Summary
The lamprey (Petromyzon marinus) undergoes developmentally programmed genome rearrangements (PGRs) that mediate deletion of ~20% of germline DNA from somatic cells during early embryogenesis. This genomic differentiation of germline and soma is intriguing, because the germline plays a unique biological role wherein it must possess the ability to undergo meiotic recombination and the capacity to differentiate into every cell type. These evolutionarily indispensible functions set the germline at odds with somatic tissues, as factors that promote recombination and pluripotency can potentially disrupt genome integrity or specification of cell fate when misexpressed in somatic cell lineages (e.g. in oncogenesis). Here, we describe the development of new genomic and transcriptomic resources for lamprey and use these to identify hundreds of genes that are targeted for programmed deletion from somatic cell lineages. Transcriptome sequencing and targeted validation studies further confirm that somatically deleted genes function both in adult (meiotic) germline and in the development of primordial germ cells during embryogenesis. Inferred functional information from deleted regions indicates that developmentally programmed rearrangement serves as a (perhaps ancient) biological strategy to ensure segregation of pluripotency functions to the germline, effectively eliminating the potential for somatic misexpression.
Results and Discussion
A Survey of Known Sequences
In lamprey, PGR events are known to occur during early stages of embryonic development (starting at approximately the midblastula transition: between day 2 and 3 of development), are inherited uniformly across all somatic tissues, and result in deletions that may individually encompass hundreds of kilobases of DNA (both single copy and repetitive) [1, 2]. To further resolve the nature of PGR, we surveyed all available lamprey germline sequence for evidence of somatic deletion using array comparative genomic hybridization (arrayCGH).
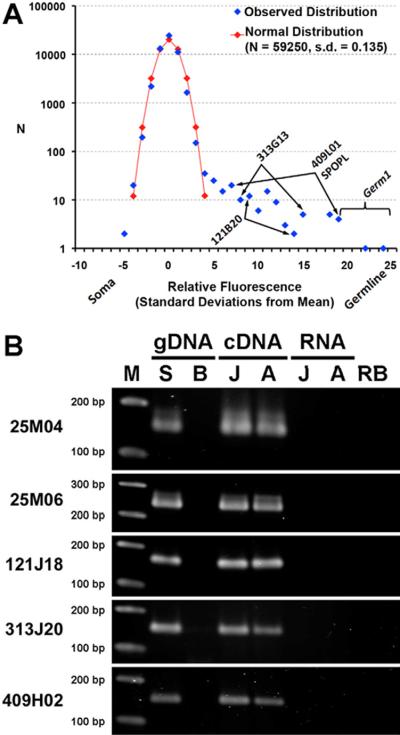
A signature of somatic rearrangement (programmed or otherwise) can be observed when a derived tissue lacks specific nucleotide sequences that were present in its progenitor cell population. To further survey for the changes that characterize lamprey PGR, we designed a customized oligonucleotide microarray to target all available germline sequence (BAC-end sequences) [2] and ~1% of the known somatic genome by arrayCGH. This microarray was used to measure the relative abundance of target sequences within an individual's germline (sperm) versus somatic (blood) DNA, using replicated, dye-swapped experiments. Analysis of relative hybridization intensities revealed that most target regions fell within the expected distribution for normalized data but also identified a substantial tail of the distribution, suggesting enrichment of several sequences within germline DNA (Figure 1A). Notably, the few sequences that had been previously classified as germline-specific [1] and that contained sufficient non-repetitive sequence to be targeted, all fell within this tail of the distribution (n = 4). No significant differences were observed in arrayCGH comparisons of DNA from somatic tissues that were derived from these same animals (Supplemental Experimental Procedures).
Figure 1. Summary of germline-specific sequence and gene discovery using arrayCGH.
