Abstract
Epigenetic dysfunctions, including DNA methylation alterations, play major roles in cancer initiation and progression. Although it is well established that gene promoter demethylation activates transcription, it remains unclear whether hypomethylation of repetitive heterochromatin similarly affects expression of non-coding RNA from these loci. Understanding how repetitive non-coding RNAs are transcriptionally regulated is important given that their established upregulation by the heat shock (HS) pathway suggests important functions in cellular response to stress, possibly by promoting heterochromatin reconstruction. We found that, although pericentromeric satellite 2 (Sat2) DNA hypomethylation is detected in a majority of cancer cell lines of various origins, DNA methylation loss does not constitutively hyperactivate Sat2 expression, and also does not facilitate Sat2 transcriptional induction upon heat shock. In melanoma tumor samples, our analysis revealed that the HS response, frequently upregulated in tumors, is probably the main determinant of Sat2 RNA expression in vivo. Next, we tested whether HS pathway hyperactivation may drive Sat2 demethylation. Strikingly, we found that both hyperthermia and hyperactivated RasV12 oncogene, another potent inducer of the HS pathway, reduced Sat2 methylation levels by up to 27% in human fibroblasts recovering from stress. Demethylation occurred locally on Sat2 repeats, resulting in a demethylation signature that was also detected in cancer cell lines with moderate genome-wide hypomethylation. We therefore propose that upregulation of Sat2 transcription in response to HS pathway hyperactivation during tumorigenesis may promote localized demethylation of the locus. This, in turn, may contribute to tumorigenesis, as demethylation of Sat2 was previously reported to favor chromosomal rearrangements.
Keywords: DNA hypomethylation, heat shock response, non-coding RNA, pericentromeric satellite DNA, Ras oncogene
Introduction
It has now become evident that cancer is a disease driven not only by genetic abnormalities, but also by epigenetic changes involving both losses and gains of DNA methylation as well as altered patterns of histone modifications. Repetitive DNA is normally highly methylated. Hypomethylation of these regions is therefore largely responsible for the global loss of DNA methylation in cancer and is believed to contribute to tumor formation and progression as malignancy grade is often associated with the level of global DNA hypomethylation.1 The mechanisms underlying cancer-linked hypomethylation are still poorly understood;2 it is also unclear whether demethylation occurs as a unique event or as successive waves. Besides, it appears that all repetitive genomic loci do not behave similarly when facing cellular demethylation processes,3-5 suggesting the involvement of locus-specific demethylation mechanisms that may differ in intensity and/or nature, according to the cellular context.
Satellite 2 DNA (Sat2), a locus mapping within the 1q12 pericentromeric region, is among the best-characterized hypomethylated sequences in cancer cells.1 Although Sat2 hypomethylation is not sufficient to induce rearrangements in this region, it is associated with pericentromeric instability.6 Accordingly, this locus is known to be a fragile site.7 In addition to its involvement in chromosomal instability, hypomethylation of CpG islands within regulatory regions reportedly activates gene transcription. Recent studies unraveled that, unlike what was previously anticipated, heterochromatic regions like Sat2 are transcriptionally competent and can produce non-coding RNAs (ncRNAs), the function of which is still unclear.8,9 The observation that satellite DNA transcription is increased by a wide range of cellular stresses including heat shock, DNA damaging agents, heavy metals, UV-C, oxidative stress and hyper-osmotic stress suggests that they may play important roles in response to stress, possibly by promoting heterochromatin reconstruction.10-14 Strong overexpression of pericentromeric satellite RNAs was also detected in experimentally-induced mouse pancreatic adenocarcinoma and in a variety of other cancer samples isolated from human patients.15 To date, however, the impact of DNA hypomethylation on Sat2 expression has remained elusive and conflicting data were reported. Eymery et al.16 showed that cell treatment with 5-aza-2’-deoxycytidine demethylating agent upregulates pericentromeric transcription. On the other hand, no increased expression of satellite III RNA was observed in HCT116 colon cancer cells deficient for DNMT3B and/or DNMT1, and a poor correlation between DNA hypomethylation and satellite DNA transcription was reported in ovarian and Wilms tumor samples.17 Similarly, cultured lymphocytes from DNMT3B-deficient patients suffering from the Immunodeficiency, Centromeric region instability and Facial anomalies (ICF) syndrome do not display strong constitutive transcription of pericentromeric Sat2 DNA sequences.17
Knowing that repetitive elements represent more than two-thirds of the human genome,18 understanding how their transcription is regulated is undoubtedly important. Here, we performed a quantitative study evaluating the influence of DNA demethylation on the transcription level of Sat2 in a variety of human cancer cells of distinct origins. In a second part of the study, we investigated the impact of heat shock response induction on Sat2 transcription and DNA methylation.
Results
Sat2 DNA hypomethylation is not associated with constitutive overexpression of Sat2 RNA in cancer cell lines
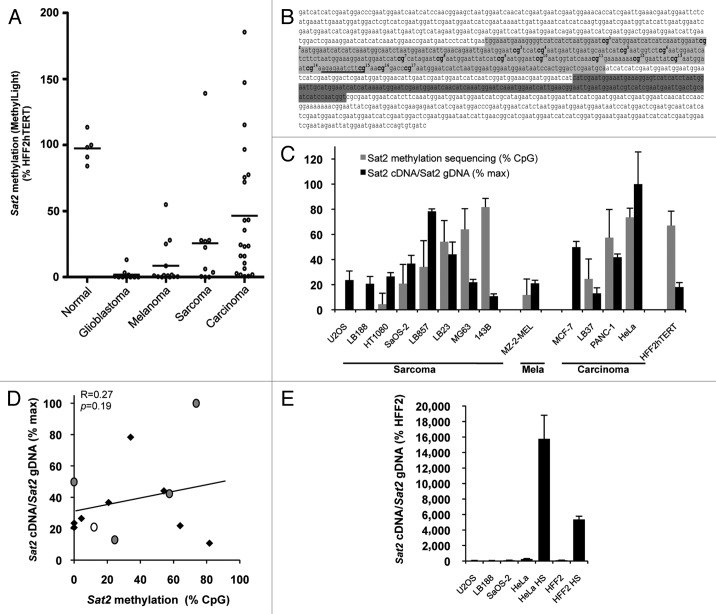
To assess the putative impact of cancer-linked Sat2 hypomethylation on transcription of the locus, we first selected a series of cancer cell lines displaying varying Sat2 methylation levels using the quantitative and rapid Sat2 MethylLight assay described previously.3 A total of 10 glioblastoma, 14 melanoma, 10 sarcoma and 22 carcinoma cell lines were included in the survey. To exclude any aberrant DNA demethylation linked to prolonged culture of cancer cell lines, Sat2 methylation levels were compared with those measured in non-tumoral cell lines obtained by immortalization with hTERT telomerase subunit of epithelial melanocytes (HNEMhTERT), foreskin fibroblasts (HFF2hTERT), human embryonic kidney cells (HEKhTERT) or mammary epithelial cells (HMEChTERT). Human mesenchymal stem cells (HMSCs) were further included in the analysis as sarcomas were previously suggested to derive from this cell type.19
Strong Sat2 hypomethylation was detected in most glioblastoma, melanoma and sarcoma cell lines, while it was less pronounced in carcinoma cell lines (Fig. 1A). Based on this survey, we selected eight sarcoma cell lines, four carcinoma cell lines and one melanoma cell line for further analyses. The non-tumoral HFF2hTERT fibroblast cell line was used as reference. A detailed analysis of Sat2 methylation was then obtained by bisulfite sequencing of the previously reported 351-bp CpG-rich region (Fig. 1B).20 Although the position of Sat2 promoter is unknown, the sequenced region contains several putative Heat Shock Factor 2 (HSF2) binding sites and one predicted HSF1 binding site (www.cbrc.jp/htbin/nph-tfsearch) (Fig. 1B), a transcription factor previously reported to promote satellite DNA transcription.12-14 Sat2 RNA levels were measured by quantitative RT-PCR, as previously described.17 In all cases, RNA was treated with DNaseI and measurements were also performed in control samples lacking reverse transcriptase. Importantly, to avoid any bias due to amplification/deletion at the locus, Sat2 cDNA levels were normalized first to ACTB cDNA levels and then to relative Sat2 genomic DNA content obtained by normalizing Sat2 gDNA to the total content in Alu repeat sequences (Fig. 1C). The Alu quantitative PCR amplifies a consensus motif conserved in ancient and more recent individual Alu repeats subfamilies (Alu-J, SluSp, AluSx, AluSq, AluSc, AluY, AluSb2, AluYb8, AluYa5 and AluYa8).3 Our results revealed no correlation between Sat2 DNA methylation and normalized Sat2 RNA levels in the 13 cancer cell lines that we tested (R = 0.27, p = 0.19) (Fig. 1C and D). As positive control of Sat2 transcriptional induction, Sat2 cDNA levels were measured in fibroblasts or HeLa cancer cells subjected to heat shock (HS), as previously reported.10,11 This revealed that Sat2 transcription remains at low basal level in cancer cell lines, even in the most hypomethylated ones (Fig. 1E). Altogether, these data show that Sat2 hypomethylation is not associated with constitutive hyperactivation of Sat2 expression in cancer cell lines.
