Abstract
We evaluated the population structure and temporal dynamics of the dominant community members within sewage influent from two wastewater treatment plants (WWTPs) in Milwaukee, WI. We generated >1.1M bacterial pyrotag sequences from the V6 hypervariable region of 16S rRNA genes from 38 influent samples and two samples taken upstream in the sanitary sewer system. Only a small fraction of pyrotags from influent samples (~15%) matched sequences from human fecal samples. The fecal components of the sewage samples included enriched pyrotag populations from Lactococcus and Enterobacteriaceae relative to their fractional representation in human fecal samples. In contrast to the large number of distinct pyrotags that represent fecal bacteria such as Lachnospiraceae and Bacteroides, only one or two unique V6 sequences represented Acinetobacter, Trichococcus and Aeromonas, which collectively account for nearly 35% of the total sewage community. Two dominant Acinetobacter V6 pyrotags (designated Acineto tag 1 and Acineto tag 2) fluctuated inversely with a seasonal pattern over a 3-year period, suggesting two distinct Acinetobacter populations respond differently to ecological forcings in the system. A single nucleotide change in the V6 pyrotags accounted for the difference in these populations and corresponded to two phylogenically distinct clades based on full-length sequences. Analysis of wavelet functions, derived from a mathematical model of temporal fluctuations, demonstrated that other abundant sewer associated populations including Trichococcus and Aeromonas had temporal patterns similar to either Acineto tag 1 or Acineto tag 2. Populations with related temporal fluctuations were found to significantly correlate with the same WWTP variables (5-day BOD, flow, ammonia, total phosphorous, and suspended solids). These findings illustrate that small differences in V6 sequences can represent phylogenetically and ecologically distinct taxa. This work provides insight into microbial community composition and dynamics within the defined environment of urban sewer infrastructure.
Introduction
Modern urban sewer infrastructure dates back only 100–150 years and encompasses a relatively new environment in which microbial communities may inhabit and propagate. Separated sewer systems with pipes for sanitary sewage and dedicated stormwater systems service most urban areas in the United States (US). Sanitary sewage from households and other buildings enters wastewater treatment plants (WWTPs), whereas stormwater runoff from rooftops, streets and other impervious surfaces discharges via a different set of pipes directly into detention ponds, rivers, or other receiving waters. However, in older cities, combined sewers convey both sanitary and stormwater into WWTPs. While this configuration captures and treats stormwater, high amounts of precipitation can inundate the system and cause it to overflow into receiving waters. In the US, most of these older systems are concentrated in the Northeast or Midwest (USEPA, 2004).
Few studies have focused on microbial populations in conveyance systems. Natural microbial communities within sewer infrastructure can exist within biofilms or in sediments but to date, studies of these communities have focused on delineating community activity or metabolic functions and have not included inventories of underlying taxa initiating degradation of nutrients and pollutants (Chen et al., 2003; Jiang et al., 2009; Mohanakrishnan et al., 2009; Pai et al., 2010). Microbial communities in sewers can play a beneficial role through pretreatment of wastewater by initiating degradation of nutrients and pollutants (Raunkjaer et al., 1995; Warith et al., 1998). They may also produce negative effects, particularly under anaerobic conditions in pressure mains or biofilms, where sulfate reducing bacteria can produce hydrogen sulfide that causes odors and contributes to corrosion of pipes (Hvitvedjacobsen et al., 1995; Norsker et al., 1995; Hao et al., 1996; Okabe et al., 2007; Satoh et al., 2009).
Release of untreated wastewater during sewage overflows contributes to a loss of biological diversity, ecosystem disruption, and degradation of water resources. Untreated wastewater contains harmful chemical and biological pollutants including nutrients, pathogens, pharmaceuticals, and cleaning products (Tong and Chen, 2002; Ternes et al., 2004; Rothenberger et al., 2009). The US Environmental Protection agency (EPA) estimates that combined sewer overflows (CSOs) discharge 3.2 trillion liters of untreated wastewater and stormwater directly to surface waters each year, while sanitary sewer overflows release an additional 11 to 37 billion liters of untreated wastewater annually (USEPA, 2004). When released into receiving waters, high densities of sewage-derived microorganisms may disrupt natural communities and alter ecosystem function or equilibrium. Untreated wastewater discharges threaten both the environment and public health (Ferguson et al., 1996; Lipp et al., 2001).
In order to evaluate microbial community structure within urban sewer systems and address the threat posed by the organisms released by overflows, it is important to understand the microbial community within sewage. We studied a large urban sewer system operated by the Milwaukee Metropolitan Sewerage District (MMSD) whose service area encompasses part or all of six watersheds and spans 1409 km2 with 28 communities and 1.1 million customers. Milwaukee’s first WWTP, Jones Island (JI), started operation in 1925 and as the city expanded, a second WWTP, South Shore (SS), was constructed in 1968. Each plant services roughly half of the service area, which in it’s entirety consists of more than 10,000 km of private laterals and local municipal pipes that converge into 545 km of a municipal interceptor system (MIS). Approximately one third of the system flow can divert to either SS or JI at a given time, but no common sections simultaneously send flow to both plants. The JI WWTP combined sewer system services the oldest and most urbanized area of the city (approximately 5% of the total service area). The SS WWTP primarily services residential neighborhoods. Figure 1S (supplemental information) outlines the conveyance system configuration for each plant. Most of the service area operates as a gravity flow system, which should promote aerobic conditions. Only short segments require pressurized mains where lift stations transport flow to higher elevations; these segments may produce anaerobic conditions. Regions to the north of the city have travel times as long as 14 hrs, whereas downtown areas have travel times as little as 1 hr. A travel time of 2–8 hrs represents the majority of the region.
Recently, we reported that microbes of non-fecal origin dominate communities in sewer infrastructure (McLellan et al., 2010). In the present study, we evaluated microbial communities at multiple time points over a three-year period using massively-parallel 16S rRNA gene pyrotag sequencing. We characterized high abundance populations in terms of seasonal fluctuations and phylogenetic relationships. This study represents one of the first detailed reports of the structure and dynamics of urban sewer infrastructure microbial communities.
Results
Diversity and taxonomic composition of sewage
We collected 19 paired sewage influent samples from JI and SS WWTPs (n=38 total) under different environmental conditions over a three-year period (Table 1S, supplemental information). Cluster analysis of the 1,085,939 bacterial pyrotags provided estimates of richness and diversity. Single-Linkage Preclustering (SLP) which overestimates richness at the 3% dissimilarity criterion (Huse et al., 2010) identified 10,545 operational taxonomic units (OTUs). The vast majority of the total number of tags (95%) formed 654 OTUs that resolved to 233 unique taxonomic descriptors, and 50% of the tags clustered into only 12 OTUs, indicating that sewage influent contains an extremely rich but uneven microbial population.
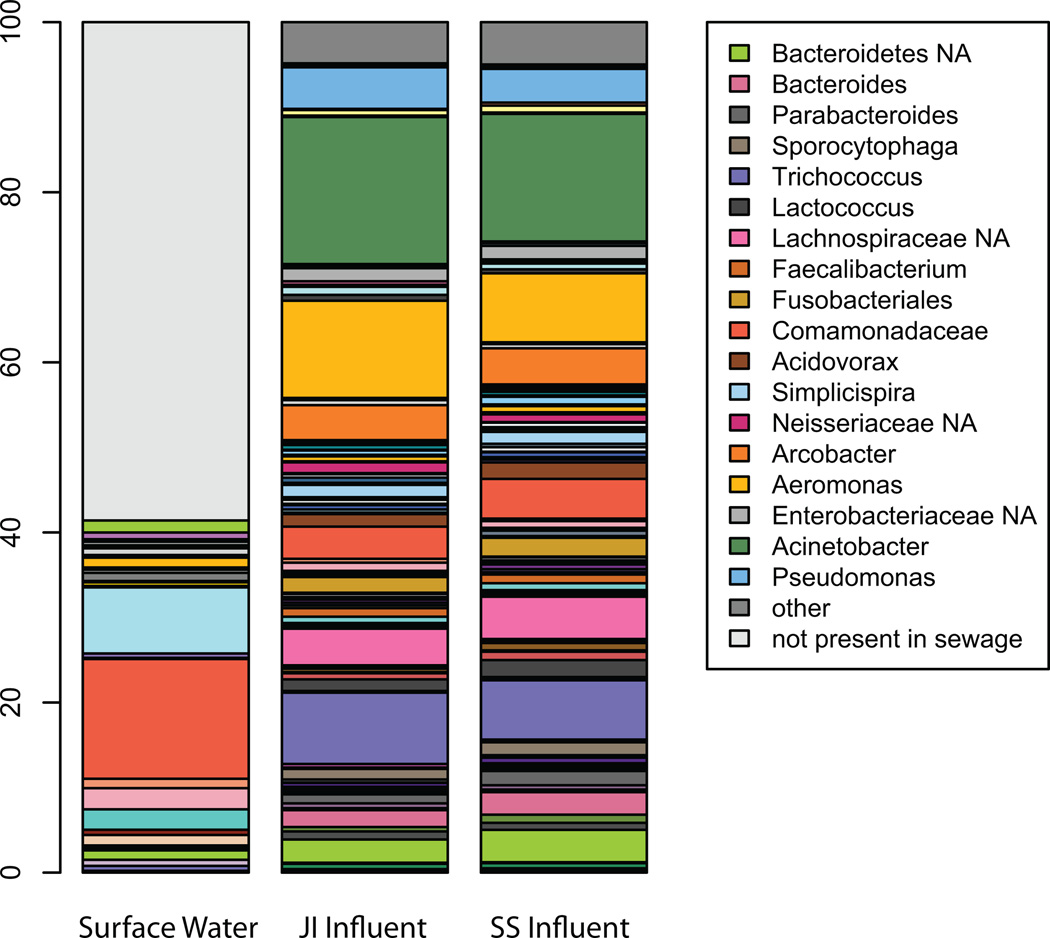
Comparisons to a reference V6 database (http://vamps.mbl.edu) identified taxonomic descriptors for the pyrotag data set. Overall, 18 taxa (e.g. V6 sequences that resolve to a unique taxonomic descriptor at either the phylum, order, family, or genus levels, see methods) comprised at least 1% of the sewage dataset. The three most dominant taxa collectively accounted for 33. 6% of all pyrotags: Acinetobacter (16.1%), Aeromonas (9.8%), and Trichococcus (7.7%) (Fig. 1). These taxa appeared to be well adapted to the sewer infrastructure environment since they occurred in low abundance in uncontaminated surface water (0.13, 0.19, and 0.01%, respectively) and were nearly absent in human fecal samples from previously published human fecal datasets (Dethlefsen et al., 2008; Turnbaugh et al., 2009). We designated six other taxa as “sewer specific” according to the criteria described in the methods. Only a small fraction of pyrotags from influent samples (~15%) matched sequences from human fecal samples. Two additional taxa that occur in low abundance in human fecal samples were enriched in the sewer environment; Lactococcus (1.7%) and Enterobacteriaceae (1.6%). Collectively, only 41% of the pyrotags recovered from surface water represented taxa that also occurred in sewage (Fig. 1), but their abundance patterns in water were distinct from sewage.
