Abstract
YAP (Yes-associated protein) is a potent oncogene and a major effector of the mammalian Hippo tumor suppressor pathway. In this review, our emphasis is on the structural basis of how YAP recognizes its various cellular partners. In particular, we discuss the role of LATS kinase and AMOTL1 junction protein, two key cellular partners of YAP that bind to its WW domain, in mediating cytoplasmic localization of YAP and thereby playing a key role in the regulation of its transcriptional activity. Importantly, the crystal structure of an amino-terminal domain of YAP in complex with the carboxy-terminal domain of TEAD transcription factor was only recently solved at atomic resolution, while the structure of WW domain of YAP in complex with a peptide containing the PPxY motif has been available for more than a decade. We discuss how such structural information may be exploited for the rational development of novel anti-cancer therapeutics harboring greater efficacy coupled with low toxicity. We also embark on a brief discussion of how recent in silico studies led to identification of the cardiac glycoside digitoxin as a potential modulator of WW domain-ligand interactions. Conversely, dobutamine was identified in a screen of known drugs as a compound that promotes cytoplasmic localization of YAP, thereby resulting in growth suppressing activity. Finally, we discuss how a recent study on the dynamics of WW domain folding on a biologically critical time scale may provide a tool to generate repertoires of WW domain variants for regulation of the Hippo pathway toward desired, non-oncogenic outputs.
Keywords: TEAD transcription factor, WW domain, PDZ domain, Nuclear localization, Digitoxin, Dobutamine
Introduction
YAP is a transcriptional co-activator and a major effector of the mammalian Hippo tumor suppressor pathway [1]. