Abstract
Innate immune activation by microbial detection receptors is a complex process involving at least a hundred proteins and multiple signaling pathways. While there continues to be a need to identify additional regulators of host-microbe interactions, a larger conceptual challenge is our lack of understanding of how the known regulators interact in space and time. This review offers a framework to explain the long appreciated (but poorly understood) observation that innate immune signaling pathways are activated from multiple organelles. Using the Toll-like receptors (TLRs) and the RIG-I like receptors (RLRs) as examples, I propose that the receptors do not necessarily define the sites of signaling. Rather, a structurally unrelated class of proteins called “sorting adaptors” functions in this capacity.
Localization of TLRs—ideally positioned for ligand binding, but not signaling
Each TLR family member is a type I transmembrane protein that contains a horseshoe-shaped ectodomain that detects molecules that are common to broad classes of microorganisms1. Classic examples of such molecules include bacterial lipopolysaccharides (LPS), lipoproteins, flagellin and double stranded RNA2. Some TLRs detect microbial ligands directly, whereas others require accessory proteins for high affinity interactions with their ligands1. In their cytosolic tail, TLRs contain a Toll Interleukin-1 receptor homology (TIR) domain3, which functions to recruit downstream TIR domain-containing adaptors to initiate signal transduction4. Despite their common structural features, different TLR family members are transported to different organelles upon translation. The evolutionary pressure to direct a receptor to a given location appears to be linked to the need for rapid responsiveness to a microbial encounter. For this reason, TLRs that detect bacterial cell surface components are often found at the surface of mammalian cells, most notably phagocytes. Examples of such receptors are TLR4 (which detects LPS)5, TLR2 (bacterial lipoproteins)6 and TLR5 (flagellin)7. Likewise, TLRs that detect nucleic acids are not found at the plasma membrane, but are located in late endosomal compartments8–11. In this location, these receptors are poised to detect microbial genomes after they are released by the hydrolytic enzymes present in these organelles. While most cells that express TLRs display them in the aforementioned locations, there are exceptions, as some cells have been reported to display nucleic acid sensing TLRs at their plasma membrane12,13. In recent years, studies of mislocalized TLRs revealed the critical importance of proper subcellular localization for the efficient binding of the microbial ligands they detect14,15. The means by which the biosynthetic trafficking machinery delivers newly synthesized TLRs to their proper subcellular destination has been reviewed elsewhere16–18 and will not be discussed further.
Upon ligand binding, TLRs are thought to self-associate to create a scaffold of TIR domains that recruit soluble cytoplasmic adaptor proteins19. MyD88, TIRAP, TRIF and TRAM are the known adaptors that initiate TLR signal transduction11,20–25. These adaptors are differentially utilized by different TLRs, and serve as the biochemical link between ligand-bound receptors and various serine/threonine kinases that induce inflammatory cytokine expression. MyD88 and TIRAP promote the expression of NF-kB dependent cytokines, whereas TRAM and TRIF lead to the expression of type I interferons (IFNs). In some instances, most notably plasmacytoid dendritic cells (pDCs), MyD88 can induce the expression of both cytokines and IFNs26–28. While these adaptors are usually considered mere intermediates in a signaling pathway, they can also be considered the cytosolic sensors of activated TLRs. In this regard, the view that adaptors are mere intermediates is altered to view these proteins as critical regulators of the earliest cytosolic events that initiate signal transduction.
For many years, it had been assumed that TLRs are immobile in the membranes where they detect their microbial ligand. As such, receptor localization would be expected to determine where within the cell signal transduction can occur. However, recent work has revealed that, in the case of both plasma membrane localized and endosomal TLRs, these receptors must be transported to new regions of the cell to initiate signal transduction29–32.
In this review I focus on where within the cell innate immune signaling first occurs, and how the specificity of signaling locale is achieved. Several examples will be given of proteins that share no primary sequence similarity, but can be grouped based on their ability to define the site where innate immune signaling occurs. I suggest that these proteins be dubbed “sorting adaptors”, the defining features of which are: 1) that they are prepositioned at the location of innate immune signal transduction; and 2) define the type of signaling pathway activated from that location.
Adaptor protein localization determines the specificity of signaling locale
The LPS receptor TLR4 has served as an excellent model to study how TLRs can be transported to new locations upon microbial detection. LPS recognition by TLR4 is facilitated by interactions with the LPS-binding cofactors CD14 and MD-233–35. When CD14 detects LPS, it transfers this microbial product to a plasma membrane-localized heterodimer of MD-2 and TLR4. Upon ligand binding TLR4 is recruited to phosphatidylinositol 4,5-bisphosphate (PIP2) rich regions of the plasma membrane, such as lipid rafts in epithelial cells and sites of phagocytosis in macrophages31,32. Interestingly, TLR2 is also thought to be mobilized into lipid rafts after engaging its various ligands36. It is within these sites that TLR4 (and perhaps TLR2) is thought to oligomerize and engage TIRAP and MyD88 to induce inflammatory cytokine expression37. Subsequent to this event, TLR4 is internalized into endosomes (or phagosomes), where it engages TRAM and TRIF to induce type I IFN expression29,30,38,39.
Analysis of how TLR4 endocytosis is regulated revealed that TLR4 does not induce its own internalization. Rather, TLR4 is cargo for an LPS-inducible endocytosis pathway that is mediated by CD1432. After transferring LPS to the MD-2/TLR4 complex, CD14 engages PLCγ2, which induces the calcium-dependent internalization of TLR432,40. The ITAM-containing transmembrane proteins DAP12 and FcεRγ facilitate this process, probably by activating the tyrosine kinase Syk. While genetic knockouts of Syk have yet to be used to examine its role in TLR4 endocytosis and TRIF signaling, RNAi- and small molecule-based studies suggest that this kinase is important for these events32,41,42.