Figure 1.Sat2 hypomethylation is not correlated with constitutive Sat2 RNA overexpression in cancer cell lines. (A) The MethyLight assay3 was used to quantify Sat2 methylation on bisulfite-treated DNA isolated from glioblastoma (n = 10), melanoma (n = 14), sarcoma (n = 10) and carcinoma (n = 22) cell lines. Non-tumoral control samples included four hTERT-immortalized cell lines (HFF2hTERT, HEKhTERT, HNEMhTERT, HMEChTERT) and HMSC mesenchymal stem cells. Methylation levels were compared with Sat2 methylation in HFF2hTERT. Bars indicate median values. (B) The Sat2 sequence of chromosome 1 corresponds to GenBank accession number X72623.1. In light gray: Sat2 fragment analyzed after bisulfite treatment of genomic DNA for methylated CpG content determination by sequencing. The 17 CpG sites are numbered. In dark gray: Sat2 fragment amplified by PCR on cDNA for quantitative RT-PCR measurement of Sat2 RNA transcripts. The same region (dark gray) was amplified in ChIP experiments. The HSF1 binding site predicted by tfsearch is underlined. (C) Sat2 methylation level in cancer cell lines derived from sarcoma, melanoma and carcinoma and in HFF2hTERT non-tumoral fibroblasts determined by bisulfite sequencing (gray bars). Data are indicated as % ± SD of total CpG. Relative Sat2 cDNA/Sat2 gDNA ratio were calculated as follows: (Sat2 cDNA/ACTB cDNA)/(Sat2 gDNA/Alu gDNA) (black bars). Data are presented as % ± SEM of the maximal expression level measured in cell lines. (D) Linear regression of data from C. Black diamonds: sarcoma; gray dots: carcinoma; white dot: melanoma. (E) Normalized Sat2 RNA levels were measured in either HFF2 or HeLa cells after 1h of heat shock (HS) at 42°C followed by 1h of recovery at 37°C. Basal expression levels in U2OS, LB188 and SaOS-2 hypomethylated cancer cell lines are also shown. Data are presented as % ± SEM of the expression measured in HFF2.
Sat2 hypomethylation in ICF fibroblasts does not exacerbate HS-induced Sat2 RNA levels
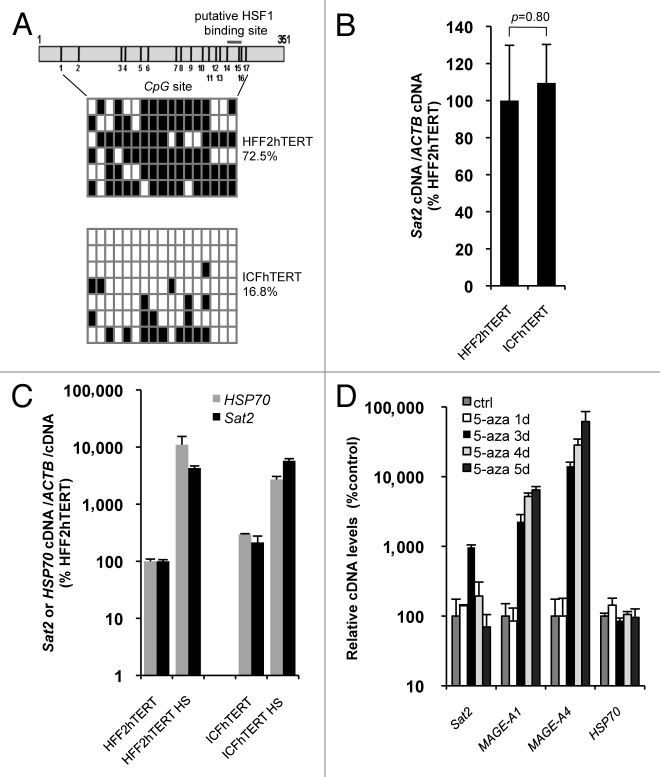
Although reduced methylated CpG content in cancer cell lines does not appear to impact on constitutive Sat2 expression, we next tested whether Sat2 hypomethylation may upregulate Sat2 RNA induction under HS conditions. To address this issue in comparable cell lines, we derived the ICFhTERT cell line from DNMT3B-deficient ICF fibroblasts and measured both basal and HS-induced Sat2 RNA levels in ICFhTERT and related HFF2hTERT fibroblasts displaying normal Sat2 methylation. As expected, Sat2 methylation levels were low in ICFhTERT, with methylated CpG content amounting to only 16.8% of total CpG, and high in HFF2hTERT, reaching 72.5% (Fig. 2A). In line with data obtained in cancer cell lines, we found that extensive hypomethylation of Sat2 sequences in ICFhTERT was not associated with constitutive hyperactivation of Sat2 transcription (Fig. 2B), which was instead triggered following exposure of ICFhTERT and HFF2hTERT fibroblasts to HS (Fig. 2C). Importantly, hypomethylation did not appear to exacerbate Sat2 transcriptional activation in heat shocked ICFhTERT cells (Fig. 2C). Further support for the lack of correlation between DNA hypomethylation and constitutive Sat2 RNA upregulation came from the demonstration that treatment of HFF2hTERT cells with 5 μM 5-aza-2’-deoxycytidine (5-aza) demethylating agent for 1 to 5 d only transiently upregulated Sat2 RNA (Fig. 2D). Consistent with a previous report,16 we found that Sat2 RNA levels were about 10-fold higher after 3 d of treatment with 5-aza with no concomitant increase in transcription of HSP70, one of the main HSF1 target genes.21 Upregulation of Sat2 RNA coincided with the transcriptional activation of MAGE-A1 and MAGE-A4 cancer germline genes (Fig. 2D) and the expected timing of 5-aza-induced genome-wide demethylation in cultured cells.22 While, as expected,22 MAGE-A1 and MAGE-A4 remained stably induced during the time-course of 5-aza treatment, Sat2 RNA had almost returned to basal levels after 4 d of treatment (Fig. 2D), unraveling a very transient induction by the demethylating agent.
