Abstract
Next-generation sequencing technologies are rapidly transforming molecular systematic studies of non-model animal taxa. The arachnid order Opiliones (commonly known as “harvestmen”) includes more than 6,400 described species placed into four well-supported lineages (suborders). Fossil plus molecular clock evidence indicates that these lineages were diverging in the late Silurian to mid-Carboniferous, with some fossil harvestmen representing the earliest known land animals. Perhaps because of this ancient divergence, phylogenetic resolution of subordinal interrelationships within Opiliones has been difficult. We present the first phylogenomics analysis for harvestmen, derived from comparative RNA-Seq data for eight species representing all suborders. Over 30 gigabases of original Illumina short-read data were used in de novo assemblies, resulting in 50–80,000 transcripts per taxon. Transcripts were compared to published scorpion and tick genomics data, and a stringent filtering process was used to identify over 350 putatively single-copy, orthologous protein-coding genes shared among taxa. Phylogenetic analyses using various partitioning strategies, data coding schemes, and analytical methods overwhelmingly support the “classical” hypothesis of Opiliones relationships, including the higher-level clades Palpatores and Phalangida. Relaxed molecular clock analyses using multiple alternative fossil calibration strategies corroborate ancient divergences within Opiliones that are possibly deeper than the recorded fossil record indicates. The assembled data matrices, comprising genes that are conserved, highly expressed, and varying in length and phylogenetic informativeness, represent an important resource for future molecular systematic studies of Opiliones and other arachnid groups.
Introduction
Animal molecular systematics is in the midst of transformation. Until very recently, studies of non-model taxa have relied upon a “PCR + Sanger Sequencing” strategy for a frustratingly small set of genes representing a minute fraction of the genome. These genes were for the most part “useable” rather than optimal (e.g., spread across the genome, evolving at different rates, easy to align, etc.). Various next-generation sequencing (NGS) technologies [1], [2] are rapidly changing this landscape, allowing for the comparative collection of partial or entire genomic data, even for non-model taxa. As proof of concept, Hittinger et al. [3] used Illumina short-read technology to generate comparative cDNA data (RNA-Seq [4]) for ten Anopheles mosquito species. These authors showed that large, highly-expressed, shared gene sets (>100 orthologous genes) could be consistently recovered using this strategy, and that these gene sets enabled robust phylogenomic analysis. This NGS-facilitated phylogenomics approach has also been extended to deeper phylogenetic levels in animals (e.g., in mollusks [5], in myzostomid worms [6]). As NGS data becomes less expensive, systematists working on non-model taxa will continue to scale up the taxonomic breadth of comparative phylogenomic datasets. Also, these comparative datasets provide glimpses of previously unknown genomes, facilitating downstream marker development, SNP discovery, and targeted NGS approaches [7]–[9].
The chelicerate arthropod class Arachnida (e.g., scorpions, spiders, mites, etc.) is rich in taxonomic diversity (eleven traditional extant orders, ∼97,000 described species [10]), but poorly characterized from a comparative genomics perspective. Complete genomes are currently only available for the Order Acari (mites and ticks; Rhipicephalus (Boophilus) microplus [11], Ixodes scapularis [12]), although various scorpions and spiders have been the subject of traditional EST or more recent RNA-Seq studies [13]–[16]. The order Opiliones is the third largest arachnid clade, with 46 families, approximately 1,500 genera, and more than 6,400 described species [17], [18]. Commonly known as harvestmen, Opiliones taxa are conspicuous members of nearly all terrestrial communities, and present compelling but generally under-appreciated opportunities for studies of ecology, sexual selection, historical biogeography, etcetera [19]. To date, large-scale studies of the nuclear genomes of harvestmen have not been conducted.
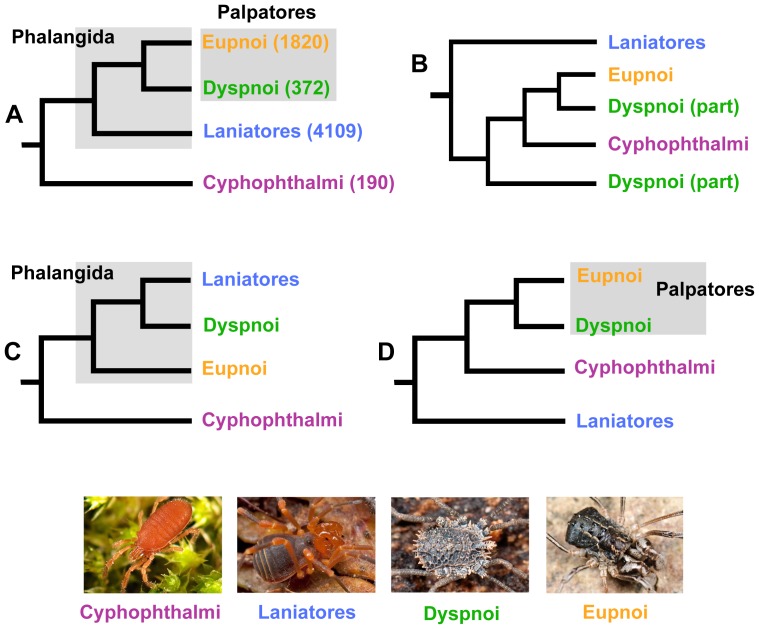
Two phylogenetic facts regarding Opiliones seem clear – the order appears monophyletic, and is comprised of four primary clades (Figure 1). Opiliones monophyly is supported by molecular phylogenetic studies [20], and several hypothesized morphological synapomorphies (e.g., prosomal defensive glands, direct sperm transfer organ, etc. [21], [22]). The four primary harvestmen clades (suborders) include the Cyphophthalmi, Eupnoi, Dyspnoi, and the Laniatores. Phylogenetic support for the monophyly of these individual clades is summarized in Giribet and Kury [20].
Figure 1. Alternative phylogenetic hypotheses for Opiliones.
A) Classical higher-level phylogeny for Opiliones. Number of described species per clade from Kury [18]. B) Cyphopalpatores hypothesis. C) Dyspnolaniatores hypothesis. D) Laniatores basal hypothesis. For details see Table 1. E-H Representative live animals: E) Siro (Cyphophthalmi) F) Sclerobunus (Laniatores) G) Ortholasma (Dyspnoi) H) Protolophus (Eupnoi). Images by M. Hedin.
There are several deep-level phylogenetic problems in harvestmen that are still unresolved to varying degrees. The arachnid sister group to Opiliones remains elusive [23]. Morphological characters suggest that harvestmen are either sister to scorpions, or sister to a larger clade that includes scorpions, pseudoscorpions, and solifugids. Recent molecular phylogenetic and phylogenomic analyses generally fail to resolve the placement of harvestmen, and do not support a close Opiliones plus Scorpiones relationship [24]–[26]. Within Opiliones there is uncertainty regarding interrelationships of the four primary lineages, particularly in placement of the root, and whether Eupnoi and Dyspnoi together form a clade called Palpatores. The “classical” hypothesis of Hansen and Sørensen [27] is that Cyphophthalmi is sister to other suborders (together called Phalangida), and that Palpatores is monophyletic (Figure 1A). This topology is the most commonly supported hypothesis in recent modern analyses (Table 1). However, the early qualitative work of Martens and co-workers [28]–[30] differs considerably from this perspective (Figure 1B, Table 1), and there are multiple subsequent quantitative phylogenetic studies that suggest alternatives to the classical view (Figure 1C, 1D; Table 1). These alternatives do not appear to strictly hinge upon data type (i.e., molecules versus morphology) or analytical method (e.g., parsimony versus model-based approaches).