The discovery that TLR4 must be delivered to PIP2-rich regions of the plasma membrane to activate MyD88 signaling31,37, and then endocytosed to activate TRIF signaling30, suggests that the initial site of receptor-ligand interaction does not determine where signal transduction occurs. Cell biological analysis of TIRAP and TRAM revealed that their localization within cells determines the sites of TLR4 signaling.
TIRAP and TRAM are peripheral membrane proteins whose localization is dictated by interactions with acidic phospholipids30,37,43. In the case of TIRAP, this protein is localized to PIP2-rich regions of the plasma membrane by a process dependent on an N-terminal lipid-binding domain that interacts selectively with this phosphoinositide.
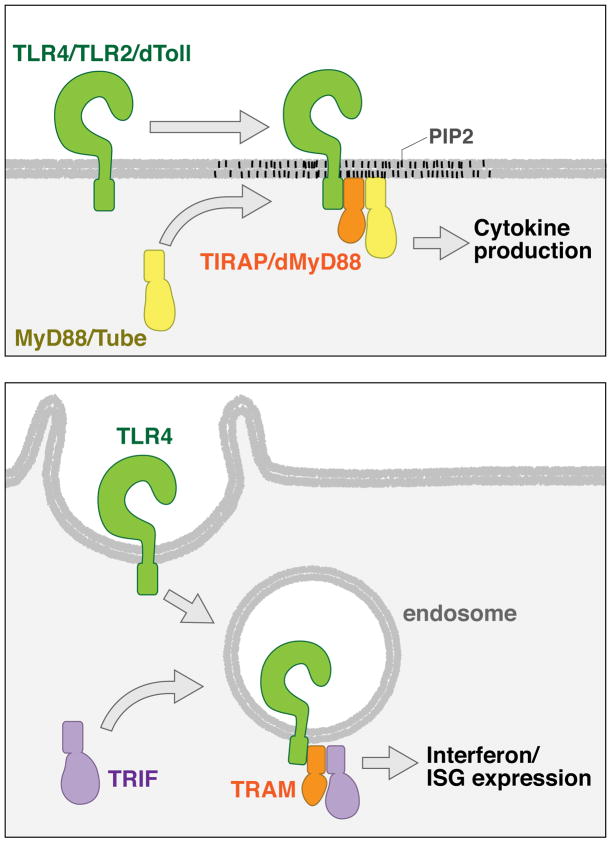
While the upstream sensory protein TLR4 and the downstream signaling adaptor MyD88 must be recruited to PIP2-rich regions of the plasma membrane to signal, TIRAP is present in this location prior to microbial encounters37 (Figure 1 top panel). Altering the localization of TIRAP by ablating the PIP2-binding domain renders this protein cytoplasmic, and MyD88-dependent signal transduction cannot occur37. The localization of TIRAP to PIP2-rich plasma membrane subdomains therefore defines where within the cell TLR4 can activate MyD88-dependent signaling.
Figure 1.
The subcellular site of innate immune signal transduction is defined by the localization of inflexible sorting adaptors. (a) Signaling by plasma membrane-bound Toll family members involves their recruitment to a new subdomain of the cell surface, where they can engage a sorting adaptor. For the Toll family members, the sorting adaptors TIRAP and dMyD88 are located in PIP2-rich regions of the cell surface. (b) TRIF-dependent signaling by TLR4 involves its delivery to endosomes, where the sorting adaptor TRAM resides.
In the case of the TRAM, similar cell biological rules apply. TRAM contains a bipartite localization domain consisting of an N-terminal myristoylation motif and a phosphoinositide binding domain30,43. This bipartite domain directs TRAM to both the plasma membrane and endosomes. The upstream receptor TLR4 and the downstream adaptor TRIF must be recruited to endosomes in order to induce type I IFN expression30,38,39, but it is only TRAM that is resident on endosomes prior to signaling 30,39 (Figure 1 bottom panel). Mislocalizing TRAM to the cytosol43, or forcing this adaptor to be mainly located at the cell surface30, diminishes the ability of TLR4 to induce type I IFN expression. In contrast, forcing TRAM to be located only on early endosomes results in very high type I IFN expression30. These data indicate that like TIRAP, TRAM defines the site in the cell where TLR4 and TRIF converge to induce signal transduction.
Interestingly, the TRAM gene encodes a splice variant called TAG that displays a distinct subcellular distribution from that of TRAM44. Whereas TRAM utilizes the above-described bipartite motif to localize to early endosomes, this domain has been replaced by a Golgi dynamics domain (GOLD) domain in TAG. The GOLD domain directs TAG to late endosomes44, where it functions to interfere with TRAM-mediated signal transduction through interactions with a newly defined protein called TMED745. Thus, the TRAM gene provides an intriguing example of how localizing an adaptor to different endosomes results in a protein with either pro-inflammatory (TRAM) or anti-inflammatory (TAG) functions.
