Abstract
Two Cryptosporidium isolates from separate infants suffering from diarrhea were obtained from a hospital in Zhengzhou, China and were genotyped by PCR amplification and sequence analysis of the small-subunit ribosomal RNA (rRNA) (SSU rRNA), 70-kDa heat shock protein (HSP70), and actin genes. Further subtyping was performed by PCR amplification and sequence analysis of the 60-kDa glycoprotein (gp60) gene. Both the isolates were identified as Cryptosporidium hominis subtype IdA21, a rare subtype previously found only in a human immunodeficiency virus-infected child in South Africa and another child in Jordan.
Introduction
Cryptosporidium, a pathogenic protozoan parasite with a worldwide distribution, causes diarrhea in humans and animals. The parasite can be transmitted from person to person through fecal-oral contact (household contact and nosocomial transmission), ingestion of contaminated food or water, and contact with infected animals. In immunocompetent hosts, the infection is typically acute and self-limiting, whereas in immunocompromised individuals, such as persons taking immunosuppressive drugs and AIDS patients, the infection is often chronic [1].
Oocysts of Cryptosporidium species that are infectious to humans share similar morphology to those that are not, which prevents differentiation by light microscopy. Therefore, prevalence surveys based on oocyst morphology fail to determine the public health contribution of animals and environment to this human disease. Recent molecular epidemiologic studies of cryptosporidiosis have helped researchers gain a better understanding of human cryptosporidiosis transmission and the public health significance of Cryptosporidium spp. found in animals and the environment. With the use of genotyping tools, at least 23 valid species and numerous genotypes of Cryptosporidium have been described; eleven Cryptosporidium species (C. hominis, C. parvum, C. meleagridis, C. felis, C. canis, C. ubiquitum, C. suis, C. muris, C. andersoni, C. cuniculus, and C. fayeri) and at least five genotypes (skunk genotype, chipmunk genotype I, horse genotype, monkey genotype, and pig genotype II) can infect humans, with C. hominis and C. parvum being the most common clinical isolates [1]–[6]. By employing highly discriminatory subtyping techniques (generally sequence analysis of the gp60 gene), researchers have been able to track the infection source and the transmission dynamics of C. hominis and C. parvum [7].
Since the first report of human cryptosporidiosis in 1987 in Nanjing by Han et al., Xuzhou, Wuhu, and at least 17 other Chinese provinces or cities have reported this disease in their populations, with an infection rate ranging from 0.917% to 9.700% [8]–[10]. However, most of these studies were conducted using microscopy, and there is little available data on the molecular epidemiology of human cryptosporidiosis in China. Cryptosporidium hominis has been found to be the dominant infectious species, and three subtype families termed Ia, Ib, and Id have been identified. In contrast, only a few clinical isolates have been identified as C. meleagridis, C. canis, and C. felis [11]–[13]. To further characterize Cryptosporidium spp. in human clinical isolates in China, we performed a genotyping and subtyping study of two isolates of C. hominis by means of multilocus sequence typing.
Materials and Methods
Cryptosporidium Samples and DNA Extraction
Fecal samples were collected from clinical patients (including infants and those suffering from cancer) attending four hospitals in Zhengzhou, China. The samples were collected with the verbal consent of patients or their guardians. Since feces testing is a standard procedure in Chinese investigating hospitals, all patients agreed to sign the informed consent form. These completed forms were submitted to the ethics committee to examine and verify, in order to ensure the protection of the rights and benefits of patients. The study and this procedure were approved by the Research Ethics Committee of Henan Agricultural University. The stool samples of two infants less than one year old with a one week history of watery diarrhea were taken. The infants were diagnosed as Cryptosporidium-positive by bright field microscopy; the oocysts were concentrated by the Sheather’s sugar flotation technique and stored in a 2.5% potassium dichromate solution at 4°C until needed. Genomic DNA was isolated as previously described [14].
Molecular Analyses
PCR amplification of the SSU rRNA, HSP70, actin and gp60 genes was performed as done in previous studies [1], [15]. RFLP analysis was performed on SSU rRNA PCR products; each sample was digested with SspI (Fermentas, USA) and VspI (Fermentas, USA), which produced a restriction pattern specific to Cryptosporidium. The PCR products of the SSU rRNA, HSP70 and gp60 genes, as well as the positive clones of actin gene, were sequenced by TaKaRa Biotechnology Co. Ltd (Dalian, China) on an ABI PRISMTM 3730 XL DNA Analyzer (Applied Biosystems, USA), using the Big Dye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA).