(A) Germline-enriched sequences were identified by comparing observed relative hybridization intensities to a normal distribution with the same number of sampled regions (N) and standard deviation (s.d.). The y-axis is plotted on log10 scale in order to magnify differences at the tails of the distribution. All previously discovered germline-specific sequences [1] (marked by arrows and brackets) and several additional germline-specific sequences were identified in this assay. (B) Examples of PCR validation of single-copy sequences eliminated from soma and their expression in the germline. Sequences are present in testes gDNA (genomic DNA) but absent from blood gDNA. These same fragments can be amplified from testes cDNA, but not from the source RNA (a control for gDNA contamination) or reagent blank. S = Sperm, B = Blood, A = Adult testes, J = Juvenile testes, RB = Reagent Blank, M = 100 bp DNA Ladder.
In addition to these previously identified sequences, our analysis identified several other germline-enriched sequences. In total, ~13% of the surveyed germline sequence (259 of 2100 fragments or 150 kb/1.08 megabases, including deleted repeats) showed evidence of somatic deletion. This percentage is consistent with previous flow cytometric estimates that compared nuclear DNA content in germline versus somatic tissues [1], confirming that programmed deletions result in extensive differentiation of germline versus somatic genomes. Candidate germline-enriched regions that were identified for the first time by arrayCGH included eight single/low-copy sequences and several tandemly-repeated sequences that appeared to be uniquely enriched in the germline. Six of eight candidate sequences were clearly validated as germline-specific, five of which were observed to be expressed in adult and juvenile testes (Figures 1, S3; Supplemental Experimental Procedures) and one of which was expressed in cells that are identical to classical anatomical descriptions of migrating primordial germ cells in lamprey embryos [3–5] (Figures 2, S2; Supplemental Experimental Procedures).
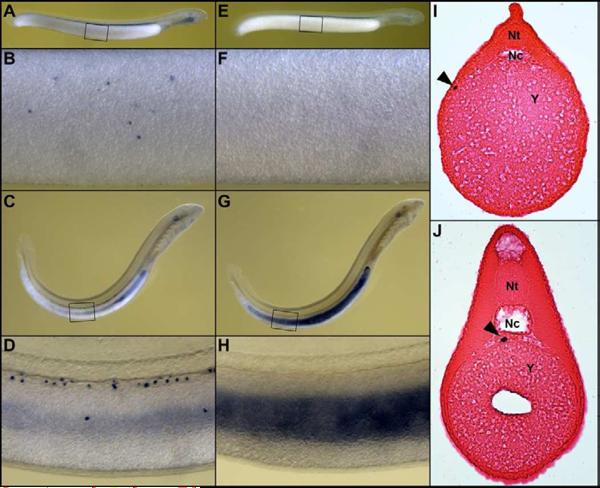
Figure 2. Expression of a germline-specific marker in embryonic germline.
In situ hybridization of an antisense probe of the germline-specific gene 25M04 (putative KRAB domain zinc finger protein) reveals expression in the developing germline cells at day 14 (A, B, I) and day 20 (C, D, J) post-fertilization. Punctate staining reveals specific expression in the presumptive primordial germ cells (PGCs). Staining of PGCs is not observed in embryos that were hybridized with the sense strand probe (E–F), but some background staining is observed due to the presence of non-cellular endogenous alkaline phosphatase activity in the developing gut, pharynx, notochord and otic capsule. Panels B, D, F, and H correspond to the circumscribed regions in A, C, E, G, respectively. Panels I and J are transverse sections of the embryos shown in panels A and C. Sections have been counterstained with eosin in order to enhance contrast; arrows mark the location of PGCs positive for the 25M04 marker. This expression pattern suggests that 25M04 is involved in some aspect of PGC differentiation and/or migration. Nc = notochord, Nt = neural tube, Y = yolk.
