Abstract
Coxsackievirus B3 (CVB3) has previously been shown to utilize autophagy in an advantageous manner during the course of infection of the host cell. However, few studies have determined whether stem cells induce autophagy in a similar fashion, and whether virus-induced autophagy occurs following infection of stem cells. Therefore, we compared the induction of autophagy following CVB3 infection of neural progenitor and stem cells (NPSCs), which we have recently shown to be highly susceptible to CVB3 infection, to HL-1 cells, a transformed cardiomyocyte cell line. As previously demonstrated for other susceptible host cells, HL-1 cells showed an increase in the activity of autophagic signaling following infection with a CVB3 expressing dsRed protein (dsRed-CVB3). Furthermore, viral titers in HL-1 cells increased in the presence of an inducer of autophagy (CCPA), while viral titers decreased in the presence of an inhibitor of autophagy (3-MA). In contrast, no change in autophagic signaling was seen in NPSCs following infection with dsRed-CVB3. Also, basal levels of autophagy in NPSCs were found to be highly elevated in comparison to HL-1 cells. Autophagy could be induced in NPSCs in the presence of rapamycin without altering levels of dsRed-CVB3 replication. In differentiated NPSC precursors, autophagy was activated during the differentiation process, and a decrease in autophagic signaling was observed within all three CNS lineages following dsRed-CVB3 infection. Hence, we conclude that the role of autophagy in modulating CVB3 replication appears cell type-specific, and stem cells may uniquely regulate autophagy in response to infection.
Keywords: coxsackievirus, enterovirus, neural stem cells, autophagy, meningitis, encephalitis, central nervous system, neurotropic virus
Introduction
Macroautophagy, hereby referred to as autophagy, is an essential process that is responsible for the breakdown of long-lived proteins and organelles within the cell. Recently, autophagy has been identified as a crucial step for the replication and survival of viral pathogens following infection of the host cell. Some viruses have been shown to manipulate the autophagic process in order to efficiently replicate within the cell, rather than fall prey to this catabolic process and be destroyed within the lysosome. Autophagy is activated downstream of class III phosphatidylinositol 3-kinase (PtdIns3K) signaling, and the growth of the autophagosome double membrane is promoted by the association of covalently conjugated autophagy proteins.1 One such protein is microtubule-associated protein 1 light chain 3 (LC3). The lipidated form of LC3, known as LC3-II, studs the inner and outer autophagosome membrane. Autophagosomes can be visualized directly by expressing LC3 as a fusion protein with green fluorescent protein (GFP-LC3), and by observing GFP-labeled vacuoles within cells. In addition, the ratio of lipidated LC3-II to unmodified LC3-I, which reflects autophagic activity, can be analyzed via western blotting.2 Once formed, the autophagosome may fuse with the lysosome and its contents can be degraded.
Autophagy can be induced by several processes, including starvation, cell damage and invading pathogens, where the autophagic engulfment of the intracellular organisms is known as xenophagy.3 The autophagic process has also been shown to play a critical role in the immune response and during inflammation.4 For example, autophagy has been shown to facilitate MHC-peptide and toll-like receptor-ligand interactions.5 While some microbes, such as Mycobacterium tuberculosis, are destroyed by autophagy, others, such as Dengue virus, use autophagy to their advantage.6,7 Interestingly, some viruses, such as herpes simplex virus-1 (HSV-1), encode proteins that inhibit autophagy during the course of infection, thus providing increased neurovirulence and pathogenicity of the cornea.8,9 For example, interaction of ICP34.5 with BECN1 modulates HSV-1 pathogenesis through control of CD4+ T cell responses. Interplay between viral infections and autophagy may occur throughout the viral life cycle and at different stages of the autophagy pathway. Hepatitis C virus utilizes autophagosomes or autophagy proteins to initiate viral replication.10,11 A complex interplay between pathogens and autophagy has evolved and reveals a process which appears to be specific to each microbe and, as we report in this study, may also be specific to each cell type.12
Enteroviruses are serious human pathogens that are responsible for a wide range of disease, ranging from simple flu-like symptoms to poliomyelitis. CVB3, an enterovirus, has been found to cause severe morbidity and mortality by contributing to myocarditis, pancreatitis and meningitis.13,14 These infections are more common in neonates and may lead to long-term sequelae later in life, including dilated cardiomyopathy, learning disabilities and demyelinating disorders.15-22 When tracking neonatal CVB3 infection in vivo using a recombinant CVB3 expressing eGFP (eGFP-CVB3), neural progenitor and stem cells (NPSCs) were identified to be highly susceptible to CVB3 infection.23 Our previous studies demonstrated the co-localization of viral protein (eGFP) expression with neural stem cell markers located in the subventricular zone, a neurogenic region of the central nervous system (CNS).24 Also, neurotropic CVB3 infection recruited nestin+ myeloid cells which became infected following their extravasation through the blood-cerebrospinal fluid-barrier.25,26
Recent publications investigating autophagy following enterovirus infection have shown that poliovirus, coxsackievirus and enterovirus-71 induce autophagy and utilize the autophagosome membrane for viral replication.27-29 Specifically, previous studies have found that the poliovirus 3A and 2BC proteins when expressed together in the absence of virus may induce autophagy. Also, 2BC viral protein can induce the lipidation of LC3.30,31 Furthermore, autophagy may play a part in the noncytolytic release of enteroviruses from the cell.32 Similar findings were established in vivo, as CVB3 infection of the pancreas induced autophagosomes, which provided a scaffold for viral replication in pancreatic acinar cells.33 In addition to the induction of autophagy following infection, CVB3 blocks the maturation of the autophagosome, thus generating megaphagosomes.33,34 Intriguingly, an inverse relationship between autophagy and apoptosis was recently identified in rat primary neurons following CVB4 infection.35 However, these previous studies have been typically performed in differentiated cell types such as HeLa cells and rat primary neurons,36 thus leaving the role of autophagy in stem cells unexplored.
We have recently demonstrated that CVB3 preferentially infects NPSCs in culture.37 Neurospheres, or free-floating spheres generated by NPSCs in culture, remain undifferentiated but also have the ability to differentiate into precursors to form all three cell lineages of the central nervous system (CNS), comprising neurons, astrocytes and oligodendrocytes.38 Differentiated NPSC precursors were found to be less susceptible to CVB3 infection than undifferentiated NPSCs, presumably due to their decreased proliferative status and other cellular changes associated with differentiation.39 The process of autophagy has yet to be explored in detail with regards to neural stem cell maintenance and during the differentiation process. Since NPSCs were found to be highly susceptible to CVB3 both in vitro and in vivo, the goal of our study was to determine whether autophagy was altered following their infection. In addition, we wished to explore the relationship of autophagy and viral replication in NPSCs. Activation of autophagic signaling following infection with a recombinant CVB3 expressing dsRed protein (dsRed-CVB3) was measured using two distinct methodologies. GFP-LC3-positive vacuoles in cells transduced using an adenovirus expressing GFP-LC3 (Adeno-GFP-LC3) were quantified by microscopy, and the ratio of LC3-II to LC3-I in cells was calculated following western blotting. Here, we observed that the role of autophagy during CVB3 infection appears cell specific. Also, NPSCs may respond uniquely to CVB3 infection with regards to autophagy and viral replication. The naturally high levels of autophagic activity in NPSCs may contribute to this phenomenon, and may also explain their relatively high susceptibility to CVB3 infection.
Results
Activation of autophagic signaling following CVB3 infection in HL-1 cells transduced with Adeno-GFP-LC3
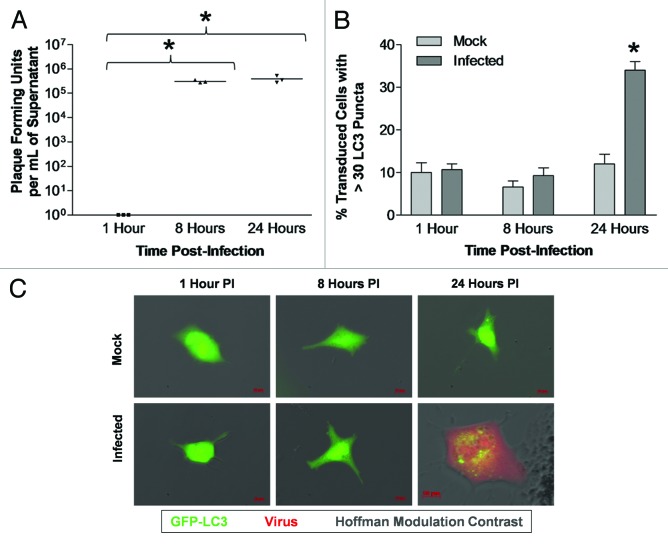
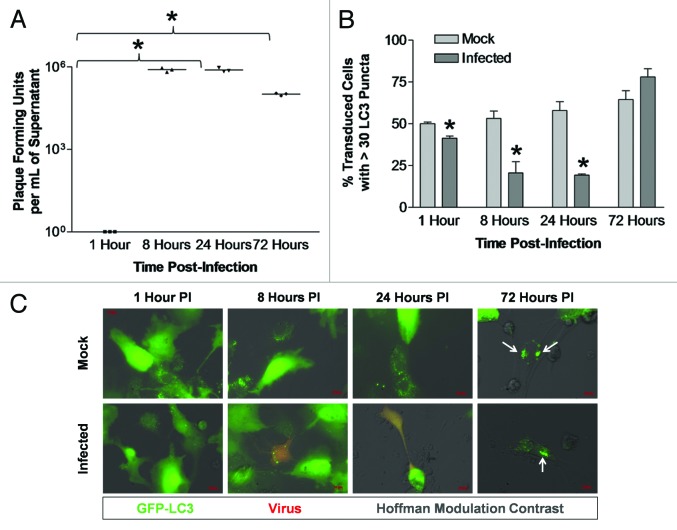
The levels of LC3 puncta formation following dsRed-CVB3 infection were first determined in a transformed cell line susceptible to infection. Although CVB3 exhibits tropism for cardiomyocytes in vivo, no previous studies have examined cardiac cells in relation to autophagy and CVB3 infection. HL-1 cells, a transformed cardiomyocyte cell line that has previously been shown to be highly susceptible to CVB3 infection,40,41 were utilized for this purpose. Following infection with dsRed-CVB3, high viral titers were observed in HL-1 cells at 8 and 24 h post-infection (PI) (Fig. 1A). We observed a statistically significant increase in LC3 puncta formation within infected HL-1 cells by 24 h PI (Fig. 1B). Fluorescence microscopy demonstrated the successful transduction of HL-1 cells with Adeno-GFP-LC3 (green), and the presence of numerous GFP-LC3+ vacuoles in CVB3-infected (red) cells at 24 h PI (Fig. 1C). High viral titers were also observed at 8 h PI although there was no increase in autophagosome abundance. The lack of correlation between virus titers and autophagosome abundance may be attributable to autophagy-independent viral replication, increased flux (clearance of autophagosomes via exosome release or lysosomal destruction increases at the same rate that autophagosome formation increases), or a modest increase in autophagy that does not rise to our threshold of > 30 puncta per cell.