The importance of adaptor protein localization for signal transduction appears to be an evolutionarily conserved aspect innate immunity. For example, the fruitfly Drosophila melanogaster encodes a Toll signaling pathway that has often been used as a reference for the mammalian network46. While Toll and its downstream serine/threonine kinases are similar to their mammalian orthologues in both structure and function46, the cytoplasmic adaptors that link the receptor to these kinases bear little similarity to the mammalian orthologues47,48. Flies do not encode any genes similar to TIRAP, TRIF or TRAM, but they do encode a protein similar to MyD88 called dMyD88 (or dmMyD88)49,50. Despite displaying strong structural similarities, dMyD88 and mammalian MyD88 exhibit distinct subcellular distributions and functional characteristics51. From a cell biological perspective, dMyD88 is more similar to TIRAP and TRAM in that it is prepositioned at the site of signaling via interactions with phosphoinositides, most notably PIP251. dMyD88 functions to recruit the cytosolic adaptor Tube to PIP2-rich regions of the plasma membrane to permit Toll signaling (Figure 1 top panel). In response to bacterial encounters, transgenic flies expressing a cytosolic dMyD88 allele in place of WT dMyD88 display low levels of Toll signaling, and cannot survive the infection51. Thus, like TIRAP and TRAM, dMyD88 probably defines the subcellular site where Toll signaling can occur.
Sorting adaptors—structurally unrelated signaling proteins that define sites of innate immune signal transduction
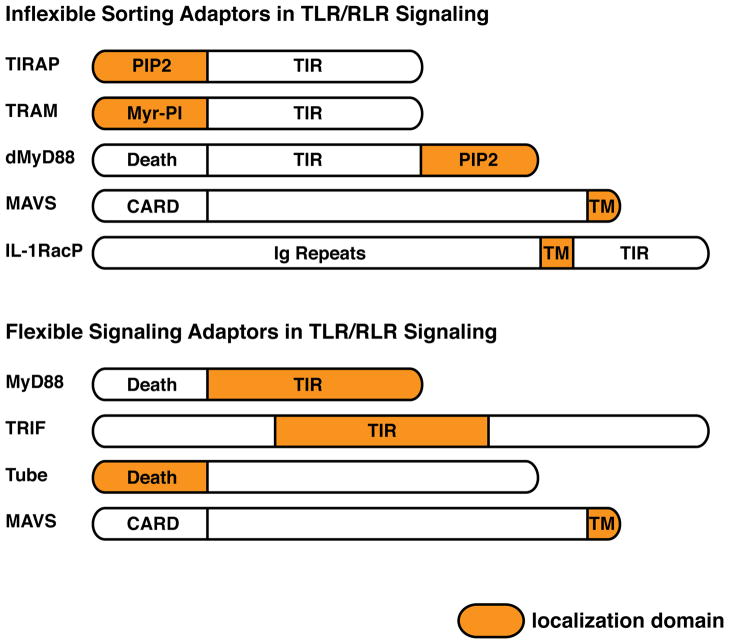
Based on our knowledge of where within the cell various signaling proteins are located, some general principles emerge. It is now clear that there are two classes of innate immune signaling factors. One class is flexible in its signaling locale, in that it can participate in signaling events that occur from multiple compartments of the cell. The second class is inflexible in its signaling locale and can only participate in signaling events that occur on a subset of organelles. The mechanism by which these flexible and inflexible factors are categorized relates directly to their means of membrane localization. The inflexible factors are localized to a given organelle by interactions with general components of the organelle itself, such as phosphoinositides in the case of TIRAP, TRAM and dMyD8830,37,51 (Figure 2). Because their localization results from interactions with a general membrane component, these factors cannot participate in signaling events that occur from a different organelle. In contrast, the flexible factors appear to be cytosolic components, whose recruitment to a given membrane occurs upon microbial detection31,37,39,51 (Figure 2). As such, these flexible factors can be recruited to any organelle harboring an active receptor.
Figure 2.
Sorting and signaling adaptors that function in innate immune pathways. (a) Shown are proteins implicated as sorting adaptors. All sorting adaptors contain a localization motif that differs from their signaling domains. Thus, all sorting adaptors should be placed at the site of signal transduction prior to the cell encountering any microbial or inflammatory stimulus. (b) Shown are the proteins implicated as signaling adaptors. Note that for TLR signaling adaptors, the TIR domains are localization domains, whereas for sorting adaptors, the TIR domains are not localization domains. Also note that while MAVS exhibits many attributes of a sorting adaptor, also functions as a signaling adaptor.
At the receptor proximal level, flexible regulators are bona fide “signaling adaptors” that interact directly with downstream enzymes (Figure 2). Examples of this include MyD88 (which interacts with IRAK family of kinases)52,53, TRIF (which interacts with TRAF family of E3 ligases)54,55 and the Drosophila Tube protein (which interacts with Pelle kinases)48. Inflexible regulators such as TIRAP, TRAM and dMyD88 bind downstream signaling enzymes indirectly, but bind directly to the activated receptors. These inflexible regulators define the site of signal transduction by recruiting signaling adaptors to the active receptor. Classically, inflexible regulators such as TIRAP and TRAM are defined as bridging adaptors. However, the very term adaptor implies a bridging function. I therefore suggest that the term “sorting adaptor” has the spatial connotation that is so important for their function in defining the locale of signal transduction (Figure 2). It is notable that in the case of TLR and Toll signaling pathways, sorting adaptors and signaling adaptors often act together (e.g. TIRAP and MyD88). Why would this be the case?