Figure 2.Sat2 hypomethylation in DNMT3B-deficient ICFhTERT fibroblasts does not exacerbate heat shock-induced Sat2 RNA expression. (A) Upper panel: schematic representation of the Sat2 fragment analyzed by sequencing. Positions of the 17 CpG sites and of the putative HSF1 binding site are indicated. Lower panel: Sat2 methylation level determined by sequencing of bisulfite-treated DNA molecules isolated from HFF2hTERT and ICFhTERT cells. White squares represent unmethylated CpG, black squares represent methylated CpG. (B) Relative Sat2 RNA levels in HFF2hTERT and ICFhTERT cells. Data are indicated as % ± SEM of the expression measured in HFF2hTERT. (C) Basal and HS-induced Sat2 and HSP70 RNA levels in HFF2hTERT and ICFhTERT. For HS, cells were incubated for 1h at 42°C followed by 1h of recovery at 37°C. Data are presented as % ± SEM of the basal expression measured in HFF2hTERT. (D) HFF2hTERT cells were incubated in the presence or absence (ctrl) of 5 μM 5-aza-2’-deoxycytidine (5-aza) for 1 to 5 d and relative expression levels of Sat2, MAGE-A1, MAGE-A4 and HSP70 were measured by qRT-PCR. Data are presented as % ± SEM of the expression measured in HFF2hTERT control cells.
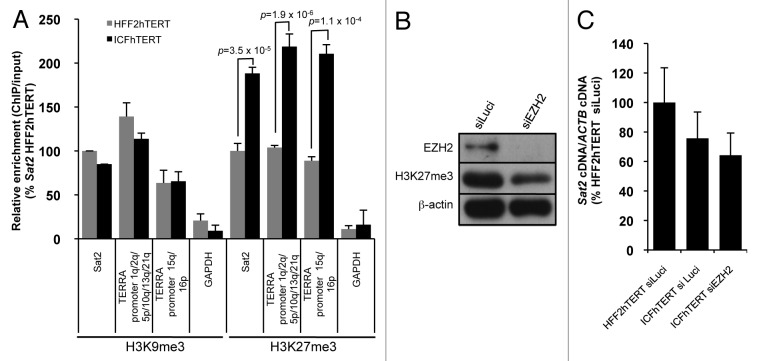
As suggested before by Eymery et al.,16 the failure of Sat2 hypomethylation to constitutively hyperactivate Sat2 transcription may be explained by the acquisition of additional repressive histone marks that would compensate for DNA methylation loss. To address this hypothesis, we performed chromatin immunoprecipitation experiments with antibodies against H3K9me3 or H3K27me3 repressive histone marks in HFF2hTERT and ICFhTERT cell lines. As expected, Sat2 was enriched in both H3K9me3 and H3K27me3 repressive histone marks when compared with the highly uncondensed GAPDH promoter (Fig. 3A). Strikingly, when compared with HFF2hTERT cells, we detected a 2-fold enrichment in H3K27me3 density at Sat2 locus of ICFhTERT cells (p = 3.5x10−5) (Fig. 3A). To test whether this enrichment in H3K27me3 density was specific to hypomethylated Sat2, we analyzed a series of subtelomeric (1q, 2q, 5p, 10q, 13q, 15q, 16p and 21q) TERRA CpG-island promoters.23 TERRA promoter methylation amounts to about 48.5% in normal telomerized fibroblasts and drops to 7.3% in ICFhTERT cells, resulting in strong upregulation of telomere transcription (data not shown). ChIP analyses revealed a similar increase in H3K27me3 density at hypomethylated TERRA promoters, showing that it is not restricted to Sat2 DNA. To test whether increased H3K27me3 density may nevertheless repress basal transcription of Sat2 in ICFhTERT, we transfected cells with siRNAs against EZH2, the histone-lysine N-methyltransferase of Polycomb Repressive Complex 2. However, although global H3K27me3 levels were reduced by 60–65%, we did not detect any upregulation of Sat2 RNA (Fig. 3B and C), suggesting that the increased density of H3K27me3 in ICFhTERT cells does not downregulate steady-state Sat2 RNA levels. On the other hand, H3K9me3 density at Sat2 repeats was similar in HFF2hTERT and ICFhTERT cell lines (Fig. 3A), in agreement with the recent observations that DNA methylation status does not impact on H3K9me3 density at several human chromosomal loci.24
Figure 3. Hypomethylated Sat2 in ICFhTERT is characterized by increased density of H3K27me3 marks. (A) Density of H3K9me3 or H3K27me3 at Sat2 locus of HFF2hTERT and ICFhTERT determined by ChIP followed by qPCR. These marks were also quantified at GAPDH and TERRA (1q, 2q, 5p, 10q, 13q, 15q, 16p and 21q) promoters as controls. ChIP/input ratios were expressed as % ± SEM of the ratios obtained for Sat2 DNA in HFF2hTERT with either H3K9me3 or H3K27me3 antibody. (B) Western blot analysis of EZH2 and H3K27me3 after treatment of ICFhTERT cells with siRNAs against either Luciferase (siLuci) or EZH2 (siEZH2) for 72h. β-actin is shown as loading control. (C) Relative Sat2 RNA levels in ICFhTERT treated as in B and expressed as % ± SEM of Sat2 RNA in HFF2hTERT siLuci control cells.
Sat2 RNA levels correlate with HSP70 expression in melanoma tissues
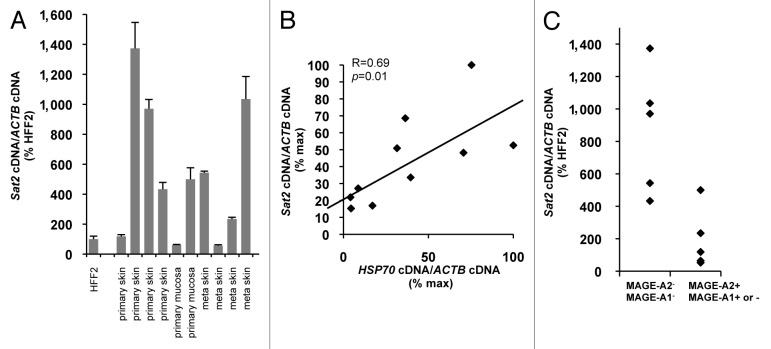
Having concluded that Sat2 hypomethylation does not constitutively upregulate Sat2 RNA levels, we next wished to get hints into Sat2 transcriptional regulation in tumors. The previous demonstration that, when activated experimentally, the HS pathway is able to trigger Sat2 transcription and the established role of HSF1 and heat shock proteins in the maintenance of cancer cell viability,25-29 led us to test whether the HS pathway may similarly regulate Sat2 expression in cancer tissues in vivo. To answer this question, we measured the expression levels of Sat2 and HSP70 in either primary (n = 6) or metastatic (n = 4) melanoma tumors. Sat2 expression levels were highly variable and not statistically different between primary and metastatic melanoma tumors (p = 0.73) (Fig. 4A). Importantly, our results indicated a good correlation between the expression levels of Sat2 and HSP70 in melanoma tissues (R = 0.69, p = 0.01) (Fig. 4B), suggesting that, in vivo, Sat2 RNA levels are correlated with activation of the HS pathway. We believe that, similarly to what was observed for cancer cell lines, DNA hypomethylation is not responsible for constitutive hyperactivation of Sat2 transcription in melanoma tissues as we found no positive correlation between Sat2 RNA and MAGE-A1/2 gene expression, previously linked to profound Sat2 and genome-wide hypomethylation (Fig. 4C).4,30,31
Figure 4.Sat2 RNA levels are correlated with HSP70 expression in melanoma tissues. (A) Relative Sat2 RNA levels measured in 6 primary melanoma tumors and 4 melanoma metastases. Data are presented as % ± SEM of the expression measured in HFF2. (B) Comparison between Sat2 and HSP70 expression levels in melanoma tissues from A. Both Sat2 and HSP70 cDNA levels were normalized to ACTB cDNA and expressed as % of the maximal value measured in the samples. (C) Melanoma tissues from A were classified into two groups according to the presence (+) or the absence (-) of MAGE-A2 or MAGE-A1 transcripts. Relative Sat2 RNA levels, as described in A, are indicated.