Figure 1. Activation of autophagic signaling following CVB3 infection in HL-1 cells transduced with Adeno-GFP-LC3. LC3 puncta formation increased following CVB3 infection in HL-1 cells transduced with Adeno-GFP-LC3. HL-1 cells were plated on gelatin/fibronectin-coated chamber slides, transduced with Adeno-GFP-LC3, infected with dsRed-CVB3, and observed by fluorescence microscopy at 63X at the indicated time points. (A) CVB3 titers over time were determined by plaque assay. No viral titers were found in mock-infected cells. Viral titers at 8 h and 24 h PI were significantly higher (*p < 0.01) than 1 h PI (with the initial inoculum subtracted) as determined by ANOVA with Newman-Keuls post-hoc analysis. (B) Quantification of cells with high levels of GFP-LC3 autophagosome vacuoles was performed by counting 50 transduced cells per well, with three wells per treatment (represented as the mean +SEM). High levels of GFP-LC3 autophagosome vacuoles were defined as greater than 30 punctate per cell. Infected cells showed a significant increase in LC3 puncta formation (*p = 0.002), as compared with mock-infected cells at 24 h PI (determined by Student’s t-test). No difference was observed between mock and infected cells at 1 h and 8 h PI. (C) Representative 63X fluorescent images are shown for each time point and treatment.
CVB3 infection and CCPA activated autophagic signaling in HL-1 cells
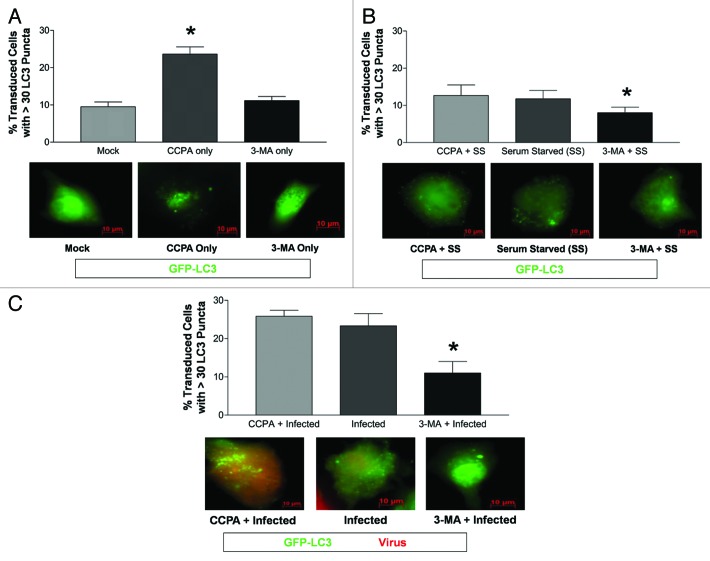
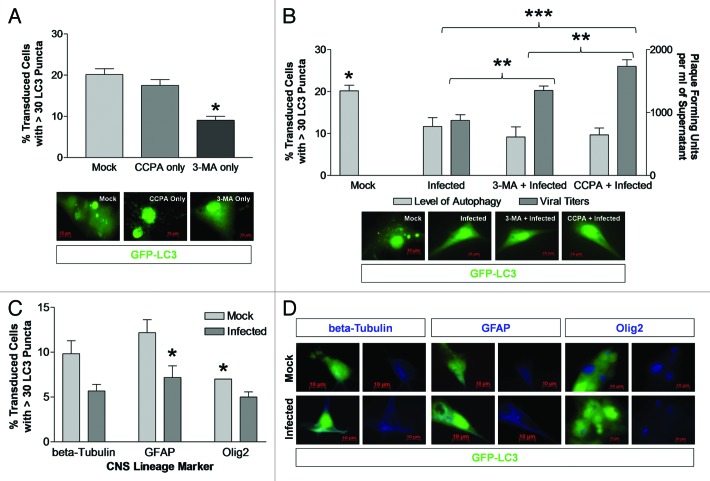
In order to determine whether an increase LC3 puncta formation following CVB3 infection influenced viral replication, the level of autophagy was altered in HL-1 cells using the autophagy inducer, CCPA, prior to infection. CCPA, an adenosine A1 agonist, activates autophagy by increasing intracellular calcium levels.42 Alternatively, HL-1 cells were treated with the autophagy inhibitor, 3-MA, prior to infection. 3-MA, a class III PtdIns3K inhibitor, halts autophagy without affecting protein synthesis.42,43 CCPA significantly increased LC3 puncta formation in HL-1 cells (Fig. 2A). Serum starvation increased LC3 puncta formation in HL-1 cells, although the increase was not statistically significant (Fig. 2B). Also, 3-MA did not reduce the low basal levels of autophagic signaling in HL-1 cells, but did suppress autophagic activation following serum starvation. Next, the effect of modulating autophagy induced by infection with dsRed-CVB3 was determined. As shown by other investigators for differentiated cell lines, CVB3 infection activated autophagic signaling; the effect of CCPA was not additive, whereas addition of 3-MA reduced LC3 puncta formation induced by viral infection (Fig. 2C).
Figure 2. CVB3 infection and CCPA increased the level of LC3 puncta formation in HL-1 cells. (A) The level of autophagy increased (*p < 0.01) following the addition of CCPA (inducer of autophagy) to HL-1 cells, as compared with mock or 3-MA (inhibitor of autophagy). (B) A significant decrease (*p < 0.05) in the percentage of transduced HL-1 cells with high levels of GFP-LC3 vacuoles was observed after serum starvation (SS) in the presence of 3-MA, as compared with serum starvation alone or serum starvation in the presence of CCPA. (C) A significant decrease (*p < 0.01) in the percent of transduced HL-1 cells with high levels of GFP-LC3 vacuoles was observed after dsRed-CVB3 infection in the presence of 3-MA, as compared with dsRed-CVB3 infection alone or in the presence of CCPA. ANOVA with Newman-Keuls post-hoc analysis was utilized to determine statistical significance for each experiment.
The activation or inhibition of autophagic signaling altered CVB3 replication and viral protein expression levels in HL-1 cells
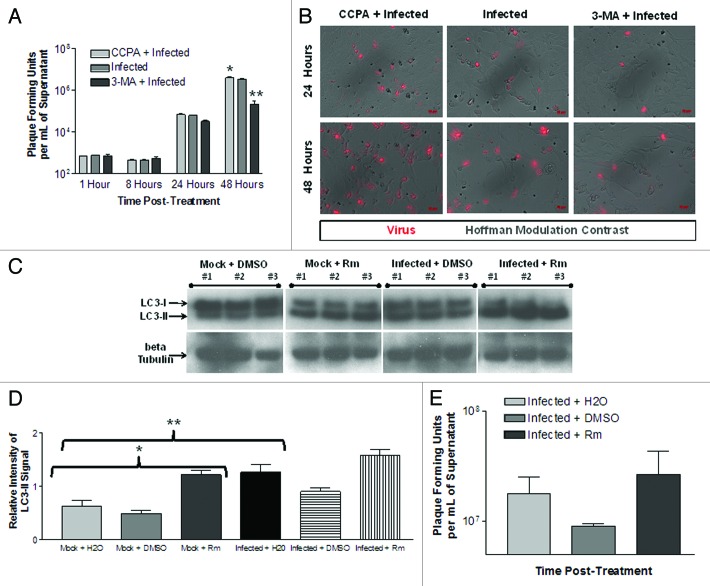
To determine if changes in autophagy modulated viral infection, CVB3 replication was evaluated in the presence of CCPA, rapamycin (Rm) or 3-MA in HL-1 cells (Fig. 3). Rm induces autophagy by inhibiting the mechanistic target of rapamycin (MTOR), a negative regulator of autophagy upstream of class III PtdIns3K.44,45 A significant decrease in viral titers was observed after 3-MA-treatment at 48 h PI (Fig. 3A). In contrast, a significant increase in viral titers was observed after CCPA-treatment at 48 h PI. A similar trend was observed for 24 h PI, yet this trend was not significant when all time points were analyzed by ANOVA with Newman-Keuls post-hoc analysis. Nonetheless when the 24 h PI time point was analyzed alone, 3-MA-treatment showed significantly lower viral titers (p < 0.05) than infected alone or CCPA-treated. Viral protein expression levels paralleled viral titers for each treatment (Fig. 3B; red signal). For example, CCPA-treated cells expressed higher levels of dsRed protein as compared with infected cells alone at 24 and 48 h PI. In contrast, 3-MA-treated cells expressed lower levels of dsRed protein than infected cells alone at 24 and 48 h PI.
Figure 3. The induction or inhibition of autophagy altered CVB3 replication and viral protein expression levels in HL-1 cells. HL-1 cells were infected with dsRed-CVB3 and overlaid with fresh media containing CCPA, 3-MA, rapamycin (Rm) or sterile water (vehicle control). Autophagy inducers (CCPA and Rm) or inhibitor (3-MA) were administered in fresh media at the time of dsRed-CVB3 infection at the following final concentrations: CCPA, 0.1 μM; Rm, 5 μM; and 3-MA, 10 mM. (A) Supernatant samples from infected cultures were taken at the indicated time points, and plaque assay was performed to determine viral titers. At 48 h post-treatment, viral titers in 3-MA-treated HL-1 cells were significantly lower (**p < 0.001) than in untreated cultures. In contrast, viral titers in CCPA-treated HL-1 cells were significantly higher (*p < 0.01) than in untreated cultures. A similar trend, although not statistically significant, was observed at 24 h post-treatment. ANOVA with Newman-Keuls post-hoc analysis was utilized to determine statistical significance. (B) Representative fluorescent images of dsRed-CVB3-infected HL-1 cells showing reduced viral protein expression levels in 3-MA-treated HL-1 cells and increased levels in CCPA-treated HL-1 cells at 24 h and 48 h post-treatment. (C) HL-1 cells were treated with H2O, dimethyl sulfoxide (DMSO, vehicle control) or DMSO + rapamycin (Rm) for 24 h. Protein extracts from lysed cells were analyzed for LC3-II levels by western blot analysis. Protein loading was determined by endogenous β-tubulin levels. (D) The relative intensity of LC3-II compared with β-tubulin levels was determined using ImageJ software. Mock + Rm showed statistically significant higher levels of relative LC3-II intensity than Mock + H20 or Mock + DMSO (*p < 0.01). These results show that Rm increased autophagic signaling in HL-1 cells. Also, CVB3 infection alone increased autophagic signaling in HL-1 cells (**p < 0.001). (E) Viral titers increased in RM-treated HL-1 cells as compared with DMSO treatment alone, although the increase was not statistically significant. ANOVA with Newman-Keuls post-hoc analysis was utilized to determine statistical significance.
We also inspected the ability of rapamycin (Rm) to induce autophagic signaling in HL-1 cells and alter levels of viral replication. Rm treatment was performed to directly compare autophagic induction of HL-1 cells with Rm-treated NPSCs. Rm was found to significantly increase levels of LC3 lipid modification in mock-infected HL-1 cells as shown by western blotting (Fig. 3C) and by quantifying the relative intensity of LC3-II signal using ImageJ software (Fig. 3D). CVB3 infection alone increased autophagosome abundance in HL-1 cells. In addition, a synergistic increase in autophagic abundance was seen in Rm-treated HL-1 cells infected with CVB3. Viral titers were performed for infected plus water (mean = 1.78 × 107 pfu/ml), infected plus DMSO (mean = 9.04 × 106 pfu/ml) and infected plus Rm (mean = 2.65 × 107 pfu/ml) (Fig. 3E). Rm treatment increased viral titers at 24 h PI as compared with DMSO treatment alone, although the level was not statistically significant.
These results are consistent with the view that autophagy facilitates CVB3 infection and/or replication in HL-1 cells. Nonetheless, the possibility remains that 3-MA treatment may affect other aspects of virus replication or virion formation and release. For example, PtdIns3K activity is beneficial for CVB3 replication by phosphorylation of AKT1/protein kinase B.46 Consequently, the inhibition of PtdIns3K by 3-MA might contribute to some of the observed reduction in viral titers following 3-MA treatment. Also, viral titers were reduced in HL-1 cells following the addition of DMSO alone. We hypothesize that viral replication in HL-1 cells may be marginally affected in the presence of DMSO, thereby slightly altering viral replication kinetics. We also observed that HL-1 cells do not respond as quickly to Rm as compared with other cell lines (data not shown). HL-1 cells might require a longer incubation time with Rm to induce similar levels of autophagy.
No change in the level of autophagic signaling in undifferentiated NPSCs following CVB3 infection
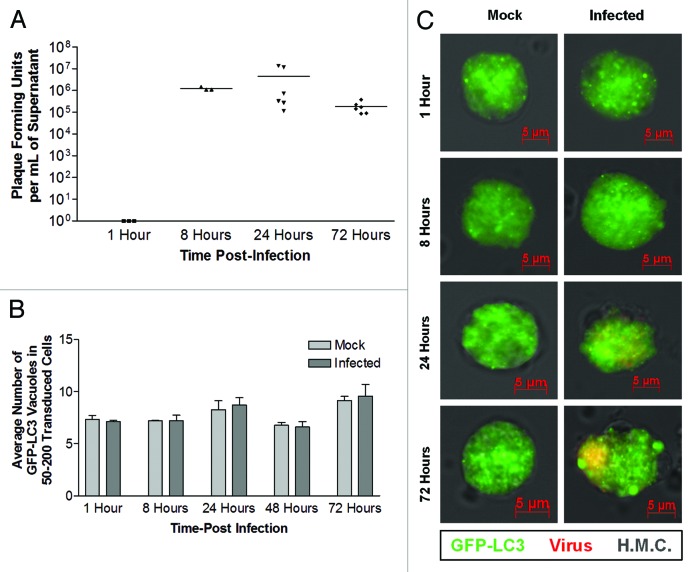
Our results for HL-1 cells paralleled previously published findings in differentiated cell types with regards to autophagy and CVB3 replication. The role of autophagy in viral infection of undifferentiated NPSCs was next examined. As previously described,37 high viral titers were observed in supernatants taken from infected undifferentiated NPCSs at 8, 24 and 72 h PI (Fig. 4A). Interestingly, basal levels of LC3 puncta formation were quite high in NPSCs, and did not increase following infection with CVB3 (Fig. 4B and C).
Figure 4. No change in the level of LC3 puncta formation in undifferentiated NPSCs following CVB3 infection. Undifferentiated NPSCs were transduced with Adeno-GFP-LC3, infected with dsRed-CVB3, and observed by fluorescence microscopy at 63X at the indicated time points. (A) Viral titers were determined by plaque assay. Viral titers increased over time, but were not significantly different due to large variance, as determined by ANOVA with Newman-Keuls post-hoc analysis. (B) Quantification of autophagosomes was performed by counting 50–200 GFP-LC3 positive cells per well, with 3–5 wells per treatment, and values are represented as the mean +SEM. No difference in the level of autophagy between mock and infected undifferentiated NPSCs was observed for any of the time points analyzed, as determined by Student’s t-test. (C) Representative 63X fluorescent images are shown for each time point and treatment.
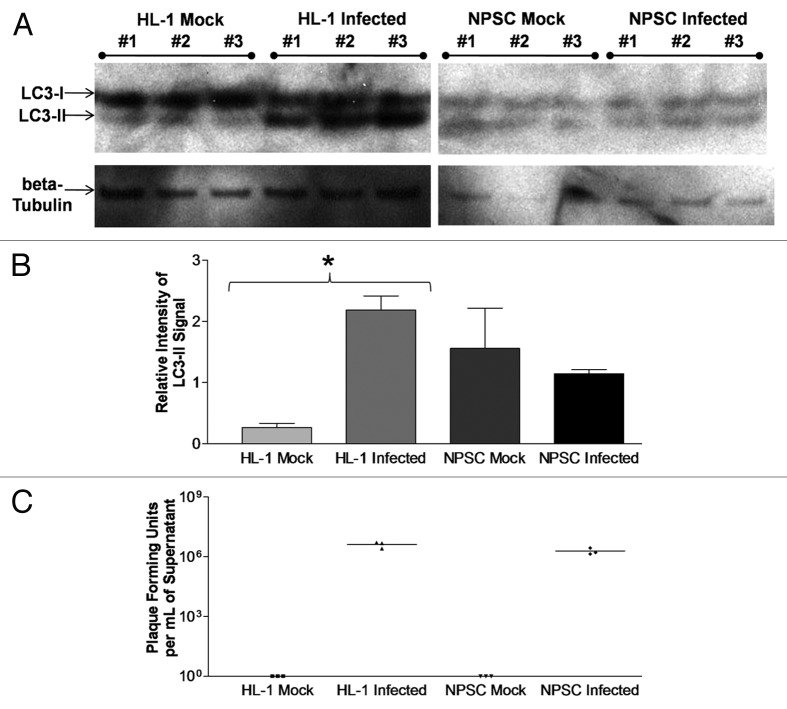
Undifferentiated NPSCs exhibit a higher basal level of autophagic signaling in comparison to HL-1 cells
We verified our GFP-LC3 vacuole quantification utilizing an independent methodology (western blotting for endogenous LC3 levels) and comparing these results to mock or infected HL-1 cells (Fig. 5A). Similar to the quantification of GFP-LC3 puncta formation shown in Figure 1C, the relative intensity of LC3-II signal compared with β-tubulin as determined by western blotting significantly increased in HL-1 cells at 24 h PI (Fig. 5B). In contrast, no increase in the relative intensity of LC3-II signal was observed in NPSCs at 24 h PI, as compared with mock-infected NPSCs. These results confirmed our GFP-LC3 vacuole quantification data shown in Figure 4. Also, the LC3-II signal in either mock or infected NPSCs was higher (although not statistically significant) than that observed for mock-infected HL-1 cells, consistent with a higher basal level of autophagic signaling in NPSCs. Of note, the relative intensity of the LC3-II signal in NPSCs was not further increased by viral infection. To ensure that differences in the level of autophagic signaling were not due to differences in viral replication between HL-1 cells and NPSCs, viral titers were determined from these experiments (Fig. 5C). No significant difference was seen in dsRed-CVB3 titers between HL-1 cells (mean = 4.08 × 106 pfu/ml) and NPSCs (mean = 1.91 × 106 pfu/ml) at 24 h PI.
Figure 5. Undifferentiated NPSCs exhibited a higher basal level of autophagic signaling, as compared with HL-1 cells. (A) HL-1 cells and undifferentiated NPSCs were infected with dsRed-CVB3 for 24 h, and protein extracts from lysed cells were analyzed for LC3-I and LC3-II levels by western blot analysis. Protein loading was determined by endogenous β-tubulin levels. (B) The relative intensity of LC3-II compared with β-tubulin levels was determined using Image J software. A statistically significant increase (*p < 0.05) in relative LC3-II intensity was observed between Infected and Mock HL-1 cells by ANOVA with Newman-Keuls post-hoc analysis. Also, a greater (although not statistically significant) level of relative LC3-II intensity was observed for Mock NPSCs and Infected NPSCs, as compared with Mock HL-1 cells. No statistical significant difference was observed between Infected HL-1 cells, Mock NPSCs and Infected NPSCs, thus indicating a higher basal level of autophagy in NPSCs regardless of viral infection. (C) No statistical difference was observed for viral titers between Infected HL-1 cells and Infected NPSCs by Student’s t-test (p = 0.0734).