FIG. 1.
Taxa comprising the top 95% of sewage influent (n=38). These taxa account for just over 41% of uncontaminated surface water; only taxa found in sewage influent are shown for comparison. Jones Island (n=19) and South Shore (n=19) are separated to highlight similarity between plants. Stacked bars are ordered by taxonomy listed alphabetically according to the phylum in which the different taxa belong. A full listing of all sewage influent taxa and the % relative abundance in JI, SS and surface water samples is provided in supplemental data (Table 2S). The 18 most dominate taxa, which accounted for at least 1% of the relative abundance individually are shown in the legend.
Sewage communities compared with uncontaminated and contaminated surface water
We compared baseflow (e.g. no rainfall within 48 hrs) conditions with water collected in the Milwaukee estuary during a CSO to determine if we could observe increases in sewage-associated organisms (e.g. of fecal origin and “sewer specific”). The CSO contaminated water contained a higher relative abundance of fecal taxa, with more than a 20 and 30-fold increase in Lachnospiraceae and Bacteroides, respectively. Acinetobacter, Trichococcus, and Aeromonas were also more abundant at a 10-, 20-, and 3-fold higher abundance compared to baseflow conditions.
Comparison of influent to WWTP from two sections of the service area
We examined whether differences in the microbial populations reaching the WWTPs reflected the system configuration and inputs. Samples taken on the same day from SS and JI had the highest Bray Curtis similarity scores and they decreased over time at each respective plant. To examine if a particular portion of the community was more homogenous between WWTPs on a given day, we performed Pearson correlation analyses for three categories of taxa: human fecal, sewage, and water (see methods). Each category highly correlated between influent samples across 19 paired dates with average correlations of 0.94, 0.96, and 0.97 for the sewage, human fecal, and water taxa, respectively. Spearman rank correlations also demonstrated high correlations across all dates. There was no significant difference in the degree of correlation considering days with rainfall in the past 24 or 48 hours vs. no rainfall (p<0.05). In contrast, the Pearson correlation for 8/21/08 samples were lower for the most abundant sewer specific taxa (0.62), the human fecal taxa (0.84) and the water associated taxa (0.82). The high abundance organisms Acinetobacter and Aeromonas displayed the most notable differences including a reciprocal relative abundance relationship between the two plant influent samples.
Upstream vs. Downstream
We compared pyrotags from two samples collected on 5 Aug 2009 upstream of each respective WWTP with pyrotag profiles for influent samples. There was a 16% decrease in the relative abundance of human fecal organisms in JI WWTP influent and a 5% decrease in SS WWTP influent compared with each respective upstream site. We also observed a 15–30 fold increase in Lactococcus and a 3–8 fold increase in Enterobacteriaceae at both plants.
Population structure of high abundance taxa
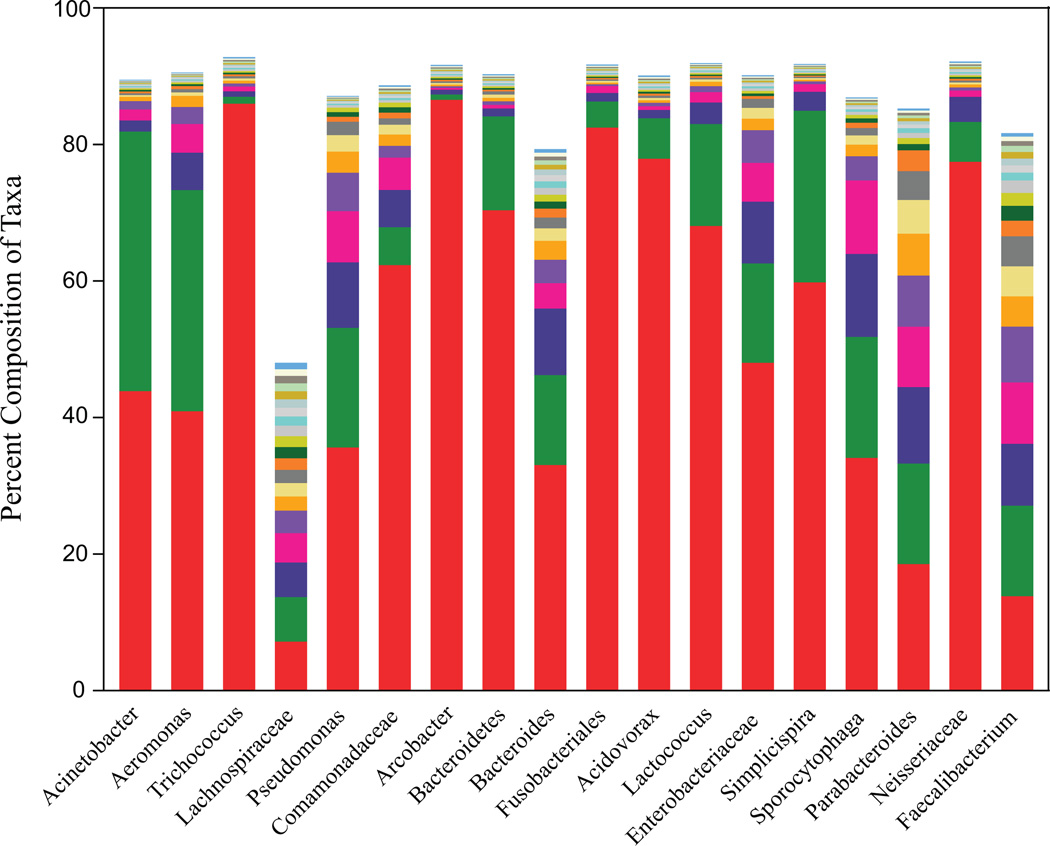
Fig. 2 shows the abundance of distinct pyrotags within each of the 18 most abundant taxa in WWTP influent samples. A single, unique pyrotag sequence represented the majority of Trichococcus, Arcobacter, Bacteroidetes (unclassified), Fusobacteriales, Acidovorax, Lactococcus, or Neisseriaceae, while two distinct pyrotag sequences represented the majority of Acinetobacter, Aeromonas, or Simplicispira. In contrast, a large number of unique pyrotag sequences represented families and genera for human associated Lachnospiraceae, Bacteroides, Parabacteroides, and Faecalibacterium. For example, the 20 most dominant Lachnospiraceae and Bacteroides pyrotag sequences accounted for only 78% and 47% of pyrotags assigned to each respective taxon.
FIG. 2.
V6 sequence composition of the 18 most abundant taxa in the WWTP influent samples. Taxa are ordered by their overall abundance. Stacked bars represent the % composition for each of the top 20 unique V6 pyrotag sequences for each taxa. The top increments are not resolved since they represent very minor fractions of the population (<1–2%). Two high abundance pyrotags represent most of the Acinetobacter and Aeromonas populations, whereas a single unique pyrotag represents nearly all Trichoccocus. Many unique sequences mapped to the fecal derived taxa such as unclassified Lachnospiraceae and Bacteroides with the 20 most abundant sequences only accounting for 47% and 78% of members within this group.