Heat shock and RasV12 oncogene induce Sat2 DNA demethylation
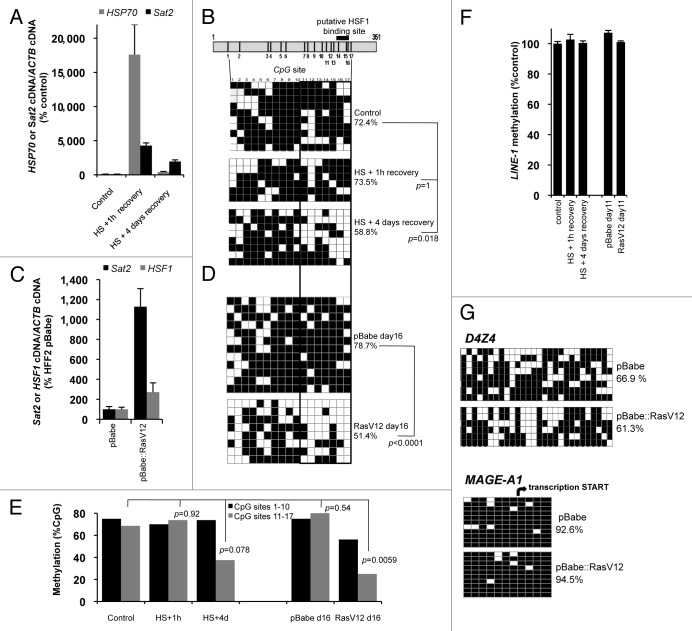
Hyperactivation of the HS pathway is likely to be a common event in the early steps of tumorigenesis, allowing cancerous cells to escape from oncogene-induced senescence.25,26,28,29,32 Given that HS pathway hyperactivation was previously reported to be associated with chromatin remodeling events,10,11,33,34 we next tested the possible role of the HS pathway in inducing Sat2 DNA demethylation. To address this possibility, we exposed HFF2hTERT cells to HS for 1h at 42°C, followed by either 1h or four days of recovery at 37°C. As expected, both HSP70 and Sat2 transcripts were strongly induced after 1h of recovery from HS (Fig. 5A), but resulting Sat2 methylation levels (73.5%) were not statistically different from the ones measured in untreated cells (72.4%) (Yates’ χ2 test: p = 1) (Fig. 5B). Strikingly, however, we detected a reduction of Sat2 methylation by about 14% in heat shocked-fibroblasts allowed to recover for four days at 37°C (p = 0.018) (Fig. 5B). These results indicate that the HS pathway induces Sat2 demethylation by a process that appears to require several rounds of DNA replication (cells underwent three population doublings within the time course of recovery).
Figure 5. Heat shock and RasV12 oncogene induce Sat2 DNA demethylation. (A) Sat2 and HSP70 expression levels in HFF2hTERT cells subjected or not to HS for 1h at 42°C followed by either 1h or 4 d of recovery at 37°C. Data are given as % ± SEM of the expression levels measured in control cells. (B) Upper panel: schematic representation of the Sat2 fragment analyzed by sequencing. The 17 CpG dinucleotides and the putative HSF1 binding site are indicated. Lower panel: analysis of Sat2 methylation by sequencing of individual molecules amplified from bisulfite-treated DNA isolated from cells described in A. White squares represent unmethylated CpG, black squares represent methylated CpG. A Yates’ χ2 test was applied to assess the statistical differences in methylated CpG content with the control. (C) Relative Sat2 and HSF1 RNA levels in HFF2 fibroblasts 16 d after transduction with either empty pBabe or pBabe::RasV12 plasmid and expressed as % ± SEM of the expression in HFF2pBabe. (D) Sat2 methylation analysis of HFF2pBabe and HFF2pBabe::RasV12 determined 16 d after transduction. See B for details. (E) Average methylation level of CpG sites 1 to 10 (black bars) or 11 to 17 (gray bars) of Sat2 determined from B and D. A Yates’ χ2 test was applied to assess the statistical differences in methylated CpG content within CpG sites 11 to 17 in all tested conditions. (F) LINE-1 methylated CpG content determined by qMS-PCR on bisulfite-treated DNA prepared from samples described in B and D. [Methylated LINE-1 /(methylated LINE-1 + unmethylated LINE-1)] ratios were expressed as % ± SEM of LINE-1 methylation in control cells. (G) Analysis of D4Z4 and MAGE-A1 promoter methylation in HFF2pBabe and HFF2pBabe::RasV12 fibroblasts 16 d after transduction.
Ras oncogene was previously reported to activate HSF1.35 Hence, we tested the impact of constitutively activated RasV12 protein, a mutant form frequently detected in tumors,36 on Sat2 methylation. HFF2 fibroblasts were transduced with either pBabe-puro::RasV12 or an empty pBabe-puro vector as control. RasV12 upregulated Sat2 RNA and the transcript was still detected 16 d after retroviral transduction (Fig. 5C). At this late time point, HSP70 mRNA was not detected (data not shown) but HSF1 transcripts were about 3-fold more abundant than in control cells (Fig. 5C). Analysis of RasV12-expressing cells 16 d after the transduction revealed a marked reduction of Sat2 methylation level, which dropped from 78.7% to 51.4% (p < 0.0001) (Fig. 5D). Interestingly, in both HS and RasV12 conditions, Sat2 demethylation appeared to occur mainly within the last 7 CpG dinucleotides of the analyzed sequence, in a region encompassing the putative HSF1 binding site (Fig. 5B, D and E). Altogether, the above results suggested that the heat shock response may be involved in Sat2 DNA demethylation. Importantly, in our experimental conditions, we did not detect any concurrent reduction of the methylation level of either LINE-1 interspersed repetitive element, D4Z4 non-satellite subtelomeric repeats located at 4q35.2 and 10q26.3 or the promoter of cancer germline gene MAGE-A1 (Fig. 5F and G and data not shown), suggesting a specific effect of HS on the demethylation of selected CpG dinucleotides within Sat2 sequence.