Table 1. Summary of subordinal phylogenetic hypotheses for Opiliones.
| Author | Data/Analysis | Comments | |
| Classical Hypothesis | Hansen and Sorensen [27] | Morphology; non-cladistic | |
| Shultz [75] | Morphology; parsimony | ||
| Giribet et al. [76] | rRNA; parsimony | ||
| Shultz and Regier [77] | EF1alpha, POL II; parsimony, maximum likelihood (ML) | No outgroups | |
| Giribet et al. [37] | 5 genes; ML | ||
| Hedin et al. [78] | EF1alpha; parsimony | No outgroups | |
| Sharma and Giribet [35] | 10 genes; ML, Bayesian | no outgroups; 4 mitochondrial genes | |
| Garwood et al. [33] | Combined rRNA and morphology; Bayesian | ||
| Cyphopalpatores Hypothesis | Martens [29], [30] | Genital morphology; non-cladistic | |
| Dyspnolaniatores Hypothesis | Giribet et al. [76] | Combined rRNA and morphology; parsimony | |
| Giribet et al. [79] | rRNA, morphology, combined; parsimony | ||
| Garwood et al. [33] | Combined rRNA and morphology; direct optimization parsimony | weakly supported (Fig. 3) | |
| Laniatores Basal Hypothesis | Giribet et al. [37] | 5 genes; direct optimization parsimony | weakly supported (Fig. 5) |
| Garwood et al. [33] | rRNA; Bayesian | weakly supported |
Opiliones suborders are ancient groups, and as such, phylogenetic resolution of subordinal interrelationships might be expected to be difficult. The oldest known fossil harvestmen (Eophalangium) from the Devonian Rhynie Chert (∼410 Myr) represents one of the earliest known land animals [31], [32]. This long-legged fossil has been classified as a Eupnoi based mostly on genital morphology, but a phylogenetic analysis has never been conducted. Recently, formal combined evidence phylogenetic analyses have placed well-preserved Carboniferous (∼305 Myr) harvestmen as members of Dyspnoi (Ameticos) and Eupnoi (Macrogyion) with reasonably high confidence [33]. The fossil record for Cyphophthalmi and Laniatores does not include Paleozoic forms [34], but molecular clock analyses suggest ancient ancestry. Relaxed clock BEAST analyses of nucleotides using an uncorrelated lognormal model estimate a most recent common ancestor for Laniatores at ∼350 Myr (95% highest posterior density (HPD) 307–382 [35]), and a most recent common ancestor for Cyphophthalmi at ∼330 Myr (95% HPD 297–362 [36]). Taken together, fossil plus molecular clock evidence indicates that land-dwelling Opiliones suborders were diverging in the late Silurian to mid-Carboniferous time interval [37].
Although previous phylogenetic studies in Opiliones have used mitochondrial, rRNA, and a handful of nuclear protein-coding genes, a phylogenomic analysis has never been conducted. Phylogenomics-scale data are expected to help resolve ancient, relatively rapid diversification events in the tree of life, although many analytical caveats exist [38]. Here we present the first phylogenomics analysis for Opiliones, derived from comparative Illumina RNA-Seq data for a taxon set representing all four primary harvestmen lineages.
Materials and Methods
Taxon Sample, RNA-Seq
None of the exemplar species used are of conservation concern, and none were collected on lands requiring special collecting permits. Our taxon sample includes representatives of all four primary harvestmen suborders: Cyphophthalmi (represented by Siro), Eupnoi (Leiobunum and Protolophus), Dyspnoi (Ortholasma, Trogulus, and Hesperonemastoma), and Laniatores (Sitalcina and Sclerobunus). The Dyspnoi sample includes representatives of the two primary Dyspnoi lineages (Ischryopsalidoidea and Troguloidea [28]), as does the Laniatores sample (“Insidiatores” and Grassatores [35]). The Eupnoi sample includes representatives of only the Phalangioidea, but not the primary sister lineage Caddoidea. Additional taxonomic information is provided in Table 2.
Table 2. Taxonomic and RNA Extraction Information.
| Suborder | Species | Geographic Origin | Sex, No. Individuals, Tissues |
| Cyphophthalmi | Siro acaroides (Ewing, 1923) | west of Wemme, Clackamas County, Oregon, USA | 8 adults (sex unknown) - entire |
| Eupnoi | Protolophus singularis Banks, 1893 | near Guatay, San Diego County, California, USA | 1 male cephalothorax |
| “ | Leiobunum verrucosum (Wood, 1870) | near Damascus, Montgomery County, Maryland, USA | 2 immatures –midgut removed |
| Dyspnoi | Ortholasma coronadense Cockerell, 1916 | San Diego, San Diego County, California, USA | 1M, 2F –midgut removed |
| “ | Trogulus martensi Chemini, 1983 | Unteruhldingen, Baden-Wurttemberg, Germany | 2 adults (sex unknown) – one entire, one midgut removed |
| “ | Hesperonemastoma modestum (Banks, 1894) | Palomar Mountain, San Diego County, California, USA | 10 adults (sex unknown) - entire |
| Laniatores | Sitalcina lobata Goodnight and Goodnight, 1942 | near Julian, San Diego County, California, USA | 3M, 4F - entire |
| “ | Sclerobunus nondimorphicus Briggs, 1971 | near Rhododendron, Clackamas County, Oregon, USA | Two adults (sex unknown) –midgut removed |
Specimens destined for RNA extractions were either sacrificed immediately prior to extraction, or preserved as fresh specimens in cold RNAlater, then extracted later. We sought “general purpose” transcriptomes, and typically extracted from whole animals, sometimes combining several small specimens in a single extraction (Table 2). Total RNA was extracted using TRIzol (Invitrogen, Carlsbad, CA) and the RNeasy Minikit (Qiagen, Valencia, CA). Protolophus total RNA was shipped to Cofactor Genomics (www.cofactorgenomics.com), where a duplex-specific nuclease (DSN) normalized library was constructed, and 60 basepair (bp) paired-end reads were generated using Illumina GAII technology. All other RNA samples were sent to the Genomic Services Lab at the HudsonAlpha Institute for Biotechnology (www.hudsonalpha.org), where non-normalized libraries were prepared using the Illumina TruSeq RNASeq kit, and sequenced as 50-bp paired-end reads using Illumina HiSeq technology.
de Novo Assembly
Data for Protolophus, Sitalcina, and Ortholasma were passed through an Illumina chastity filter, but were otherwise not filtered prior to de novo assembly. The program FastQC [39] was used to confirm data quality prior to assembly for these taxa. Fastq files for all other taxa were filtered for low-quality reads prior to de novo assembly using a custom Python script. De novo assemblies of paired-end data were constructed with Oases v 0.1.18 [40], which uses as input preliminary assemblies produced by Velvet v1.1.05 [41]. A k-mer length of 31 was used for all assemblies. Gene transcripts (contigs) resulting from assemblies were given taxon-specific names in text files, then all transcripts were combined into a single “eight harvestmen transcript” file. Oases sometimes provides alternative assemblies for the same putative transcript; these redundant transcripts were retained at this stage of analysis.
SCORP - Matrices with Scorpion & Ixodes as Outgroups
Nucleotide data from three different scorpion venom EST projects (Heterometrus, [14]; Hadrurus [13]; Lychas [42]) and a targeted Sanger sequencing phylogenomics project (Hadrurus [24]) were retrieved from GenBank. FASTA files from these projects were combined into a single file, which was searched locally against the eight harvestmen transcript file using tblastx (max hits = 20, max e value 1e-10) in Geneious Pro v5.4 [43]. Searches resulting in short regions of overlap (<200 bp) or apparent paralogy (multiple divergent gene transcripts from the same harvestmen taxon, for one or more taxa) were discarded. Genes were only retained if Siro, and at least one taxon representing Laniatores, Dyspnoi and Eupnoi, were present for the target gene region. For promising candidate genes, the scorpion and Siro sequences were searched locally against the tick (Ixodes) peptide set (blastx, max hits = 2, max e value 1e-10), downloaded from VectorBase (http://iscapularis.vectorbase.org/). Ixodes sequences were retained if both Siro and scorpion matched the same sequence. Nucleotide data were initially aligned using the Translation alignment tool in Geneious Pro (12∶3 gap open to gap extension penalty ratio). Non-coding data 5′ of start codons, and 3′ of stop codons (not always present in partial scorpion gene fragments) were trimmed from matrices, and matrices were re-aligned using the Geneious Pro MAFFT v6.814b plug-in [44] with default parameter settings. Assembled transcriptomes for some taxa (Protolophus in particular, see Results) were fragmented for certain genes – i.e., represented by multiple transcripts arranged in an adjacent or minimally overlapping manner for a single putative ortholog. If orthology seemed apparent (e.g., for Protolophus we expected high similarity to Leiobunum sequences and identical Protolophus sequence in regions of transcript overlap), these multiple different transcripts were merged into a single contiguous “super-transcript” for that taxon [3]. Matrices were exported to EXCEL to check that harvestmen transcripts were not represented more than once in the SCORP gene set.