Temporal variability of highly abundant sewer specific pyrotags
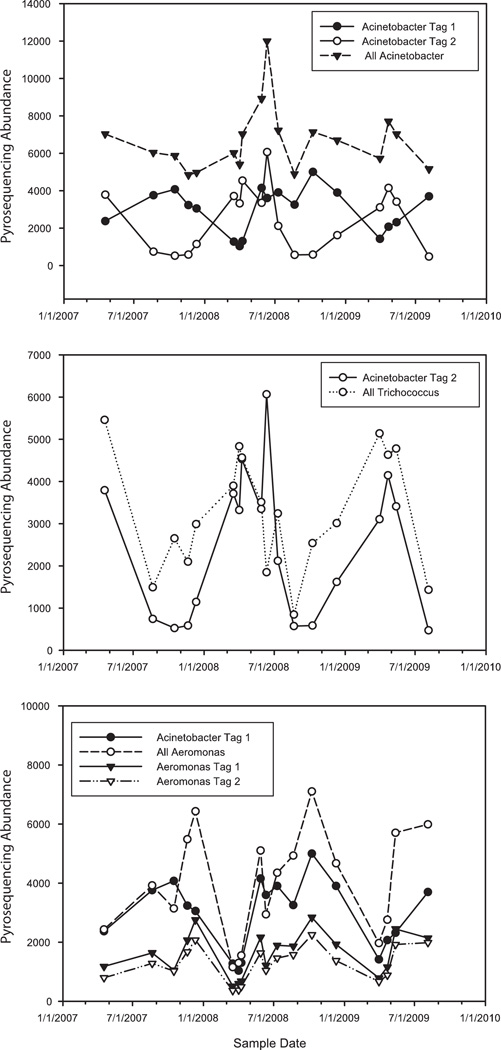
The relative abundance of all Acinetobacter varied on average by only 20% across the 38 samples. However, the two dominant Acinetobacter V6 pyrotag sequences (designated Acineto tag 1 and Acineto tag 2) had reciprocal seasonal patterns (Fig. 3, top panel). We noted Acineto tag 2 increased in winter and spring relative to Acineto tag 1, which increased in summer and fall. Other high abundance sewage-specific taxa also followed seasonal patterns, with Trichococcus following a pattern similar to Acineto tag 2 (Fig. 3, middle panel) and both dominant Aeromonas tags following a pattern similar to Acineto tag 1 (Fig. 3, bottom panel).
FIG. 3.
Seasonal patterns of high abundance taxa in urban infrastructure. Numbers of pyrotags in a normalized sample of 41,453 tags are shown for each taxonomic group. Top panel shows total Acinetobacter with Acineto tag 1 and Acineto tag 2, which are inversely related. Seasonal fluctuations reoccurred in spring 2007, 2008, and 2009. Middle panel shows total Trichococcus fluctuated seasonally similar to Acineto tag 2. Bottom panel shows total Aeromonas which was primarily represented by Aero tag 1 and Aero tag 2 followed a pattern similar to Acineto tag 2.
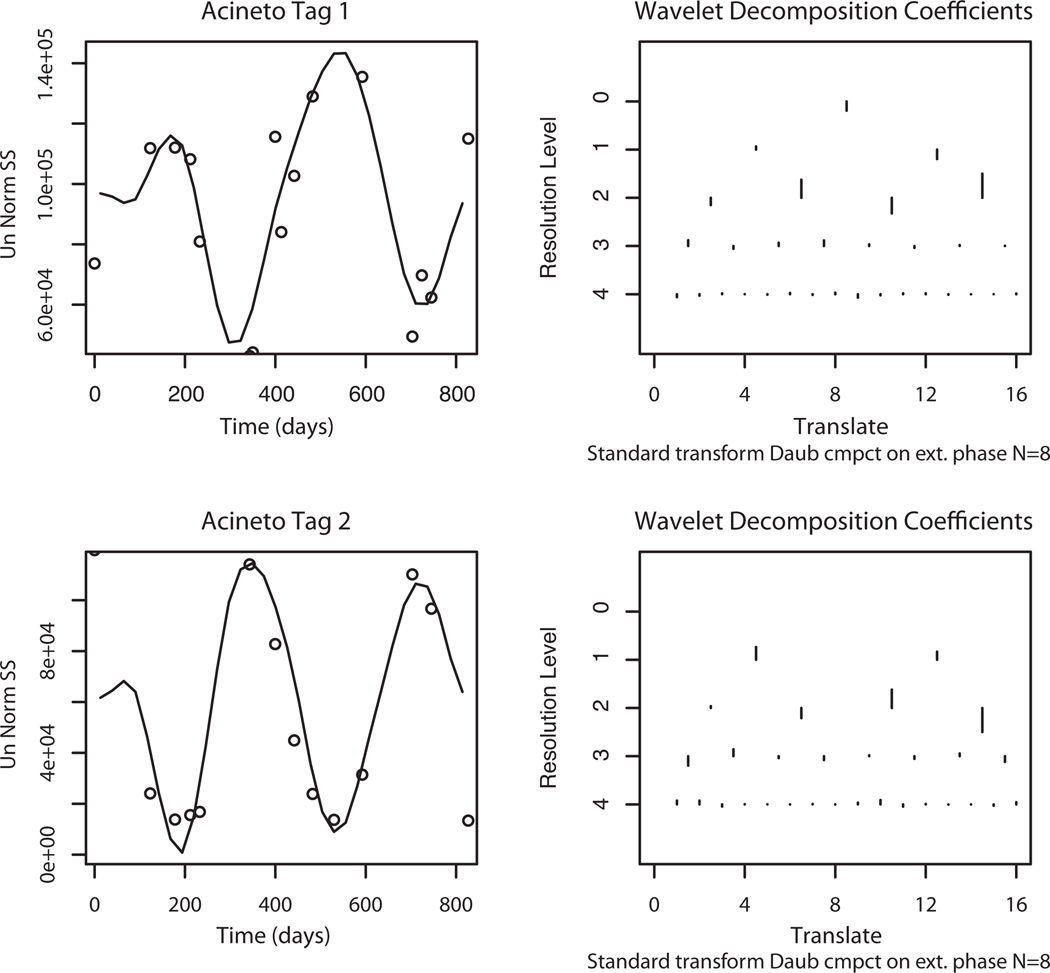
We modeled the temporal patterns using Daubechies’ wavelets to investigate whether temporal patterns of the other sewer specific abundant taxa were related to each other or were similar to either Acineto tag 1 or Acineto tag 2. The Daubechies’ wavelets provide a simple mathematical model for the temporal pattern that can be can be clustered into similar and dynamic groups. The coefficients of the Daubechies’ wavelets give the magnitude for each term of the wavelet decomposition, where different levels of the coefficients represent the fineness of the terms that are needed to represent the temporal pattern. The wavelet patterns and associated coefficients of Acineto tag 1 and Acineto tag 2 in Fig. 4 provide an example of this analysis. The differences in the location (the x axis) and size (the vertical bars) of the level 0 and level 1 coefficients indicate an inverse abundance relationship for the two Acineto time series.
FIG. 4.
Wavelet Smoothed Curve (Filter level is 8) and Data Points for Acinetobacter Tag 1 and Acinetobacter Tag 2 showing the peaks and valleys occur at different seasons. Daubechies Wavelet Coefficients showing the different temporal patterns lead to different sets of coefficients for clustering. For the wavlet decomposition graphs, the vertical axis is desginated “Resolution Level” and it represents the size of the wavelet, were the scale begins with the very low frequency wavelets and the frequency is doubled at each resolution level. The horizontal axis is designated “Translate” and it represents the shift along the time axis for the center of different size wavelets. For the largest wavelet (resolution 0), there is no translation. For the next level, there are two (orthogonal) wavelets and two translations to cover the whole time period, etc. The size of the line in the graph is the magnitude of the coefficient of the wavelet. Large lines correspond to dominant frequencies and small ones correspond to minor frequencies.
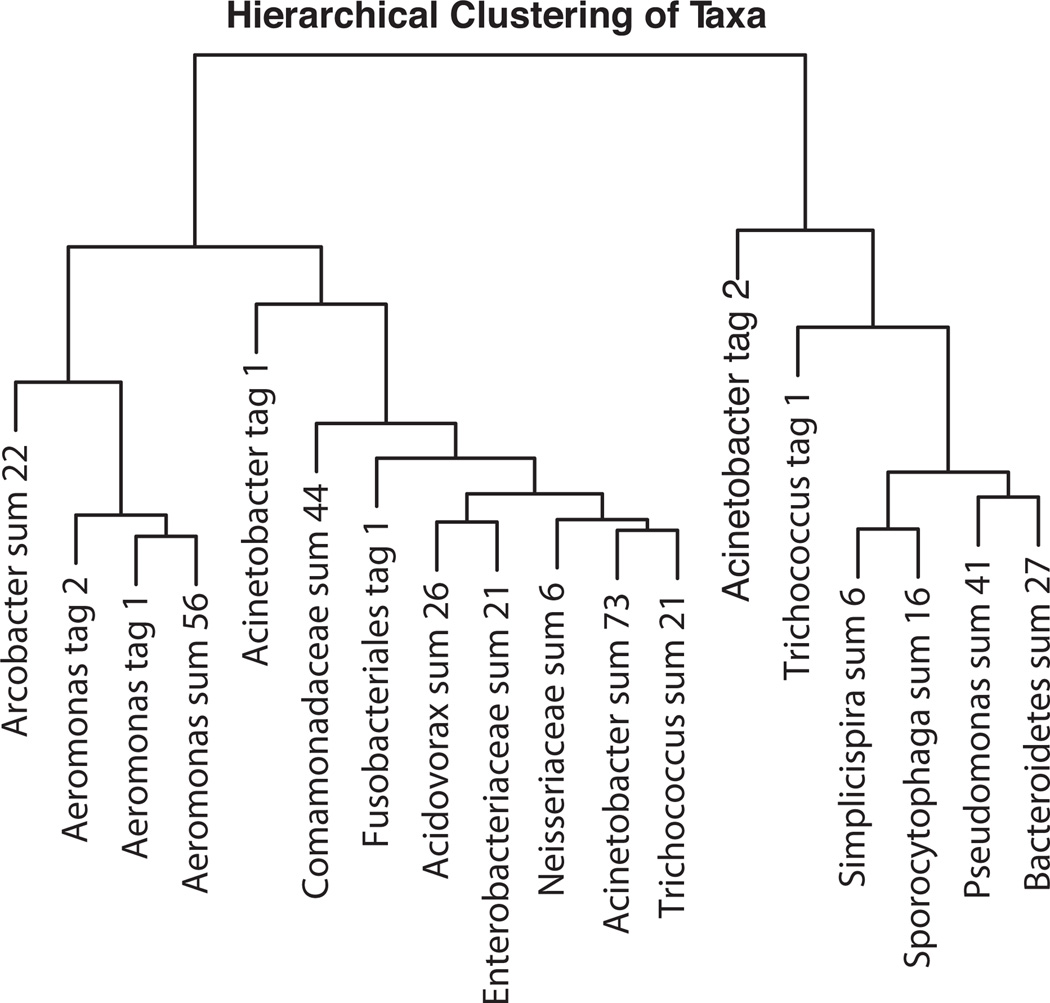
To group wavelet coefficient patterns based on temporal abundance patterns we used hierarchical cluster analysis. We found several dominant pyrotags had similar dynamics and clustered together (Fig. 5). The dendrogram shows two main groups, the Acineto tag 1 pattern and the Acineto tag 2 pattern, with a subgroup representing the Aeromonas patterns that clustered close to Acineto tag 1.