Sequencing and Analysis of Lamprey Germline DNA
Our hybridization-based assays and earlier computational studies [1] hold the capacity to identify candidate deletion regions, yet both methods carry the same limitation in that they can only identify differences when sequences are known a priori. To address this limitation, a single 454 Titanium shotgun sequencing run was performed on lamprey germline (sperm) DNA. This sequence set consisted of 554,979 sequence reads with a minimum quality score of Q20 and a minimum trimmed length of 300 bp (median length: 484 bp, mean length: 428 bp, total length: 230 megabases), representing ~10% of the germline genome. The availability of a large whole-genome shotgun (WGS) dataset from the lamprey genome project (liver DNA) [6] enabled us to develop a relatively simple analytical pipeline in order to discover novel sequences and architectures present in the germline and absent from soma. This pipeline involved aligning all germline reads to all somatic (liver) reads then computationally examining alignments to search for signatures of rearrangement (Figure 3). Different alignment patterns were considered indicative of: 1) “normal” single-copy or repetitive DNA, 2) WGS coverage gaps, 3) candidate deletion regions, and 4) candidate recombination sites. Numerous putative deletion and recombination regions were identified (41,996 “deletion” and 18,842 “recombination” reads: Table S1). Importantly, alignment of “deletion” reads to the human RefSeq dataset identified 246 non-redundant gene hits for the deleted fraction (E <1e−20 with a total of 2265 homology-informative reads, including several redundant alignments to zinc finger genes, which may resent multiple independent loci). This suggests that a substantial fraction of the somatically deleted DNA corresponds to single-copy and protein-coding DNA.
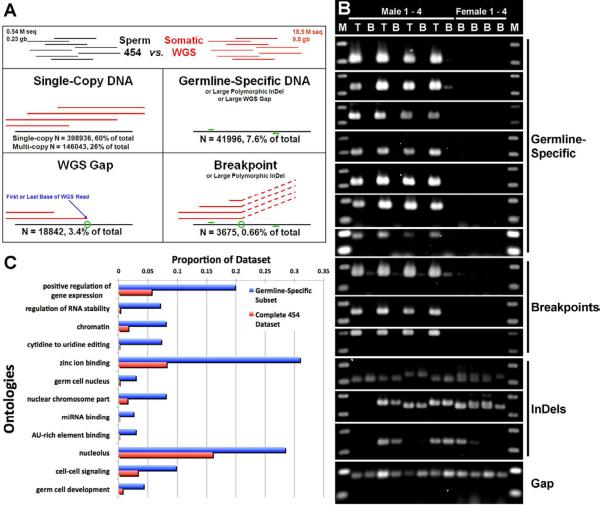
Figure 3. Analysis of pilot 454 sequencing data.
(A) All 454 reads were categorized on the basis of alignment patterns with the complete lamprey WGS dataset (liver DNA). A majority (82%) of reads appeared as “normal” DNA (multicopy or single-copy). Other alignment patterns were consistent with coverage gaps in the WGS dataset (3.4%), germline-specific DNA (7.6%) or recombination breakpoints (0.66%). Green circles depict the positions of alignment breaks and green arrows depict the generic locations of primer binding sites for validation PCRs. (B) Results of PCR validations of germline-specific/gene-containing (BLAST hit) reads and breakpoint-flanking reads provided positive validation of members of both rearrangement classes and identified segregating (in the population) insertion/deletion (InDel) polymorphisms and WGS coverage gaps, which mimic programmed rearrangement outcomes. Note, the “Germline-Specific” and “Breakpoint” classes result in similar PCR validation patterns because one primer (breakpoint) or both primers (germline-specific) are designed to germline-specific regions. T – template is testes DNA, B – template is blood DNA, M = 100 bp DNA ladder. (C) Overrepresented gene ontologies from 234 predicted germline-specific genes, relative to the entire 454 dataset (p>1e-8, corrected using false discovery rate control, as implemented by Blast2Go [9]).