There are two major benefits of utilizing sorting and signaling adaptor pairs to promote immune signaling. The first benefit is that sorting adaptor localization facilitates the reliable activation of signal transduction, probably because these adaptors are the first cytosolic proteins to detect activated receptors. In mammals, evidence in support of the need for reliable responsiveness comes from studies showing that cytosolic alleles of TIRAP or TRAM cannot detect active receptors efficiently, and therefore TLR4 signaling occurs with diminished efficacy37,43. Studies in Drosophila further underscored the importance of sorting adaptor localization to execute a reliable innate immune response. For example, flies encoding cytosolic alleles of dMyD88 exhibit a highly variable ability to control bacterial replication during infections51. This vastly different ability of flies of the same genotype to control bacterial replication is in contrast to the situation with flies expressing WT dMyD88, that nearly always control bacterial growth49,51. These examples indicate that the reliability of signaling pathway activation is achieved through the proper subcellular positioning of sorting adaptors prior to any microbial encounter.
In principle, one could achieve a rapid and reliable innate immune response by prepositioning the signaling adaptors in sites of signal transduction. For example, MyD88 could have evolved the ability to bind to PIP2 directly. In this case, MyD88 would be in an ideal position for rapid responsiveness to TLR4 activation. However, the benefit of rapid responsiveness to activated TLR4 would come at the cost of restricting its movement to a single subcellular site. Thus, the second major benefit of utilizing sorting and signaling adaptor pairs is to permit signaling adaptors and downstream enzymes to be recruited to multiple organelles. By placing distinct sorting adaptors in distinct locations, the same downstream enzymes can function from many locations in the cell. In the case of Toll signaling, the localization of TIRAP to PIP2-rich regions of the plasma membrane facilitates MyD88 recruitment to this location37, but MyD88 also retains the flexibility to be recruited to other locations in the cell. For example, MyD88 can be recruited to the interleukin 1 receptor (IL-1R)56, which signals from a region of the plasma membrane that contains low levels of PIP237. In this regard, the “sorting adaptor” model would mandate the existence of a sorting adaptor that is prepositioned in PIP2-poor regions of the cell surface to recruit MyD88.
Although never discussed in this context, the IL-1RacP may function as a sorting adaptor for the recruitment of MyD88 to PIP2-poor regions of the plasma membrane. IL-1RacP is a transmembrane protein that is a central component of the signaling complex induced by IL-1β57. IL-1R signaling appears to occur from caveolae, and the receptor is subsequently internalized into caveosomes58,59. Relative to lamellipodia, where TIRAP is located37, caveolae contain low amounts of PIP260. Similar to the function of TIRAP, the primary function of IL-1RacP is to recruit MyD88 to IL-1R, although it modestly contributes to ligand binding as well57,61,62.
I predict that while TIRAP serves as a sorting adaptor to recruit MyD88 to plasma membrane subdomains that are enriched in PIP2, IL-1RacP serves as a sorting adaptor to recruit MyD88 to plasma membrane subdomains that are devoid of PIP2 (Figure 2). This prediction may help explain the reliance of some receptors on TIRAP for inducing MyD88-dependent responses (e.g. TLR4, TLR2/1 and TLR2/6)63,64 and other receptors on IL-1RacP for inducing MyD88-dependent responses (e.g. IL-1R, IL-33R)61,65. It is important to note however, that the ability to implicate IL-1RacP as a bona fide sorting adaptor awaits detailed cell biological analysis of the subcellular localization of this protein and its signaling receptor.
Expanding the use of sorting adaptors to intracellular organelles
The aforementioned properties of sorting-signaling adaptor pairs would not only benefit an organelle containing diverse subdomains such as the cell surface, but should also benefit the diverse organelles present in the cytosol of mammalian cells. In this regard, I speculate that sorting adaptors exist in non-plasma membrane compartments. Some obvious organelles to consider in this discussion are endosomal vesicles, peroxisomes and mitochondria. The ability of TRAM to function as an endosomal sorting adaptor for TLR4 suggests that signaling pathways operating from endosomes can indeed benefit from the use of sorting-signaling adaptor pairs. But are there others?
Nucleic acid sensing TLRs such as TLRs 3, 7, 8 and 9 induce innate immune signaling from endosomes16, and in the case of the latter three receptors, they signal through MyD88. I speculate that a dedicated set of sorting adaptors exists that are uniquely found on endosomes. The localization of these endosomal sorting adaptors would permit the recruitment of MyD88 to a subset of endosomes and endow these organelles with unique signaling capabilities. It is possible that in the case of TLR9, the ability of MyD88 to be recruited to distinct populations of endosomes (e.g. PI(3,5)P2-rich endosomes and Lysosome-related organelles) results from the actions of differentially localized sorting adaptors. The diversification of sorting adaptors on different subsets of endosomes also has the advantage of diversifying the downstream effectors that can be recruited to such compartments, thus explaining the specificity of the signaling response induced by TLR9 in these organelles66.
Another biological process where these principles may apply is in the antiviral immune responses induced by RIG-I like receptors (RLRs)67. Despite their distinct structural characteristics, RLRs and TLRs have much in common. Like the TLRs, RLRs recognize microbial infections, utilize downstream adaptors to activate cytokine and IFN expression, and can induce signal transduction from multiple organelles67.
There are two RLRs that have well-established proinflammatory functions—RIG-I and MDA526,68,69. Both RLRs are RNA helicases that survey the cytosol for the presence of viral RNA. The distinction between self and viral RNA is thought to occur through the recognition of specific molecular features that are found within viral nucleic acids. For example, RIG-I signaling occurs when it encounters RNA containing 5′ triphosphate groups, short double stranded regions and/or uridine-rich 3′ regions70–72. MDA5 signaling, in contrast, occurs when it encounters long stretches of double stranded RNA73,74. The RNA detected by RLRs can be of microbial origin, such as viral genomes, mRNAs or replication intermediates, but can also be produced by the host. For example, the host-encoded RNA polymerase III can transcribe the DNA of Epstein-Barr Virus to produce RNA that activates RIG-I signaling75,76. Thus, like the TLRs, RLRs have a widespread role in detecting multiple types of viral infections.