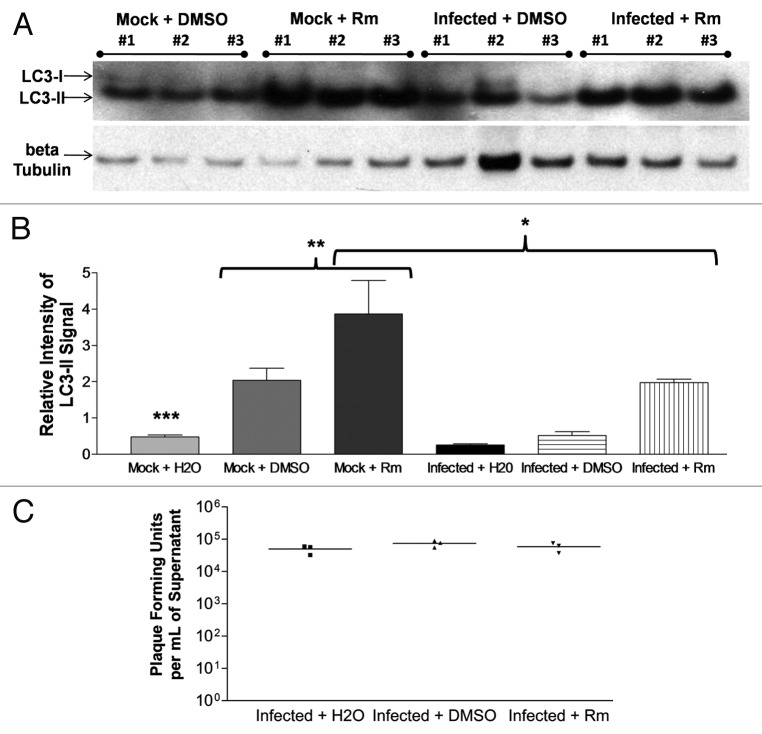
Rapamycin induced the activation of autophagic signaling in undifferentiated NPSCs, yet did not alter CVB3 replication
In order to determine whether undifferentiated NPSCs were able to increase autophagic signaling, rapamycin was utilized to induce autophagy, and the levels of LC3 lipid modification were analyzed by western blotting for endogenous LC3. Of note, CCPA failed to activate autophagic signaling in NPSCs (data not shown), presumably due to the absence of a functional adenosine A1 receptor in NPSCs.47 Also, 3-MA was shown to be toxic in undifferentiated NPSCs, as shown previously by other investigators.48,49 Rm was found to significantly increase levels of LC3 lipid modification in mock-infected NPSCs as shown by western blotting (Fig. 6A) and by quantifying the relative intensity of LC3-II signal using ImageJ software (Fig. 6B). Also, CVB3 infection appeared to suppress Rm-induced autophagic signaling in NPSCs. Viral titers were performed for infected plus water (mean = 50,000 pfu/ml), infected plus DMSO (mean = 74,167 pfu/ml) and infected plus Rm (mean = 58,333 pfu/ml) (Fig. 6C). No differences (p = 0.2983) in viral titers were observed between any treatments. Thus, CVB3 replication did not appear to be altered by rapamycin-induced autophagy in undifferentiated NPSCs, although this effect could be due to inhibition of MTOR and possible secondary effects on viral replication unrelated to autophagy.50
Figure 6. Rapamycin induced autophagic signaling in undifferentiated NPSCs but did not alter CVB3 replication. (A) Undifferentiated NPSCs were treated with dimethyl sulfoxide (DMSO, vehicle control) or DMSO + rapamycin (Rm) for 24 h. Protein extracts from lysed cells were analyzed for LC3-II levels by western blot analysis. Protein loading was determined by endogenous β-tubulin levels. (B) The relative intensity of LC3-II compared with β-tubulin levels was determined using ImageJ software. Mock + Rm showed statistically significant higher levels of relative LC3-II intensity than Mock + H20 (***p < 0.001) or Mock + DMSO (**p < 0.001). These results show that Rm could further increase autophagic signaling in NPSCs. Also, CVB3 infection suppressed Rm-induced autophagic signaling in NPSCs (*p < 0.05). (C) No statistically significant difference (p = 0.2983) was found in viral titers between Infected + H2O, Infected + DMSO or Infected + Rm at 24 h PI. These results demonstrated that rapamycin-induced increases in autophagy within undifferentiated NPSCs failed to alter CVB3 replication levels. ANOVA with Newman-Keuls post-hoc analysis was utilized to determine statistical significance.
Decreased autophagic signaling in differentiated NPSCs following CVB3 infection
We hypothesized that autophagy may be altered upon NPSC differentiation, and that CVB3 infection may also act in a unique fashion with regards to autophagy in differentiated NPSCs. Therefore, NPSCs were differentiated for 5 d in differentiation media on gelatin/fibronectin-coated chamber slides. Differentiated NPSC begin to express markers for all three CNS lineages after 5 d in differentiation media, although these cells continue to divide in culture.37 Following infection of differentiated NPSC with dsRed-CVB3, viral titers were observed at 8, 24 and 72 h PI (Fig. 7A). Also, a statistically significant increase in viral titers was observed over time in these cultures (*p < 0.05). Intriguingly, differentiated NPSC precursors showed high basal levels LC3 puncta formation which decreased in a statistically significant manner following dsRed-CVB3 infection at 1, 8 and 24 h PI (Fig. 7B). The relative intensity of LC3-II signal indicated that differentiated NPSCs in the absence of infection showed elevated levels of autophagic signaling (mean = 2.05) as compared with the levels observed in undifferentiated NPSCs (mean = 1.56; data not shown). By 72 h PI, both mock and infected cultures exhibited similar high levels of LC3 puncta formation. Also, very large autophagy-related structures were observed at 72 h PI in both mock and infected cultures, perhaps due to the extended amount of time without media replenishment (Fig. 7C, white arrows).
Figure 7. Decreased levels of LC3 puncta formation in differentiated NPSCs following CVB3 infection. NPSCs were differentiated for 5 d on gelatin/fibronectin coated chamber slides, transduced with Adeno-GFP-LC3, infected with dsRed-CVB3, and observed by fluorescence microscopy at 63X at the indicated time points. (A) Viral titers were determined by plaque assay. Viral titers in infected cells were significantly higher at 8 h and 24 h PI (*p < 0.05), as compared with 1 h PI. No viral titers were found in mock infected cells. (B) At 1 h, 8 h and 24 h PI, a significant decrease in GFP-LC3 autophagosome vacuoles were observed following infection, as compared with mock-infected cells (p = 0.008, p = 0.0154 and p = 0.0019, respectively). (C) Representative 63X images depicted for each time point and treatment showing decreased GFP-LC3 autophagosome vacuoles in differentiated NPSCs infected with dsRed-CVB3. Possible megaphagosomes are depicted (white arrows) in 72 h PI images. Quantification of cells with high levels of GFP-LC3 vacuoles (defined as greater than 30 punctate per cell) was performed by counting 200 transduced cells per well, with three wells per treatment (represented as the mean +SEM). ANOVA with Newman-Keuls post-hoc analysis was utilized to determine statistical significance for each experiment.
Differentiated NPSCs treated with both 3-MA and CCPA showed an increase in viral titers following CVB3 infection
CCPA and 3-MA were utilized to modulate the levels of autophagy in differentiated NPSCs. Although neurons, astrocytes and oligodendrocytes have been previously shown to express the adenosine A1 receptor,51-53 CCPA failed to increase the high basal levels of autophagy observed within differentiated NPSCs. Given that basal levels of autophagic signaling were high in differentiated NPSCs, we examined the effect of 3-MA on autophagy and viral replication. 3-MA decreased LC3 puncta formation in differentiated NPSCs in comparison to mock-treated cultures (Fig. 8A). In contrast to the findings with undifferentiated NPSCs, CVB3 infection of differentiated NPSCs reduced the abundance of autophagosomes; also, 3-MA did not have an additive effect (Fig. 8B). Surprisingly, viral titers significantly increased in differentiated NPSCs treated with either 3-MA or CCPA. Since these adenine derivatives did not modulate autophagic signaling in infected differentiated NPSCs, the effect on viral replication could be due to off-target activities.
Figure 8. Differentiated NPSCs treated with both 3-MA and CCPA showed an increase in viral titers following CVB3 infection. (A) The level of autophagy in differentiated NPSCs decreased (*p < 0.01) in the presence of 3-MA (inhibitor of autophagy), as compared with mock or CCPA treatment (inducer of autophagy). (B) No change in the level of autophagy (left y-axis) was observed in infected differentiated NPSCs following 3-MA or CCPA treatment. Mock-treated differentiated NPSCs were significantly different (*p < 0.05) than all other treatments. Viral titers (right y-axis) were significantly different (**p < 0.01) between 3-MA + Infected vs. Infected alone and CCPA + Infected and a highly significantly different (***p < 0.001) between Infected and CCPA + Infected, despite the similar level of autophagy in all of these treatments. (C) No bias in the level of autophagy was observed in the three downstream cell lineages of differentiated NPSCs following CVB3 infection. All three cell lineages showed a trend toward a reduction in the level of autophagy following infection, as compared with mock-infected. However, only GFAP+ cells were significantly different (*p < 0.05) in the level of autophagy following infection, as compared with mock-infected. Mock-infected OLIG2+ cells showed a significantly lower (*p < 0.05) level of autophagy, as compared with mock-infected GFAP+ cells. (D) Representative images of transduced cells (green) expressing each CNS lineage marker (blue). Quantification of cells with high levels of GFP-LC3 autophagosome vacuoles was performed by counting 200 transduced cells per well, with three wells per treatment (represented as the mean +SEM). ANOVA with Newman-Keuls post-hoc analysis was utilized to determine statistical significance.
We immunostained infected differentiated NPSCs using three well-known markers of neuronal and glial differentiation to determine whether a reduction of LC3 puncta formation preferentially occurred in one type of CNS precursor. Neuronal class III β-tubulin-labeled neuronal precursor cells, GFAP-labeled astrocyte precursor cells and OLIG2-labeled oligodendrocyte precursor cells. We analyzed the level of autophagy in each cell type by evaluating the abundance of punctate GFP-LC3 (Fig. 8C and D). The activation of autophagic signaling varied among the three cell lineages, with oligodendrocyte (OLIG2+) precursor cells showing the lowest level. Infection with CVB3 resulted in a decrease in the level of autophagic signaling in all cell lineages, although only GFAP+ cells showed difference that reached the level of statistically significance.