It should be noted that different individuals were used for the somatic WGS and germline 454 projects. This is because a female was selected for the lamprey WGS project whilst pure germline DNA is much more readily accessible from sperm. Therefore, apparent deletion and recombination signatures could also reflect polymorphic insertion/deletion events that segregate in the lamprey population and were differentially inherited by the sequenced individuals. In order to address this potential issue, we performed further analyses on a subset of predicted gene deletions (n = 20) and recombination events (n = 28). We used PCR to test several candidate regions, focusing on predicted genes and recombination sites (Figure 3). These validation experiments revealed seven sites of programmed deletion, three recombination breakpoints, three segregating insertion/deletion polymorphisms and five WGS sequence coverage gaps. Comparison of PCR validated breakpoint sequences reveals the presence of short 5′/3′ palindromes near the predicted breakpoint position, but no defined consensus sequence (Figure 4). The potential functionality of these is as-yet unclear, although the presence of such sequences is considered strong evidence that site-specific recombination events facilitate the elimination of DNA from the lamprey genome (though chromosome loss cannot be ruled out as a contributing factor). Genes present within validated deletions included: APOBEC-1 Complementation Factor, RNA Binding Motif 46 (cancer/testis antigen 68) and 47, KRAB Zinc Fingers 79 and 180, Lysophosphatidic Acid Receptor 1, and WNT7A/B. Summaries of functional information for homologs of these genes (NCBI gene: http://www.ncbi.nlm.nih.gov/gene/) indicate functional roles in maintenance of cell fate, cell proliferation and oncogenesis/tumorigenesis.
Figure 4. Sequence of PCR validated breakpoint regions.
Breakpoints contain short 5′/3′ palindromes (green) at the junction between somatically retained (blue) and germline-specific (red) sequence. The breakpoint of junction 2 contains an imperfect 5′/3′ palindrome. It is as-yet unclear if these are functionally related to programmed genome rearrangement.
To gain a better perspective on gene functions within the larger predicted deletion dataset, we compared homology-derived ontology information [7] for all candidate deletions to ontology information for the remainder of the 454 shotgun dataset (Table S2). Several ontologies were statistically overrepresented among predicted deletions, including categories related to regulation of gene expression, chromatin organization, and development of germ/stem cells (Figure 3, Table S2). A subset of these regions, with validated expression in meiotic testes, was also similarly enriched in transcriptional regulatory and germline developmental functions (Table S2). Coupled with the above studies, ontology analyses seem to indicate that the genomic differentiation of germline vs. somatic lineages leads to differentiation in their capacities to deploy specific transcriptional programs, thereby regulating germline vs. somatic cell fate.
Analyses of our germline 454 corroborate previous findings that lamprey deletes ~20% of its genome through PGR [1, 2], though the method does not identify deletions of repetitive sequences when one or more members are retained in the soma. It known that repetitive elements constitute a substantial fraction of lamprey germline-specific (and somatically retained) DNA [1, 2] although these and other non-functional single-copy regions do not necessarily contribute to the development or maintenance of germline. More importantly, our studies indicate that the deleted fraction contains a substantial complement of functional or potentially functional genes: 7.6% of germline reads and 3.8% of germline gene homologies that are completely absent from the somatic WGS dataset (Table S1). When interpreting these results, it is also important to note that homology information cannot identify all functional components within deleted regions. For example, the current analyses do not specifically identify recent gene duplicates, lamprey-specific genes, or functional non-coding sequences that are deleted via PGR.
Although the current dataset does not identify the entire set of germline-specific genes, it seems clear that the developmentally-regulated segregation of a few thousand protein-coding genes and associated regulatory elements should substantially limit the functional capacities of somatic cell lineages, relative to the germline. On the basis of gene homology, ontology, and gene expression data (Figures 1–3), we hypothesize that DNA loss may be critical for segregating “totipotency” gene functions into the germline, thereby preventing the dysregulated deployment of germline-specific gene functions in somatic cell lineages. Notably though, several genes identified within the germline-specific fraction possess vertebrate homologs that are not currently known to function in either the development or maintenance of germline. We reason that their restriction to the germline-specific fraction of the lamprey genome, in itself, provides insight into their biological function. Specifically, the physical restriction of these genes to the lamprey germline genome implies that they 1) contribute to the development or maintenance of totipotent germline and 2) are dispensable (or deleterious) with respect to the maintenance and development of soma.
