Abstract
Shiga toxin-producing Escherichia coli (STEC) serotypes, particularly E. coli O157:H7, possess a variety of fimbrial and afimbrial adhesins which have emerged as important contributors to intestinal colonization. E. coli O157:H7 possesses two chromosomal operons encoding long polar fimbriae (Lpf), which have been found to influence adherence in vitro and colonization in vivo. In a recent Infection and Immunity paper, we further explored the role of Lpf in E. coli O157:H7 intestinal colonization by using the infant rabbit model of STEC infection. We found that an E. coli O157:H7 Lpf-deficient mutant was outcompeted in the rabbit intestine by its parental strain, which may suggest that Lpf contributes to colonization. In contrast, the Lpf-deficient mutant showed an increased adherence to cultured intestinal epithelial cells, and we discovered that this strain overexpressed curli fibers. In this addendum article, we provide a continued perspective on the predicted roles of Lpf and curli, both in vivo and in vitro.
Keywords: long polar fimbriae, curli, E. coli O157:H7, enterohemorrhagic E. coli, fimbria, colonization
Food-borne pathogens are able to survive in a diverse range of environmental niches, including those encountered in the mammalian intestine or on the surfaces of different vegetables. The ability to adhere is a vital first step in the successful colonization of these environments. Thus, organisms have acquired an array of fimbrial and afimbrial adhesins that mediate attachment to biotic and/or abiotic surfaces. A determination of the factors that are important in adhesion and the conditions under which these factors operate and interact will enable us to develop more effective approaches to prevent bacterial attachment to surfaces, and thereby reduce the chance of human infection.
Shiga toxin-producing E. coli (STEC) O157:H7 is an important cause of human gastrointestinal disease and the best-studied STEC serotype associated with large outbreaks worldwide. E. coli O157:H7 strains are common in the intestines of livestock and can be transmitted to humans following the consumption of fecal-contaminated meat, fruits and vegetables.1 Like many food-borne pathogens, E. coli O157:H7 contains a multitude of putative adhesion factors (reviewed in ref. 2). However, the outer membrane protein, intimin, and the long polar fimbriae (Lpf) are the only two E. coli O157:H7 adhesins that have been demonstrated to play a role in the colonization of human intestinal epithelial cells.3,4 While the role for intimin in E. coli O157:H7 attachment to human intestinal explants is well-established,4,5 the role of Lpf is less clear.
E. coli O157:H7 possesses two lpf operons, lpf1 and lpf2, both of which contain genes closely related to the long polar fimbriae of Salmonella enterica serovar Typhimurium.6 Expression of lpf1 and lpf2 is induced during the late exponential-phase growth in tissue culture media at pH 6.5 and 37°C7,8 or under iron restricted conditions,9 and has been found to influence E. coli O157:H7 adherence to cultured epithelial monolayers.10,11 We have shown that a mutation of the lpf1 operon results in a significant reduction in E. coli O157:H7 adherence to, as well as a reduction in microcolony formation on, cultured epithelial cells.7 Similarly, an lpf2 mutation showed partial reduction in adherence, and a possible role for Lpf2 in early adherence to intestinal cells has been suggested.9
Studies in a variety of animal models have supported the role of Lpf in intestinal colonization and persistence. For example, a report on in vivo colonization assays using samples from sheep and pigs demonstrated that the absence of both lpf1 and lpf2 loci impair E. coli O157:H7 colonization.11 Yet in contrast to the in vitro study results, the single lpf1 mutant was found to have performed as well as the parent strain in colonizing the intestines of sheep and pigs.11 Contrasting results were obtained from a different set of experiments that used a collection of lpf mutants constructed in a Shiga toxin-negative E. coli O157:H7 strain and were conducted in 6-week-old cross-bred lambs. In this case, the recovery of both the lpf1 mutant and the double lpf1 lpf2 mutant in fecal material was significantly attenuated when compared with the findings in the wild type.10 Further complicating matters are findings from studies using tissue explants. For example, Lpf did not influence the ability of E. coli O157:H7 to adhere to intestinal explants obtained from lambs.10 Similar studies in human tissues showed that E. coli O157:H7 strains with lpf mutations colonized intestinal regions that were not normally bound by a wild-type strain.3 Taken all together, all of these studies indicate environmental conditions may dictate which mechanisms control E. coli O157:H7 adherence and that there is redundancy between some of these systems examined.