Cancer cell lines display epigenetic scars of possible heat shock pathway activation during tumorigenesis
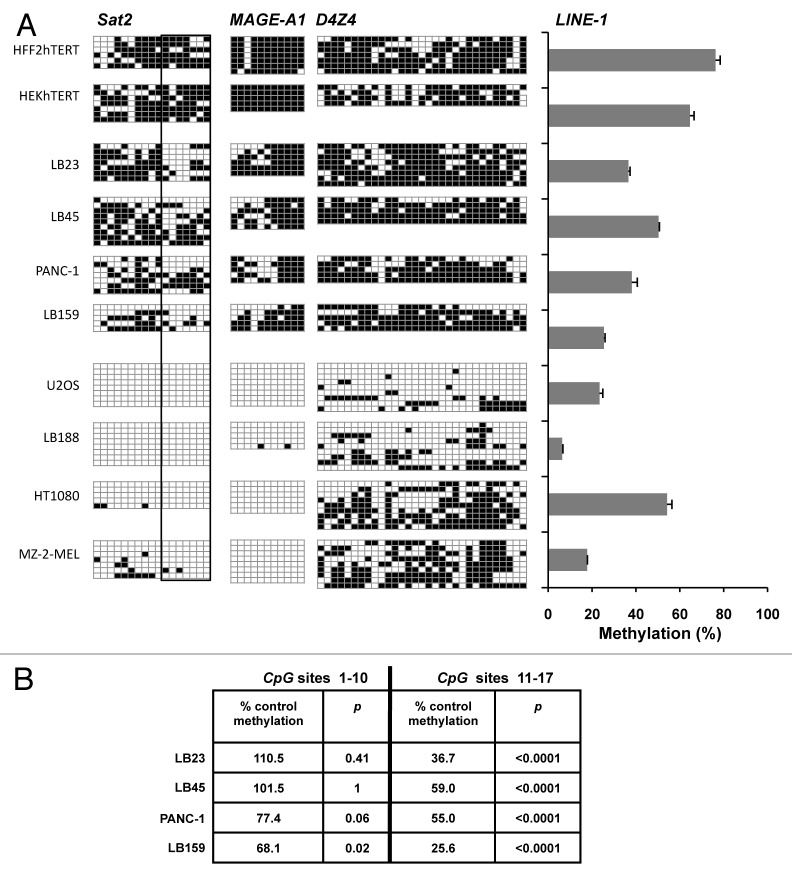
Our in vitro assay showed that HS pathway activation in fibroblasts is able to induce localized demethylation events within the last 7 CpG dinucleotides of Sat2 sequence, giving rise to a HS-dependent demethylation signature. In order to provide evidence that the HS response may be involved in tumorigenesis-associated Sat2 demethylation processes, we analyzed the methylation profile of Sat2 in eight cancer cell lines, using HFF2hTERT and HEKhTERT as non-tumoral reference cell lines. Methylation levels within the MAGE-A1 promoter, D4Z4 and LINE-1 sequences were also tested in these samples. As shown in Figure 6A, four cancer cell lines (LB23, LB45, PANC-1 and LB159) displayed an intermediate level of hypomethylation, as all four tested genomic regions retained a significant proportion of methylated CpGs. The other four cancer cell lines (U2OS, LB188, HT1080 and MZ-2-MEL), instead, were characterized by extensive genome-wide hypomethylation as evidenced by the complete or nearly complete demethylation of the tested loci (Fig. 6A). Closer analysis of cancer cell lines with intermediate genome-wide hypomethylation revealed preferential demethylation of the last 7 CpG dinucleotides within the Sat2 sequence (Fig. 6A and B), similarly to what we observed following experimentally induced HS.
Figure 6. Heat shock pathway-dependent signature of Sat2 demethylation is also observed in cancer cell lines. (A) Left panel: methylation profile of Sat2, D4Z4 and MAGE-A1 promoter determined by sequencing of bisulfite-treated DNA molecules isolated from sarcoma (LB23, U2OS, LB188, HT1080), melanoma (LB45, MZ-2-MEL) and carcinoma (PANC-1, LB159) cell lines and from HFF2hTERT and HEKhTERT non-tumoral cell lines; right panel: LINE-1 methylation level determined by qMS-PCR on bisulfite-treated DNA with [methylated LINE-1 /(methylated LINE-1 + unmethylated LINE-1)] ratios expressed as % ± SEM (B) Relative methylation levels of CpG sites 1–10 or 11–17 of Sat2 in LB23, LB45, PANC-1 and LB159 cancer cell lines compared with the methylated CpG content of Sat2 DNA from HFF2hTERT and HEKhTERT control cells together. Yates’ χ2p-values are indicated.
Discussion
Hypomethylation of repetitive DNA is largely responsible for the global loss of DNA methylation in cancer cells and pericentromeric Sat2 locus is frequently hypomethylated.1 Given the possible important role of repetitive ncRNAs in cancer cell biology, understanding how these ncRNAs are transcriptionally regulated is important. Our quantitative analysis of various cancer cell lines revealed that Sat2 transcription is not constitutively hyperactivated by DNA hypomethylation and, in agreement with a previous study,17 we did not detect constitutive overexpression of Sat2 RNA in DNMT3B-deficient ICFhTERT fibroblasts either. We further showed that Sat2 hypomethylation does not exacerbate Sat2 RNA induction upon heat shock. Because we do not know where Sat2 transcription starts, we cannot however exclude the possibility that, as suggested earlier,17 dispersed chromosomal copies of Sat2, with undefined methylation profiles, may be transcribed and contribute to cellular Sat2 RNA. The previous demonstration that HS induces the production of long molecules of satellite III transcripts emanating mostly—if not entirely—from chromosome 9 pericentromeric region suggests, however, that, if any, short transcripts from dispersed copies of Sat2 are unlikely to contribute significantly to the total amount of Sat2 RNA.10,11
A previous study suggested the existence of two distinct pathways for Sat2 transcriptional induction, one relying on the HS pathway, through HSF1 activation, and the other one induced by treatment with 5-azadeoxycytidine demethylating agent independently of HS pathway activation.16 We found that 5-aza only transiently upregulated Sat2 RNA after three days of treatment without any detectable increase in HSP70 transcription. Two possible explanations may account for this observation: (1) the heat shock pathway has been only transiently activated in response to genome-wide DNA damage37 and, because HSP70 mRNA may be much less stable than Sat2 RNA, we did not detect it, or (2) Sat2 transcription was hyperactivated by DNA demethylation but rapidly shut down by a still unknown mechanism, possibly linked to the immediate acquisition of additional and undefined repressive marks. Although this did not appear to counteract basal Sat2 expression, we found an increase of H3K27me3 density at hypomethylated Sat2 locus in ICFhTERT cells that was also detected at hypomethylated subtelomeric TERRA promoters. The reason why DNA hypomethylation is correlated with increased H3K27me3 density is unknown but fits with the observation that demethylation of the mouse imprinted Rasgrf1 locus is similarly associated with increased H3K27me3 density, and suggests the existence of a—still unexplained—antagonism between these two repressive marks.38 Other repressive marks, like Heterochromatin Protein 1 (HP1), may account for Sat2 transcriptional repression. In support of this, mouse satellite ncRNAs were shown to promote the recruitment of HP1α to pericentric heterochromatin through direct interaction between RNA and the hinge domain of HP1α.39 In the future, it would be interesting to test this hypothesis further.
Overexpression of satellite RNAs was previously detected in a variety of tumors.15 Our analysis of Sat2 expression in primary and metastatic melanoma tumor samples suggested that the level of HS pathway activation in cancer tissues may be the main determinant of Sat2 expression in vivo, irrespective of DNA methylation status. The HS pathway is indeed frequently activated in cancerous lesions that experience oxidative stress, oncogene activation or proteotoxic damages and, not surprisingly, HSF1 is required for cancer cell survival, both in mouse and in human.25-29,32 While this manuscript was in preparation, a study similarly reported that, although frequently detected in prostate cancer tissues, rRNA (rRNA) overexpression is not linked to rDNA promoter hypomethylation, and authors provided evidences that rRNA levels are instead correlated with MYC mRNA levels, a transcriptional activator of rDNA.40 In addition to being required for survival of fully transformed cells, HSF1 also acts as a powerful driver of oncogenic transformation, notably by ensuring escape from oncogene-induced senescence.28,29,32 Hence, we can reasonably assume that the HS pathway is strongly hyperactivated during the first steps of tumorigenesis. We found that both HS treatment and RasV12 oncogene overexpression reduced Sat2 methylation level by, respectively, 13 and 27%, suggesting that hyperactivation of HS pathway may be causally involved in tumorigenesis-driven Sat2 demethylation. In support of this, we observed similar patterns of Sat2 demethylation in cancer cell lines with moderate genome-wide hypomethylation. Interestingly, we found that HS pathway-induced demethylation of Sat2 mainly occurred within a region encompassing the putative HSF1 binding site. These observations are reminiscent to the demonstration that recruitment of DNA-binding factors to chromatin is sufficient to locally induce DNA methylation loss,41 and fits with a more recent base-pair-resolution methylome analysis in mouse stem cells, which unraveled an overlap between low-methylated regions and DNA-binding factor occupancy.42 It is likely that, in light of the role of ncRNA in heterochromatin formation, HS-induced transcriptional upregulation, although leading to localized DNA demethylation, may be associated with the acquisition of other repressive marks to maintain pericentromeric heterochromatin.