Tick - Matrices with Ixodes only as Outgroup
Annotations available at VectorBase were used to classify Ixodes proteins into the following categories: single copy, single exon proteins (SCSE, n = 507); proteins with introns but no annotated paralogs (IntNoPar, n = 3286); proteins with a single annotated paralog (SPar, n = 1160). This reduced Ixodes protein set was searched locally against the eight harvestmen transcript file (tblastn, max hits = 20, max e value 1e-10). A strict filter was applied to retain candidate proteins: 1) only proteins represented in all 8 harvestmen transcriptomes were retained; 2) genes or gene regions less than 300 bp were discarded; 3) proteins with evidence for paralogy (multiple divergent transcripts for the same region for a single taxon) in one or more taxa were discarded; 4) harvestmen proteins with high similarity, but conspicuously longer reading frames than in Ixodes, were discarded; 5) difficult-to-align proteins (>5% gaps in tblastn alignments) were discarded; 6) complete proteins with both start and stop codons in harvestmen were preferred.
As for the SCORP matrices, nucleotide data were initially aligned using the Translation alignment tool in Geneious Pro, terminal non-coding data were trimmed, and matrices were re-aligned using MAFFT. In some cases “super-transcripts” were merged (see Results). Matrices were exported to EXCEL to check that harvestmen transcripts were not represented more than once in the TICK gene set, and were not also found in the SCORP gene set. We discontinued the above process after finding 300 suitable and unique TICK proteins.
Phylogenomics
The 300 TICK proteins were subdivided into 3 separate 100-gene bins based on relative rates of evolution [45], using average pairwise nucleotide divergence as a measure of such rate variation (calculated in Geneious Pro). Five total concatenated nucleotide data matrices (SCORP, TICKFAST, TICKMED, TICKSLOW, TICKALL) were analyzed unpartitioned and with codon partitioning (i.e., 3 total partitions), using RAxML version 7.2.8 [46]. A GTRGAMMA model was used for each partition, as the GTRCAT model is not recommended for matrices with so few taxa [47]. Multiple ML searches (100) were conducted per matrix, each using a randomized stepwise addition parsimony tree to initiate an ML tree inference. Non-parametric bootstrap replicates (500) were conducted on each matrix using a partitioned GTRGAMMA model. Bootstrap values were visualized on the best-scoring ML tree derived from multiple ML searches.
The five DNA matrices were also translated to amino acids for phylogenetic analysis, with stop codons recoded as missing data. For each concatenated amino acid matrix a single best-fit model of protein evolution was chosen using ProtTest v2.4 [48], for a subset of models also available in RAxML. As above, multiple ML searches (100) and non-parametric bootstrap replicates (500) were conducted per amino acid matrix. Bootstrap values were visualized on the best-scoring ML tree from multiple inferences.
We also conducted a Bayesian phylogenetic analysis of the TICKALL protein matrix using PhyloBayes version 3.3 [49]. PhyloBayes can implement a site-heterogeneous Bayesian mixture model (CAT) that allows site-specific rates for different amino acid positions in a protein alignment [50]. The CAT model has been shown to better capture the complexities of protein evolution, and is more robust to long-branch attraction than standard site-homogeneous protein models [50], [51]. Using a default CAT-Poisson model, two Monte Carlo Markov chains were run in parallel until the largest discrepancy observed across all bipartitions (maxdiff) was lower or equal to 0.3, and effective sizes for all summary variables exceeded 50. A majority-rule posterior consensus tree was reconstructed from a set of post burn-in trees (discarding 20% as burn-in).
Divergence Time Estimation
The MCMCtree Bayesian program in the PAML v. 4.4c package [52], [53] was used to estimate harvestmen divergence times using a relaxed molecular clock. Analyses were conducted on nucleotide data from the TICKALL matrix, partitioned by codon position, without Ixodes outgroup sequences. The tree topology was constrained to ingroup relationships recovered in RAxML and PhyloBayes analyses of TICKALL nucleotides and amino acids (see Results). Priors for lineage-specific substitution rates were specified using autocorrelated and independent rates models [54]–[56]. Both of these analyses were run with and without sequence data. We estimated the parameters (shape and scale) of the gamma distribution of the overall substitution rate prior (μ) using Baseml estimates of the substitution rate for each partition. The substitution rate prior (μ) was estimated under three calibration schemes (see below); each calibration scheme used three fixed calibration points. The shape and scale for μ were 0.42 and 1.31, 0.34 and 0.13, and 0.40 and 1.17, under calibrations schemes 1, 2, and 3, respectively. The gamma distributed prior for σ2, which specifies how variable the substitution rate is among branches, was set to shape = 1 and scale = 1. The analysis was run with birth rate, death rate, and species sampling priors of 2, 2, and 0.1, respectively. The HKY sequence model was used and gamma priors for κ (the transition/transversion ratio) and α (shape parameter for among site rate variation) were left as default [54]. Calibrations (see below) were treated as soft boundaries (i.e., 2.5% chance date falls beyond boundary [55], [57]). The first 20,000 iterations were discarded as burnin, followed by 50,000 iterations sampled every five iterations. Analyses including sequence data were run twice to assess MCMC convergence.
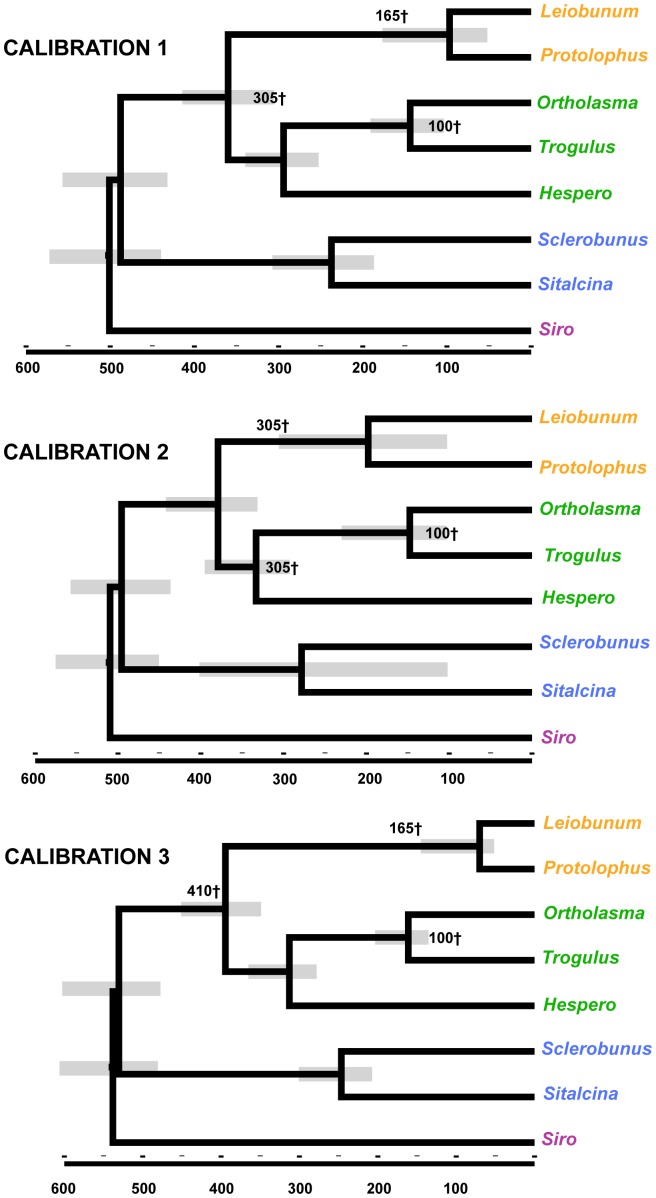
Three different fossil calibration strategies were used as follows: 1) We treated the Garwood et al. [33] fossils as stem group members of Eupnoi (Macrogyion) and Dyspnoi (Ameticos) respectively, and constrained the minimum age of the common ancestor of the group including these fossils ( = Palpatores) at 305 Myr. We treated the Huang et al. [58] Mesobunus fossil (165 Myr) as a crown group phalangioid, and the Giribet and Dunlop [59] Halitherses fossil (100 Myr) as a crown group troguloid, constraining the minimum age for these clades. 2) Garwood et al. [33] fossils were treated as crown group members of Eupnoi and Dyspnoi, constraining minimum ages for these clades at 305 Myr; Halitherses (100 Myr) was treated as a crown group troguloid, constraining the minimum age for this clade. 3) Eophalangium was treated as a stem group Eupnoi, constraining the minimum age of the common ancestor of the group including these fossils ( = Palpatores) at 410 Myr [31], [32]. Mesobunus (165 Myr [58]) was treated as a crown group phalangioid, and Halitherses (100 Myr [59]) as a crown group troguloid, constraining minimum ages for these clades. All calibration strategies required “soft” maximum ages for Opiliones, which we set at 500 Myr. This age is older than the oldest known paleontological evidence for terrestrial arachnids (∼430 Myr [60], [61]), and is arguably too old. However, recent molecular clock studies have estimated Cambrian or Pre-Cambrian divergence dates for the common ancestor of arachnids [26], [62], [63]. Schaefer et al. [62] suggest that arachnids were colonizing terrestrial habitats approximately 150 Myr before the earliest known terrestrial animal fossils.