Given the relatively subtle effects of lpf mutations on adherence in vitro, coupled with the divergent findings from in vivo or organ culture experiments, the precise role of Lpf in E. coli O157:H7 adherence remains somewhat unclear. Therefore, in our recent study published in Infection and Immunity,12 the role of the E. coli O157:H7 lpf loci was further tested in an infant rabbit model which mimics the diarrhea and gut pathology, including the histopathological attaching and effacing lesions, seen in patients with STEC infection.13 We assessed the importance of Lpf for intestinal colonization by performing competition experiments between E. coli O157:H7 and an isogenic lpf1 lpf2 double mutant and found that the mutant was outcompeted in the ileum, cecum, and mid-colon of rabbits, confirming that Lpf contributes to intestinal colonization.12 Unexpectedly, we observed that the lpf1 lpf2 double mutant showed an increased adherence to colonic epithelial cells in vitro, and transmission electron microscopy revealed curli-like structures on the surface of this mutant, as confirmed by immunoblotting and Congo red binding assays, immunogold-labeling electron microscopy, and real-time RT-PCR measuring csgA (encoding the major curli subunit) expression. Interestingly, deletion of csgA per se did not appear to affect intestinal colonization. Therefore, in addition to conclusively demonstrating that Lpf contribute to E. coli O157:H7 intestinal colonization, our observations indicated that the regulatory mechanisms controlling expression of Lpf and curli are interconnected.
Curli fibers are commonly thought to be involved in the colonization of abiotic surfaces and the development of biofilms (reviewed in ref. 14). Curli are thin (2 to 5 nm wide), coiled fibers of varying lengths that self-assemble outside the cell and aggregate to form an amorphous matrix.15–18 Curli fibers bind Congo red dye and certain host proteins including fibronectin, laminin, and plasminogen.19,20 The production of curli polymers is environmentally regulated and RpoS-dependent, which means that the transcription of csgA is under the control of an environmental program that responds positively to low temperature, low osmolarity, and stationary-phase growth conditions.21,22
The role of curli in the pathogenesis associated with different diarrheagenic E. coli pathogroups is not clear. For example, in a collection of 49 bovine and human E. coli O157:H7 strains, only two human isolates were found to produce curli and exhibit Congo red binding under the conditions tested.23,24 It has been proposed that natural E. coli isolates can be either “on” or “off” for the program controlling curli expression and that different growth conditions may select for these variants in both commensal and pathogenic E. coli.21,25,16 Therefore, the low percentage of curli-positive strains found in the study above may mean that curli production in the human intestine is not common.