Whether HS response hyperactivation may induce demethylation of other repetitive DNA sequences remains to be established as transcription of a variety of other centromeric and pericentric sequences was reported to occur upon HS.16 However, the observation that, instead of being localized within the last 7 CpG sites, demethylated CpG sites spread over the entire Sat2 sequence in cancer cell lines displaying strong genome-wide hypomethylation, suggests the existence of additional and still unexplained demethylation processes that may also be responsible for MAGE-A1 and D4Z4 demethylation. Cancer-testis gene promoter demethylation was reported to be a late event in prostate cancer progression43 and it is assumed that, while cancer-linked DNA hypomethylation is generally found early during carcinogenesis, more extensive DNA hypomethylation can appear later during tumor progression.1 Hence, we propose that HS pathway activation may contribute to initial demethylation events of Sat2, which may, in some instances, be followed by additional undefined genome-wide demethylation events. We do not exclude the possibility that the latter events may be facilitated by—and spread from—early-induced localized demethylation events.
In summary, our results indicate that cancer-linked Sat2 hypomethylation is not associated with constitutive hyperactivation of Sat2 but that HS pathway activation in cancer tissues is likely to be the main determinant of Sat2 expression. Our study further suggests that strong HS response activation during tumorigenesis may play a role in localized demethylation of pericentromeric Sat2. In the future, it would be interesting to address further the role of the HS pathway and its downstream effectors in cancer-linked DNA demethylation.
Materials and Methods
Non-tumoral or cancer cell lines and melanoma tissues
HFF2 normal human foreskin fibroblasts were purchased from ATCC (SCRC-1042). HMEC human mammary epithelial cells and ICF fibroblasts were purchased from Coriell. HNEM normal human epidermal melanocytes and HMSC human mesenchymal stem cells were kindly provided by E De Plaen (Ludwig Institute for Cancer Research) and E Sokal (UCL) respectively. HFF2hTERT,44 ICFhTERT, HMEChTERT and HNEMhTERT cell lines were obtained by retroviral transduction with pBMN::hTERT plasmid (kindly provided by C Heirman, VUB). Cancer cell lines used in this study are listed in Table S1. Cell culture media were from GIBCO (Life Technologies) and FBS was from HyClone (Thermo Fisher Scientific, SV30160.03). HEKhTERT cDNA and gDNA samples were provided by A Londoño-Vallejo (Institut Curie). Melanoma tumor samples were obtained from patients undergoing surgery or tumor resection and immediately frozen in liquid nitrogen. Experimental procedures involving the use of biological material were approved by the Ludwig Institute for Cancer Research Institutional Review Board. All patients gave informed consent mentioning that part of the tumor samples could be used for research purposes. Most tumor biopsies were obtained as part of screening procedures for participation in clinical immunotherapy trials.
Cell treatment with 5-aza-2’-deoxycytidine
HFF2hTERT cells were incubated for 1 to 5 d in culture medium containing 5 μM 5-aza-2’-deoxycytidine (Sigma-Aldrich).
Retroviral transductions with RasV12 oncogene
150 000 HFF2 cells were incubated with retroviral supernatant containing either pBABE-puro or pBABE::Ha-RasV12-puro plasmid (kindly provided by F d’Adda di Fagagna, IFOM foundation, Milan, Italy) and 4 μM polybrene (Sigma-Aldrich, H9268) for 24h. Transduction procedure was repeated once and cells were then selected with 0.8 μg/ml puromycin (Sigma-Aldrich, P7255). The day following the second transduction was designated as day 1.
siRNA transfections
Cells were transfected with Lipofectamine 2000 (Life Technologies, 11668019) and siRNAs (Eurogentec) against either Luciferase (5′CUUACGCUGAGUACUUCGA) or EZH2 (5′GACUCUGAAUGCAGUUGCU).45 650 000 cells were transfected with 0.7 nmole siRNA and collected for RNA or protein extraction 72h post-transfection.
Genomic DNA extraction and sodium bisulfite treatment
Genomic DNA was extracted with the Wizard Genomic DNA Purification Kit following manufacturer’s instructions (Promega, A1125). Sodium bisulfite treatment of 10 μg gDNA was performed as described earlier and resuspended in a final volume of 100 μl.46
Sat2 and LINE-1 methylation measurement by qPCR
When indicated, quantification of Sat2 methylation was achieved using the MethylLight assay on bisulfite-treated gDNA and values were normalized by the amount of CpG-depleted Alu sequences as described previously.3 LINE-1 methylation was quantified as follows. First, a semi-quantitative methylation-independent PCR was performed on a 1:100 dilution of bisulfite-modified gDNA (30 sec at 95°C, 30 sec at 58°C and 30 sec at 72°C for 25 cycles). Next, two independent qPCR were performed on a 1:100 dilution of the first PCR with primers specific for either the methylated or the unmethylated LINE-1 sequence. Annealing temperature was of 60°C for both Sat2 MethylLight and LINE-1 qMS-PCR. Primer and probe sequences are listed in Table S2.
Methylation analysis of Sat2, D4Z4 and MAGE-A1 promoter by sequencing
Methylation CpG content determination of Sat2,20 D4Z4,31 and MAGE-A1 promoter,47 was performed by sequencing of bisulfite-treated DNA using primers listed in Table S2. In all cases, the final PCR product was cloned into pJET 1.2/blunt vector using CloneJET PCR Cloning kit (Fermentas, Thermo Fischer Scientific, K1232) before sequencing (Macrogen Inc.).
Quantitative RT-PCR
Total RNA was extracted from cell lines using TriPure Isolation Reagent (Roche Applied Science) and treated with DNaseI (Ambion, Life Technologies, AM2238) according to manufacturer’s instructions. Reverse transcription was performed as described previously.31 Sat2/ACTB cDNA ratios were normalized to Sat2/Alu gDNA ratios determined by qPCR on a 1:1000 dilution of gDNA. The Alu sequences that were amplified correspond to an Alu consensus motif determined from the sequences of ancient and more recent individual Alu repeats subfamilies (Alu-J, SluSp, AluSx, AluSq, AluSc, AluY, AluSb2, AluYb8, AluYa5 and AluYa8).3 Annealing temperature was of 60°C for all qPCR except for ACTB (β-actin) (62°C). qPCR reactions were performed using either TaqMan or SYBR Green technology. Primers and probes (listed in Table S2), qPCR Core kit and SYBR Green were purchased from Eurogentec.
Western blot
Cell lysates were obtained by resuspending cell pellets into 2X Laemmli sample buffer (25% glycerol, 2% SDS, 0.01% bromophenol blue, 0.0625 M TRIS-HCl, pH 6.8) with Complete mini protease inhibitor cocktail (Roche Applied Science, 11836153001). The following antibodies were used: EZH2 (Millipore, 17–662, 1:2000), H3K27me3 (Active Motif, 39156–39158, 1:10000) and β-Actin (Sigma-Aldrich, A5441, 1:50000).