FIG. 5.
Hierarchical clustering of the temporal patterns based on the wavelet coefficients. Single dominant tags or a sum of minor tags were used in the analysis. The total number of unique tags summed to represent the group is shown as the “sum” (see methods).
Correlations of dominant pyrotags with environmental data
Several sewer specific taxa had strong correlations to measured parameters at the WWTPs. Acineto tag 1, Aeromonas tags 1 and 2 as well as the remainder of tags within the genus, and pyrotags belonging to Arcobacter, Enterobacteriaceae, Fusobacteriales and Acidovorax, positively correlated with ammonia (median correlation 0.481 with interquartile range (IQR) 0.315–0.662, BOD (Biological Oxygen Demand) (median 0.541, IQR .412 to 0.616), total phosphorous (median 0.276, IQR 0.226 to 0.693), and suspended solids (median 0.269, IQR 0.121 to 0.426), and negatively correlated to flow (median −.459, IQR −.425 to −.520). In contrast, two other high abundance sewer specific pyrotags, Acineto tag 2, and the one dominant Trichococcus pyrotag had the reciprocal relationship, with significant negative correlations to nutrients, BOD and solids and positive correlation to flow. These correlations were consistent with the coordinated temporal patterns observed with these sewer specific taxa and clustering of wavelet coefficients (Fig. 3 and Fig. 5). The sewer specific taxa did not correlate with meteorological data, namely, three of the abundant human taxa (Bacteroides, Faecalibacterium and Lachnospiraceae) had a non-significant negative correlation to daily low temperatures for JI WWTP influent (r = −0.448, −0.304, −0.349, respectively with p<0.06, 0.22 and 0.16). We observed a similar non-significant relationship for SS WWTP influent (r = −0.085, −0.203, −0.217, respectively, with p<0.74, 0.42 and 0.39, respectively).
Quantitative PCR targeting dominant taxa
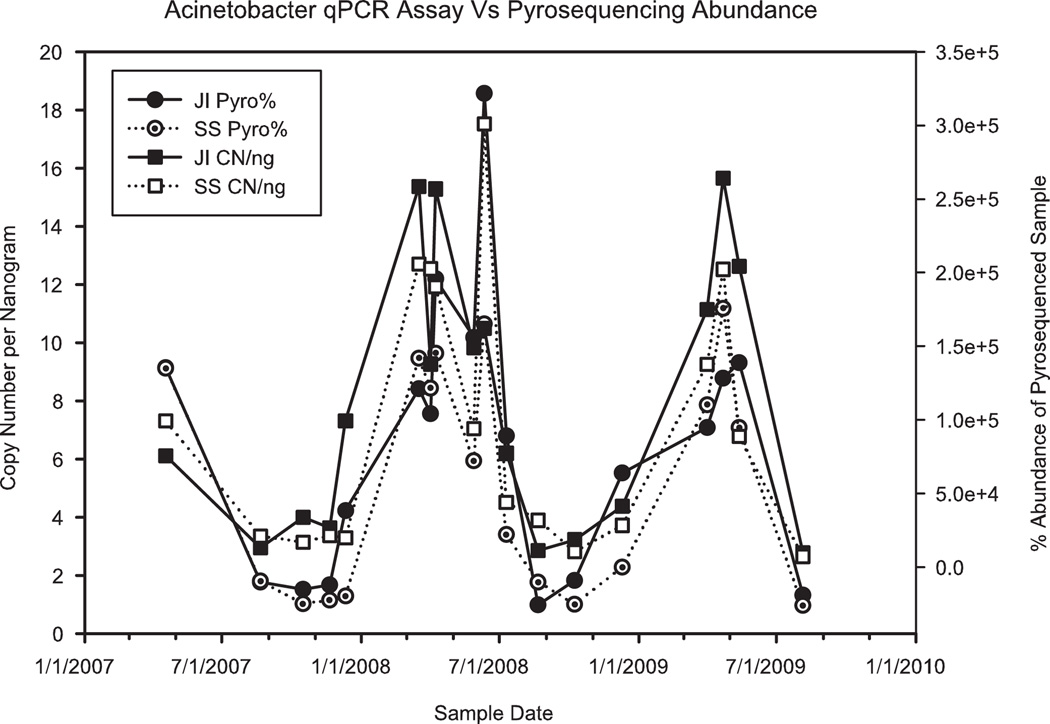
qPCR targeting Acinetobacter, Aeromonas, and Trichococcus served as an independent measure to validate the relative abundance data derived from the sequencing method. After normalization of DNA concentration, the temporal fluctuation in qPCR patterns very closely resembled shifts in the pyrosequencing data (Fig. 6).
FIG. 6.
Results of qPCR assay targeting Acinetobacter pyrotag 2. The secondary Y-axis shows the percent abundance in the pyrosequencing data. Patterns of temporal variation produced by qPCR follow the same fluctuation patterns produced by pyrosequencing. This demonstrates that temporal variation was accurately represented in the pyrosequencing data.
Phylogenetic analysis of full-length clone libraries of sewage WWTP samples
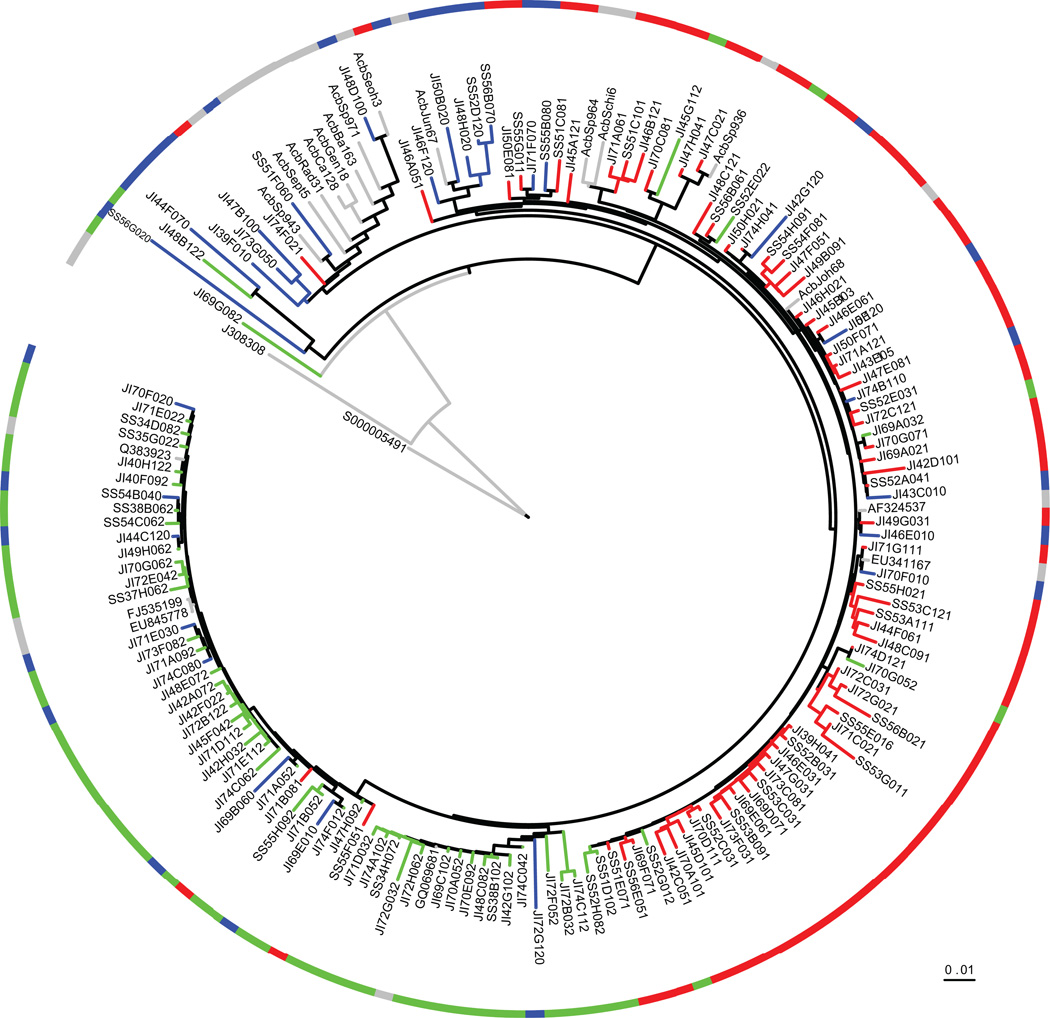
We generated clone libraries of near full-length sequences of the 16S rRNA gene to characterize the most abundant organisms for phylogenetic analysis. Clones identified as Acinetobacter were binned according to their match to specific V6 pyrotags (Table 1). The V6 pyrotag data demonstrated Acineto tag 1 and Acineto tag 2 differed at a single position across the V6 region (Table 2). A phylogenetic tree revealed that Acineto tag 1 and Acineto tag 2 were in separate clades, demonstrating that each tag represents phylogenetically distinct populations (Fig. 7). None of the WWTP Acinetobacter sequences perfectly matched SILVA reference sequences. We designated the major assemblage containing Acineto tag 1 as the summer clade (shown in red), which also contained SILVA reference sequences, AcbJoh68 (DQ911549), Acbsp964 (NZ_ABYN01000133) and AcbSchi6 (AJ278311). Two other major clades that we designated the spring clades (shown in green) included sequences that matched Acineto tag 2. Other Acinetobacter clones that did not contain Acineto tag 1 or Acineto tag 2 V6 sequences generally distributed across the tree.