The results of several studies have led to a suggestion that curli play an important role in mediating attachment to surfaces other than those found in the intestine. For example, expression of the E. coli O157:H7 csg operon in laboratory E. coli strains increased the ability of these strains to bind to alfalfa sprouts and seed coats.26 Furthermore, transcriptional analyses revealed that genes involved in curli production were significantly upregulated during E. coli O157:H7 attachment to lettuce leaves.27 Finally, curli-expressing E. coli O157:H7 strains appeared to develop stronger associations bound in higher numbers to the surface of spinach leaves than did isogenic curli-deficient mutants.28 Examination of the inoculated leaves revealed that curli-expressing E. coli O157:H7 were embedded in extracellular material that immunostained with anti-curli antibodies, possibly meaning that these structures may offer protection against the harsh desiccating environment found on the surface of leaves.28
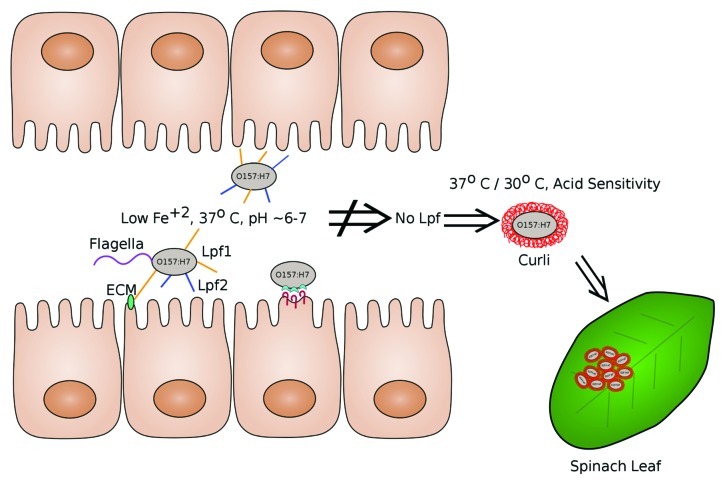
The prevailing dogma is that curli expression in most E. coli strains is inhibited at mammalian body temperatures (i.e., 37°C) (reviewed in ref. 14), conditions which instead favor the transcription of most classes of fimbriae genes (reviewed in refs. 29 and 2). Our findings lead us to suggest that the regulation of these two adherence mechanisms may be linked. In our model, we propose that conditions in the intestine (37°C, pH 6–7, and/or low iron) favor the expression of Lpf, which in turn mediates bacterial colonization (see Fig. 1). In the absence of Lpf1 and Lpf2, the bacterium expresses curli, thus providing the organism with an alternative adhesion mechanism. Yet curli is known to be expressed better in low-salt and at 30°C,30 and to mediate adhesion to surfaces other than the intestine.26 Thus, deletion of csgA does not further attenuate E. coli O157:H7 intestinal colonization. It is clear that E. coli O157:H7 has evolved multiple adherence mechanisms to maximize bacterial survival fitness in different environments.
Figure 1.
Model of intestinal expression of Lpf and environmental expression of curli. Long polar fimbriae (Lpf1 in yellow and Lpf2 in blue) are induced in conditions that mimic the intestine (pH 6.5, 37°C and low iron) and are proposed to participate in early attachment to intestinal cells and binding to extracellular matrix proteins (ECM). Intimin (light blue) and Tir (dark red), along with other Type III secreted effector proteins mediate the pedestal formation and the intimate attachment (attaching and effacing lesion, A/E) characteristic of E. coli O157:H7 and other A/E-producing bacteria. Curli (bright red) is expressed in the absence of Lpf at 30°C and 37°C, and curliated variants of E. coli O157:H7 are known to display acid sensitivity. Curli was shown not to contribute to intestinal colonization12 and thus is thought to play a role in attachment to plant surfaces, such as in the case of spinach leaves, or other environmental substrates.
In other pathogenic scenarios, curli has been implicated in the pathogenesis of sepsis and uropathogenic E. coli (UPEC) infections.31,32 However, in vivo curli production or an in vivo role for curliated E. coli O157:H7 has not yet been demonstrated, although some strains of E. coli O157:H7 do produce curli at 37°C and have been shown to adhere to or invade cultured cells or bovine intestinal explants.33,34 Thus, we can only speculate that curli may act as an alternative adhesin in vivo in the Lpf-deficient mutant. Instead, curliated strains of E. coli O157:H7 exhibit characteristics that may provide a better fitness advantage in soils, sediments and on plant tissues.35 A recent report indicates environmental stages of the life history of E. coli are more important than previously thought and that the mammalian intestine may not be the primary or even preferred environment for some E. coli strains.36,37 For example, White et al. found curli production was more prevalent in host-generalist strains of E. coli (strains that are likely to be better adapted to the environment) than in predicted host-adapted strains.36 In addition, Carter et al. found that curli-producing variants of E. coli O157:H7 were more sensitive to acid than were curli-negative variants, while the curli-negative variants were less fit in nutrient-limited conditions.35 These results support the idea that curliated E. coli O157:H7 variants are more fit in conditions that mimic the environment, while non-curliated E. coli O157:H7 are better adapted to the host.