Phylogenetic Informativeness of Genes
Gene-specific measures of phylogenetic informativeness (PI [64]) were calculated using the online PhyDesign application [65]. PI values attempt to measure the informativeness of a character set (“genes”, either nucleotides or amino acids) in comparison to other character sets, over time intervals relevant to the clade of interest. Informativeness values can be visualized as PI profile curves, where total area and peak curve values provide evidence for differential informativeness over time [64]–[67]. Net and per-site PI values were calculated for 300 individual genes in the TICKALL matrix for both nucleotides and amino acids, excluding Ixodes sequences. The chronogram from MCMCtree calibration analysis 1 (replicate 1) was used as an input tree. Nucleotide site rate models were estimated using HyPhy [68], with empirical base frequencies and a time reversible mode of substitution (transitions = 2, transversions = 1). Amino acid site rate models were estimated using rate4site [69] using a JTT model and ML inference method. Similar model settings were recently used in a phylogenetic informativeness analysis of vertebrate genes [70]; crown group vertebrates are likely younger than Opiliones, sharing a common ancestor ∼420 MYA [71].
Results
Assemblies
Between 2.5–6 Gb of post-filter Illumina data were used per taxon assembly (Table 3). For non-normalized libraries (all taxa except for Protolophus), de novo assemblies resulted in approximately 50–80,000 Oases transcripts over 100 bp in length, with mean transcript lengths ranging from 471–694 bp (Table 3). Assembled sequences have been deposited in the NCBI Transcriptome Shotgun Assembly (TSA) database. Assembly of the normalized Protolophus library data resulted in many more transcripts of shorter average length (i.e., a more fragmented transcriptome; Table 3). The number of Oases transcripts per taxon is an overestimate of harvestmen transcriptome sizes (because of exogenous sequences, “redundant” Oases transcripts, multiple non-overlapping or partially overlapping transcripts for any single gene, etc.), but full characterization of single harvestmen transcriptomes was not a focus of this paper.
Table 3. Raw data and Assembly Information.
| Taxon | # Illumina PE Reads (post-QCfilter) | # Oases Transcripts | Mean, Max Transcript Length | No. “Super Transcripts” in Phylogenetic Matrices |
| Siro | 50341482×50 bp = 2.52 Gb | 53,006 | 477, 6979 | 12 in 367 (0.0327) |
| Sitalcina | 85228254×50 bp = 4.26 Gb | 79,075 | 471, 12417 | 16 in 367 (0.0436) |
| Sclerobunus | 88356268×50 bp = 4.42 Gb | 77,342 | 675, 17875 | 8 in 367 (0.0218) |
| Ortholasma | 80744734×50 bp = 4.04 Gb | 50,665 | 570, 11074 | 18 in 366 (0.0492) |
| Trogulus | 54675768×50 bp = 2.73 Gb | 77,444 | 567, 9614 | 5 in 366 (0.0137) |
| Hesperonemastoma | 120052764×50 bp = 6.00 Gb | 68,378 | 613, 8952 | 23 in 365 (0.0630) |
| Leiobunum | 62470530×50 bp = 3.12 Gb | 56,048 | 694, 11393 | 12 in 365 (0.0329) |
| Protolophus | 74989348×60 bp = 4.50 Gb | 282,910 | 231, 8979 | 85 in 363 (0.2341) |
Data Matrices & Phylogenomics
Summary information for all individual gene matrices (e.g., gene identities, nucleotide alignment lengths, PI values, average pairwise divergence values, taxon composition, etc.) is available in Table S1; aligned nucleotide matrices are available in Text S1. The final SCORP concatenated matrix included data for 67 unique protein-coding genes, with a total nucleotide alignment length of approximately 32 kb. Nucleotide alignment lengths of TICKFAST, TICKMED and TICKSLOW 100-gene concatenated matrices were approximately 72, 67 and 74 kb, respectively (Text S1). The proportion of gaps and missing data for individual nucleotide matrices is very low (2.8%, 1.8%, 1.3%, and 5.24% for TICKFAST, TICKMED, TICKSLOW and SCORP respectively). When present, we found that alternative Oases transcripts for the same taxon differed only in UTR regions (which were trimmed from matrices); alternative transcripts were thus randomly chosen for inclusion in phylogenetic matrices. The number of super-transcripts used per taxon is low (<5%) for all taxa except for Protolophus; approximately 23% of individual gene matrices include a Protolophus super-transcript (Table 3, Table S1).
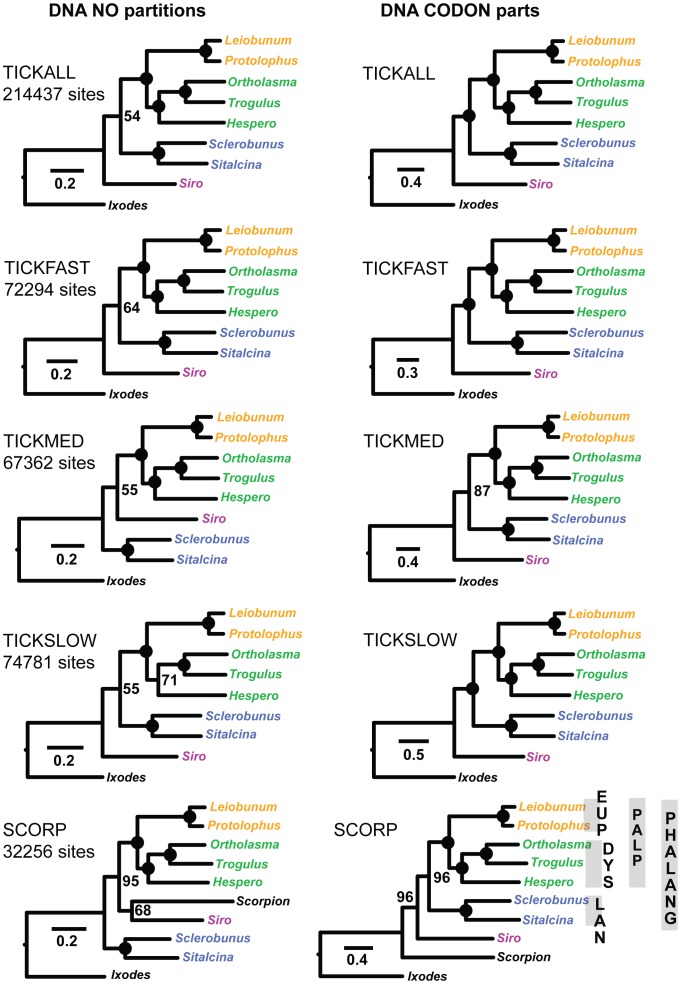
RAxML analyses with codon partitioning result in identical topologies, with proportionally similar branch lengths, for all concatenated nucleotide matrices (Figure 2). Across all trees, all but three nodes are supported by bootstrap proportion values of 100. The recovered tree topology includes a monophyletic Eupnoi, Dyspnoi, Palpatores, Laniatores, and Phalangida, consistent with the classical view of Opiliones relationships (Figure 1A). Rooting of the SCORP matrix with Ixodes supports Opiliones monophyly (bootstrap = 96). The phylogenetic placement of Protolophus is as expected in all analyses (sister to Leiobunum in Eupnoi), suggesting that the inclusion of super-transcripts did not negatively impact phylogenetic analyses.
Figure 2. RAxML trees inferred from analyses of nucleotide data.
Analyses without codon partitioning on left, analyses with codon partitioning on right. Circles at nodes indicate bootstrap values of 100. Abbreviation for Hesperonemastoma = “Hespero”.
Phylogenetic analyses of unpartitioned concatenated nucleotide matrices strongly support Eupnoi, Dyspnoi, Palpatores and Laniatores (bootstrap = 100), but are less resolved at deeper phylogenetic levels. First, although analyses of TICK matrices recover the classical Opiliones tree, these analyses do not strongly support Phalangida monophyly (bootstrap values less than 70; Figure 2). Second, analysis of the SCORP matrix does not recover Opiliones monophyly, instead suggesting a scorpion + Siro relationship (Figure 2). Analyses of the unpartitioned nucleotide matrices with third position sites removed result in trees consistent with the classical view, with bootstrap proportion values at or near 100 for all nodes (Figure S1).