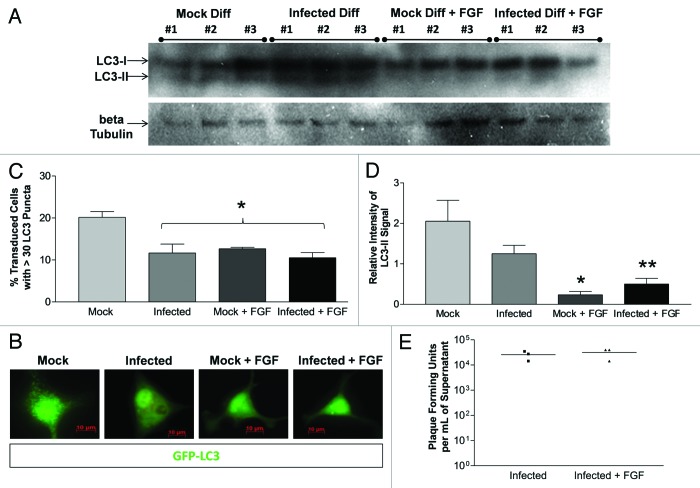
The addition of fibroblast growth factor (FGF) decreased the level of autophagic signaling in differentiated NPSCs without affecting viral replication
FGF, a component of undifferentiated NPSC media, is typically removed to assist in the differentiation process. However, a recent study has shown that the addition of FGF to mesencephalic neural progenitor cell cultures may inhibit autophagy-mediated cell death.49 Therefore, we examined the effect of basic fibroblast growth factor (FGF) on differentiated NPSCs. LC3 western blotting was performed on mock and infected differentiated NPSCs in the presence and absence FGF at 24 h PI (Fig. 9A). The addition of FGF suppressed autophagic signaling in differentiated NPSCs based on fluorescence microscopy of LC3 puncta formation (Fig. 9B), by GFP-LC3 vacuole quantification (Fig. 9C), and by quantification of LC3 western blotting (Fig. 9D). Infection with dsRed-CVB3 also decreased autophagic signaling to a similar extent, while the addition of FGF to infected cells did not additively suppress LC3 puncta formation. Also, the addition of FGF to infected cells did not reduce the relative intensity of LC3-II, as compared with Mock + FGF cultures. Although FGF treatment reduced autophagic signaling in differentiated NPSCs, no change in virus production was observed in FGF-treated differentiated NPSCs (Fig. 9E).
Figure 9. The addition of fibroblast growth factor decreased autophagic signaling in differentiated NPSCs without affecting CVB3 replication. NPSCS were differentiated for 5 d on gelatin/fibronectin coated chamber slides and transduced [for (B and C) only] with Adeno-GFP-LC3. Differentiated NPSCs were infected with dsRed-CVB3 and/or treated with fibroblast growth factor (FGF). (A) Protein extracts from lysed cells were analyzed after 24 h for LC3-II levels by western blot analysis. (B) Representative 63X fluorescent images are depicted for each treatment. (C) Cells were fixed after 8 h and observed by fluorescence microscopy at 63X. Quantification of autophagosomes was performed by counting 200 transduced cells per well, with three wells per treatment, and represented as the mean +SEM. Mock treatment was statistically higher than all other treatments (*p < 0.01). No statistical difference was observed between any other treatments. (D) The relative intensity of LC3-II compared with β-tubulin levels was determined from (A) using ImageJ software. A statistically significant decrease in the relative intensity of LC3-II was observed between Mock and Mock + FGF (*p < 0.01), or Mock and Infected + FGF (**p < 0.05), indicating that FGF significantly decreased the level of autophagy in differentiated NPSCs. Also, the relative intensity of LC3-II was lower in Infected, as compared with Mock, although this difference was not statistically significant. ANOVA with Newman-Keuls post-hoc analysis was utilized to determine statistical significance for each experiment. (E) No difference (p = 0.6047) in viral titers was observed between Infected and Infected + FGF, as determined by Student’s t-test. These results indicate that altering autophagy levels in differentiated NPSCs did not affect CVB3 replication.
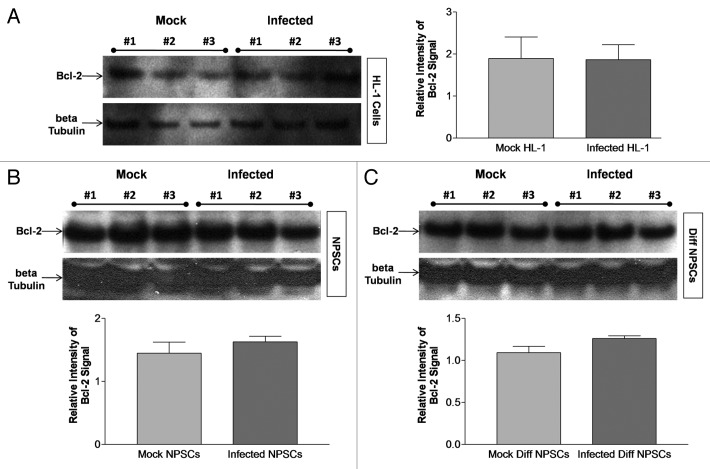
BCL2 levels remained unchanged in HL-1 cells, undifferentiated NPSCs and differentiated NPSCs following CVB3 infection
Similar to the effects of FGF, BCL2, an antiapoptotic protein, has been found to prevent autophagy-mediated cell death in neural progenitor cell cultures when overexpressed.49 BCL2 expression may be induced downstream of FGF signaling, though a previous study did not see a difference in endogenous levels when FGF was added.54 BCL2 levels were analyzed before and after infection in all three cultures (HL-1 cells, undifferentiated and differentiated NPSCs) included in our study. No change was observed in BCL2 expression levels by western blot analysis at 24 h PI for HL-1 cells (Fig. 10A), undifferentiated NPSCs (Fig. 10B) or differentiated NPSCs (Fig. 10C) after normalization to β-tubulin expression levels. These results indicate that the disparity in the role of autophagy following CVB3 infection in undifferentiated or differentiated NPSCs does not appear to be mediated by BCL2. The differential role of autophagy after CVB3 infection has been summarized for each cell type in Table 1, which also described the relationship between autophagy and CVB3 replication within each cell type.
Figure 10. Bcl-2 levels remain unchanged in HL-1 cells, undifferentiated NPSCs and differentiated NPSCs following CVB3 infection. (A) HL-1 cells, (B) undifferentiated NPSCs and (C) differentiated (Diff) NPSCs were infected with dsRed-CVB3 for 24 h. Protein extracts from lysed cells were utilized for BCL2 western blot analysis. Protein loading levels were determined by endogenous β-tubulin levels. The quantification of BCL2 signal was performed using ImageJ software and normalized to β-tubulin levels for each cell type analyzed. No statistical difference in the level of BCL2 was observed following infection of HL-1 cells (p = 0.9648), NPSCs (p = 0.3877) or Diff NPSCs (p = 0.1138), as compared with mock-infected cells by Student’s t-test.
Table 1. Summary of the role of autophagy in HL-1 cells, undifferentiated NPSCs and differentiated NPSCs.
| HL-1 Cells | Undifferentiated NPSCs | Differentiated NPSCs | |
|---|---|---|---|
| Does the level of autophagy change after CVB3 infection? |
Yes—An increase in the level of autophagy |
No change |
Yes—A decrease in the level of autophagy |
| Are inducers of autophagy able to induce autophagy in this cell type? |
Yes |
Yes |
No—But differentiation itself induces autophagy |
| Is there a relationship between autophagy and CVB3 replication? | Yes—Direct relationship | No—Increasing the level of autophagy with rapamycin does not alter viral titers | No—Decreasing the level of autophagy with FGF does not alter viral titers |
Results are summarized by cell type in regards to whether activation of autophagic signaling is induced following CVB3 infection, if autophagy can be induced in the particular cell type, and how autophagy affects viral replication. While HL-1 cells followed previous results published by other groups, a divergent relationship regarding autophagy and CVB3 in both undifferentiated and differentiated NPSCs was observed.
Discussion
CVB3 is a common childhood enterovirus that can infect and kill NPSCs,37 thus leading to neurodevelopmental delays in the newborn.55 Antibodies against enteroviruses have been detected in approximately 75% of the population, and previous infection has been linked to a number of long-term diseases, including schizophrenia and neurological diseases causing damage to the white matter of the CNS.56-58 Several publications have shown that enterovirus infection induced autophagy, and that this induction enhanced viral replication, either by facilitating virus spread/cell invasion, intracellular virus production and/or virus release. These previous findings highlight the possibility of using autophagy inhibitors as potential antiviral treatment strategies.30 However, these studies were performed in differentiated cell types, thus bringing into question whether autophagy may function similarly during viral replication in stem cells. The role of autophagy in neural stem cells in response to infection or during CNS development remains largely uncharacterized. To further inspect the consequences of CVB3 infection in this cell population, primary neonatal NPSCs were isolated and alterations in autophagic activation were evaluated following their infection. Hence, the goal of this study was to determine the role of autophagy following CVB3 infection of NPSCs.
In order to confirm previous results in differentiated cell types, HL-1 cells were analyzed side-by-side with NPSCs in our study. Autophagic activation was induced in HL-1 cells following dsRed-CVB3 infection, and this induction was a direct result of infection. However, autophagic activation was not induced until 24 h PI. In HeLa cells and HEK293A cells, induction was seen as early as 7 h post-CVB3 infection, which corresponds to roughly one round of viral replication.28 Since dsRed-CVB3 may replicate more slowly than wild-type CVB3, the slower viral kinetics associated with this recombinant virus may be responsible for the delayed activation of autophagic signaling following infection of HL-1 cells. Conversely, in primary rat neurons the level of autophagy began to increase by 16 h post-CVB3 infection and continued to increase at 24 h.36 While 16 h PI was not analyzed in our study, the induction of autophagy in rat primary neurons may be more similar to that observed in HL-1 cells, in comparison to HeLa and HEK293A cells. Therefore, cell type, which may also influence the rate of viral replication, may influence the onset of autophagy following infection.39 Our study is also the first to analyze autophagy in a cardiomyocyte cell line following CVB3 infection. Since CVB3 may be one of the most common infectious causes of myocarditis,59 understanding the factors contributing to virus replication in cardiomyocytes may be extremely important in designing potential antiviral treatment regimes.60 Consequently, novel autophagy-regulating treatments to control CVB3 replication in the heart may be of great clinical importance. Further investigation of the role of autophagy during CVB3 replication in additional cell types, as well as in vivo, may be of particular importance.