Heterogeneity of adhesin expression within a genetically homogeneous population during infection is thought to be important for niche colonization and immune evasion.38 E. coli O157:H7 strains grown in vitro show heterogeneity of curli production, and subpopulations of curliated E. coli O157:H7 may also be present during host colonization or in the environment. In the study by Carter et al., the curli-producing phenotype was found to be stable at the population level in vitro and in all conditions tested, with infrequent conversion to a non-curliated phenotype.35 The Lpf-deficient mutant described in our paper also exhibited stable curli production. At both 30°C and 37°C on LB plates containing Congo red and no-salt, following 48 h incubation, the majority of the Lpf-deficient colonies produced curli, while the wild-type strain showed a heterogeneous population with a majority of white colonies (data not shown). There is some evidence that both lpf1 and lpf2 are capable of phase variation,39 so it may be that the wild-type colonies that produce curli are in the “off” phase for Lpf production. An example of such mutually exclusive expression has been demonstrated in UPEC in the regulation of Ag43 and type 1 fimbriae.40 Ag43 (flu) mRNA levels increased in the absence of type 1 fimbriae (fim) and decreased when type 1 fimbriae were overexpressed.40 A similar relationship in E. coli O157:H7 in which curli production is dependent on the absence of Lpf would provide strong support for our model. A possible mechanism for interdependent regulation of Lpf and curli may be through the differential action of transcriptional activators that regulate the global repressor, histone-like nucleoid structuring protein (H-NS). In E. coli O157:H7, both curli and Lpf expression is silenced by H-NS.41–44 In order for lpf1 transcription to occur, the E. coli O157:H7 Ler regulator anti-silences H-NS, activating the lpf1 locus as well as inducing the expression of other virulence factors, such as the intimin adhesin.44,8 In contrast, activation of the stress/stationary phase response and RpoS de-represses H-NS, resulting in transcription of the curli operons.21,42 Ler is activated by conditions that mimic the intestine, such as pH, carbon source and temperature (37°C), while RpoS is activated by conditions found outside the host such as low temperature and nutrient limitation.45 However, the highly complex regulatory network involving H-NS, RpoS and Ler may obscure the effects of any one regulator, much as the effect of particular fimbriae may be masked by the compensatory and/or interdependent expression of other fimbriae. Therefore, the Lpf-deficient mutant may serve as a useful tool in determining whether interdependence occurs between Lpf and curli expression.
Finally, E. coli are ubiquitous in the intestines of mammals and other animals, and an environmental role has been recently brought to light in the wake of produce-associated outbreaks of infection by STEC O157:H7 and other STEC serotypes.46 Thus it is not surprising to discover that E. coli possess redundant and/or compensatory mechanisms for colonization both in and out of the intestine. Although we propose that Lpf is preferred over curli in the colonization of the intestine, and the likely primary role for curli is in the environment, we may well discover that, true to the plasticity of E. coli, curli fibers are able to mediate adherence and/or survival in multiple host and environmental niches.
Acknowledgments
The authors are grateful to Katie Johnston for designing the figure and Mardelle Susman for editorial advice. The paper discussed in this addendum was supported by NIH/ NIAID grant AI079154 to A.G.T. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the NIAID, or NIH.