Conclusions
Genetic conflicts between germline and soma that are evident in our analyses of the PGR are conceptually similar to the definition of cancer/testes genes (or cancer/testes antigens) [8, 9], although such conflicts are not necessarily limited to the development of cancer. Cancer/testes genes are diverse in evolutionary origin, but share a common feature in that they normally exhibit testes-restricted expression and are only observed in somatic tissues in the context of oncogenesis [8, 9]. From a biological standpoint, it seems plausible that misregulation of genes with germline-specific functions (recombination, unlimited proliferation and a capacity for genomic reprogramming) could contribute to oncogenesis or other disease states [10]. Indeed, it has been shown empirically that ectopic expression of germline-specific genes can drive tumor growth in Drosophila [11] and Hydractinia [12]. In light of the differential and conflicting requirements of germline and soma, we hypothesize that PGR events might serve, in part, to segregate totipotency functions into the germline, thereby alleviating such untoward effects of these genes in the soma. Intriguingly, the conceptual similarities between cancer/testes and PGR models are seemingly further bolstered by our detection of cancer/testis antigen 68 within the germline specific fraction of the lamprey genome. As such, the lamprey genome appears to present a large, readily identifiable, and evolutionarily informative collection of germline-limited genes that can be leveraged to understand the unique genetic requirements and pleiotropic liability of vertebrate germline.
Future studies aimed at dissecting the functionality of deleted lamprey genes and other molecular details of PGR should provide novel insights into molecular mechanisms of germline totipotency, somatic recombination and biological tradeoffs between germline and soma. Notably, both extant lineages of jawless vertebrates (agnathans: lampreys and hagfish) are known to undergo PGR [1, 13], which would seem to indicate that the phenomenon is common to all extant agnathans and potentially represents an ancestral condition [14]. Thus PGR may represent an ancient mechanism for moderating genetic conflict between germline and soma that evolved within an ancestral vertebrate lineage (alternately, repeated evolution of PGR in lamprey, hagfish and numerous invertebrate and protist lineages [13, 15–21], may reflect recurrent selective advantages for PGR). Under either scenario, lamprey PGR holds the potential to fill an important gap in our understanding of the cause and consequence of dysregulated rearrangement of vertebrate genomes (e.g., in oncogenesis) [22–28] and the capacity for tight regulation of genome rearrangements in phylogenetically disparate lineages [13, 15–21, 29]. The availability of a draft genome [6] and established gene knockdown/replacement methods [30–32] for lamprey should promote future progress in resolving the causes, consequences and evolutionary relevance of PGR.
Experimental Procedures
Microarray Design, Processing, Analysis and Validation
We designed a customized NimbleGen (Roche) microarray consisting of 385,000 oligonucleotides targeted to lamprey germline and somatic sequences. DNA samples were prepared from agarose embedded nuclei (for sperm versus blood comparisons) or whole tissues by standard phenol/chloroform extraction [33]. Soma-germline array comparative genomic hybridization experiments were performed using DNA that originated from the same individual (blood versus sperm). Additional comparisons were performed between somatic tissues to assess the extent of somatic variation (Supplemental Experimental Procedures, Figure S1). Candidate regions identified by arrayCGH or from the analysis of 454 data were further evaluated by PCR and rtPCR. Detailed methods and validation procedures are provided as supplementary material (Supplemental Experimental Procedures). Array data were deposited in the NCBI gene expression omnibus (Table S3, http://www.ncbi.nlm.nih.gov/geo/).
454 Sequencing and Analysis
We isolated sperm DNA from a single individual and outsourced 454 sequencing to the Duke IGSP Sequencing Core Facility. Sequences were trimmed to Q20 using phred [34, 35] and any sequences less than 300 bp in length were removed from the dataset prior to analysis. The remaining 554,979 sequences were aligned (BLAST) [36] to a local database of 18,506,949 trimmed sequences from the lamprey genome project (liver DNA, average trimmed read length = 529 bp at Q20) [1, 6]. Alignments were parsed using custom scripts to identify various patterns that are indicative of rearrangement, insertion/deletion polymorphisms, or sampling artifacts (Figure 3A). Alignments to human RefSeq datasets were also performed using BLAST.