Phylogenetic trees were constructed using the software PHYLIP (version 3.69). Neighbor-joining trees were constructed on the basis of evolutionary distances calculated using the Kimura two-parameter model. Sequence identity analysis was performed using the MegAlign program in the DNAStar software package.
The partial nucleotide sequences of the SSU rRNA, HSP70, actin and gp60 genes obtained in this study were deposited in the GenBank database under accession numbers EF570921 to EF570922 and EF591783 to EF591788.
Results
DNA nucleotides of 837, 1951 and 1068 base pairs (bp) were obtained by PCR for the SSU rRNA, HSP70 and actin genes, respectively, from the two human isolates (CHZF1 and CHZF2). RFLP analysis of the SSU rRNA gene product with SspI and VspI showed restriction patterns identical to C. hominis [16]. The two Cryptosporidium isolates did not show any sequence differences in each gene. In the SSU rRNA gene, the sequence obtained was identical to AF093489 (from a USA isolate), AF108865 (from an Austria isolate), AJ849464 (from a Slovenia isolate), and DQ286403 (from a Chile isolate). However, it had 1–6 nucleotide differences with other C. hominis isolates (DQ523506, AF093492, AJ849462, EU331242 and AF093491), and the sequence identity varied from 99.3% to 99.9%. Similarly, in the HSP70 gene, three nucleotide changes (two C to T at nucleotide 4 and 1900, and a T to A at nucleotide 1903) and one nucleotide change (a C to T change at nucleotide 1912) were respectively observed for two different C. hominis isolates (AF401506 and AF401504). The nucleotide similarities with these two isolates were 99.8% and 99.9%, respectively. Interestingly, however, no nucleotide change was found in the actin gene when compared to other C. hominis isolates.
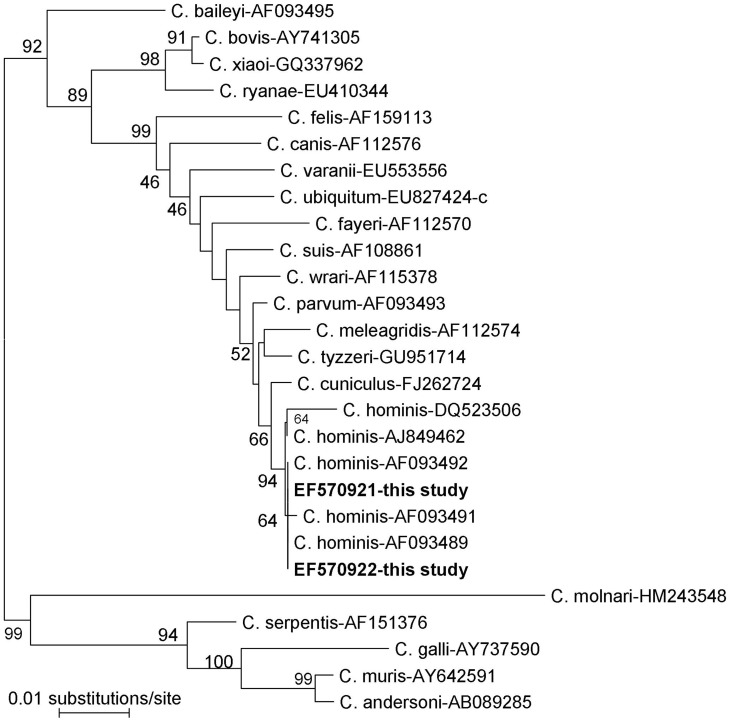
Neighbor-joining trees were constructed from the aligned partial SSU rRNA, HSP70 and actin sequences of these two C. hominis isolates, as well as those downloaded from the GenBank database. In the actin locus, the two isolates formed a cluster with C. hominis, and this was supported by bootstrap analysis. However, in the SSU rRNA and HSP70 genes, the two isolates clustered together and formed a sister clade with C. hominis (a neighbor-joining tree of the SSU rRNA gene is shown in Figure 1).
Figure 1. Phylogenetic relationship of Cryptosporidium parasites inferred by neighbor-joining analysis of the SSU rRNA based on evolutionary distances calculated using the Kimura two-parameter model.
Bootstrap values were obtained using 1,000 pseudoreplicates.