Glossary
- Abbreviations: STEC
Shiga toxin-producing Escherichia coli
- Lpf
long polar fimbriae
- A/E
attaching and effacing
Footnotes
Previously published online: www.landesbioscience.com/journals/gutmicrobes/article/20661
References
- 1.Caprioli A, Morabito S, Brugère H, Oswald E. Enterohaemorrhagic Escherichia coli: emerging issues on virulence and modes of transmission. Vet Res. 2005;36:289–311. doi: 10.1051/vetres:2005002. [DOI] [PubMed] [Google Scholar]
- 2.Farfan MJ, Torres AG. Molecular mechanisms that mediate colonization of Shiga toxin-producing Escherichia coli strains. Infect Immun. 2012;80:903–13. doi: 10.1128/IAI.05907-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fitzhenry R, Dahan S, Torres AG, Chong Y, Heuschkel R, Murch SH, et al. Long polar fimbriae and tissue tropism in Escherichia coli O157:H7. Microbes Infect. 2006;8:1741–9. doi: 10.1016/j.micinf.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 4.Mundy R, Schüller S, Girard F, Fairbrother JM, Phillips AD, Frankel G. Functional studies of intimin in vivo and ex vivo: implications for host specificity and tissue tropism. Microbiology. 2007;153:959–67. doi: 10.1099/mic.0.2006/003467-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fitzhenry RJ, Pickard DJ, Hartland EL, Reece S, Dougan G, Phillips AD, et al. Intimin type influences the site of human intestinal mucosal colonisation by enterohaemorrhagic Escherichia coli O157:H7. Gut. 2002;50:180–5. doi: 10.1136/gut.50.2.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bäumler AJ, Heffron F. Identification and sequence analysis of lpfABCDE, a putative fimbrial operon of Salmonella typhimurium. J Bacteriol. 1995;177:2087–97. doi: 10.1128/jb.177.8.2087-2097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torres AG, Giron JA, Perna NT, Burland V, Blattner FR, Avelino-Flores F, et al. Identification and characterization of lpfABCC’DE, a fimbrial operon of enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 2002;70:5416–27. doi: 10.1128/IAI.70.10.5416-5427.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rojas-López M, Arenas-Hernández MM, Medrano-López A, Martínez de la Peña CF, Puente JL, Martínez-Laguna Y, et al. Regulatory control of the Escherichia coli O157:H7 lpf1 operon by H-NS and Ler. J Bacteriol. 2011;193:1622–32. doi: 10.1128/JB.01082-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torres AG, Kanack KJ, Tutt CB, Popov V, Kaper JB. Characterization of the second long polar (LP) fimbriae of Escherichia coli O157:H7 and distribution of LP fimbriae in other pathogenic E. coli strains. FEMS Microbiol Lett. 2004;238:333–44. doi: 10.1016/j.femsle.2004.07.053. [DOI] [PubMed] [Google Scholar]
- 10.Torres AG, Milflores-Flores L, Garcia-Gallegos JG, Patel SD, Best A, La Ragione RM, et al. Environmental regulation and colonization attributes of the long polar fimbriae (LPF) of Escherichia coli O157:H7. Int J Med Microbiol. 2007;297:177–85. doi: 10.1016/j.ijmm.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 11.Jordan DM, Cornick N, Torres AG, Dean-Nystrom EA, Kaper JB, Moon HW. Long polar fimbriae contribute to colonization by Escherichia coli O157:H7 in vivo. Infect Immun. 2004;72:6168–71. doi: 10.1128/IAI.72.10.6168-6171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lloyd SJ, Ritchie JM, Rojas-Lopez M, Blumentritt CA, Popov VL, Greenwich JL, et al. A double, long polar fimbria mutant of Escherichia coli O157:H7 expresses Curli and exhibits reduced in vivo colonization. Infect Immun. 2012;80:914–20. doi: 10.1128/IAI.05945-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ritchie JM, Thorpe CM, Rogers AB, Waldor MK. Critical roles for stx2, eae, and tir in enterohemorrhagic Escherichia coli-induced diarrhea and intestinal inflammation in infant rabbits. Infect Immun. 2003;71:7129–39. doi: 10.1128/IAI.71.12.7129-7139.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barnhart MM, Chapman MR. Curli biogenesis and function. Annu Rev Microbiol. 2006;60:131–47. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prigent-Combaret C, Prensier G, Le Thi TT, Vidal O, Lejeune P, Dorel C. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: role of flagella, curli and colanic acid. Environ Microbiol. 2000;2:450–64. doi: 10.1046/j.1462-2920.2000.00128.x. [DOI] [PubMed] [Google Scholar]