TABLE 1.
Summary of full length clones corresponding to specific tag sequences for 16S rRNA gene libraries from spring and summer WWTP samples.
| Total no. clones |
Acinetobacter | Aeromonas | Trichococcus | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Taxa specific |
Tag 1 | Tag 2 | Taxa specific |
Tag 1 | Tag 2 | Taxa specific |
Tag 1 | ||
| Spring library | 1541 | 107 | 30 | 49 | 80 | 44 | 23 | 41 | 35 |
| (% total) | (6.9%) | (2.0%) | (3.2%) | (7.6%) | (4.2%) | (2.2%) | (3.9%) | (3.3%) | |
| Summer library | 895 | 110 | 57 | 15 | 82 | 36 | 31 | 8 | 8 |
| % total | (12.3%) | (6.4%) | (1.7%) | (13.5%) | (5.9%) | (5.1%) | (1.3%) | (1.3%) | |
TABLE 2.
Dominant V6 tag sequences in Acinetobacter, Trichococcus, and Aeromonas populations.
| V6 tag designation |
Sequence |
|---|---|
| Acineto tag 1 | |
| Acineto tag 2 | |
| Trich tag 1 | |
| Aero tag 1 | |
| Aero tag 2 |
FIG. 7.
Neighbor joining tree of sewage Acinetobacter clones, select top BLAST matches to clones, and divergent Acinetobacter SILVA reference sequences. SILVA sequences are designated Acb and accession numbers correspond to top BLAST matches, both are coded as grey. Clones corresponding to Acineto tag 1 are designated red, Acineto tag 2 as green, and other, lower abundance tags as blue. A tree constructed using maximum likelihood recovered similar topography with Acineto tag 1 and Acineto tag 2 in distinct clades.
Unifrac analysis of near full-length sequences demonstrated that Acineto tag 1 and Acineto tag 2 populations differed significantly from each other and from other sewage Acinetobacter (p-test values from <0.0001 to 0.0400). We found no significant year to year variation for Acineto tag 1 sequences for the spring libraries, or between spring and summer libraries. Similarly, the Acinetobacter sequence corresponding to Acineto tag 2 did not significantly vary between years or libraries (data not shown).
Two different V6 pyrotags comprised the majority of Aeromonas pyrotags (Fig. 2), however, Aeromonas full-length clones matching either Aero tag 1 or Aero tag 2 did not appear as distinct clades; this agrees with the Unifrac analysis. Similarity between the sewage Trichococcus full-length clone sequences always exceeded 98%, demonstrating little sequence variation in the 16S rRNA gene of this population. These observations are consistent with recovering a single dominant V6 sequence (Trich tag 1) in our sewage dataset. Overall this V6 tag sequence could not track Trichococcus subpopulations, but did represent Trichococcus as a genus.
Discussion
Urban infrastructure as a defined environment for microbial communities
Sewer infrastructure provides a relatively new environment in which bacteria can colonize and propagate. Despite the large distance between WWTPs (Figure 1S, supplemental information) and separate pipes dedicated to each plant, the entire service area acts as a single, defined system with fairly dynamic changes in the influent microbial community on a weekly, monthly and annual time scale. These changes were mirrored at JI and SS WWTPS. We noted this phenomenon in an earlier study with a limited number of samples (McLellan et al., 2010), and designed the present study to collect 19 paired samples at different temporal scales. These findings provide valuable insight into variability across space and time and provide important guidance for future sampling aimed at understanding differences in municipal systems.
Since a portion of the JI service area includes combined sewers that collect stormwater, we expected to observe differences in JI and SS WWTP communities during rainfall because of the influx of stormwater (and associated bacteria) into the combined system. However, JI and SS WWTP influent communities were highly similar on almost all days regardless of rainfall. Stormwater runoff carries with it soil and other bacteria from land surfaces. Yet, the 95% most abundant pyrotags did not include sequences from common soil taxa such as Sphingomonas and Bradyrhizobium (T. Schmidt, personal communication). Instead, the dominant populations in the JI community (Fig. 1 and Table 2S, supplemental information) included fecal bacteria and sewer-specific bacteria. Since combined sewers only represent 10% of the service area for JI, the relatively minor input of bacteria from stormwater relative to the total sewage community agrees with expectations.
Functional roles of high abundant sewage taxa
Treatment of wastewater can begin in the sewer conveyance systems, particularly with gravity flow systems, which are primarily aerobic (Norsker et al., 1995; Chen et al., 2003). Bacterial activity in sewers is thought to occur most often in sediments and biofilms (Raunkjaer et al., 1995; Chen et al., 2003; Leung et al., 2005; Jiang et al., 2009). Evidence of microbial communities associated with sediments and biofilms may be found in WWTP influent as this material is resuspended or detached. However, other sources of suspended solids, such as fecal material or soils make it difficult to identify community members originating from sediment and biofilm communities. Specific sampling of pipe biofilms or sediment would be necessary to make this distinction.
Gammaproteobacteria, which utilize a wide range of carbon sources, dominate influent from the Milwaukee conveyance system and correspond to several of the “sewer specific” bacteria. Acinetobacter commonly lives in soil and water (Warskow and Juni, 1972) and can utilize a wide range of carbon sources (Baumann et al., 1968). Members of this genus degrade a wide range of compounds including fuel oil (Barberio and Fani, 1998), and estradiol (Pauwels et al., 2008). Acinetobacter also contribute to activated sludge communities in WWTPs (Snaidr et al., 1997). Aeromonas spp. commonly occur in sewage (Brenner et al., 2004). Pseudomonas and Aeromonas spp. thrive in a mesophilic environment, can grow both aerobically and anaerobically and utilize a wide range of carbohydrate sources (Farmer et al., 2006; Moore et al., 2006). Sporocytophaga and Cytophaga were among the 18 most dominant taxa and these organisms can breakdown cellulose in aquatic systems (Madigan and Martinko, 2005).
V6 sequence tags represent specific populations within high abundant taxa
Next generation DNA sequencing provides in depth descriptions of microbial communities. The complexity of these data sets challenges our ability to place these data into an ecological context. An average recovery of ~28,000 pyrotags for each sample allowed us to identify major and minor populations for most genera. The V6 pyrotag data identified two dominant groups of Acinetobacter that differed by one nucleotide; these tag sequences represent distinct populations based on full-length 16S rRNA gene sequencing and demonstrated different seasonal fluctuations. For Trichococcus, we identified more than 1000 unique pyrotags matching this genus, but one sequence represented 86% of all Trichococcus. This high abundance Trichococcus pyrotag corresponded to a collection of full-length sequences with very limited diversity, where identity was >98%.
We did not observe distinct clades in Aeromonas based on full-length sequences, even though we found two different highly abundant pyrotags. The two tags most likely represent the sequence heterogeneity of a single Aeromonas population that responds coordinately to environmental influences, however, these tags could also represent variation in the rRNA operons of a single organism. However, genomic sequence information for Aeromonas hydrophila shows that its 10 copies of the 16S rRNA gene are absolutely conserved in the V6 region, with the exception of a C to T transition in one operon. Similarly, eight of the nine Aeromonas salmonilca rRNA operons show no sequence variation. The ninth operon has two divergent positions in the V6 region that exactly match the same region in A. hydrophilia. Given the conserved nature of the operons of the two Aeromonas genomes that we examined, it is most likely the two pyrotags identified in sewage represent different organisms in the population.
Temporal fluctuations of urban infrastructure microbial communities
Our pyrotag data revealed clear seasonal fluctuations in two V6 tag sequences that represent two distinct Acinetobacter populations (Fig. 5), as well as seasonal variation for a V6 sequence from Trichococcus and two V6 sequences from Aeromonas. Samples taken over nearly three years allowed us to discern temporal cycling patterns and model these patterns as a wavelet function. Certain populations had similar wavelet functions (as shown in Fig. 5), suggesting that they were modulated by similar forces. A multiple linear regression using variables such as temperature, rainfall, and data collected at the plant (BOD, ammonia, etc.) demonstrated that the populations (defined either by a single tag, or a sum of the pyrotags within a taxa) with similar wavelet functions significantly correlated with the same group of variables. These two independent analyses show that the observed relative abundance fluctuations represent ecological forcings upon the bacteria, and that these forces can impact bacterial populations in different ways.