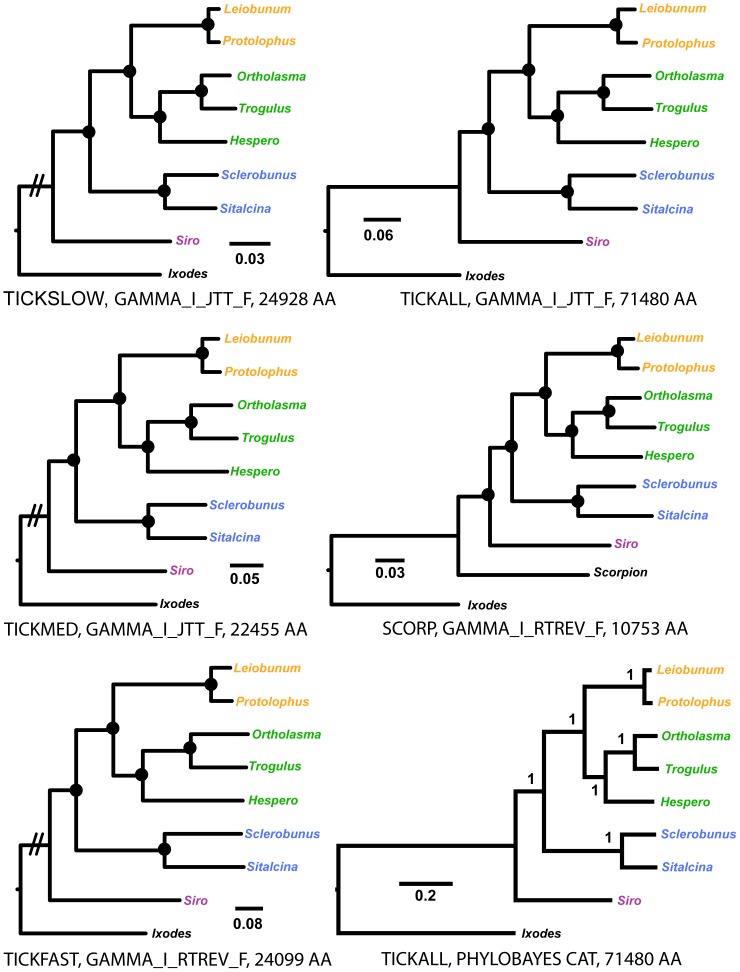
Figure 3 shows best-fit models of protein evolution for individual concatenated amino acid matrices. For any given matrix, a single best-fit model was consistently preferred using the multiple criteria available in ProtTest (e.g., AIC, BIC, etc. [48]). RAxML analyses of the amino acid matrices result in identical topologies, with similar branch lengths, for all data partitions (Figure 3). All nodes for all trees are supported by bootstrap values of 100. The recovered tree topology is consistent with codon-partitioned nucleotide analyses, and the classical view of Opiliones relationships (Figure 1A). Rooting of the SCORP matrix with Ixodes supports Opiliones monophyly (bootstrap = 100). PhyloBayes analysis of the TICKALL amino acid matrix implementing the CAT model recovers the classical tree, with posterior probability values of 1.0 at all nodes (Figure 3).
Figure 3. RAxML and PhyloBayes trees inferred from analyses of amino acid data.
Root branch for TICKFAST, TICKMED and TICKSLOW trees not drawn to scale. Trees resulting from analysis of SCORP matrix rooted presuming Scorpion plus Opiliones relationship. Circles at nodes indicate bootstrap values of 100. Abbreviation for Hesperonemastoma = “Hespero”.
Comparative Gene Ontologies
Representative nucleotide sequences (usually Ixodes, scorpion for 9 SCORP matrices where Ixodes was unavailable) for four separate matrices (SCORP, TICKFAST, TICKMED, TICKSLOW) were imported into Blast2GO V.2.5.0 (http://www.blast2go.com [72]) for bioinformatic functional annotation (see Text S2). Matrix-specific GO terms for biological process and molecular function are provided in Table S2. Consistent with the observations of Hittinger et al. [3], our RNA-Seq derived phylogenomic matrices appear dominated by core metabolic genes.
Divergence Time Estimation
Here we focus on results of MCMCtree analyses under the autocorrelated rates model; results from independent rates model analyses are provided in Text S3. In general, these alternative analyses imply generally similar nodal times estimates (Text S3). Autocorrelated rates chronograms resulting from alternative calibration strategies are shown in Figure 4. Although replicate MCMCtree analyses result in similar most recent common ancestor (MRCA) time estimates for calibration strategies 1 and 3, this was not the case for calibration strategy 2, where MRCA divergence time estimates were more variable over replicates, particularly for the MRCA of Eupnoi (Table 4). Confidence intervals for the MRCA of Eupnoi and Laniatores are also conspicuously large for calibration strategy 2 (Figure 4, Table 4). Time estimates for nodes that are unconstrained in two or more analyses are reasonably consistent. Calibration strategies 1 and 3 suggest a MRCA for Dyspnoi at ∼300 Myr. All calibration strategies suggest a mean MRCA estimate for Laniatores at 220–280 Myr, which is younger than implied by relaxed clock BEAST analyses of Sharma and Giribet (mean estimate of ∼350 Myr, HPD 307–382 [35]). Old fossil calibration minimums within or at the base of Palpatores (for all calibration strategies), in combination with relatively long branches to Laniatores (e.g., Figure 2), imply ancient origins for Phalangida (MRCA mean estimates of 480–530 Myr). These time estimates for Phalangida are older than in previous relaxed molecular clock analyses (mean estimate of ∼415 Myr [37]; mean estimate of ∼427 Myr, HPD 417–438 [35]). Analyses of Giribet et al. [37] were conducted with a fixed root age for Opiliones of 420 Myr.
Figure 4. MCMCtree chronograms.
MCMCtree chronograms resulting from three alternative calibration schemes (replicate one) using autocorrelated rates model. Scale in millions of years. Fossil calibrations (treated as soft minimums) indicated by cross symbols. Soft maximum of 500 Myr for the root used in each analysis (see Material and Methods). Abbreviation for Hesperonemastoma = “Hespero”.
Table 4. MCMC tree results for the autocorrelated rates model.
| Calibration | Root | Phalan | Lan | Palp | Eupnoi | Dyspnoi | Trog |
| Setting 1, run 1 | 501, 440–567 | 488, 424–556 | 237, 185–309 | 360, 310–413 | 97, 51–174 | 294, 253–345 | 144, 106–186 |
| Setting 1, run 2 | 499, 438–566 | 485, 420–554 | 224, 147–317 | 360, 310–412 | 103, 51–178 | 295, 254–344 | 146, 106–191 |
| Setting 2, run 1 | 510, 457–578 | 496, 440–565 | 278, 101–398 | 380, 334–440 | 198, 97–303 | 333, 289–398 | 148, 110–235 |
| Setting 2, run 2 | 515, 464–584 | 503, 448–572 | 267, 191–375 | 378, 336–432 | 149, 55–279 | 319, 280–377 | 157, 105–223 |
| Setting 3, run 1 | 538, 483–611 | 530, 474–603 | 246, 206–300 | 394, 351–447 | 70, 53–145 | 313, 277–358 | 161, 136–199 |
| Setting 3, run 2 | 538, 482–611 | 530, 472–601 | 250, 206–317 | 394, 350–447 | 75, 53–156 | 314, 277–362 | 162, 134–204 |
Notes – time in Myr. Phalan = MRCA of Phalangida, Lan = MRCA of Laniatores, Palp = MRCA of Palpatores, Trog = MRCA of Troguloidea.