The activity of autophagic signaling following CVB3 infection in NPSCs was investigated to determine if similar alterations in autophagy may occur in these cells. Although CVB3 infection did not increase autophagy in NPSCs, their basal level of LC3 puncta formation was higher than in HL-1 cells, perhaps due to unique features associated with stem cells. The naturally elevated levels of autophagic signaling in NPSCs may be related to their particular high susceptibility to CVB3 infection.37 Even so, production of infectious virus progeny reached similar levels in both NPSCs and HL-1 cells in our experiments. Technical considerations may artificially reduce infectious virus production in NPSCs, as compared with HL-1. The free-floating cell aggregate structure characteristic of neurospheres may limit initial viral infection and lower the effective multiplicity of infection following inoculation, unless these neurospheres are completed dissociated prior to infection.
Autophagic signaling in NPSCs was further upregulated by addition of Rm. Also, no cell death was observed following the induction of autophagy in NPSCs by Rm (data not shown), indicating that autophagic cell death was not occurring. In fact, inhibition of autophagy with 3-MA was cytotoxic in NPSCs, as previously reported.48,49 In contrast to HL-1 cells, the induction of autophagy in NPSCs by Rm did not alter levels of viral replication. CCPA was also utilized in our study, although this compound was not successful at inducing autophagy in NPSCs. The lack of autophagic induction by CCPA may be due to the absence of a functional adenosine A1 receptor in NPSCs.47
While basal levels of autophagic signaling differed among the three CNS precursor cell types, CVB3 infection suppressed LC3 puncta formation in all three cell lineages. NPSCs may exhibit a predisposition to differentiate into astrocytes based on plating density, and this effect could potentially reflect changes in autophagy and viral replication in these cultures.49,61 The addition of FGF was found to decrease the activation of autophagic signaling in differentiated NPSCs, both in the presence and absence of infection. Nonetheless, levels of viral replication were not altered in FGF-treated differentiated NPSCs. Hence, the addition of FGF decreased autophagic activation without affecting CVB3 replication. FGF might be considered essential growth factor included in complete NPSC media. FGF assists in retaining the undifferentiated status of neurospheres and is removed to assist in the differentiation process. Also, autophagy may be highly active during the process of differentiation and development.62 Other aspects associated with cellular differentiation may be a contributing factor in the relatively low level of viral production seen in differentiated NPSCs despite the high levels of autophagic signaling during differentiation.37 For example cellular proliferation may be substantially reduced during differentiation, or differentiated cell types such as astrocytes may naturally restrict CVB3 replication. The observation that viral titers increased in differentiated NPSCs despite inhibition of autophagy with 3-MA might suggest that the basal levels of autophagy in these cells are sufficient for viral replication/infection.
In order to further investigate why autophagic signaling was not induced in NPSCs, BCL2 was examined for its ability to inhibit autophagy-mediated cell death in these cultures, similar to what has been shown for FGF.63 BCL2, an anti-apoptotic protein, has been shown to alter autophagy in NPSCs and HL-1 cells.49 BCL2 has also been shown to be induced following CVB3 infection.64 However, BCL2 expression remained unchanged following CVB3 infection in HL-1 cells, undifferentiated NPSCs or differentiated NPSCs. Nonetheless, signaling pathways involved in modulating autophagy within NPSCs following CVB3 infection merit further investigation.
The activity of viral proteins 3A and 2BC, which have been shown to induce autophagy in other studies, may be ineffective in NPSCs for reasons yet to be determined. For example, cellular proteins influencing autophagy may be downregulated or nonfunctional in NPSCs due to differences in post-translational processing or altered subcellular localization. Expressing fluorescent tagged versions of the CVB3 viral proteins 3A and 2BC within NPSCs may assist in answering these questions. Such studies may also shed light on the currently unclear mechanism of autophagy induction by viral proteins.
Our study examined only extracellular viral titers, yet a decrease in extracellular vs. intracellular poliovirus was previously observed when autophagy was inhibited. Also, exocytosis of poliovirus protein and LC3-positive vesicles was previously shown by electron microscopy.30 Because of these previous findings, autophagy has been implicated as a mechanism for the noncytolytic release of virus from the cell, and may thus provide a means for enteroviruses to persist. Since persistent CVB3 infection has been documented in both the myocardium and in the CNS, the role of autophagy during this process may be of great importance.65,66 Also, the cell type in the CNS supporting viral persistence remains unknown. Neural progenitor cells may be attractive candidates due to their ability to support persistent infection in culture.37
In the CNS, relatively high basal levels of autophagy in neural progenitor cells may provide CVB3 with receptive targets for infection. However upon differentiation of neural progenitor cells, CVB3 may direct the differentiated cells to reduce basal levels of autophagy for reasons yet to be determined. Alterations in autophagy following infection of neural progenitor cells, and within their differentiated counterparts, may play a role in the observed neuropathology following infection which may include loss of progenitor cells, neuronal apoptosis, inflammation and the recruitment of immune cells into the CNS.23,24,66,67 Given the unusual autophagic response seen in NPSCs following CVB3 infection, the role of autophagy and its contribution to viral persistence and pathogenesis in the host CNS may be of particular significance, and should be inspected in more detail.
Materials and Methods
Ethics statement
This study was performed in strict accordance with the requirements pertaining to animal subjects protections within the Public Health Service Policy and USDA Animal Welfare Regulations. All experimental procedures with mice were approved by the San Diego State University Institutional Animal Care and Use Committee (Animal Protocol Form #10-05-013F), and all efforts were made to minimize suffering.
Isolation and production of a recombinant coxsackievirus
The generation of a recombinant coxsackievirus expressing foreign proteins has been described previously.39,68 Briefly, the CVB3 infectious clone (pH3) (obtained from Dr. Kirk Knowlton at University of California at San Diego) was engineered to contain a unique SfiI site which facilitates the insertion of any foreign sequence into the CVB3 genome. The generation of recombinant coxsackievirus expressing dsRed protein (dsRed-CVB3) has been described previously.37 The gene encoding dsRed was amplified from a dsRed expression plasmid using dsRed sequence-specific primers with flanking Sfi1 sequences. The PCR product was cloned into pMKS1. Following transfection of HeLa RW cells with dsRed-CVB3 plasmid, infectious virus was generated. Virus stocks were grown on HeLa RW cells. Virus titrations were performed as described previously.39 Viral stocks were prepared on HeLa RW cells maintained in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, 12491-015) supplemented with 10% fetal bovine serum (FBS) which has been heat-treated to inactivate complement. Viral stocks were diluted in DMEM before inoculation.
Isolation and culture of neurospheres
C57BL/6 mice were obtained from the Scripps Research Institute animal facilities or Harlan Laboratories. Breeding pairs were checked every day. NPSCs were derived from isolated cortices of newborn mice, mechanically and enzymatically dissociated, and then plated as a single cell suspension in DMEM/F12 media supplemented with 2% B27 Supplement (Invitrogen, 17504-044), 20 ng/mL EGF (Invitrogen, PHG0311L), 20 ng/mL bFGF (Preprotech, 450-33), 5 µg/mL Heparin (Sigma, H3149) and 0.5% penicillin/streptomycin. Free-floating neurospheres were separated and transferred into new flasks every two days. Neurospheres were vigorously triturated and resuspended in culture medium to a concentration of 105 cells/mL in a T-25 flask (BD Falcon, 353109). Neurospheres were differentiated using gelatin/fibronectin coated chamber slides in DMEM media supplemented with 1% N1 supplement, 1% FBS and 0.5% pen/strep for 5 d.
Transfection, infection and treatment of NPSCs and HL-1 cells
Undifferentiated NPSCs, differentiated NPSCs and HL-1 cells cultured as previously described,40 were transfected with GFP-LC3 adenovirus for 40 h then infected with dsRed-CVB3 at an MOI of 10. Autophagy inducers (m-chlorocyclopentyladenosine, CCPA and rapamycin, Rm) or inhibitor (3-methyladenine, 3-MA) were administered in fresh media at the time of dsRed-CVB3 infection at the following final concentrations: CCPA, 0.1 μM; Rm, 5 μM; and 3-MA, 10 mM. CCPA (C7938) and 3-MA (M9281) were both obtained from Sigma-Aldrich Co. Rm (553211-500) was obtained from EMD Chemicals Inc. Supernatants were harvested at the indicated time points and analyzed by plaque assay to determine viral titers.
GFP-LC3 vacuole quantification
For time-course experiments, NPSCs and HL-1 cells were imaged live for GFP-LC3 vacuoles at the indicated time points and 50–200 cells per well were counted in triplicate for mock and infected cultures. For non-time-course experiments, cells were fixed in 4% paraformaldehyde, washed three times in 1× PBS and 200 cells per well were counted in triplicate for all treatments. For differentiated NPSC precursors and HL-1 cells, a bimodal distribution of GFP-LC3 vacuoles was observed. Therefore, transduced cells were scored as having less than or greater than 30 GFP-positive vacuoles, as previously described.63 For undifferentiated NPSCs, a bimodal distribution was not observed. Therefore, the average number of GFP-positive vacuoles per transduced cell was counted.
Immunofluorescence microscopy
NPSCs were fixed in 4% paraformaldehyde, washed three times in 1× PBS and permeabilized with 0.5% Triton X-100. Viral protein expression was determined by native dsRed (red) expression. Fixed cells were blocked with 10% normal goat serum and immunostained using the following antibodies: neuronal class III β-tubulin (Covance, PRB-435P) at 1:1000, GFAP (Sigma, G9269) at 1:500 and OLIG2 (Abcam, ab33427) at 1:1000. Secondary antibodies were labeled with Alexa Fluor 594 or Alex Fluor 350 at (1:1000). Live and fixed cultures were imaged using a Zeiss Axio Observer D.1 inverted fluorescence microscope. Three to five representative images of the cultures were taken for each time point.