CHIP assay
About 20x106 cells were cross-linked with 1% formaldehyde (Sigma-Aldrich, 344198) at RT for 10 min before addition of 125 mM glycine (Sigma-Aldrich, G7126) followed by resuspension into lysis buffer [1% SDS, 10 mM EDTA, 50 mM Tris, pH 8.1, 1x Complete mini complete protease inhibitor cocktail (Roche, 11836153001)] and sonication with Bioruptor (Diagenode) for 9x30s. Immunoprecipitations were performed with OneDay ChIP kit (Diagenode, Kch-oneDIP-180) following manufacturer’s instructions and with antibodies purchased from Millipore (H3K9me3, 07–442; H3K27me3, 17–622). Immunoprecipitated Sat2 DNA was quantified by qPCR, together with GAPDH and subtelomeric TERRA (1q, 2q, 5p, 10q, 13q, 15q, 16p and 21q) promoter DNA as controls. Primers are described in Table S2.
Statistical analyses
Results obtained from experiments relying on either qRT-PCR or qPCR on bisulfite-treated gDNA are shown as means ± standard error of the mean (SEM). SEM was obtained by dividing the standard deviation by the square root of the number of measurements performed (SD/√n). Results obtained after sequencing of bisulfite-treated gDNA molecules are given as means ± standard deviations. p-values were calculated with Student t-tests. Linear regression analyses and Yates’ chi-square (χ2) were performed on http://faculty.vassar.edu/lowry/VassarStats.html.
Supplementary Material
Acknowledgments
This work was supported by a Mandat d’Impulsion Scientifique from the Fonds National de la Recherche Scientifique (MIS/FNRS) and by the Special Funds for Research (FSR) from the Catholic University of Louvain. GT was supported by the Télévie/FNRS and the de Duve Institute. AD, NA, SL and AL were supported by the FNRS. We are grateful to M Swinarska, F Brasseur and E De Plaen (Ludwig Institute for Cancer Research, Brussels), E Sokal (UCL, Brussels), C Heirman (VUB, Brussels), F Fuks, F Journe and H Id Boukfer (ULB, Brussels), J Piette (ULg, Liège), J Lingner (ISREC, Lausanne, Switzerland), A Londoño-Vallejo (Institut Curie, Paris, France), C Hill (Cancer Research UK, London) and F d’Adda di Fagagna (IFOM foundation, Milan, Italy) for sharing reagents, plasmids, melanoma tumor RNA samples and cell lines. We thank all members of the GEPI group for helpful discussions and the de Duve Institute for constant support.
Glossary
Abbreviations:
- Sat2
satellite 2
- HS
heat shock
- ICF
immunodeficiency, centromeric region instability and facial anomalies
- ncRNAs
non-coding RNAs
- HSF1
heat shock factor 1
- DNMT
DNA methyltransferase
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/21107
References
- 1.Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics. 2009;1:239–59. doi: 10.2217/epi.09.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Smet C, Loriot A. DNA hypomethylation in cancer: Epigenetic scars of a neoplastic journey. Epigenetics. 2010;5:206–13. doi: 10.4161/epi.5.3.11447. [DOI] [PubMed] [Google Scholar]
- 3.Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, et al. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33:6823–36. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cadieux B, Ching TT, VandenBerg SR, Costello JF. Genome-wide hypomethylation in human glioblastomas associated with specific copy number alteration, methylenetetrahydrofolate reductase allele status, and increased proliferation. Cancer Res. 2006;66:8469–76. doi: 10.1158/0008-5472.CAN-06-1547. [DOI] [PubMed] [Google Scholar]
- 5.Choi SH, Worswick S, Byun HM, Shear T, Soussa JC, Wolff EM, et al. Changes in DNA methylation of tandem DNA repeats are different from interspersed repeats in cancer. Int J Cancer. 2009;125:723–9. doi: 10.1002/ijc.24384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ehrlich M, Hopkins NE, Jiang G, Dome JS, Yu MC, Woods CB, et al. Satellite DNA hypomethylation in karyotyped Wilms tumors. Cancer Genet Cytogenet. 2003;141:97–105. doi: 10.1016/S0165-4608(02)00668-4. [DOI] [PubMed] [Google Scholar]
- 7.Sawyer JR, Tian E, Thomas E, Koller M, Stangeby C, Sammartino G, et al. Evidence for a novel mechanism for gene amplification in multiple myeloma: 1q12 pericentromeric heterochromatin mediates breakage-fusion-bridge cycles of a 1q12 approximately 23 amplicon. Br J Haematol. 2009;147:484–94. doi: 10.1111/j.1365-2141.2009.07869.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eymery A, Callanan M, Vourc’h C. The secret message of heterochromatin: new insights into the mechanisms and function of centromeric and pericentric repeat sequence transcription. Int J Dev Biol. 2009;53:259–68. doi: 10.1387/ijdb.082673ae. a. [DOI] [PubMed] [Google Scholar]
- 9.Probst AV, Okamoto I, Casanova M, El Marjou F, Le Baccon P, Almouzni G. A strand-specific burst in transcription of pericentric satellites is required for chromocenter formation and early mouse development. Dev Cell. 2010;19:625–38. doi: 10.1016/j.devcel.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 10.Jolly C, Metz A, Govin J, Vigneron M, Turner BM, Khochbin S, et al. Stress-induced transcription of satellite III repeats. J Cell Biol. 2004;164:25–33. doi: 10.1083/jcb.200306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rizzi N, Denegri M, Chiodi I, Corioni M, Valgardsdottir R, Cobianchi F, et al. Transcriptional activation of a constitutive heterochromatic domain of the human genome in response to heat shock. Mol Biol Cell. 2004;15:543–51. doi: 10.1091/mbc.E03-07-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valgardsdottir R, Chiodi I, Giordano M, Rossi A, Bazzini S, Ghigna C, et al. Transcription of Satellite III non-coding RNAs is a general stress response in human cells. Nucleic Acids Res. 2008;36:423–34. doi: 10.1093/nar/gkm1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eymery A, Souchier C, Vourc’h C, Jolly C. Heat shock factor 1 binds to and transcribes satellite II and III sequences at several pericentromeric regions in heat-shocked cells. Exp Cell Res. 2010;316:1845–55. doi: 10.1016/j.yexcr.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 14.Vourc’h C, Biamonti G. Transcription of satellite DNAs in mammals. Prog Mol Subcell Biol. 2011;51:95–118. doi: 10.1007/978-3-642-16502-3_5. [DOI] [PubMed] [Google Scholar]
- 15.Ting DT, Lipson D, Paul S, Brannigan BW, Akhavanfard S, Coffman EJ, et al. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science. 2011;331:593–6. doi: 10.1126/science.1200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eymery A, Horard B, El Atifi-Borel M, Fourel G, Berger F, Vitte AL, et al. A transcriptomic analysis of human centromeric and pericentric sequences in normal and tumor cells. Nucleic Acids Res. 2009;37:6340–54. doi: 10.1093/nar/gkp639. b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alexiadis V, Ballestas ME, Sanchez C, Winokur S, Vedanarayanan V, Warren M, et al. RNAPol-ChIP analysis of transcription from FSHD-linked tandem repeats and satellite DNA. Biochim Biophys Acta. 2007;1769:29–40. doi: 10.1016/j.bbaexp.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7:e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lafferty-Whyte K, Cairney CJ, Will MB, Serakinci N, Daidone MG, Zaffaroni N, et al. A gene expression signature classifying telomerase and ALT immortalization reveals an hTERT regulatory network and suggests a mesenchymal stem cell origin for ALT. Oncogene. 2009;28:3765–74. doi: 10.1038/onc.2009.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hassan KM, Norwood T, Gimelli G, Gartler SM, Hansen RS. Satellite 2 methylation patterns in normal and ICF syndrome cells and association of hypomethylation with advanced replication. Hum Genet. 2001;109:452–62. doi: 10.1007/s004390100590. [DOI] [PubMed] [Google Scholar]