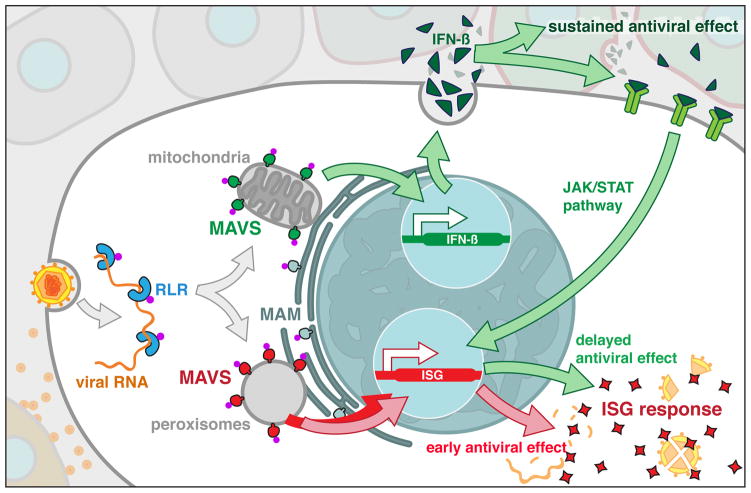
Upon detecting viral RNA, RLRs engage the adaptor protein MAVS (also known as IPS-1, Cardif or VISA) to induce the expression of IFNs and other inflammatory mediators77–80. MAVS is located on the limiting membranes of peroxisomes, mitochondria and mitochondria-associated membrane (MAM)81,82 (Figure 3). These organelles share several homeostatic functions in lipid synthesis and metabolism83, but have only recently been implicated the control of antiviral immunity. MAVS signaling from mitochondria induces the expression of Type I IFNs and antiviral factors called interferon stimulated genes (ISGs)80. In contrast, MAVS signaling from peroxisomes induces ISG expression without inducing the expression of Type I IFNs81. Recently, it was shown that mitochondria and peroxisomes interact at the MAM during viral infection82. This coalescence of organelles on the MAM has been deemed the “intracellular immune synapse”, where the actions of peroxisomes and mitochondrial MAVS are coordinated to ensure effective antiviral immunity (Figure 3).
Figure 3.
RLR-mediated detection of viral RNAs leads to receptor transport to the sorting-signaling adaptor hybrid MAVS. MAVS is located on mitochondria, peroxisomes and the MAM. The docking of these organelles at the MAM creates an innate immune synapse that maximizes antiviral innate immunity.
Once detecting a viral infection, RLRs are recruited to an inflexible regulator (MAVS) at sites of signaling (Figure 3). Altering MAVS localization to the cytosol renders this protein unable to induce antiviral signaling80,81, despite the fact that the signaling domains of this protein remain intact. Thus, similar to the TLR system, the subcellular localization of a downstream adaptor protein, not the site of RLR-virus interaction, determines the initial site of signal transduction.
MAVS therefore has properties that are common to other sorting adaptors, such as defining the site of RLR signaling, and recruiting downstream signaling enzymes to these sites (see examples below). However, this protein also has properties of a signaling adaptor. For example MAVS can interact with downstream enzymes that facilitate RLR signaling, such as TBK1, TRAF3, TRAF2 and TRAF684. Thus, MAVS appears to be a hybrid sorting-signaling adaptor (Figure 2 top and bottom panels), which perhaps explains why RLR signaling only requires this single adaptor to engage downstream enzymes.
It remains to be determined how RLRs are delivered to MAVS to promote antiviral signal transduction. It is possible that the interactions between MAVS and RLRs are facilitated by an intermediate protein, whose function would be to recruit RLRs to the site of MAVS residence. In this regard, this intermediate protein would serve a function analogous to CD14, which delivers TLR4 to endosomes to activate TRAM-TRIF dependent signaling32.