Germline Transcriptome
Germline transcripts were obtained from a single male lamprey collected in the early in the 2009 spawning run. RNA was extracted from testes using Trizol extraction, purified using Qiagen RNeasy midi kit, and polyA selected via two rounds affinity purification (Dynabeads mRNA Direct purification kit, Invitrogen). Sequencing was performed by the UC Davis Genome Center using a single flow cell lane on a GAIIx machine. Reads were assembled using ABySS 1.2.0 [37] and aligned to local databases of germline and somatic reads using BLAST.
In situ Hybridization
Whole-embryo in situ hybridization was performed following the methods of Nikitina et al. [32]. A fragment of the 3′ untranslated region from 25M04 (KRAB domain zinc finger gene) was amplified by PCR then cloned into the plasmid vector pCRII-TOPO (Invitrogen). Following sequencing to verify orientation, the one amplicons were generated using M13 primer and of two gene-specific primers, 25M04_insitu.F or 25M04_insitu.R (Table S4), to generate templates for synthesis of sense and antisense probes, respectively. Digoxigenin-labeled riboprobes were synthesized from these templates using the SP6 polymerase and the MAXIscript (Ambion) in vitro transcription kit.
Ontology Classification
All ontology analyses were performed using a custom ontology database, using Blast2GO [7]. The frequency of each ontology category within the predicted germline-specific subset was compared to the overall frequency of that ontology in the entire 454 dataset, using best blast hits to SwissProt as a source of ontology information.
Supplementary Material
Highlights
-
1)
The lamprey (Petromyzon marinus) undergoes developmentally-programmed gene deletions
-
2)
Analysis of lamprey germline shotgun sequence identifies germline-specific genes
-
3)
Deleted genes are specifically enriched in pluripotency-related functions
-
4)
Deleted genes function in meiotic germline and in developing primordial germ cells
Acknowledgments
We thank M. Bronner-Fraser and T. Sauka-Spengler for access to their lamprey husbandry facilities at the California Institute of Technology. We also thank M.C. Yao for his valuable discussions regarding programmed genome rearrangement. This work was supported by the National Institutes of Health (grant numbers GM079492, GM090049, and RR014085) and the National Science Foundation (grant number MCB-0719558) to C.T.A., the National Institutes of Health (grant numbers T32-HG00035, F32-GM087919) to J.J.S. and the National Institutes of Health (grant number GM58815) to E.E.E. E.E.E. is an Investigator of the Howard Hughes Medical Institute. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of NIH. Microarray data for this study were deposited with the NCBI Gene Expression omnibus under accession numbers GSE23757, GPL10847 and GSM586211 - 6. 454 reads from sperm genomic DNA were deposited in the NCBI short read archive under accession number SRA023537.3. Transcriptomic reads were deposited in the NCBI short read archive under accession numbers SRA047838.2.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Smith JJ, Antonacci F, Eichler EE, Amemiya CT. Programmed loss of millions of base pairs from a vertebrate genome. Proc.Natl.Acad.Sci.U.S.A. 2009;106:11212–11217. doi: 10.1073/pnas.0902358106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith JJ, Stuart AB, Sauka-Spengler T, Clifton SW, Amemiya CT. Development and analysis of a germline BAC resource for the sea lamprey, a vertebrate that undergoes substantial chromatin diminution. Chromosoma. 2010;119:381–389. doi: 10.1007/s00412-010-0263-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okkelberg P. The early history of the germ cells in the brook lamprey, entosphenus wilderi (GAGE), up to and including the period of sex differentiation. J.Morphol. 1921;35:1–151. [Google Scholar]