Western blotting
Undifferentiated NPSCs, differentiated NPSCs and HL-1 cells were washed with 1× PBS 24 h after infection/treatment. Cells were scraped and/or centrifuged at 7,000 rpm for 1 min, based on adherence, in 1× PBS and resuspended in cell extraction buffer (Invitrogen; FNN0011) with protease inhibitors for 30 min on ice with periodic vortexing. Cells were then centrifuged for 10 min at 4°C and supernatants were transferred to a new tube. Samples were prepared for SDS-PAGE by adding 19.5 μl protein extract, 7.5 µl lithium dodecyl sulfate sample buffer and 3 µl NuPAGE reducing agent (Invitrogen, NP0004), and by heating for 3 min at 70°C. Gel electrophoresis was performed for 1 h at 150V with 15 μl of each sample. Proteins from the gel were transferred to a PVDF membrane for 1 h at 30V. The membrane was blocked with 5% skimmed milk in Tris-Buffered Saline Tween 20 (TBST) for 1 h at room temperature. The membrane was cut in half just under the 40-kDa marker. The top half was probed for rabbit-anti β-Tubulin (50 kDa; Abcam, ab6046) at (1:500) in 1% Bovine Serum Albumin (BSA, G-Biosciences, 786-193) in TBST overnight at 4°C. The bottom half of the membrane was probed for either endogenous LC3 (16 and 18 kDa) or BCL2 (26kDa). Rabbit anti-LC3A/B (Cell Signaling Technologies, 4108) was used at (1:1000) in 5% BSA in TBST overnight at 4°C. Mouse anti-BCL2 (Invitrogen, 33-6100) was used at (1:500) in 1% BSA in TBST overnight at 4°C. For all primary antibodies, HRP-conjugated secondary was applied at (1:2000–5000) for 1 h at room temperature. Washes were performed in triplicate for 5 min in 1× TBST between all incubations. ECL substrate (Invitrogen, WP20005) was applied for 1 min and membranes were exposed to film. Quantification of band density was performed using Image J software.
Statistical analyses
Statistical analyses were performed using Graphpad Prism 3.0 software. One-way ANOVA with Newman-Keuls post-hoc analysis was used to analyze three or more groups, while Student’s t-test was utilized to compare two groups. Significance was determined by a p value of 0.05 or lower.
Acknowledgments
This work was supported by National Institutes of Health (NIH) R01 Award NS054108 (to R.F.), R01 Award HL092136 (to R.A.G.), an NIH Research Supplement to Promote Diversity in Health-Related Research Award 3R01NS054108-01A2S1 (to R.F.) and a National Institutes of Mental Health (NIMH) Minority Research Infrastructure Support Program (M-RISP) R24 Faculty Fellow Award MH065515 (to R.F.). J.M.T.-G. and G.T. are recipients of an Achievement Rewards for College Scientists Foundation Scholarship. G.T. is a recipient of the Inamori Fellowship and the Gen-Probe Fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. No conflicts of interest exist between the subject matter and the authors included in the manuscript.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/19781
References
- 1.Gustafsson AB, Gottlieb RA. Recycle or die: the role of autophagy in cardioprotection. J Mol Cell Cardiol. 2008;44:654–61. doi: 10.1016/j.yjmcc.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–75. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kudchodkar SB, Levine B. Viruses and autophagy. Rev Med Virol. 2009;19:359–78. doi: 10.1002/rmv.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–35. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deretic V. Multiple regulatory and effector roles of autophagy in immunity. Curr Opin Immunol. 2009;21:53–62. doi: 10.1016/j.coi.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–66. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 7.Lee YR, Lei HY, Liu MT, Wang JR, Chen SH, Jiang-Shieh YF, et al. Autophagic machinery activated by dengue virus enhances virus replication. Virology. 2008;374:240–8. doi: 10.1016/j.virol.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alexander DE, Leib DA. Xenophagy in herpes simplex virus replication and pathogenesis. Autophagy. 2008;4:101–3. doi: 10.4161/auto.5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orvedahl A, Levine B. Autophagy and viral neurovirulence. Cell Microbiol. 2008;10:1747–56. doi: 10.1111/j.1462-5822.2008.01175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dreux M, Gastaminza P, Wieland SF, Chisari FV. The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci U S A. 2009;106:14046–51. doi: 10.1073/pnas.0907344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dreux M, Chisari FV. Autophagy proteins promote hepatitis C virus replication. Autophagy. 2009;5:1224–5. doi: 10.4161/auto.5.8.10219. [DOI] [PubMed] [Google Scholar]
- 12.Lin LT, Dawson PW, Richardson CD. Viral interactions with macroautophagy: a double-edged sword. Virology. 2010;402:1–10. doi: 10.1016/j.virol.2010.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berger JR, Chumley W, Pittman T, Given C, Nuovo G. Persistent Coxsackie B encephalitis: Report of a case and review of the literature. J Neurovirol. 2006;12:511–6. doi: 10.1080/13550280601090546. [DOI] [PubMed] [Google Scholar]
- 14.Whitton JL, Cornell CT, Feuer R. Host and virus determinants of picornavirus pathogenesis and tropism. Nat Rev Microbiol. 2005;3:765–76. doi: 10.1038/nrmicro1284. [DOI] [PubMed] [Google Scholar]
- 15.Romero JR. Pediatric group B coxsackievirus infections. Curr Top Microbiol Immunol. 2008;323:223–39. doi: 10.1007/978-3-540-75546-3_10. [DOI] [PubMed] [Google Scholar]
- 16.Wikswo ME, Khetsuriani N, Fowlkes AL, Zheng X, Peñaranda S, Verma N, et al. Increased activity of Coxsackievirus B1 strains associated with severe disease among young infants in the United States, 2007-2008. Clin Infect Dis. 2009;49:e44–51. doi: 10.1086/605090. [DOI] [PubMed] [Google Scholar]
- 17.Chamberlain RN, Christie PN, Holt KS, Huntley RM, Pollard R, Roche MC. A study of school children who had identified virus infections of the central nervous system during infancy. Child Care Health Dev. 1983;9:29–47. doi: 10.1111/j.1365-2214.1983.tb00301.x. [DOI] [PubMed] [Google Scholar]
- 18.Kühl U, Pauschinger M, Noutsias M, Seeberg B, Bock T, Lassner D, et al. High prevalence of viral genomes and multiple viral infections in the myocardium of adults with “idiopathic” left ventricular dysfunction. Circulation. 2005;111:887–93. doi: 10.1161/01.CIR.0000155616.07901.35. [DOI] [PubMed] [Google Scholar]
- 19.Romero JR. Pediatric group B coxsackievirus infections. Curr Top Microbiol Immunol. 2008;323:223–39. doi: 10.1007/978-3-540-75546-3_10. [DOI] [PubMed] [Google Scholar]
- 20.Wikswo ME, Khetsuriani N, Fowlkes AL, Zheng X, Peñaranda S, Verma N, et al. Increased activity of Coxsackievirus B1 strains associated with severe disease among young infants in the United States, 2007-2008. Clin Infect Dis. 2009;49:e44–51. doi: 10.1086/605090. [DOI] [PubMed] [Google Scholar]
- 21.Chamberlain RN, Christie PN, Holt KS, Huntley RM, Pollard R, Roche MC. A study of school children who had identified virus infections of the central nervous system during infancy. Child Care Health Dev. 1983;9:29–47. doi: 10.1111/j.1365-2214.1983.tb00301.x. [DOI] [PubMed] [Google Scholar]
- 22.Kühl U, Pauschinger M, Noutsias M, Seeberg B, Bock T, Lassner D, et al. High prevalence of viral genomes and multiple viral infections in the myocardium of adults with “idiopathic” left ventricular dysfunction. Circulation. 2005;111:887–93. doi: 10.1161/01.CIR.0000155616.07901.35. [DOI] [PubMed] [Google Scholar]
- 23.Feuer R, Mena I, Pagarigan RR, Harkins S, Hassett DE, Whitton JL. Coxsackievirus B3 and the neonatal CNS: the roles of stem cells, developing neurons, and apoptosis in infection, viral dissemination, and disease. Am J Pathol. 2003;163:1379–93. doi: 10.1016/S0002-9440(10)63496-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feuer R, Pagarigan RR, Harkins S, Liu F, Hunziker IP, Whitton JL. Coxsackievirus targets proliferating neuronal progenitor cells in the neonatal CNS. J Neurosci. 2005;25:2434–44. doi: 10.1523/JNEUROSCI.4517-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tabor-Godwin JM, Ruller CM, Bagalso N, An N, Pagarigan RR, Harkins S, et al. A novel population of myeloid cells responding to coxsackievirus infection assists in the dissemination of virus within the neonatal CNS. J Neurosci. 2010;30:8676–91. doi: 10.1523/JNEUROSCI.1860-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhoades RE, Tabor-Godwin JM, Tsueng G, Feuer R. Enterovirus infections of the central nervous system. Virology. 2011;411:288–305. doi: 10.1016/j.virol.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suhy DA, Giddings TH, Jr., Kirkegaard K. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J Virol. 2000;74:8953–65. doi: 10.1128/JVI.74.19.8953-8965.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong J, Zhang J, Si X, Gao G, Mao I, McManus BM, et al. Autophagosome supports coxsackievirus B3 replication in host cells. J Virol. 2008;82:9143–53. doi: 10.1128/JVI.00641-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang SC, Chang CL, Wang PS, Tsai Y, Liu HS. Enterovirus 71-induced autophagy detected in vitro and in vivo promotes viral replication. J Med Virol. 2009;81:1241–52. doi: 10.1002/jmv.21502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jackson WT, Giddings TH, Jr., Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, et al. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor MP, Kirkegaard K. Modification of cellular autophagy protein LC3 by poliovirus. J Virol. 2007;81:12543–53. doi: 10.1128/JVI.00755-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor MP, Kirkegaard K. Potential subversion of autophagosomal pathway by picornaviruses. Autophagy. 2008;4:286–9. doi: 10.4161/auto.5377. [DOI] [PubMed] [Google Scholar]