Concluding remarks
In this review, I highlighted how cell biological and biochemical analysis of proteins can reveal common biological functions that could not have been predicted by structural and genetic analysis alone. This appears to be the case for TIRAP, TRAM, dMyD88, MAVS and perhaps IL-1RacP (Figure 2). These proteins share little structural similarity, but all share common biological features of being “hard-wired” to the site in the cell where innate immune signaling occurs. The unifying features of these proteins that define them as sorting adaptors are: 1) they are localized to specific organelles at steady state; 2) they recruit signaling proteins to their site of residence to initiate signal transduction; and 3) their mislocalization to the cytosol results in a deficient signaling response. Since these proteins share no specific structural features, it is difficult to predict additional sorting adaptors bioinformatically. Rather, detailed biochemical and cell biological analysis is necessary to expand the list of sorting adaptors to include regulators of additional signaling pathways.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kang JY, Lee JO. Structural biology of the Toll-like receptor family. Annu Rev Biochem. 2011;80:917–941. doi: 10.1146/annurev-biochem-052909-141507. [DOI] [PubMed] [Google Scholar]
- 2.Barton GM, Medzhitov R. Toll-like receptors and their ligands. Curr Top Microbiol Immunol. 2002;270:81–92. doi: 10.1007/978-3-642-59430-4_5. [DOI] [PubMed] [Google Scholar]
- 3.Gay NJ, Keith FJ. Drosophila Toll and IL-1 receptor. Nature. 1991;351 (6325):355–356. doi: 10.1038/351355b0. [DOI] [PubMed] [Google Scholar]
- 4.O’Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. 2007;7 (5):353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 5.Akashi S, et al. Cutting edge: cell surface expression and lipopolysaccharide signaling via the toll-like receptor 4-MD-2 complex on mouse peritoneal macrophages. J Immunol. 2000;164 (7):3471–3475. doi: 10.4049/jimmunol.164.7.3471. [DOI] [PubMed] [Google Scholar]
- 6.Underhill DM, et al. The Toll-like receptor 2 is recruited to macrophage phagosomes and discriminates between pathogens. Nature. 1999;401 (6755):811–815. doi: 10.1038/44605. [DOI] [PubMed] [Google Scholar]
- 7.Gewirtz AT, et al. Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J Immunol. 2001;167 (4):1882–1885. doi: 10.4049/jimmunol.167.4.1882. [DOI] [PubMed] [Google Scholar]
- 8.Latz E, et al. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat Immunol. 2004;5 (2):190–198. doi: 10.1038/ni1028. [DOI] [PubMed] [Google Scholar]
- 9.Ewald SE, et al. The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature. 2008;456 (7222):658–662. doi: 10.1038/nature07405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barton GM, et al. Intracellular localization of Toll-like receptor 9 prevents recognition of self DNA but facilitates access to viral DNA. Nat Immunol. 2006;7 (1):49–56. doi: 10.1038/ni1280. [DOI] [PubMed] [Google Scholar]
- 11.Oshiumi H, et al. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat Immunol. 2003;4 (2):161–167. doi: 10.1038/ni886. [DOI] [PubMed] [Google Scholar]
- 12.Lee J, et al. Maintenance of colonic homeostasis by distinctive apical TLR9 signalling in intestinal epithelial cells. Nat Cell Biol. 2006;8 (12):1327–1336. doi: 10.1038/ncb1500. [DOI] [PubMed] [Google Scholar]
- 13.Matsumoto M, et al. Establishment of a monoclonal antibody against human Toll-like receptor 3 that blocks double-stranded RNA-mediated signaling. Biochem Biophys Res Commun. 2002;293 (5):1364–1369. doi: 10.1016/S0006-291X(02)00380-7. [DOI] [PubMed] [Google Scholar]
- 14.Mouchess ML, et al. Transmembrane mutations in Toll-like receptor 9 bypass the requirement for ectodomain proteolysis and induce fatal inflammation. Immunity. 2011;35 (5):721–732. doi: 10.1016/j.immuni.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukui R, et al. Unc93B1 restricts systemic lethal inflammation by orchestrating Toll-like receptor 7 and 9 trafficking. Immunity. 2011;35 (1):69–81. doi: 10.1016/j.immuni.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 16.Marek LR, Kagan JC. Deciphering the function of nucleic acid sensing TLRs one regulatory step at a time. Front Biosci. 2012;17:2060–2068. doi: 10.2741/3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ewald SE, Barton GM. Nucleic acid sensing Toll-like receptors in autoimmunity. Curr Opin Immunol. 2011;23 (1):3–9. doi: 10.1016/j.coi.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barton GM, Kagan JC. A cell biological view of Toll-like receptor function: regulation through compartmentalization. Nat Rev Immunol. 2009;9 (8):535–542. doi: 10.1038/nri2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Latz E, et al. Ligand-induced conformational changes allosterically activate Toll-like receptor 9. Nat Immunol. 2007;8 (7):772–779. doi: 10.1038/ni1479. [DOI] [PubMed] [Google Scholar]
- 20.Yamamoto M, et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301 (5633):640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto M, et al. TRAM is specifically involved in the Toll-like receptor 4-mediated MyD88-independent signaling pathway. Nat Immunol. 2003;4 (11):1144–1150. doi: 10.1038/ni986. [DOI] [PubMed] [Google Scholar]
- 22.Oshiumi H, et al. TIR-containing adapter molecule (TICAM)-2, a bridging adapter recruiting to toll-like receptor 4 TICAM-1 that induces interferon-beta. J Biol Chem. 2003;278 (50):49751–49762. doi: 10.1074/jbc.M305820200. [DOI] [PubMed] [Google Scholar]
- 23.Medzhitov R, et al. MyD88 is an adaptor protein in the hToll/IL-1 receptor family signaling pathways. Mol Cell. 1998;2 (2):253–258. doi: 10.1016/s1097-2765(00)80136-7. [DOI] [PubMed] [Google Scholar]
- 24.Horng T, et al. TIRAP: an adapter molecule in the Toll signaling pathway. Nat Immunol. 2001;2 (9):835–841. doi: 10.1038/ni0901-835. [DOI] [PubMed] [Google Scholar]
- 25.Fitzgerald KA, et al. Mal (MyD88-adapter-like) is required for Toll-like receptor-4 signal transduction. Nature. 2001;413 (6851):78–83. doi: 10.1038/35092578. [DOI] [PubMed] [Google Scholar]