- 4.Butcher EO. The origin of the germ cells in the lake lamprey (Petromyzon marinus unicolor) Biol Bull-Us. 1929;56:87–99. [Google Scholar]
- 5.Hardisty MW, Cosh J. Primordial germ cells and fecundity. Nature. 1966;210:1370–1371. doi: 10.1038/2101370a0. [DOI] [PubMed] [Google Scholar]
- 6.Washington University Genome Sequencing, C. Washington University School of Medicine; 2007. http://genome.wustl.edu/pub/organism/Other_Vertebrates/Petromyzon_marinus/assembly/Petromyzon_marinus-3.0. [Google Scholar]
- 7.Gotz S, Garcia-Gomez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talon M, Dopazo J, Conesa A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat.Rev.Cancer. 2005;5:615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 9.Hofmann O, Caballero OL, Stevenson BJ, Chen YT, Cohen T, Chua R, Maher CA, Panji S, Schaefer U, Kruger A, et al. Genome-wide analysis of cancer/testis gene expression. Proc.Natl.Acad.Sci.U.S.A. 2008;105:20422–20427. doi: 10.1073/pnas.0810777105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 11.Janic A, Mendizabal L, Llamazares S, Rossell D, Gonzalez C. Ectopic expression of germline genes drives malignant brain tumor growth in Drosophila. Science. 2010;330:1824–1827. doi: 10.1126/science.1195481. [DOI] [PubMed] [Google Scholar]
- 12.Millane RC, Kanska J, Duffy DJ, Seoighe C, Cunningham S, Plickert G, Frank U. Induced stem cell neoplasia in a cnidarian by ectopic expression of a POU domain transcription factor. Development. 2011;138:2429–2439. doi: 10.1242/dev.064931. [DOI] [PubMed] [Google Scholar]
- 13.Nakai Y, Kubota S, Kohno S. Chromatin diminution and chromosome elimination in four Japanese hagfish species. Cytogenet.Cell Genet. 1991;56:196–198. doi: 10.1159/000133087. [DOI] [PubMed] [Google Scholar]
- 14.Smith JJ, Saha NR, Amemiya CT. Genome biology of the cyclostomes and insights into the evolutionary biology of vertebrate genomes. Integr Comp Biol. 2010;50:130–137. doi: 10.1093/icb/icq023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yao MC, Yao CH, Halasz LM, Fuller P, Rexer CH, Wang SH, Jain R, Coyne RS, Chalker DL. Identification of novel chromatin-associated proteins involved in programmed genome rearrangements in Tetrahymena. J.Cell Sci. 2007;120:1978–1989. doi: 10.1242/jcs.006502. [DOI] [PubMed] [Google Scholar]
- 16.Nowacki M, Higgins BP, Maquilan GM, Swart EC, Doak TG, Landweber LF. A functional role for transposases in a large eukaryotic genome. Science. 2009;324:935–938. doi: 10.1126/science.1170023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bachmann-Waldmann C, Jentsch S, Tobler H, Muller F. Chromatin diminution leads to rapid evolutionary changes in the organization of the germ line genomes of the parasitic nematodes A. suum and P. univalens. Mol.Biochem.Parasitol. 2004;134:53–64. doi: 10.1016/j.molbiopara.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 18.Rasch EM, Wyngaard GA, Connelly BA. Heterochromatin endoreduplication prior to gametogenesis and chromatin diminution during early embryogenesis in Mesocyclops edax (Copepoda: Crustacea) J.Morphol. 2008;269:387–397. doi: 10.1002/jmor.10576. [DOI] [PubMed] [Google Scholar]
- 19.Boveri T. Uber Differenzierung der Zellkerne wahrend der Furchung des Eies von Ascaris megalocephala. Anat Anz. 1887;2:688–693. [Google Scholar]
- 20.Yao MC, Gorovsky MA. Comparison of the sequences of macro- and micronuclear DNA of Tetrahymena pyriformis. Chromosoma. 1974;48:1–18. doi: 10.1007/BF00284863. [DOI] [PubMed] [Google Scholar]