Gene Informativeness
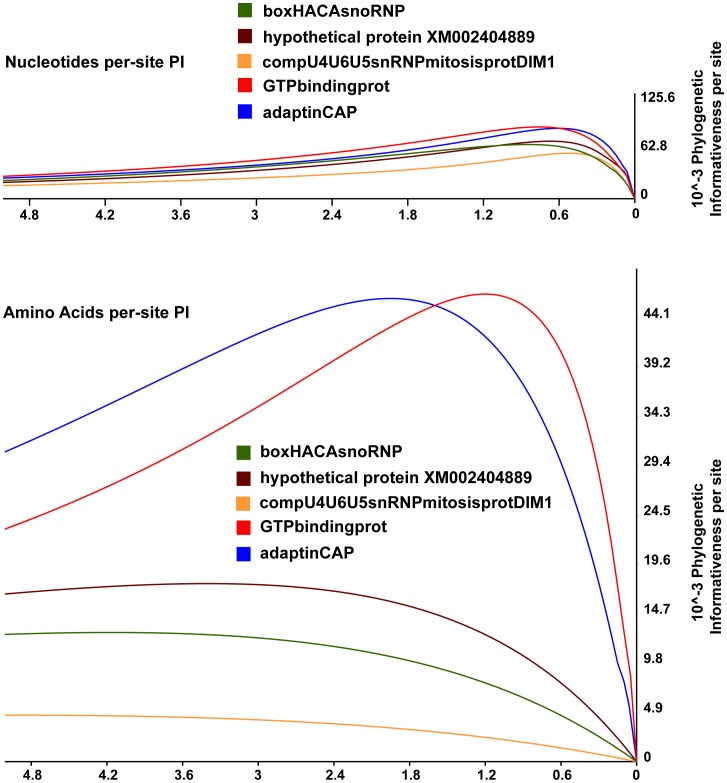
Nucleotide and amino acid PI values for 300 TICKALL genes are provided in Table S1. Summary information for each gene includes the sum of PI values over the entire chronogram time interval (for both net and per-site PI), and the time interval at which PI value is maximized (same for net and per-site PI). Net PI values are confounded by gene length, i.e., longer genes have generally higher net PI values (Table S1), so we focus on per-site values. Nucleotide max PI values are universally smaller (values ranging from 0.50–1.10, i.e., 50–110 Myr) than corresponding amino acid PI values (values ranging from 1.20–5.01), suggesting that amino acids are more informative for older divergences, which is an expected result. The narrow range of nucleotide max PI values in comparison to amino acid values also suggests a greater breadth of informativeness in amino acid characters for the ancient divergences considered here. This greater breadth of informativeness is also suggested by summed PI values for nucleotides versus amino acids (nucleotide ranges 3.3–9.91, amino acid ranges 0.16–12.62). Figure 5 illustrates this difference in informativeness breadth for five representative genes.
Figure 5. Phylogenetic informativeness profiles.
Per-site nucleotide and amino acid phylogenetic informativeness profiles for five exemplar TICKALL genes.
Discussion
One of the primary goals of this phylogenetic study was to resolve the interrelationships of Opiliones suborders. We successfully used comparative RNA-Seq data to assemble large matrices of putatively orthologous genes, and unlike many other phylogenomic studies [73], these matrices include a very low percentage of missing data. Phylogenetic analyses using various partitioning strategies, data coding schemes, and analytical methods overwhelmingly support the “classical” hypothesis of Opiliones relationships (Figures 2 and 3). This hypothesis includes the higher-level clades Palpatores and Phalangida, and is inconsistent with Cyphopalpatores and Dyspnolaniatores hypotheses (Figure 1B, 1C).
As in many phylogenomic studies, our analysis includes many genes and few taxa, and increasing the taxonomic sampling breadth within Opiliones should be a goal for future phylogenomic studies. A potentially more serious shortcoming relates to use of only a single outgroup (tick) in many analyses, and the fact that this outgroup is perhaps too distant from Opiliones. A potential phylogenetic artifact when using a distant outgroup is long branch attraction, as divergent ingroup taxa (e.g., Siro) may be pulled to a basal position [38]. We attempted to accommodate this possibility by including scorpion sequences in some analyses, working from the Opiliones plus Scorpiones sister relationship inferred from morphology [23]. Tree topologies within Opiliones are identical for SCORP and TICK amino acid matrices. Even so, it is not clear that Scorpiones represents the best taxon choice for rooting harvestmen trees, as published molecular phylogenetic and phylogenomic studies with sufficient taxon sampling fail to support a close relationship of Opiliones and Scorpiones [24]–[26], [74]. Given the general lack of genomics resources currently available for Arachnida, and the fact that the sister group of Opiliones is effectively unknown at present, we argue that our rooting approach was justified. It could also be argued that all Opiliones outgroups are effectively equivalent given the deep and apparently rapid divergences near the base of Arachnida. A large-scale phylogenomics analysis of all arachnid orders, an obvious target for comparative RNA-Seq phylogenomics, would help to resolve the issues discussed here.
The assembled SCORP and TICK data matrices represent an important resource for future molecular systematic studies of Opiliones and other arachnid groups. The 300 TICK genes were present in all harvestmen transcriptomes, suggesting conservation and high expression levels, a result supported by GO analyses (Table S2). The 300 TICK gene set includes apparently single copy-genes (or if members of gene families, including easily recognizable divergent paralogs) that vary in length and phylogenetic informativeness (Table S1), and are easily aligned. We envision two strategies for future use of this gene set in Opiliones and other arachnid groups. First, retrieving complete or partial gene sets in newly available transcriptome data will greatly streamline downstream phylogenomic analysis. Because of conserved function and high expression levels, we hypothesize that a majority of these genes will be present in most arachnid transcriptomes. Second, some members of these gene sets can be used as resources for a traditional Sanger sequencing strategy, or NGS targeted amplicon sequencing. Proteins that are single copy, single exon genes in Ixodes are highlighted in Table S1, and several of these genes are long (over 1 kb). Although this intronless condition is not expected to apply in all arachnids, these proteins are clear candidates for PCR primer design. Overall, we view the gene set assembled here as a resource which allows us to resolve a higher-level phylogenetic framework, and subsequently move towards the tips of the Opiliones tree. This general strategy should be reproducible within and across all arachnid orders.
The early diversification history of Opiliones is enigmatic. The fossil taxon Eophalangium is both remarkably old (∼410 Myr) but also surprisingly modern in several aspects of morphology. Authors of the original description suggest that this taxon represents a crown group Eupnoid [31], [32]. At the same time, the earliest known terrestrial arachnid fossils are only about 20 million years older (∼430 Myr [60], [61]), and are among the first known land animals. This paradox implies that Opiliones diversification may have been remarkably rapid, with the radiation of multiple divergent morphological clades (i.e., Cyphophthalmi, Eupnoi, Dyspnoi, Laniatores) occurring over a very short time interval. However, estimated branch lengths from codon-partitioned nucleotide analyses (Figure 2), and all amino acid analyses (Figure 3), appear inconsistent with a rapid radiation scenario at the base of Phalangida or Palpatores.
An alternative explanation is that Opiliones history is deeper than the recorded fossil record indicates, and/or that Eophalangium is not as phylogenetically nested as previously hypothesized. Relaxed clock analyses conservatively treating Eophalangium as a stem group Eupnoid (Figure 4, calibration 3; Text S3) imply a deep cryptic history for Phalangida and Opiliones, and branch length compression at the base of all chronograms (e.g., Figure 4) suggests that a soft max of 500 Myr for the root of Opiliones may be too recent. Additional analyses treating Eophalangium as a stem group Palpatores or Phalangida would be worthwhile, but imply perhaps untenable patterns of morphological character evolution within Opiliones. As hypothesized for mites [62], small omnivorous harvestmen may have been transitioning from marine to terrestrial habitats during the late Cambrian and Ordovician, but are currently unrecorded in the fossil record. An increased phylogenomic sample for Opiliones and other arachnids, in the context of a more densely sampled fossil calibration analysis, is needed to further explore this possibility.
Supporting Information
Results of analyses of unpartitioned nucleotide matrices with third position sites removed.
(TIF)
Summary information for individual gene matrices (e.g., gene identities, nucleotide alignment lengths, PI values, average pairwise divergence values, taxon composition, etc.).
(XLSX)
Gene ontologies.
(DOCX)
Individual gene matrices.
(DOCX)
Acknowledgments
Jordan Satler, Shahan Derkarbetian and S. Huber provided specimens. Technical support was provided by Jon Armstrong at CoFactor Genomics (www.cofactorgenomics.com), and Braden Boone, Liz Dye, and Shawn Levy from the Genomic Services Lab at Hudson Alpha Institute for Biotechnology (www.hudsonalpha.org/gsl). We thank Cheryl Hayashi at UC Riverside for use of lab facilities, and Rob Edwards at SDSU for use of computer resources. Comments of three anonymous reviews helped to improve the manuscript.