We noted a relative decrease of human fecal bacteria comparing upstream and downstream sites. Decay of fecal anaerobes in aerobic conditions has been noted for Bacteroides, a major member of human fecal biota (Okabe and Shimazu, 2007; Balleste and Blanch, 2010). For influent samples, statistical analysis revealed that common human fecal bacterial taxa negatively correlated to temperature suggesting that these taxa can persist longer in the system at lower temperatures. Seasonal dynamics linked to temperature shifts have been reported previously for members of the wastewater microbial community, including sulfate reducing bacteria (Ben-Dov et al., 2009) and Aeromonas spp. (Monfort and Baleux, 1990). The seasonal nature of the cyclic pattern did not always correlate with temperature. For urban sewer infrastructure, ambient air temperature may only be a proxy for seasonality; the temperature within the pipes likely has a narrower range than air temperatures (P. Topchewski, personal communication). Several other seasonal factors, including rainfall, which introduces stormwater runoff and hence nutrients into the system, and the influx of road salt could also be modulating factors.
While there were notable shifts in the relative abundance of Acinetobacter, Aeromonas, and Trichococcus, phylogenetic analysis of full-length clones constructed from multiple time-points did not reveal shifts in genotypes over time. These findings suggest that changes in the relative abundance may reflect reoccurring cycles of individual taxon dominance rather than seasonal replacement by new populations within a genus (e.g. populations remain consistent, but levels change). Further, the qPCR results reflected what we observed in relative abundance measures (e.g. sequencing).
Conclusions and significance
Overall, these findings illustrate the usefulness of next generation sequencing for tracking phylogenetically and ecologically distinct populations. Three pieces of evidence emerged to support this statement: (i) wavelet functions divided the community into groups that modulate in a coordinate manner; (ii) multiple regression revealed the same variables significantly correlated with each individual V6 sequence within wavelet defined sequence groups; and (iii) phlyogenetic analysis of full length clones identified distinct clades corresponding to V6 tags that had different temporal patterns and correlating environmental variables (e.g. Acineto tag 1 and Acineto tag 2). In some cases, the relative abundance of multiple V6 pyrotags from the same taxon had similar temporal variation and ecological patterns. For example, the two abundant Aeromonas V6 tag sequences had similar temporal patterns and were correlated to the same WWTP variables. Phylogenetic analysis of full-length 16S rRNA genes associated with these Aeromonas tags revealed a monophyletic clade despite the presence of the two V6 tags.
We used next generation sequencing to describe population dynamics of microbial communities in a previously uncharacterized environment, urban sewer infrastructure. Understanding these communities has important implications toward advancing our knowledge of microbes associated with sewage and potential ways treatment might be enhanced by microbes resident to sewer systems. Urban sewer infrastructure also imposes a major impact to the environment (USEPA, 2004). Combined and sanitary sewer overflows release high loads of non-indigenous bacteria into waterways each year. These discharges contain approximately 109 to 1010 organisms per ml (McLellan et al., 2010) which outnumber bacterial densities in surface waters by three to four orders of magnitude (Amann et al., 1999; Lemke and Leff, 1999; Yokomaku et al., 2000). Some of the high abundance sewer taxa represent minor members of surface water communities, and their introduction may disrupt natural communities at the base of the foodweb or the biogeochemical functions they serve. Finally, microorganisms that thrive within the sewer system may serve as useful adjuncts to fecal indicators for tracking sewage contamination because their population levels exceed traditional indicator species and they could provide a complex signature of sewage pollution in surface waters. Understanding microbial signatures for urban sewer systems, beyond the fecal organisms that are derived from human sources, will improve our ability to identify signatures of pollution sources and assess the ecological implications of sewage release.
Methods
Sewage treatment plant influent sample collection
We obtained WWTP influent samples of the two major facilities servicing Metropolitan Milwaukee, WI on multiple dates in 2005, 2007, 2008, and 2009 (Table 1S, supplemental information). The entire service area is 1065 km2. JI WWTP is located in downtown Milwaukee and receives sewage from 122 km of metropolitan interceptor sewer (MIS), with 10% of these (5% of total service area) comprised of combined sewers that capture sanitary sewage and stormwater from Milwaukee’s most densely urban area. SS WWTP services primarily residential areas of the metropolitan area and receives sewage from 201 km of MIS. An additional 143 km of MIS can be diverted to either SS or JI. Samples consisted of 1 L of 24-hour flow-weighted samples collected from 6 am on the preceding day until 6 am on the stated collection day. Flow into the WWTP was averaged between measurements taken at the 6 am time points each day. Meteorological data accompanying the samples included high/low temperatures (five day average of collection day and four days previous) and precipitation totals (the day of and for 48 hrs previous). Ancillary data collected at the WWTPs included flow, ammonia determined by SM(20) 4500-NH3D, BOD (5 day total) SM(20) 5210B, total phosphorous USEPA method 365.1, and suspended solids SM(20) 2540D (American Public Health Association) (Table 1S, supplemental information).
Sewage samples were filtered through a 47 mm, 0.45 µm nitrocellulose membrane (Millipore, Billerica, MA) at 100 ml or until the filter clogged. Surface water was collected at the Milwaukee estuary on 9 June 2008 during a CSO and 24 June 2008 during baseflow conditions using a 5 L grab sampler. Three samples were pooled prior to filtering 200 ml as described above. All samples were stored at −80°C until processing for DNA extraction.
DNA Extractions
DNA extractions were carried out using the Fast DNA spin kit for Soil (MP Biomedicals, Solon, OH), and purified using MO BIO Powerclean DNA clean-up kit (MO BIO Laboratories Inc., Carlsbad, CA). DNA concentrations were measured using NanoDrop® ND-1000 (Thermo Fisher Scientific Inc., Pittsburg, PA).
454 pyrosequencing
We amplified the V6 hypervariable region of the 16S rRNA coding region using a mixture of five fused primers at the 5′ end of the V6 region (E. coli positions 967- 985) and four primers at the 3’ end (E. coli positions 1046-1028) to capture the breadth of diversity of rRNA sequences represented in molecular databases (Sogin et al., 2006; Huber et al., 2007). We amplified libraries from at least three independent PCR cocktails for each sample to minimize the impact of potential early-round PCR errors. Amplicons were prepared and sequenced using the Roche Genome Sequencer GS-FLX according to the Roche standard protocols. Sequencing provided approximately 800,000 reads per run.
Quality trimming and taxonomic assignments
We trimmed the resulting sequences as described previously (Huse et al., 2007). We used Global Assignment of Sequence Taxonomy (GAST) (Huse et al., 2008) to assign taxonomy through comparison to a reference database of full-length 16S rRNA sequences based on the SILVA database (Pruesse et al., 2007). We compared pyrotags at two levels of resolution, first, grouped according to their taxonomic assignment. In some cases, the pyrotags could not be resolved to a genus or species level and the highest taxa to which it mapped was assigned. For example, the taxa designated as Bacteroidetes includes only pyrotags that could not be resolved further, other Bacteroidetes such as Bacteroides are listed as a separate taxonomic designation. We also compared pyrotags at the sequence level using the “TaxbyRef” dataset in the web-based data analysis platform Visualization and Analysis of Microbial Populations (VAMPS) (http://vamps.mbl.edu). Here, pyrotags are matched to their closest reference sequence, which eliminates many sequences that are slightly different length and some singlets, while providing a refined grouping that preserves single base pair changes between tags.
Data grouping for WWTP comparison
Samples were grouped by WWTP (i.e. 19 samples from JI were considered as a whole and the same for the 19 samples from SS). Data was trimmed to include only the 95% most abundant organisms, which included 102 unique taxonomic designations. By comparing to surface water samples and the previously published human fecal dataset, the 102 taxa were categorized as human fecal (n=22), water (n=13), or sewage (n=39). Some fecal taxa were categorized as “enriched” (n=10). Taxa not clearly associates with a particular environment were uncategorized (n=20).
To bin WWTP influent taxa and the corresponding pyrotag sequences into these groups, we used a tiered approach. We utilized a normalized dataset consisting of V6 pyrotags for 38 sewage samples, and comparison samples that included two estuary samples and 48 individual fecal samples. The “TaxbyTax” dataset in VAMPS was used, meaning that the V6 pyrotags were grouped according to their taxonomic assignment in GAST. First, the probable fecal organisms were identified by comparison to human fecal samples from two previously published datasets (Dethlefsen et al., 2008; Turnbaugh et al., 2009). Taxa that were at an equal or higher abundance in fecal samples compared with sewage (>1:1) were assigned to the “human fecal” group; two additional taxa, unclassified Clostridiales and Bacteroidales that were at lower ratios (0.72:1 and 0.43:1, respectively) were also assigned to the human fecal group. All of these taxa occurred in sewage samples at >0.25% abundance. Second, fecal taxa that were present in human fecal samples, but higher in sewage influent (3–80 fold higher depending on the taxa) were designated as “enriched”. We did not include the enriched organisms in the comparisons of the WWTPs, but used this category for comparing upstream and downstream samples.
Organisms that were “sewer specific” were identified as taxa that were very low (< or equal to 0.01% relative abundance) or absent from fecal samples and occurred at a 10 fold higher abundance in sewage influent compared to surface water. To identify taxa that commonly occur in surface water, the non-fecal associated taxa in sewage influent were compared with their occurrence in uncontaminated surface water. There was a distinguishable group of taxa that were present at >0.2% relative abundance in sewage and were >2 fold higher in surface water in comparison; these taxa were designated as “surface water”; other surface water taxa were at very low abundance in sewage. Finally, the remaining taxa that did not clearly fall into one of the four groups were designated as “not catagorized” and were not used in the group specific comparisons; there were minor members and accounted for 9% of the total pyrotags. The group assignment for each taxa found in sewage influent is shown in Table 2S, supplemental information.