- 16.Brown PK, Dozois CM, Nickerson CA, Zuppardo A, Terlonge J, Curtiss R., 3rd MlrA, a novel regulator of curli (AgF) and extracellular matrix synthesis by Escherichia coli and Salmonella enterica serovar Typhimurium. Mol Microbiol. 2001;41:349–63. doi: 10.1046/j.1365-2958.2001.02529.x. [DOI] [PubMed] [Google Scholar]
- 17.Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, et al. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295:851–5. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammar M, Bian Z, Normark S. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc Natl Acad Sci U S A. 1996;93:6562–6. doi: 10.1073/pnas.93.13.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olsén A, Jonsson A, Normark S. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature. 1989;338:652–5. doi: 10.1038/338652a0. [DOI] [PubMed] [Google Scholar]
- 20.Olsén A, Wick MJ, Mörgelin M, Björck L. Curli, fibrous surface proteins of Escherichia coli, interact with major histocompatibility complex class I molecules. Infect Immun. 1998;66:944–9. doi: 10.1128/iai.66.3.944-949.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olsén A, Arnqvist A, Hammar M, Sukupolvi S, Normark S. The RpoS sigma factor relieves H-NS-mediated transcriptional repression of csgA, the subunit gene of fibronectin-binding curli in Escherichia coli. Mol Microbiol. 1993;7:523–36. doi: 10.1111/j.1365-2958.1993.tb01143.x. [DOI] [PubMed] [Google Scholar]
- 22.Pratt LA, Silhavy TJ. Crl stimulates RpoS activity during stationary phase. Mol Microbiol. 1998;29:1225–36. doi: 10.1046/j.1365-2958.1998.01007.x. [DOI] [PubMed] [Google Scholar]
- 23.Uhlich GA, Keen JE, Elder RO. Mutations in the csgD promoter associated with variations in curli expression in certain strains of Escherichia coli O157:H7. Appl Environ Microbiol. 2001;67:2367–70. doi: 10.1128/AEM.67.5.2367-2370.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uhlich GA, Keen JE, Elder RO. Variations in the csgD promoter of Escherichia coli O157:H7 associated with increased virulence in mice and increased invasion of HEp-2 cells. Infect Immun. 2002;70:395–9. doi: 10.1128/IAI.70.1.395-399.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Provence DL, Curtiss R., 3rd Role of crl in avian pathogenic Escherichia coli: a knockout mutation of crl does not affect hemagglutination activity, fibronectin binding, or Curli production. Infect Immun. 1992;60:4460–7. doi: 10.1128/iai.60.11.4460-4467.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torres AG, Jeter C, Langley W, Matthysse AG. Differential binding of Escherichia coli O157:H7 to alfalfa, human epithelial cells, and plastic is mediated by a variety of surface structures. Appl Environ Microbiol. 2005;71:8008–15. doi: 10.1128/AEM.71.12.8008-8015.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fink RC, Black EP, Hou Z, Sugawara M, Sadowsky MJ, Diez-Gonzalez F. Transcriptional responses of Escherichia coli K-12 and O157:H7 associated with lettuce leaves. Appl Environ Microbiol. 2012;78:1752–64. doi: 10.1128/AEM.07454-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macarisin D, Patel J, Bauchan G, Giron JA, Sharma VK. Role of curli and cellulose expression in adherence of Escherichia coli O157:H7 to spinach leaves. Foodborne Pathog Dis. 2012;9:160–7. doi: 10.1089/fpd.2011.1020. [DOI] [PubMed] [Google Scholar]
- 29.Torres AG, Zhou X, Kaper JB. Adherence of diarrheagenic Escherichia coli strains to epithelial cells. Infect Immun. 2005;73:18–29. doi: 10.1128/IAI.73.1.18-29.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jubelin G, Vianney A, Beloin C, Ghigo JM, Lazzaroni JC, Lejeune P, et al. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J Bacteriol. 2005;187:2038–49. doi: 10.1128/JB.187.6.2038-2049.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, et al. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295:851–5. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bian Z, Brauner A, Li Y, Normark S. Expression of and cytokine activation by Escherichia coli curli fibers in human sepsis. J Infect Dis. 2000;181:602–12. doi: 10.1086/315233. [DOI] [PubMed] [Google Scholar]