Funding Statement
Research funding was provided by a collaborative National Science Foundation grant awarded to JWS (award no. 0640179) and MH (award no. 0640173), and a California State University Program for Education and Research in Biotechnology (CSUPERB) grant to MH. JWS was also supported by the Maryland Agricultural Experiment Station, and AS was supported by a postdoc grant from the Deutsche Forschungsgemeinschaft (DFG). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Metzker ML (2010) Sequencing technologies – the next generation. Nat Reviews 11: 31–46. [DOI] [PubMed] [Google Scholar]
- 2. Mardis ER (2011) A decade's perspective on DNA sequencing technology. Nature 470: 198–203. [DOI] [PubMed] [Google Scholar]
- 3. Hittinger CT, Johnston M, Tossberg JT, Rokas A (2010) Leveraging skewed transcript abundance by RNA-Seq to increase the genomic depth of the tree of life. Proc Natl Acad Sci 107: 1476–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kocot KM, Cannon JT, Todt C, Citarella MR, Kohn AB, et al. (2011) Phylogenomics reveals deep molluscan relationships. Nature 477: 452–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hartmann S, Helm C, Nickel B, Meyer M, Struck TH, et al. (2012) Exploiting gene families for phylogenomic analysis of myzostomid transcriptome data. PLoS One 7 1:e29843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bybee SM, Bracken-Grissom H, Haynes BD, Hermansen RA, Byers RL, et al. (2011) Target amplicon sequencing (TAS): A scalable Next-Gen approach to multilocus, multitaxa phylogenetics. Genome Biol Evol 3: 1312–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, et al. (2011) Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Reviews 12: 499–510. [DOI] [PubMed] [Google Scholar]
- 9. McCormack JE, Hird SM, Zellmer AJ, Carstens BC, Brumfield RT (2012) Applications of next-generation sequencing to phylogeography and phylogenetics. Mol Phylo Evol in press. [DOI] [PubMed] [Google Scholar]
- 10. Harvey MS (2002) The neglected cousins: What do we know about the smaller arachnid orders? J Arachnology 30: 357–372. [Google Scholar]
- 11. Bellgard MI, Moolhuijzen PM, Guerrero FD, Schibeci D, Rodriguez-Valle M, et al. (2012) CattleTickBase: An integrated Internet-based bioinformatics resource for Rhipicephalus (Boophilus) microplus . Int J Parasitology 42: 161–169. [DOI] [PubMed] [Google Scholar]
- 12. Pagel Van Zee J, Geraci NS, Guerrero FD, Wikel SK, Stuart JJ, et al. (2007) Tick genomics: The Ixodes genome project and beyond. Int J Parasitology 37: 1297–1305. [DOI] [PubMed] [Google Scholar]
- 13. Schwartz EF, Diego-Garcia E, Rodríguez de la Vega RC, Possani LD (2007) Transcriptome analysis of the venom gland of the Mexican scorpion Hadrurus gertschi (Arachnida: Scorpiones). BMC Genomics 8: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ma Y, Zhao Y, Zhao R, Zhang W, He Y, et al. (2010) Molecular diversity of toxic components from the scorpion Heterometrus petersii venom revealed by proteomic and transcriptome analysis. Proteomics 10: 2471–2485. [DOI] [PubMed] [Google Scholar]
- 15. Prosdocimi F, Bittencourt D, Rodrigues da Silva F, Kirst M, Motta PC, et al. (2011) Spinning gland transcriptomics from two main clades of spiders (Order Araneae) – Insights on their molecular, anatomical and behavioral evolution. PLoS ONE 6 6:e21634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mattila TM, Bechsgaard JS, Hansen TT, Schierup MH, Bilde T (2012) Orthologous genes identified by transcriptome sequencing in the spider genus Stegodyphus . BMC Genomics 13: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Machado G, Pinto-da-Rocha R, Giribet G (2007) What are harvestmen? In: Pinto da Rocha R, Machado G, Giribet G, editors. Harvestmen: The Biology of Opiliones. Harvard University Press. Cambridge, Massachusetts, and London, England, pp. 1–13.
- 18.Kury AB (2011) Classification of Opiliones. Available: http://www.museunacional.ufrj.br/mndi/Aracnologia/opiliones.html. Accessed 2012 Apr 15.
- 19.Pinto-da-Rocha R, Machado G, Giribet G, editors. (2007) Harvestmen: The Biology of Opiliones. Harvard Univ. Press, Cambridge, Massachusetts, and London, England, 597 p.
- 20.Giribet G, Kury AB (2007) Phylogeny and Biogeography. In: Pinto da Rocha R, Machado G, Giribet G, editors. Harvestmen. The Biology of Opiliones. Harvard University Press, Cambridge, Massachusetts, and London, England, pp. 62–87.
- 21. Shultz JW (1990) Evolutionary morphology and phylogeny of Arachnida. Cladistics 6: 1–38. [DOI] [PubMed] [Google Scholar]
- 22.Shultz JW, Pinto-da-Rocha R (2007) Morphology and Functional Anatomy. In: Pinto da Rocha R, Machado G, Giribet G, editors. Harvestmen. The Biology of Opiliones. Harvard University Press, Cambridge, Massachusetts, and London, England, pp. 14–61.
- 23. Shultz JW (2007) A phylogenetic analysis of the arachnid orders based on morphological characters. Zool J Linnean Soc 150: 221–265. [Google Scholar]
- 24. Regier JC, Shultz JW, Zwick A, Hussey A, Ball B, et al. (2010) Arthropod relationships revealed by phylogenomic analysis of nuclear protein-coding sequences. Nature 463: 1079–1084. [DOI] [PubMed] [Google Scholar]
- 25. Masta SE (2010) Mitochondrial rRNA secondary structures and genome arrangements distinguish chelicerates: comparisons with a harvestman (Arachnida: Opiliones: Phalangium opilio). Gene 449: 1–9. [DOI] [PubMed] [Google Scholar]
- 26. Sanders KL, Lee MSY (2010) Arthropod molecular divergence times and the Cambrian origin of pentastomids. Syst Biodiv 8: 63–74. [Google Scholar]
- 27.Hansen HJ, Sørensen W (1904) On two orders of Arachnida – Opiliones, especially the suborder Cyphophthalmi, and Ricinulei, namely the family Cryptostemmatoidae. University Press, Cambridge. 178 p.
- 28. Martens J (1976) Genitalmorphologie, system und phylogenie der Weberknechte (Arachnida: Opiliones). Entomol Germanica 3: 51–68. [Google Scholar]
- 29. Martens J (1980) Versuch eines phylogenetischen Systems der Opiliones. Verh 8 Intern Arachn Kong Wien 355–360. [Google Scholar]
- 30. Martens J (1986) Die Grossgliederung der Opiliones und die Evolution der Ordung (Arachnida). Actas X Congr Int. Arachnol Jaca/Espana I: 289–310. [Google Scholar]
- 31. Dunlop JA, Anderson LI, Kerp H, Hass H (2003) Preserved organs of Devonian harvestmen. Nature 425: 916. [DOI] [PubMed] [Google Scholar]
- 32. Dunlop JA, Anderson LI, Kerp H, Hass H (2004) A harvestmen (Arachnida: Opiliones) from the early Devonian Rhynie cherts, Aberdeenshire, Scotland. Trans Royal Soc Edinburgh: Earth Sciences 94: 341–354. [Google Scholar]
- 33. Garwood RJ, Dunlop JA, Giribet G, Sutton MD (2011) Anatomically modern Carboniferous harvestmen demonstrate early cladogenesis and stasis in Opiliones. Nature Communications 2: 444. [DOI] [PubMed] [Google Scholar]
- 34.Dunlop JA (2007) Paleontology. In: Pinto da Rocha R, Machado G, Giribet G, editors. Harvestmen. The Biology of Opiliones. Harvard University Press, Cambridge, Massachusetts, and London, England, pp. 247–265.