- 21.Abravaya K, Phillips B, Morimoto RI. Heat shock-induced interactions of heat shock transcription factor and the human hsp70 promoter examined by in vivo footprinting. Mol Cell Biol. 1991;11:586–92. doi: 10.1128/mcb.11.1.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weber J, Salgaller M, Samid D, Johnson B, Herlyn M, Lassam N, et al. Expression of the MAGE-1 tumor antigen is up-regulated by the demethylating agent 5-aza-2′-deoxycytidine. Cancer Res. 1994;54:1766–71. [PubMed] [Google Scholar]
- 23.Nergadze SG, Farnung BO, Wischnewski H, Khoriauli L, Vitelli V, Chawla R, et al. CpG-island promoters drive transcription of human telomeres. RNA. 2009;15:2186–94. doi: 10.1261/rna.1748309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeng W, de Greef JC, Chen YY, Chien R, Kong X, Gregson HC, et al. Specific loss of histone H3 lysine 9 trimethylation and HP1γ/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD) PLoS Genet. 2009;5:e1000559. doi: 10.1371/journal.pgen.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calderwood SK, Khaleque MA, Sawyer DB, Ciocca DR. Heat shock proteins in cancer: chaperones of tumorigenesis. Trends Biochem Sci. 2006;31:164–72. doi: 10.1016/j.tibs.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 26.Dai C, Whitesell L, Rogers AB, Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130:1005–18. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Min JN, Huang L, Zimonjic DB, Moskophidis D, Mivechi NF. Selective suppression of lymphomas by functional loss of Hsf1 in a p53-deficient mouse model for spontaneous tumors. Oncogene. 2007;26:5086–97. doi: 10.1038/sj.onc.1210317. [DOI] [PubMed] [Google Scholar]
- 28.Meng L, Gabai VL, Sherman MY. Heat-shock transcription factor HSF1 has a critical role in human epidermal growth factor receptor-2-induced cellular transformation and tumorigenesis. Oncogene. 2010;29:5204–13. doi: 10.1038/onc.2010.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meng L, Hunt C, Yaglom JA, Gabai VL, Sherman MY. Heat shock protein Hsp72 plays an essential role in Her2-induced mammary tumorigenesis. Oncogene. 2011;30:2836–45. doi: 10.1038/onc.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Smet C, De Backer O, Faraoni I, Lurquin C, Brasseur F, Boon T. The activation of human gene MAGE-1 in tumor cells is correlated with genome-wide demethylation. Proc Natl Acad Sci U S A. 1996;93:7149–53. doi: 10.1073/pnas.93.14.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tilman G, Loriot A, Van Beneden A, Arnoult N, Londoño-Vallejo JA, De Smet C, et al. Subtelomeric DNA hypomethylation is not required for telomeric sister chromatid exchanges in ALT cells. Oncogene. 2009;28:1682–93. doi: 10.1038/onc.2009.23. [DOI] [PubMed] [Google Scholar]
- 32.Gabai VL, Yaglom JA, Waldman T, Sherman MY. Heat shock protein Hsp72 controls oncogene-induced senescence pathways in cancer cells. Mol Cell Biol. 2009;29:559–69. doi: 10.1128/MCB.01041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Corey LL, Weirich CS, Benjamin IJ, Kingston RE. Localized recruitment of a chromatin-remodeling activity by an activator in vivo drives transcriptional elongation. Genes Dev. 2003;17:1392–401. doi: 10.1101/gad.1071803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inouye S, Fujimoto M, Nakamura T, Takaki E, Hayashida N, Hai T, et al. Heat shock transcription factor 1 opens chromatin structure of interleukin-6 promoter to facilitate binding of an activator or a repressor. J Biol Chem. 2007;282:33210–7. doi: 10.1074/jbc.M704471200. [DOI] [PubMed] [Google Scholar]
- 35.Stanhill A, Levin V, Hendel A, Shachar I, Kazanov D, Arber N, et al. Ha-ras(val12) induces HSP70b transcription via the HSE/HSF1 system, but HSP70b expression is suppressed in Ha-ras(val12)-transformed cells. Oncogene. 2006;25:1485–95. doi: 10.1038/sj.onc.1209193. [DOI] [PubMed] [Google Scholar]
- 36.Macara IG, Lounsbury KM, Richards SA, McKiernan C, Bar-Sagi D. The Ras superfamily of GTPases. FASEB J. 1996;10:625–30. doi: 10.1096/fasebj.10.5.8621061. [DOI] [PubMed] [Google Scholar]
- 37.Palii SS, Van Emburgh BO, Sankpal UT, Brown KD, Robertson KD. DNA methylation inhibitor 5-Aza-2′-deoxycytidine induces reversible genome-wide DNA damage that is distinctly influenced by DNA methyltransferases 1 and 3B. Mol Cell Biol. 2008;28:752–71. doi: 10.1128/MCB.01799-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindroth AM, Park YJ, McLean CM, Dokshin GA, Persson JM, Herman H, et al. Antagonism between DNA and H3K27 methylation at the imprinted Rasgrf1 locus. PLoS Genet. 2008;4:e1000145. doi: 10.1371/journal.pgen.1000145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maison C, Bailly D, Roche D, Montes de Oca R, Probst AV, Vassias I, et al. SUMOylation promotes de novo targeting of HP1α to pericentric heterochromatin. Nat Genet. 2011;43:220–7. doi: 10.1038/ng.765. [DOI] [PubMed] [Google Scholar]
- 40.Uemura M, Zheng Q, Koh CM, Nelson WG, Yegnasubramanian S, De Marzo AM. Overexpression of ribosomal RNA in prostate cancer is common but not linked to rDNA promoter hypomethylation. Oncogene. 2012;31:1254–63. doi: 10.1038/onc.2011.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsieh CL. Dynamics of DNA methylation pattern. Curr Opin Genet Dev. 2000;10:224–8. doi: 10.1016/S0959-437X(00)00064-2. [DOI] [PubMed] [Google Scholar]
- 42.Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Schöler A, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–5. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 43.Yegnasubramanian S, Haffner MC, Zhang Y, Gurel B, Cornish TC, Wu Z, et al. DNA hypomethylation arises later in prostate cancer progression than CpG island hypermethylation and contributes to metastatic tumor heterogeneity. Cancer Res. 2008;68:8954–67. doi: 10.1158/0008-5472.CAN-07-6088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mattiussi M, Tilman G, Lenglez S, Decottignies A. Human telomerase represses ROS-dependent cellular responses to Tumor Necrosis Factor-α without affecting NF-κB activation. Cell Signal. 2012;24:708–17. doi: 10.1016/j.cellsig.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 45.Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–35. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Smet C, Loriot A, Boon T. Promoter-dependent mechanism leading to selective hypomethylation within the 5′ region of gene MAGE-A1 in tumor cells. Mol Cell Biol. 2004;24:4781–90. doi: 10.1128/MCB.24.11.4781-4790.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loriot A, De Plaen E, Boon T, De Smet C. Transient down-regulation of DNMT1 methyltransferase leads to activation and stable hypomethylation of MAGE-A1 in melanoma cells. J Biol Chem. 2006;281:10118–26. doi: 10.1074/jbc.M510469200. [DOI] [PubMed] [Google Scholar]
- 48.Ding XZ, Smallridge RC, Galloway RJ, Kiang JG. Rapid assay of HSF1 and HSF2 gene expression by RT-PCR. Mol Cell Biochem. 1996;158:189–92. doi: 10.1007/BF00225845. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.