Data Grouping for Correlations to Environmental Conditions
Pyrotags were grouped according to GAST reference sequence (Huse et al., 2008). The thirty-eight sewage samples were trimmed to 95% of the total sewage dataset which corresponded to tags grouped to a reference sequence with at least 30 occurrences overall. Correlations were calculated for the 18 taxa that comprised at least 1% of the total sewage dataset. Pyrotags assigned to a reference sequence with identical taxonomic designations were grouped. However, Acinetobacter, Aeromonas, and Trichococcus were split further. For these taxa, the dominant high abundance pyrotag or pyrotags were calculated separate from remaining tags within the taxa, which was grouped together as a sum of the remaining pyrotags.
Statistical Analysis
For each taxa comprising more than 1% of the sewage dataset (n=18), the mean occurrence and average % difference from the mean were calculated to determine temporal stability. This calculation was also performed for individual tag sequences within each taxa. Because the data was not normal, Mann-Whitney tests were used to determine if there was a significant difference between top taxa at the Jones Island versus South Shore plants. Bray Cutis similarity scores were calculated for paired samples over time at each WWTP. Spearman rank and Pearson correlations of taxanomic groups at SS and JI WWTPs on a single day were also calculated. Multivariable linear regression was used to examine the taxa relationships with the meteorological data (high, low, and average daily temperatures, and rainfall in past 24 hours) and the metadata from the WWTP (ammonia, BOD, total phosphorous, and suspended solids).
Wavelet decomposition with Daubechies’ wavelets was used to model the temporal pattern in the bacterial abundance (Silverman, 1999). The Daubechies’ wavelets provide a method to smooth the data without losing the fidelity of the representation, which occurs with methods such as moving averages, which result in substantially reduced peaks and valleys. Wavelets are an alternative to sine and cosine (Fourier modeling) of time series that have the advantage that (1) they are much superior if the time points are irregular, (2) they are only affected locally if there is an abrupt change, whereas Fourier modeling is affected globally and (3) they require fewer terms to model the time series. The equation for a wavelet model is
where djk is the kth wavelet coefficient at level j and φjk( ) is the corresponding wavelet function.
The WaveThresh library of the R statistical package was used for the modeling (Nason, 2008). Daubechies’ wavelets were chosen because they are smooth and can be differentiated, unlike the Haar wavelets. The coefficients of the Daubechies’ wavelets give the magnitude of each of the terms of the wavelet decomposition. Different levels of the coefficients represent the fineness of the terms that are needed to represent the temporal pattern and different coefficients in the same level represents where the peaks and valleys of the temporal pattern occur. As the level of the wavelet decomposition increases (0, 1, 2 …), the fineness of the description of the temporal pattern increases. Clustering of the wavelet coefficients of pyrotags (or collections of pyrotags) divides them into groups with similar temporal patterns. Clustering used the R library cluster with hierarchical clustering and the complete linkage method. We attempted to use Fourier models; however the irregular sampling points and the need to model abrupt changes in the pattern over time, led to very poor fit and no commonality of the sine-cosine coefficients.
qPCR Assays
Quantitative PCR assays were developed for Acinetobacter, Aeromonas, and Trichococcus. For each assay, full length clones were characterized using NCBI Blast. Full length clones were sorted and selected based on greater than 90% identity to the target taxa. Related sequences were downloaded from RDP within the target family and order. These sequences along with the most abundant sequence tag were aligned using Vector NTI. Primers were designed to match full length clones containing the sequence tag at regions that excluded related sequences (Table 3). RDP probe match was used to check for primer cross reactivity. Standard curves with SYBR reagents were developed for each target organism using linearized DNA from a full-length sewage clone with 100% identity to the primer set. For total bacteria, primers used were 331F and 797R as described by (Nadkarni et al., 2002). Standard curves were generated using a mixture of full length sewage clones matching several different taxa.
TABLE 3.
List of oligonucleotide primers used for qPCR. The 16s rRNA gene was targeted for primer design.
| Trichococcus | Trich555F | 5′GTCTTGACATCCTTTGACAAT3′ |
| Trich2723R | 5′TTGACCTCGCGGTCTTGCT3′ | |
| Aeromonas | Aero710F | 5′GCTGTGTCCTTGAGACGTGGC3′ |
| Aero510R | 5′TTCTGATTCCCGAAGGCACTCC3′ | |
| Acinetobacter | Acineto668F | 5′GCATTCGATACTGGGAGA3′ |
| Acineto1079R | 5′CAGCACCTGTATGTAAGC3′ |
All samples were run in duplicate 25 µl reactions: 12.5 µl 2x ABI SYBR master mix, 5 µl template DNA, and 1 µl forward and reverse primer for a reaction concentration of 100 nM each. The thermocycler program was as follows: 95°C for 10 minutes followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute.
16S rRNA clone library construction
Sample DNA from three spring WWTP samples (JI 4/20/05, SS 4/18/07, JI 4/18/07) and two summer samples (SS 8/21/07 and JI 8/21/07) was amplified using universal 16S rRNA primers 8F and 1492R (Crump et al., 1999). PCR product was purified using QIAGEN PCR purification kit (Qiagen Inc., Valencia, CA). Products were cloned into pCR2.1 vector using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA). Plasmid DNA was isolated using a manual method adapted to a 96-well microtiter plate format (Sambrook and Russell, 2001). Sequencing was carried out from the 8F and 1492R primers using the ABI Big Dye Terminator Kit (Applied Biosystems, Foster City, CA) on an ABI Prism 3700xi (Applied Biosystems, Foster City, CA), which generated approximately 800 bp reads. Sequences were trimmed for quality using PHRED (Ewing and Green, 1998) and sequence reads assembled using Cap3 (Huang and Madan, 1999). The near full-length sequences were analyzed for chimeras using Mallard (Ashelford et al., 2006). Sequences flagged by Mallard were analyzed using Chimera Check (Cole et al., 2003). Table 1 shows the fraction of nearly full-length clone sequences that perfectly match tags from Acinetobacter, Trichococcus or Aeromonas.
Phylogenetic trees and Unifrac analysis
All clone sequences with taxonomic identity to Acinetobacter at 90% or better by the RDP classifier (Wang et al., 2007) were compiled for analysis. An alignment and neighbor-joining tree were constructed in ClustalW (Thompson et al., 1994). One or two sequences representing each clade were put into NCBI BLAST, and the top blast hit for each sequence was downloaded to include in a second alignment. All Acinetobacter sequences available on the SILVA (Pruesse et al., 2007) database were downloaded as an aligned file and used to construct a tree in ARB (Ludwig et al., 2004). Based on major clades, thirteen sequences were chosen to represent the range of diversity within the Acinetobacter branch, and these sequences were included in the Acinetobacter sewage clone alignment. E. coli (X80723) and Pseudomonas fluorescens (AJ308308) were used as outgroup sequences. The final alignment was trimmed to equal sequence length and hand aligned prior to construction of neighbor-joining and maximum likelihood trees in ARB. The tree was then uploaded onto iTOL (Letunic and Bork, 2007, 2011) in order to illustrate sequence tag and seasonal relationships via color-coding. This same procedure was repeated for Aeromonas clones, and Trichococcus clones using outgroup P. fluorescens (AJ308308) and Enterococcus faecalis (AF039902), respectively.
Aligned full-length sewage clone libraries were uploaded onto Unifrac (Lozupone et al., 2006). Unifrac significance and P-tests were used to calculate significant differences among clones matching tag 1, tag 2, or neither (in the case of Trichococcus clones, analysis consisted of clones matching tag 1 and those that did not match tag 1) and to determine if there was a significant difference in clones corresponding to a specific tag sequence between the two dates of the spring library, and between the spring versus summer libraries.
Sequence diversity of Aeromonas 16S rRNA genes
Two fully sequenced Aeromonas genomes are available on NCBI (A. hydrophila subsp. hydrophila ATCC 7966 gi_117617447 and A. salmonicida subsp. salmonicida A449 gi_145297124). All copies of the 16s rRNA gene from each species (10 for A. hydrophila and 9 for A. salmonicida) were downloaded as fastas and aligned in Vector NTI (Invitrogen, Carlsbad, CA) and compared with Aeromonas high abundance pyrotags.
Sequence data submission
Clone library sequences corresponding to Acinetobacter, Trichococcus, and Aeromonas have been submitted to GenBank under accession numbers GU356036 through GU356392. Eight of the 40 samples have been previously deposited under the study accession ID SRP000905. Pyrotag sequences are available through VAMPS (http://www.vamps.mbl.edu).
Supplementary Material
Acknowledgments
This work was funded by NIH grant 1 R21 AI076970-01A1. This work was also funded in part by the Woods Hole Center for Oceans and Human Health (COHH), NIH award no. ES012742 and NSF award no. 0430724. We thank Ryan Newton for assistance in constructing comprehensive trees in ARB, selection of reference sequences, comments on previous manuscript versions, and insightful discussion. We also thank the Milwaukee Metropolitan Sewerage District for cooperation and assistance with this study, in particular Peter Topchewski and Thomas Petrie for providing information on the conveyance system, and Jeff MacDonald for facilitating sample coordination.