- 35. Sharma PP, Giribet G (2011) The evolutionary and biogeographic history of the armoured harvestmen – Laniatores phylogeny based on ten molecular markers, with description of two new families of Opiliones (Arachnida). Invert Syst 25: 106–142. [Google Scholar]
- 36. Giribet G, Sharma PP, Benavides LR, Boyer SL, Clouse RM, et al. (2012) Evolutionary and biogeographical history of an ancient and global group of arachnids (Arachnida: Opiliones: Cyphophthalmi) with a new taxonomic arrangement. Biol J Linnean Soc 105: 92–130. [Google Scholar]
- 37. Giribet G, Vogt L, Perez Gonzalez A, Sharma P, Kury AB (2010) A multilocus approach to harvestman (Arachnida: Opiliones) phylogeny with emphasis on biogeography and the systematics of Laniatores. Cladistics 26: 408–437. [DOI] [PubMed] [Google Scholar]
- 38. Phillipe H, Brinkmann H, Lavrov DV, Littlewood DTJ, Manuel M, et al. (2011) Resolving difficult phylogenetic questions. Why more sequences are not enough. PLoS Biology 9 3:e1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andrews S (2011) FastQC. A quality control tool for high throughput sequence data. Available: www.bioinformatics.bbsrc.ac.uk/projects/fastqc. Accessed 2012 15 April.
- 40. Shulz MH, Zerbino DR, Vingron M, Birney E (2012) Oases: Robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics advance access. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zerbino DR, Birney E (2008) Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Research 18: 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ruiming Z, Yibao M, Yawen H, Zhiyong D, Yingliang W, et al. (2010) Comparative venom gland transcriptome analysis of the scorpion Lychas mucronatus reveals intraspecific toxic gene diversity and new venomous components. BMC Genomics 11: 452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, et al. (2011) Geneious v5.4, Available: http://www.geneious.com/. Accessed 2012 15 April.
- 44. Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nuc Acids Res 30: 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pick C, Ebersberger I, Spielmann T, Bruchhaus I, Burmester T (2011) Phylogenomic analyses of malaria parasites and evolution of their exported proteins. BMC Evol Biol 11: 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stamatakis A (2006) RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- 47.Stamatakis A (2008) The RAxML 7.0.4 manual. Available: sco.h-its.org/exelixis/countManual7.0.4.php. Accessed 2012 15 April.
- 48. Abascal F, Zardoya R, Posada D (2005) ProtTest: Selection of best-fit models of protein evolution. Bioinformatics 21: 2104–2105. [DOI] [PubMed] [Google Scholar]
- 49.Lartillot N, Blanquart S, Lepage T (2008) PhyloBayes 2.3—a Bayesian software for phylogenetic reconstruction using mixture models. Available: http://www.phylobayes.org. Accessed 2012 15 April.
- 50. Lartillot N, Philippe H (2004) A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol 21: 1095–1109. [DOI] [PubMed] [Google Scholar]
- 51. Lartillot N, Brinkmann H, Philippe H (2007) Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. BMC Evol Biol 7: S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yang Z (1997) PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13: 555–56. [DOI] [PubMed] [Google Scholar]
- 53. Yang Z (2007) PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- 54.Yang Z (2006) Computational Molecular Evolution. Oxford University Press, Oxford, England. 376 p.
- 55. Yang Z, Rannala B (2006) Bayesian estimation of species divergence times under a molecular clock using multiple fossil calibrations with soft bounds. Mol Biol Evol 23: 212–226. [DOI] [PubMed] [Google Scholar]
- 56. Rannala B, Yang Z (2007) Inferring speciation times under an episodic molecular clock. Syst Biol 56: 453–466. [DOI] [PubMed] [Google Scholar]
- 57. Inoue J, Donoghue PCJ, Yang Z (2010) The impact of the representation of fossil calibrations on Bayesian estimation of species divergence times. Syst Biol 59: 74–89. [DOI] [PubMed] [Google Scholar]
- 58. Huang D, Selden PA, Dunlop JA (2009) Harvestmen (Arachnida: Opiliones) from the middle Jurassic of China. Naturwissenschaften 96: 955–962. [DOI] [PubMed] [Google Scholar]
- 59. Giribet G, Dunlop JA (2005) First identifiable Mesozoic harvestman (Opiliones: Dyspnoi) from Cretaceous Burmese amber. Proc Royal Soc Series B, Biol Sciences 272: 1007–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Dunlop JA, Selden PA (2009) Calibrating the chelicerate clock: a paleontological reply to Jeyaprakash and Hoy. Exp Appl Acarol 48: 183–197. [DOI] [PubMed] [Google Scholar]
- 61. Dunlop JA (2010) Geological history and phylogeny of Chelicerata. Arthropod Struct Devel 39: 124–142. [DOI] [PubMed] [Google Scholar]
- 62. Schaefer I, Norton RA, Scheu S, Maraun M (2010) Arthropod colonization of land – Linking molecules and fossils in oribatid mites (Acari, Oribatida). Mol Phylo Evol 57: 113–121. [DOI] [PubMed] [Google Scholar]
- 63. Rehm P, Borner J, Meusemann K, von Reumont BM, Simon S, et al. (2011) Dating the arthropod tree based on large-scale transcriptome data. Mol Phylo Evol 61: 880–887. [DOI] [PubMed] [Google Scholar]
- 64. Townsend JP (2007) Profiling phylogenetic informativeness. Syst Biol 56: 222–231. [DOI] [PubMed] [Google Scholar]
- 65. López-Giráldez F, Townsend JP (2011) PhyDesign: an online application for profiling phylogenetic informativeness. BMC Evol Biol 11: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Townsend JP, López-Giráldez F, Friedman R (2008) The phylogenetic informativeness of nucleotide and amino acid sequences for reconstructing the Vertebrate tree. J Mol Evol 67: 437–447. [DOI] [PubMed] [Google Scholar]
- 67. Moeller AH, Townsend JP (2011) Phylogenetic informativeness profiling of 12 genes for 28 taxa without divergence dates. Mol Phylo Evol 60: 271–272. [DOI] [PubMed] [Google Scholar]
- 68. Pond SL, Frost SD, Muse SV (2005) HyPhy: hypothesis testing using phylogenies. Bioinformatics 21: 676–679. [DOI] [PubMed] [Google Scholar]
- 69. Mayrose I, Graur D, Ben-Tal N, Pupko T (2004) Comparison of site-specific rate-inference methods for protein sequences: empirical Bayesian methods are superior. Mol Biol Evol 21: 1781–1791. [DOI] [PubMed] [Google Scholar]
- 70. Fong JJ, Fujita MK (2011) Evaluating phylogenetic informativeness and data-type usage for new protein-coding genes across Vertebrata. Mol Phylo Evol 61: 300–307. [DOI] [PubMed] [Google Scholar]
- 71. Benton MJ, Donoghue PCJ (2006) Paleontological evidence to date the Tree of Life. Mol Biol Evol 24: 26–53. [DOI] [PubMed] [Google Scholar]
- 72. Conesa A, Götz S, Garcia-Gomez JM, Terol J, Talon M, et al. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- 73. Sanderson MJ, McMahon MM, Steel M (2010) Phylogenomics with incomplete taxon coverage: the limits to inference. BMC Evol Biol 10: 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Giribet G, Edgecombe GD, Wheeler WC (2001) Arthropod phylogeny based on eight molecular loci and morphology. Nature 413: 157–161. [DOI] [PubMed] [Google Scholar]
- 75. Shultz JW (1998) Phylogeny of Opiliones (Arachnida): An assessment of the “Cyphopalpatores” concept. J Arachnology 26: 257–272. [Google Scholar]
- 76. Giribet G, Rambla M, Carranza S, Riutort M, Baguñà J, et al. (1999) Phylogeny of the arachnid order Opiliones (Arthropoda) inferred from a combined approach of complete 18S, partial 28S ribosomal DNA sequences and morphology. Mol Phylo Evol 11: 296–307. [DOI] [PubMed] [Google Scholar]
- 77. Shultz JW, Regier JC (2001) Phylogenetic analysis of Phalangida (Arachnida, Opiliones) using two nuclear protein-encoding genes supports monophyly of Palpatores. J Arachnology 29: 189–200. [Google Scholar]
- 78. Hedin M, Derkarabetian S, McCormack M, Richart C, Shultz JW (2010) The phylogenetic utility of the nuclear protein-coding gene EF-1a for resolving recent divergences in Opiliones, emphasizing intron evolution. J Arachnology 38: 9–20. [Google Scholar]
- 79. Giribet G, Edgecombe GD, Wheeler WC, Babbitt C (2002) Phylogeny and systematic position of Opiliones: a combined analysis of chelicerate relationships using morphological and molecular data. Cladistics 18: 5–70. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Results of analyses of unpartitioned nucleotide matrices with third position sites removed.
(TIF)
Summary information for individual gene matrices (e.g., gene identities, nucleotide alignment lengths, PI values, average pairwise divergence values, taxon composition, etc.).
(XLSX)
Gene ontologies.
(DOCX)
Individual gene matrices.
(DOCX)