- 26.Kato H, et al. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23 (1):19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 27.Lund JM, et al. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc Natl Acad Sci U S A. 2004;101 (15):5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diebold SS, et al. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303 (5663):1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 29.Husebye H, et al. The Rab11a GTPase controls Toll-like receptor 4-induced activation of interferon regulatory factor-3 on phagosomes. Immunity. 2010;33 (4):583–596. doi: 10.1016/j.immuni.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kagan JC, et al. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nat Immunol. 2008;9 (4):361–368. doi: 10.1038/ni1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Triantafilou M, et al. Mediators of innate immune recognition of bacteria concentrate in lipid rafts and facilitate lipopolysaccharide-induced cell activation. J Cell Sci. 2002;115 (Pt 12):2603–2611. doi: 10.1242/jcs.115.12.2603. [DOI] [PubMed] [Google Scholar]
- 32.Zanoni I, et al. CD14 controls the LPS-induced endocytosis of Toll-like receptor 4. Cell. 2011;147 (4):868–880. doi: 10.1016/j.cell.2011.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore KJ, et al. Divergent response to LPS and bacteria in CD14-deficient murine macrophages. J Immunol. 2000;165 (8):4272–4280. doi: 10.4049/jimmunol.165.8.4272. [DOI] [PubMed] [Google Scholar]
- 34.Gioannini TL, et al. Isolation of an endotoxin-MD-2 complex that produces Toll-like receptor 4-dependent cell activation at picomolar concentrations. Proc Natl Acad Sci U S A. 2004;101 (12):4186–4191. doi: 10.1073/pnas.0306906101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.da Silva Correia J, et al. Lipopolysaccharide is in close proximity to each of the proteins in its membrane receptor complex. transfer from CD14 to TLR4 and MD-2. J Biol Chem. 2001;276 (24):21129–21135. doi: 10.1074/jbc.M009164200. [DOI] [PubMed] [Google Scholar]
- 36.Triantafilou M, et al. Membrane sorting of toll-like receptor (TLR)-2/6 and TLR2/1 heterodimers at the cell surface determines heterotypic associations with CD36 and intracellular targeting. J Biol Chem. 2006;281 (41):31002–31011. doi: 10.1074/jbc.M602794200. [DOI] [PubMed] [Google Scholar]
- 37.Kagan JC, Medzhitov R. Phosphoinositide-mediated adaptor recruitment controls Toll-like receptor signaling. Cell. 2006;125 (5):943–955. doi: 10.1016/j.cell.2006.03.047. [DOI] [PubMed] [Google Scholar]
- 38.Tseng PH, et al. Different modes of ubiquitination of the adaptor TRAF3 selectively activate the expression of type I interferons and proinflammatory cytokines. Nat Immunol. 2010;11 (1):70–75. doi: 10.1038/ni.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanimura N, et al. Roles for LPS-dependent interaction and relocation of TLR4 and TRAM in TRIF-signaling. Biochem Biophys Res Commun. 2008;368 (1):94–99. doi: 10.1016/j.bbrc.2008.01.061. [DOI] [PubMed] [Google Scholar]
- 40.Chiang CY, et al. Phospholipase Cgamma-2 and intracellular calcium are required for lipopolysaccharide-induced Toll-like receptor 4 (TLR4) endocytosis and interferon regulatory factor 3 (IRF3) activation. J Biol Chem. 2012;287 (6):3704–3709. doi: 10.1074/jbc.C111.328559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chevrier N, et al. Systematic discovery of TLR signaling components delineates viral-sensing circuits. Cell. 2011;147 (4):853–867. doi: 10.1016/j.cell.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dang O, et al. Inhibition of lipopolysaccharide-induced interferon regulatory factor 3 activation and protection from septic shock by hydroxystilbenes. Shock. 2004;21 (5):470–475. doi: 10.1097/01.shk.0000123513.13212.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rowe DC, et al. The myristoylation of TRIF-related adaptor molecule is essential for Toll-like receptor 4 signal transduction. Proc Natl Acad Sci U S A. 2006;103 (16):6299–6304. doi: 10.1073/pnas.0510041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palsson-McDermott EM, et al. TAG, a splice variant of the adaptor TRAM, negatively regulates the adaptor MyD88-independent TLR4 pathway. Nat Immunol. 2009;10 (6):579–586. doi: 10.1038/ni.1727. [DOI] [PubMed] [Google Scholar]
- 45.Doyle SL, et al. The GOLD domain-containing protein TMED7 inhibits TLR4 signalling from the endosome upon LPS stimulation. Nat Commun. 2012;3:707. doi: 10.1038/ncomms1706. [DOI] [PubMed] [Google Scholar]
- 46.Hoffmann JA. The immune response of Drosophila. Nature. 2003;426 (6962):33–38. doi: 10.1038/nature02021. [DOI] [PubMed] [Google Scholar]
- 47.Dunne A, et al. Structural complementarity of Toll/interleukin-1 receptor domains in Toll-like receptors and the adaptors Mal and MyD88. J Biol Chem. 2003;278 (42):41443–41451. doi: 10.1074/jbc.M301742200. [DOI] [PubMed] [Google Scholar]
- 48.Sun H, et al. A heterotrimeric death domain complex in Toll signaling. Proc Natl Acad Sci U S A. 2002;99 (20):12871–12876. doi: 10.1073/pnas.202396399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tauszig-Delamasure S, et al. Drosophila MyD88 is required for the response to fungal and Gram-positive bacterial infections. Nat Immunol. 2002;3 (1):91–97. doi: 10.1038/ni747. [DOI] [PubMed] [Google Scholar]
- 50.Horng T, Medzhitov R. Drosophila MyD88 is an adapter in the Toll signaling pathway. Proc Natl Acad Sci U S A. 2001;98 (22):12654–12658. doi: 10.1073/pnas.231471798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marek LR, Kagan JC. Phosphoinositide binding by the Toll adaptor dMyD88 controls antibacterial responses in Drosophila. Immunity. 2012;36 (4):612–622. doi: 10.1016/j.immuni.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Motshwene PG, et al. An oligomeric signaling platform formed by the Toll-like receptor signal transducers MyD88 and IRAK-4. J Biol Chem. 2009;284 (37):25404–25411. doi: 10.1074/jbc.M109.022392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin SC, et al. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465 (7300):885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hacker H, et al. Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature. 2006;439 (7073):204–207. doi: 10.1038/nature04369. [DOI] [PubMed] [Google Scholar]