The 533 bp nucleotide sequence of the gp60 gene was amplified by nested PCR, and the two isolates yielded the same sequence and the subtype identity as established by sequence alignment and phylogenetic analysis: both isolates belonged to the subtype IdA21. In comparison with the previous reported Chinese C. hominis isolate IdA14 (AF403169), seven more TCA repeats was found, with the exception of a T to C change at nucleotide 11, and the isolates shared a nucleotide similarity of 95.6%.
Discussion
SSU rRNA, HSP70, and actin genes are common molecular markers for genotyping and have been very valuable when used in conjunction with morphological, biological, or host specificity studies [1]. In this study, although there were no nucleotide changes found between the two isolates and other C. hominis isolates at the actin gene, sequence diversity was observed in the SSU rRNA and HSP70 genes when compared to other C. hominis isolates. Five copies of the SSU rRNA gene are present in the Cryptosporidium genome, and previous studies have suggested that there is slight sequence heterogeneity in some of these copies [17]. Therefore, some of the sequence differences might be attributed to the intragenomic variation of the multiple rRNA loci. However, the sequence differences found in the HSP70 gene between isolates could be due to a single nucleotide polymorphism (SNP).
In the present study, two Cryptosporidium-positive isolates were both found to belong to C. hominis based on sequence analyses of three genes. Previous studies suggest C. parvum and C. hominis are responsible for greater than 90% of human cases of cryptosporidiosis in most areas; however, the contribution of each species varies among different geographic regions [7]. In the United Kingdom, other parts of Europe, and New Zealand, C. parvum is responsible for slightly more infections than C. hominis. In contrast, C. hominis is responsible for far more infections than C. parvum in the United States, Australia, and Japan, as well as developing countries where genotyping studies have been conducted. Major differences in the transmission routes may be responsible for these observed differences in the distribution of Cryptosporidium species [7], [18]. In China, to the best of our knowledge, more than 100 Cryptosporidium isolates have been genotyped, and most of them were identified as C. hominis [11]–[13]. Thus, the predominant infectious C. hominis found in humans indicated human-to-human transmissions of Cryptosporidium spp. might play an important role in China. However, since exposure information was not gathered, it is difficult to determine the infection sources.
Ia, Ib and Id are the three common C. hominis subtype families, and each has been found in humans in many areas. Nevertheless, there are geographic differences in the distribution of subtype families. For example, Ib was a predominant C. hominis subtype family in humans in C. parvum-endemic areas such as Portugal, United Kingdom, and Australia [18], [19]. Both Ib and Id were common in developing countries such as Malawi, South Africa, India and Peru [18], [20], [21]. In New Orleans, Ib and Ia were both common, while Id subtypes were absent [18]. In the present study, the two C. hominis isolates were identified as IdA21, a relatively rare subtype that has only been detected in South Africa and Jordan [22], [23]. Id is one of the predominant subtype families found in China: Ia (35/95), Id (40/95) and Ib (7/95) have been identified in humans in Shanghai and Zhengzhou [12], [13]. However, due to the absence of genetic data of clinical C. hominis isolates in other areas, the population genetic structure of C. hominis subtypes is still unclear.
Further dissection of the infection sources and transmission dynamics of human cryptosporidiosis in China remains an important area of future investigation.
Funding Statement
This study was supported in part by funds for the Key National Science and Technology Specific Projects (no. 2012ZX10004220-001,http://www.nmp.gov.cn/zxjs/crb/201012/t20101208_2127.htm), the National Natural Science Foundation of China (no. 31172311, http://www.nsfc.gov.cn/Portal0/default152.htm), and the Program for Innovative Research Team (in Science and Technology) in University of Henan Province (012IRTSTHN005, http://est.haedu.gov.cn/Article/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Xiao L, Fayer R, Ryan U, Upton SJ (2004) Cryptosporidium taxonomy: recent advances and implications for public health. Clin Microbiol Rev 17: 72–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Leoni F, Amar C, Nichols G, Pedraza-Diaz S, McLauchlin J (2006) Genetic analysis of Cryptosporidium from 2,414 humans with diarrhoea in England between 1985 and 2000. J Med Microbiol 55: 703–707. [DOI] [PubMed] [Google Scholar]
- 3. Feltus DC, Giddings CW, Schneck BL, Monson T, Warshauer D, et al. (2006) Evidence supporting zoonotic transmission of Cryptosporidium spp. in Wisconsin. J Clin Microbiol 44: 4303–4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Morse TD, Nichols RA, Grimason AM, Campbell BM, Tembo KC, et al. (2007) Incidence of cryptosporidiosis species in paediatric patients in Malawi. Epidemiol Infect 135: 1307–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Robinson G, Elwin K, Chalmers RM (2008) Unusual Cryptosporidium genotypes in human cases of diarrhea. Emerg Infect Dis 14: 1800–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ren X, Zhao J, Zhang L, Ning C, Jian F, et al. (2012) Cryptosporidium tyzzeri n. sp. (Apicomplexa: Cryptosporidiidae) in domestic mice (Mus musculus). Exp Parasitol 130: 274–281. [DOI] [PubMed] [Google Scholar]
- 7. Xiao L (2010) Molecular epidemiology of cryptosporidiosis: An update. Exp Parasitol 124: 80–89. [DOI] [PubMed] [Google Scholar]
- 8. Chen YG, Yao FB, Li HS, Shi WS, Dai MX (1992) Cryptosporidium infection and diarrhea in rural and urban areas of Jiangsu, People’s Republic of China. J Clin Microbiol 30: 492–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang KX, Li CP, Wang J, Pan BR (2002) Epidemiological survey of cryptosporidiosis in Anhui Province China. World J Gastroenterol 8: 371–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen XW (2004) Cryptosporidium. In: Wu GL, editor. Human Parasitology. Beijing: People’s Medical Publishing House. 269–275.
