [PubMed] [Google Scholar]
- 53. Kuo CY, Huang CT (2008) A reliable transformation method and heterologous expression of beta-glucuronidase in Lentinula edodes . J Microbiol Methods 72: 111–115. [DOI] [PubMed] [Google Scholar]
- 54. Chen J, Zheng W, Zheng S, Zhang D, Sang W, et al. (2008) Rac1 is required for pathogenicity and Chm1-dependent conidiogenesis in rice fungal pathogen Magnaporthe grisea . PLoS Pathog 4: e1000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 56. Geer LY, Marchler-Bauer A, Geer RC, Han L, He J, et al. (2010) The NCBI BioSystems database. Nucleic Acids Res 38: D492–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, et al. (2006) Pfam: clans, web tools and services. Nucleic Acids Res 34: D247–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Protein domains analysis of G. lucidum DCL-1 homologue. This figure shows the 4 domains that characterize the Dicer-2 proteins that were present in the G. lucidum DCL-1 homologue fragment. The domains were identified using the NCBI Conserved Domain Database. The domains in the 1544 amino acid fragment were: DEXDc (DEAD Like), HELIC_c (helicase domain), dsRNA binding, and the 2 RIBOc domains.
(PDF)
DNA and Amino acid sequence GLDCL-1. The partial DNA and derived amino acid sequence of the gldcl-1 gene. coding regions and amino acids are given in upper case letters. The DEAD domain is shadowed in blue-green, the helicase domain is shadowed in purple, the dsRNA binding domain is shadowed in green and the RNAse 3 domain is shadowed in red and gray.
(PDF)
The predicted amino acid sequence of G. lucidum DCL-1 and DCL-1 homologues from other fungi. In the alignment, blue shading to red shading indicates BAD identity to GOOD identity. Important domains are highlighted in colored boxes. The DEAD domain, the helicase domain, dsRNA binding domain and the two RNAse III domains are highlighted in blue-green, purple, green, red and gray boxes, respectively. blue-green, purple, green, red and gray.
(PDF)