- 33.Saldaña Z, Xicohtencatl-Cortes J, Avelino F, Phillips AD, Kaper JB, Puente JL, et al. Synergistic role of curli and cellulose in cell adherence and biofilm formation of attaching and effacing Escherichia coli and identification of Fis as a negative regulator of curli. Environ Microbiol. 2009;11:992–1006. doi: 10.1111/j.1462-2920.2008.01824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uhlich GA, Gunther NW, 4th, Bayles DO, Mosier DA. The CsgA and Lpp proteins of an Escherichia coli O157:H7 strain affect HEp-2 cell invasion, motility, and biofilm formation. Infect Immun. 2009;77:1543–52. doi: 10.1128/IAI.00949-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carter MQ, Brandl MT, Louie JW, Kyle JL, Carychao DK, Cooley MB, et al. Distinct acid resistance and survival fitness displayed by Curli variants of enterohemorrhagic Escherichia coli O157:H7. Appl Environ Microbiol. 2011;77:3685–95. doi: 10.1128/AEM.02315-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White AP, Sibley KA, Sibley CD, Wasmuth JD, Schaefer R, Surette MG, et al. Intergenic sequence comparison of Escherichia coli isolates reveals lifestyle adaptations but not host specificity. Appl Environ Microbiol. 2011;77:7620–32. doi: 10.1128/AEM.05909-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo C, Walk ST, Gordon DM, Feldgarden M, Tiedje JM, Konstantinidis KT. Genome sequencing of environmental Escherichia coli expands understanding of the ecology and speciation of the model bacterial species. Proc Natl Acad Sci U S A. 2011;108:7200–5. doi: 10.1073/pnas.1015622108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holden NJ, Gally DL. Switches, cross-talk and memory in Escherichia coli adherence. J Med Microbiol. 2004;53:585–93. doi: 10.1099/jmm.0.05491-0. [DOI] [PubMed] [Google Scholar]
- 39.Low AS, Holden N, Rosser T, Roe AJ, Constantinidou C, Hobman JL, et al. Analysis of fimbrial gene clusters and their expression in enterohaemorrhagic Escherichia coli O157:H7. Environ Microbiol. 2006;8:1033–47. doi: 10.1111/j.1462-2920.2006.00995.x. [DOI] [PubMed] [Google Scholar]
- 40.Schembri MA, Klemm P. Coordinate gene regulation by fimbriae-induced signal transduction. EMBO J. 2001;20:3074–81. doi: 10.1093/emboj/20.12.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ogasawara H, Yamada K, Kori A, Yamamoto K, Ishihama A. Regulation of the Escherichia coli csgD promoter: interplay between five transcription factors. Microbiology. 2010;156:2470–83. doi: 10.1099/mic.0.039131-0. [DOI] [PubMed] [Google Scholar]
- 42.Lee JH, Kim YG, Cho MH, Wood TK, Lee J. Transcriptomic analysis for genetic mechanisms of the factors related to biofilm formation in Escherichia coli O157:H7. Curr Microbiol. 2011;62:1321–30. doi: 10.1007/s00284-010-9862-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Torres AG, López-Sánchez GN, Milflores-Flores L, Patel SD, Rojas-López M, Martínez de la Peña CF, et al. Ler and H-NS, regulators controlling expression of the long polar fimbriae of Escherichia coli O157:H7. J Bacteriol. 2007;189:5916–28. doi: 10.1128/JB.00245-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Torres AG, Slater TM, Patel SD, Popov VL, Arenas-Hernández MMP. Contribution of the Ler- and H-NS-regulated long polar fimbriae of Escherichia coli O157:H7 during binding to tissue-cultured cells. Infect Immun. 2008;76:5062–71. doi: 10.1128/IAI.00654-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dong T, Schellhorn HE. Global effect of RpoS on gene expression in pathogenic Escherichia coli O157:H7 strain EDL933. BMC Genomics. 2009;10:349. doi: 10.1186/1471-2164-10-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eppinger M, Mammel MK, Leclerc JE, Ravel J, Cebula TA. Genomic anatomy of Escherichia coli O157:H7 outbreaks. Proc Natl Acad Sci U S A. 2011;108:20142–7. doi: 10.1073/pnas.1107176108. [DOI] [PMC free article] [PubMed] [Google Scholar]