- 21.Lauth MR, Spear BB, Heumann J, Prescott DM. DNA of ciliated protozoa: DNA sequence diminution during macronuclear development of Oxytricha. Cell. 1976;7:67–74. doi: 10.1016/0092-8674(76)90256-7. [DOI] [PubMed] [Google Scholar]
- 22.Kolomietz E, Meyn MS, Pandita A, Squire JA. The role of Alu repeat clusters as mediators of recurrent chromosomal aberrations in tumors. Genes Chromosomes.Cancer. 2002;35:97–112. doi: 10.1002/gcc.10111. [DOI] [PubMed] [Google Scholar]
- 23.Flores M, Morales L, Gonzaga-Jauregui C, Dominguez-Vidana R, Zepeda C, Yanez O, Gutierrez M, Lemus T, Valle D, Avila MC, et al. Recurrent DNA inversion rearrangements in the human genome. Proc.Natl.Acad.Sci.U.S.A. 2007;104:6099–6106. doi: 10.1073/pnas.0701631104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strout MP, Marcucci G, Bloomfield CD, Caligiuri MA. The partial tandem duplication of ALL1 (MLL) is consistently generated by Alu-mediated homologous recombination in acute myeloid leukemia. Proc.Natl.Acad.Sci.U.S.A. 1998;95:2390–2395. doi: 10.1073/pnas.95.5.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elliott B, Richardson C, Jasin M. Chromosomal translocation mechanisms at intronic alu elements in mammalian cells. Mol.Cell. 2005;17:885–894. doi: 10.1016/j.molcel.2005.02.028. [DOI] [PubMed] [Google Scholar]
- 26.Raphael BJ, Volik S, Yu P, Wu C, Huang G, Linardopoulou EV, Trask BJ, Waldman F, Costello J, Pienta KJ, et al. A sequence-based survey of the complex structural organization of tumor genomes. Genome Biol. 2008;9:R59. doi: 10.1186/gb-2008-9-3-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, Stebbings LA, Leroy C, Edkins S, Mudie LJ, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell PJ, Stephens PJ, Pleasance ED, O'Meara S, Li H, Santarius T, Stebbings LA, Leroy C, Edkins S, Hardy C, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat.Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nemetschke L, Eberhardt AG, Hertzberg H, Streit A. Genetics, chromatin diminution, and sex chromosome evolution in the parasitic nematode genus Strongyloides. Curr Biol. 2010;20:1687–1696. doi: 10.1016/j.cub.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 30.Sauka-Spengler T, Meulemans D, Jones M, Bronner-Fraser M. Ancient evolutionary origin of the neural crest gene regulatory network. Dev.Cell. 2007;13:405–420. doi: 10.1016/j.devcel.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 31.Nikitina N, Sauka-Spengler T, Bronner-Fraser M. Dissecting early regulatory relationships in the lamprey neural crest gene network. Proc.Natl.Acad.Sci.U.S.A. 2008;105:20083–20088. doi: 10.1073/pnas.0806009105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nikitina N, Bronner-Fraser M, Sauka-Spengler T. Emerging Model Organisms: A Laboratory Manual. Volume 1. CSHL Press; Cold Spring Harbor, NY, USA: 2009. The Sea Lamprey Petromyzon marinus: A Model for Evolutionary and Developmental Biology; pp. 405–429. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Russell DW. Molecular Cloning: A laboratory Manual. 3rd Eds Volume 3. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2001. [Google Scholar]
- 34.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 35.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 36.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J.Mol.Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 37.Birol I, Jackman SD, Nielsen CB, Qian JQ, Varhol R, Stazyk G, Morin RD, Zhao Y, Hirst M, Schein JE, et al. De novo transcriptome assembly with ABySS. Bioinformatics. 2009;25:2872–2877. doi: 10.1093/bioinformatics/btp367. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.