References
- Amann R, Glockner FO, Fuchs BM. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl Environ Microbiol. 1999;65:3721–3726. doi: 10.1128/aem.65.8.3721-3726.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Public Health Association. Standard Methods for the Examination of Wastwater. 20th edition [Google Scholar]
- Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl Environ Microbiol. 2006;72:5734–5741. doi: 10.1128/AEM.00556-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balleste E, Blanch AR. Persistence of Bacteroides Species Populations in a River as Measured by Molecular and Culture Techniques. Applied and Environmental Microbiology. 2010;76:7608–7616. doi: 10.1128/AEM.00883-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberio C, Fani R. Biodiversity of an Acinetobacter population isolated from activated sludge. Res Microbiol. 1998;149:665–673. doi: 10.1016/s0923-2508(99)80014-x. [DOI] [PubMed] [Google Scholar]
- Baumann P, Doudoroff M, Stanier RY. A study of the Moraxella group. II. Oxidative-negative species (genus Acinetobacter) J Bacteriol. 1968;95:1520–1541. doi: 10.1128/jb.95.5.1520-1541.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Dov E, Kushmaro A, Brenner A. Long-term surveillance of sulfate-reducing bacteria in highly saline industrial wastewater evaporation ponds. Saline Systems. 2009;5:2. doi: 10.1186/1746-1448-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner DJ, Krieg NR, Staley JT. Bergey's Manual of Systematic Bacteriology: Proteobacteria. New York: Springer; 2004. [Google Scholar]
- Chen GH, Leung DH, Hung JC. Biofilm in the sediment phase of a sanitary gravity sewer. Water Res. 2003;37:2784–2788. doi: 10.1016/S0043-1354(03)00083-6. [DOI] [PubMed] [Google Scholar]
- Cole JR, Chai B, Marsh TL, Farris RJ, Wang Q, Kulam SA, et al. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 2003;31:442–443. doi: 10.1093/nar/gkg039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump BC, Armbrust EV, Baross JA. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia river, its estuary, and the adjacent coastal ocean. Appl Environ Microbiol. 1999;65:3192–3204. doi: 10.1128/aem.65.7.3192-3204.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Research. 1998;8:186–194. [PubMed] [Google Scholar]
- Farmer JJ, Arduino MJ, Hickman-Brenner FW. The Genera Aeromonas and Plesiomonas. In: Dworkin M, editor. The Prokaryotes. Singapore: Springer; 2006. pp. 564–645. (ed in chief) [Google Scholar]
- Ferguson CM, Coote BG, Ashbolt NJ, Stevenson IM. Relationships between indicators, pathogens and water quality in an estuarine system. Water Res. 1996;30:2045–2054. [Google Scholar]
- Hao OJ, Chen JM, Huang L, Buglass RL. Sulfate-reducing bacteria. Crit Rev Environ Sci Technol. 1996;26:155–187. [Google Scholar]
- Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber JA, Welch DB, Morrison HG, Huse SM, Neal PR, Butterfield DA, Sogin ML. Microbial population structures in the deep marine biosphere. Science. 2007;318:97–100. doi: 10.1126/science.1146689. [DOI] [PubMed] [Google Scholar]
- Huse SM, Welch DM, Morrison HG, Sogin ML. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol. 2010;12:1889–1898. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 2007;8:R143. doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse SM, Dethlefsen L, Huber JA, Welch DM, Relman DA, Sogin ML. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000255. e1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hvitvedjacobsen T, Raunkjaer K, Nielsen PH. Volatile fatty-acids and sulfide in pressure mains. Water Sci Tech. 1995;31:169–179. [Google Scholar]
- Jiang F, Leung DH, Li S, Chen GH, Okabe S, van Loosdrecht MC. A biofilm model for prediction of pollutant transformation in sewers. Water Res. 2009;43:3187–3198. doi: 10.1016/j.watres.2009.04.043. [DOI] [PubMed] [Google Scholar]
- Lemke MJ, Leff LG. Bacterial Populations in an Anthropogenically Disturbed Stream: Comparison of Different Seasons. Microb Ecol. 1999;38:234–243. doi: 10.1007/s002489900173. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011;39:W475–W478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung HD, Chen G, Sharma K. Effect of detached/re-suspended solids from sewer sediment on the sewage phase bacterial activity. Water Sci Technol. 2005;52:147–152. [PubMed] [Google Scholar]
- Lipp EK, Farrah SA, Rose JB. Assessment and impact of microbial fecal pollution and human enteric pathogens in a coastal community. Mar Pollut Bull. 2001;42:286–293. doi: 10.1016/s0025-326x(00)00152-1. [DOI] [PubMed] [Google Scholar]
- Lozupone C, Hamady M, Knight R. UniFrac--an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madigan M, Martinko J. Brock Biology of Microorganisms. 11th edn. New York, USA: Pearson Higher Education; 2005. p. 404. [Google Scholar]
- McLellan SL, Huse SM, Mueller-Spitz SR, Andreishcheva EN, Sogin ML. Diversity and population structure of sewage-derived microorganisms in wastewater treatment plant influent. Environ Microbiol. 2010;12:378–392. doi: 10.1111/j.1462-2920.2009.02075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanakrishnan J, Gutierrez O, Sharma KR, Guisasola A, Werner U, Meyer RL, et al. Impact of nitrate addition on biofilm properties and activities in rising main sewers. Water Res. 2009;43:4225–4237. doi: 10.1016/j.watres.2009.06.021. [DOI] [PubMed] [Google Scholar]
- Monfort P, Baleux B. Dynamics of Aeromonas hydrophila, Aeromonas sobria, and Aeromonas caviae in a sewage treatment pond. Appl Environ Microbiol. 1990;56:1999–2006. doi: 10.1128/aem.56.7.1999-2006.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore ERB, Tindall BJ, Santos VAPMd, Pieper DH, Ramos JL, Palleroni NJ. Nonmedical Pseudomonas. In: Dworkin M, editor. The Prokaryotes. Singapore: Springer; 2006. pp. 646–703. (ed in chief). [Google Scholar]
- Nadkarni MA, Martin FE, Jacques NA, Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology-Sgm. 2002;148:257–266. doi: 10.1099/00221287-148-1-257. [DOI] [PubMed] [Google Scholar]
- Nason GP, editor. Wavelet Methods in Statistics with R. New York: Springer; 2008. [Google Scholar]
- Norsker NH, Nielsen PH, Hvitvedjacobsen T. Influence of oxygen on biofilm growth and potential sulfate reduction in gravity sewer biofilm. Wat Sci Technol. 1995;31:159–167. [Google Scholar]
- Okabe S, Odagiri M, Ito T, Satoh H. Succession of sulfur-oxidizing bacteria in the microbial community on corroding concrete in sewer systems. Appl and Environ Microbiol. 2007;73:971–980. doi: 10.1128/AEM.02054-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe S, Shimazu Y. Persistence of host-specific Bacteroides-Prevotella 16S rRNA genetic markers in environmental waters: effects of temperature and salinity. Appl Microbiol Biotechnol. 2007;76:935–944. doi: 10.1007/s00253-007-1048-z. [DOI] [PubMed] [Google Scholar]
- Pai TY, Chen CL, Chung H, Ho HH, Shiu TW. Monitoring and assessing variation of sewage quality and microbial functional groups in a trunk sewer line. Environ Monit and Assess. 2010;171:551–560. doi: 10.1007/s10661-009-1299-5. [DOI] [PubMed] [Google Scholar]
- Pauwels B, Wille K, Noppe H, De Brabander H, van de Wiele T, Verstraete W, Boon N. 17 alpha-ethinylestradiol cometabolism by bacteria degrading estrone, 17 beta-estradiol and estriol. Biodegradation. 2008;19:683–693. doi: 10.1007/s10532-007-9173-z. [DOI] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raunkjaer K, Hvitvedjacobsen T, Nielsen PH. Transformation of organic-matter in a gravity sewer. Wat Environ Res. 1995;67:181–188. [Google Scholar]
- Rothenberger MB, Burkholder JM, Brownie C. Long-term effects of changing land use practices on surface water quality in a coastal river and lagoonal estuary. Environ Manage. 2009;44:505–523. doi: 10.1007/s00267-009-9330-8. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell D. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Satoh H, Odagiri M, Ito T, Okabe S. Microbial community structures and in situ sulfate-reducing and sulfur-oxidizing activities in biofilms developed on mortar specimens in a corroded sewer system. Water Research. 2009;43:4729–4739. doi: 10.1016/j.watres.2009.07.035. [DOI] [PubMed] [Google Scholar]
- Silverman BW. Wavelets in statistics: beyond the standard assumptions. Philosophical Transactions of the Royal Society of London Series a-Mathematical Physical and Engineering Sciences. 1999;357:2459–2473. [Google Scholar]
- Snaidr J, Amann R, Huber I, Ludwig W, Schleifer KH. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl Environ Microbiol. 1997;63:2884–2896. doi: 10.1128/aem.63.7.2884-2896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, et al. Microbial diversity in the deep sea and the underexplored "rare biosphere". Proc Natl Acad Sci U S A. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ternes TA, Joss A, Siegrist H. Scrutinizing pharmaceuticals and personal care products in wastewater treatment. Environ Sci Technol. 2004;38:392A–399A. doi: 10.1021/es040639t. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong ST, Chen W. Modeling the relationship between land use and surface water quality. J Environ Manage. 2002;66:377–393. doi: 10.1006/jema.2002.0593. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- United States Environmental Protection Agency (USEPA) Washington, D.C., USA: Office of Water; 2004. Report to Congress: Impacts and Control of CSOs and SSOs. EPA 833-R-04-001. [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl and Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warith MA, Kennedy K, Reitsma R. Use of sanitary sewers as wastewater pre-treatment systems. Waste Manage. 1998;18:235–247. [Google Scholar]
- Warskow AL, Juni E. Nutritional requirements of Acinetobacter strains isolated from soil, water, and sewage. J Bacteriol. 1972;112:1014–1016. doi: 10.1128/jb.112.2.1014-1016.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokomaku D, Yamaguchi N, Nasu M. Improved direct viable count procedure for quantitative estimation of bacterial viability in freshwater environments. Appl Environ Microbiol. 2000;66:5544–5548. doi: 10.1128/aem.66.12.5544-5548.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.