- 33.Kemball CC, Alirezaei M, Flynn CT, Wood MR, Harkins S, Kiosses WB, et al. Coxsackievirus infection induces autophagy-like vesicles and megaphagosomes in pancreatic acinar cells in vivo. J Virol. 2010;84:12110–24. doi: 10.1128/JVI.01417-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dreux M, Gastaminza P, Wieland SF, Chisari FV. The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci U S A. 2009;106:14046–51. doi: 10.1073/pnas.0907344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoon SY, Ha YE, Choi JE, Ahn J, Lee H, Kim DH. Autophagy in coxsackievirus-infected neurons. Autophagy. 2009;5:388–9. doi: 10.4161/auto.5.3.7723. [DOI] [PubMed] [Google Scholar]
- 36.Yoon SY, Ha YE, Choi JE, Ahn J, Lee H, Kweon HS, et al. Coxsackievirus B4 uses autophagy for replication after calpain activation in rat primary neurons. J Virol. 2008;82:11976–8. doi: 10.1128/JVI.01028-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsueng G, Tabor-Godwin JM, Gopal A, Ruller CM, Deline S, An N, et al. Coxsackievirus preferentially replicates and induces cytopathic effects in undifferentiated neural progenitor cells. J Virol. 2011;85:5718–32. doi: 10.1128/JVI.02261-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eriksson C, Björklund A, Wictorin K. Neuronal differentiation following transplantation of expanded mouse neurosphere cultures derived from different embryonic forebrain regions. Exp Neurol. 2003;184:615–35. doi: 10.1016/S0014-4886(03)00271-1. [DOI] [PubMed] [Google Scholar]
- 39.Feuer R, Mena I, Pagarigan R, Slifka MK, Whitton JL. Cell cycle status affects coxsackievirus replication, persistence, and reactivation in vitro. J Virol. 2002;76:4430–40. doi: 10.1128/JVI.76.9.4430-4440.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Claycomb WC, Lanson NA, Jr., Stallworth BS, Egeland DB, Delcarpio JB, Bahinski A, et al. HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc Natl Acad Sci U S A. 1998;95:2979–84. doi: 10.1073/pnas.95.6.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan J, Cheung PK, Zhang H, Chau D, Yanagawa B, Cheung C, et al. A phosphorothioate antisense oligodeoxynucleotide specifically inhibits coxsackievirus B3 replication in cardiomyocytes and mouse hearts. Lab Invest. 2004;84:703–14. doi: 10.1038/labinvest.3700083. [DOI] [PubMed] [Google Scholar]
- 42.Yitzhaki S, Huang C, Liu W, Lee Y, Gustafsson AB, Mentzer RM, Jr., et al. Autophagy is required for preconditioning by the adenosine A1 receptor-selective agonist CCPA. Basic Res Cardiol. 2009;104:157–67. doi: 10.1007/s00395-009-0006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seglen PO, Gordon PB. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci U S A. 1982;79:1889–92. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–95. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 45.Carreira RS, Lee Y, Ghochani M, Gustafsson AB, Gottlieb RA. Cyclophilin D is required for mitochondrial removal by autophagy in cardiac cells. Autophagy. 2010;6:6. doi: 10.4161/auto.6.4.11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Esfandiarei M, Luo H, Yanagawa B, Suarez A, Dabiri D, Zhang J, et al. Protein kinase B/Akt regulates coxsackievirus B3 replication through a mechanism which is not caspase dependent. J Virol. 2004;78:4289–98. doi: 10.1128/JVI.78.8.4289-4298.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stafford MR, Bartlett PF, Adams DJ. Purinergic receptor activation inhibits mitogen-stimulated proliferation in primary neurospheres from the adult mouse subventricular zone. Mol Cell Neurosci. 2007;35:535–48. doi: 10.1016/j.mcn.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 48.Yu SW, Baek SH, Brennan RT, Bradley CJ, Park SK, Lee YS, et al. Autophagic death of adult hippocampal neural stem cells following insulin withdrawal. Stem Cells. 2008;26:2602–10. doi: 10.1634/stemcells.2008-0153. [DOI] [PubMed] [Google Scholar]
- 49.Ćrdenas-Aguayo MdelC, Santa-Olalla J, Baizabal JM, Salgado LM, Covarrubias L. Growth factor deprivation induces an alternative non-apoptotic death mechanism that is inhibited by Bcl2 in cells derived from neural precursor cells. J Hematother Stem Cell Res. 2003;12:735–48. doi: 10.1089/15258160360732759. [DOI] [PubMed] [Google Scholar]
- 50.Nichols LA, Adang LA, Kedes DH. Rapamycin blocks production of KSHV/HHV8: insights into the anti-tumor activity of an immunosuppressant drug. PLoS One. 2011;6:e14535. doi: 10.1371/journal.pone.0014535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanz JM, Vendite D, Ferńndez M, Andrés A, Ros M. Adenosine A1 receptors in cultured cerebellar granule cells: role of endogenous adenosine. J Neurochem. 1996;67:1469–77. doi: 10.1046/j.1471-4159.1996.67041469.x. [DOI] [PubMed] [Google Scholar]
- 52.D’Alimonte I, Ballerini P, Nargi E, Buccella S, Giuliani P, Di Iorio P, et al. Staurosporine-induced apoptosis in astrocytes is prevented by A1 adenosine receptor activation. Neurosci Lett. 2007;418:66–71. doi: 10.1016/j.neulet.2007.02.061. [DOI] [PubMed] [Google Scholar]
- 53.Othman T, Yan H, Rivkees SA. Oligodendrocytes express functional A1 adenosine receptors that stimulate cellular migration. Glia. 2003;44:166–72. doi: 10.1002/glia.10281. [DOI] [PubMed] [Google Scholar]
- 54.Désiré L, Courtois Y, Jeanny JC. Endogenous and exogenous fibroblast growth factor 2 support survival of chick retinal neurons by control of neuronal neuronal bcl-x(L) and bcl-2 expression through a fibroblast berowth factor receptor 1- and ERK-dependent pathway. J Neurochem. 2000;75:151–63. doi: 10.1046/j.1471-4159.2000.0750151.x. [DOI] [PubMed] [Google Scholar]
- 55.Euscher E, Davis J, Holzman I, Nuovo GJ. Coxsackie virus infection of the placenta associated with neurodevelopmental delays in the newborn. Obstet Gynecol. 2001;98:1019–26. doi: 10.1016/S0029-7844(01)01625-8. [DOI] [PubMed] [Google Scholar]
- 56.Sawyer MH. Enterovirus infections: diagnosis and treatment. Semin Pediatr Infect Dis. 2002;13:40–7. doi: 10.1053/spid.2002.29756. [DOI] [PubMed] [Google Scholar]
- 57.Rantakallio P, Jones P, Moring J, Von Wendt L. Association between central nervous system infections during childhood and adult onset schizophrenia and other psychoses: a 28-year follow-up. Int J Epidemiol. 1997;26:837–43. doi: 10.1093/ije/26.4.837. [DOI] [PubMed] [Google Scholar]
- 58.Verboon-Maciolek MA, Utrecht FG, Cowan F, Govaert P, van Loon AM, de Vries LS. White matter damage in neonatal enterovirus meningoencephalitis. Neurology. 2008;71:536. doi: 10.1212/01.wnl.0000324706.94229.88. [DOI] [PubMed] [Google Scholar]
- 59.Daley AJ, Isaacs D, Dwyer DE, Gilbert GL. A cluster of cases of neonatal coxsackievirus B meningitis and myocarditis. J Paediatr Child Health. 1998;34:196–8. doi: 10.1046/j.1440-1754.1998.00176.x. [DOI] [PubMed] [Google Scholar]
- 60.Knowlton KU. CVB infection and mechanisms of viral cardiomyopathy. Curr Top Microbiol Immunol. 2008;323:315–35. doi: 10.1007/978-3-540-75546-3_15. [DOI] [PubMed] [Google Scholar]
- 61.Adachi T, Takanaga H, Sakurai Y, Ishido M, Kunimoto M, Asou H. Influence of cell density and thyroid hormone on glial cell development in primary cultures of embryonic rat cerebral hemisphere. J Neurosci Res. 2002;69:61–71. doi: 10.1002/jnr.10279. [DOI] [PubMed] [Google Scholar]
- 62.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–30. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brady NR, Hamacher-Brady A, Yuan H, Gottlieb RA. The autophagic response to nutrient deprivation in the hl-1 cardiac myocyte is modulated by Bcl-2 and sarco/endoplasmic reticulum calcium stores. FEBS J. 2007;274:3184–97. doi: 10.1111/j.1742-4658.2007.05849.x. [DOI] [PubMed] [Google Scholar]
- 64.Colston JT, Chandrasekar B, Freeman GL. Expression of apoptosis-related proteins in experimental coxsackievirus myocarditis. Cardiovasc Res. 1998;38:158–68. doi: 10.1016/S0008-6363(97)00323-4. [DOI] [PubMed] [Google Scholar]
- 65.Kim KS, Tracy S, Tapprich W, Bailey J, Lee CK, Kim K, et al. 5′-Terminal deletions occur in coxsackievirus B3 during replication in murine hearts and cardiac myocyte cultures and correlate with encapsidation of negative-strand viral RNA. J Virol. 2005;79:7024–41. doi: 10.1128/JVI.79.11.7024-7041.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Feuer R, Ruller CM, An N, Tabor-Godwin JM, Rhoades RE, Maciejewski S, et al. Viral persistence and chronic immunopathology in the adult central nervous system following Coxsackievirus infection during the neonatal period. J Virol. 2009;83:9356–69. doi: 10.1128/JVI.02382-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Feuer R, Whitton JL. Preferential coxsackievirus replication in proliferating/activated cells: implications for virus tropism, persistence, and pathogenesis. Curr Top Microbiol Immunol. 2008;323:149–73. doi: 10.1007/978-3-540-75546-3_7. [DOI] [PubMed] [Google Scholar]
- 68.Slifka MK, Pagarigan R, Mena I, Feuer R, Whitton JL. Using recombinant coxsackievirus B3 to evaluate the induction and protective efficacy of CD8+ T cells during picornavirus infection. J Virol. 2001;75:2377–87. doi: 10.1128/JVI.75.5.2377-2387.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]