- 11.Peng MM, Matos O, Gatei W, Das P, Stantic-Pavlinic M, et al. (2001) A comparison of Cryptosporidium subtypes from several geographic regions. J Eukaryot Microbiol Suppl: 28–31. [DOI] [PubMed]
- 12. Wang RJ, Zhang XS, Zhu HL, Zhang LX, Feng YY, et al. (2011) Genetic characterizations of Cryptosporidium spp. and Giardia duodenalis in humans in Henan, China. Exp Parasitol 127: 42–45. [DOI] [PubMed] [Google Scholar]
- 13. Feng YY, Wang L, Duan LP, Gomez-Puerta LA, Zhang LX, et al. (2012) Extended Outbreak of Cryptosporidiosis in a Pediatric Hospital, China. Emerg Infect Dis 18: 312–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang R, Zhang L, Ning C, Feng Y, Jian F, et al. (2008) Multilocus phylogenetic analysis of Cryptosporidium andersoni (Apicomplexa) isolated from a bactrian camel (Camelus bactrianus) in China. Parasitol Res 102: 915–920. [DOI] [PubMed] [Google Scholar]
- 15. Alves M, Xiao L, Sulaiman I, Lal AA, Matos O, et al. (2003) Subtypes analysis of Cryptosporidium isolates from humans, cattle, and ruminants in Portugal. J Clin Microbiol 41: 2744–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Das P, Roy SS, MitraDhar K, Dutta P, Bhattacharya MK, et al. (2006) Molecular characterization of Cryptosporidium spp. in children in Kolkata, India. J Clin Microbiol 44: 4246–4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xiao L, Escalante L, Yang C, Sulaiman I, Escalante AA, et al. (1999) Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl Environ Microbiol 65: 1578–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao L, Ryan UM (2008) Molecular epidemiology. In: Fayer R, Xiao L, editors. Cryptosporidium and Cryptosporidiosis. Boca Raton: CRC Press and IWA Publishing. 119–171.
- 19. Alves M, Xiao L, Antunes F, Matos O (2006) Distribution of Cryptosporidium subtypes in humans and domestic and wild ruminants in Portugal. Parasitol Res 99: 287–292. [DOI] [PubMed] [Google Scholar]
- 20.Peng MM, Meshnick SR, Cunliffe NA, Thindwa BD, Hart CA, et al.. (2003) Molecular epidemiology of cryptosporidiosis in children in Malawi. J Eukaryot Microbiol Suppl: 557–559. [DOI] [PubMed]
- 21. Cama VA, Bern C, Roberts J, Cabrera L, Sterling CR, et al. (2008) Cryptosporidium species and subtypes and clinical manifestations in children, Peru. Emerg Infect Dis 14: 1567–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Leav BA, Mackay MR, Anyanwu A, O’Connor RM, Cevallos AM, et al. (2002) Analysis of sequence diversity at the highly polymorphic Cpgp40/15 locus among Cryptosporidium isolates from human immunodeficiency virus-infected children in South Africa. Infect and Immun 70: 3881–3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hijjawi N, Ng J, Yang R, Atoum MFM, Ryan U (2010) Identification of rare and novel Cryptosporidium gp60 subtypes in human isolates from Jordan. Exp Parasitol 125: 161–164. [DOI] [PubMed] [Google Scholar]