- 55.Sato S, et al. Toll/IL-1 receptor domain-containing adaptor inducing IFN-beta (TRIF) associates with TNF receptor-associated factor 6 and TANK-binding kinase 1, and activates two distinct transcription factors, NF-kappa B and IFN-regulatory factor-3, in the Toll-like receptor signaling. J Immunol. 2003;171 (8):4304–4310. doi: 10.4049/jimmunol.171.8.4304. [DOI] [PubMed] [Google Scholar]
- 56.Muzio M, et al. IRAK (Pelle) family member IRAK-2 and MyD88 as proximal mediators of IL-1 signaling. Science. 1997;278 (5343):1612–1615. doi: 10.1126/science.278.5343.1612. [DOI] [PubMed] [Google Scholar]
- 57.Brikos C, et al. Mass spectrometric analysis of the endogenous type I interleukin-1 (IL-1) receptor signaling complex formed after IL-1 binding identifies IL-1RAcP, MyD88, and IRAK-4 as the stable components. Mol Cell Proteomics. 2007;6 (9):1551–1559. doi: 10.1074/mcp.M600455-MCP200. [DOI] [PubMed] [Google Scholar]
- 58.Blanco AM, et al. Ethanol mimics ligand-mediated activation and endocytosis of IL-1RI/TLR4 receptors via lipid rafts caveolae in astroglial cells. J Neurochem. 2008;106 (2):625–639. doi: 10.1111/j.1471-4159.2008.05425.x. [DOI] [PubMed] [Google Scholar]
- 59.Liu P, Anderson RG. Compartmentalized production of ceramide at the cell surface. J Biol Chem. 1995;270 (45):27179–27185. doi: 10.1074/jbc.270.45.27179. [DOI] [PubMed] [Google Scholar]
- 60.Watt SA, et al. Subcellular localization of phosphatidylinositol 4,5-bisphosphate using the pleckstrin homology domain of phospholipase C delta1. Biochem J. 2002;363 (Pt 3):657–666. doi: 10.1042/0264-6021:3630657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Korherr C, et al. A critical role for interleukin-1 receptor accessory protein in interleukin-1 signaling. Eur J Immunol. 1997;27 (1):262–267. doi: 10.1002/eji.1830270139. [DOI] [PubMed] [Google Scholar]
- 62.Radons J, et al. Identification of essential regions in the cytoplasmic tail of interleukin-1 receptor accessory protein critical for interleukin-1 signaling. J Biol Chem. 2002;277 (19):16456–16463. doi: 10.1074/jbc.M201000200. [DOI] [PubMed] [Google Scholar]
- 63.Horng T, et al. The adaptor molecule TIRAP provides signalling specificity for Toll-like receptors. Nature. 2002;420 (6913):329–333. doi: 10.1038/nature01180. [DOI] [PubMed] [Google Scholar]
- 64.Yamamoto M, et al. Essential role for TIRAP in activation of the signalling cascade shared by TLR2 and TLR4. Nature. 2002;420 (6913):324–329. doi: 10.1038/nature01182. [DOI] [PubMed] [Google Scholar]
- 65.Chackerian AA, et al. IL-1 receptor accessory protein and ST2 comprise the IL-33 receptor complex. J Immunol. 2007;179 (4):2551–2555. doi: 10.4049/jimmunol.179.4.2551. [DOI] [PubMed] [Google Scholar]
- 66.Sasai M, et al. Bifurcation of Toll-like receptor 9 signaling by adaptor protein 3. Science. 2010;329 (5998):1530–1534. doi: 10.1126/science.1187029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nakhaei P, et al. RIG-I-like receptors: sensing and responding to RNA virus infection. Semin Immunol. 2009;21 (4):215–222. doi: 10.1016/j.smim.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 68.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441 (7089):101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 69.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5 (7):730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 70.Pichlmair A, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314 (5801):997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 71.Saito T, et al. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454 (7203):523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Uzri D, Gehrke L. Nucleotide sequences and modifications that determine RIG-I/RNA binding and signaling activities. J Virol. 2009;83 (9):4174–4184. doi: 10.1128/JVI.02449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kato H, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205 (7):1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pichlmair A, et al. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J Virol. 2009;83 (20):10761–10769. doi: 10.1128/JVI.00770-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ablasser A, et al. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10 (10):1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chiu YH, et al. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138 (3):576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Meylan E, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437 (7062):1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 78.Xu LG, et al. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19 (6):727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 79.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6 (10):981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 80.Seth RB, et al. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122 (5):669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 81.Dixit E, et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141 (4):668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Horner SM, et al. Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc Natl Acad Sci U S A. 2011;108 (35):14590–14595. doi: 10.1073/pnas.1110133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hettema EH, Motley AM. How peroxisomes multiply. J Cell Sci. 2009;122 (Pt 14):2331–2336. doi: 10.1242/jcs.034363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Belgnaoui SM, et al. Orchestrating the interferon antiviral response through the mitochondrial antiviral signaling (MAVS) adapter. Curr Opin Immunol. 2011;23 (5):564–572. doi: 10.1016/j.coi.2011.08.001. [DOI] [PubMed] [Google Scholar]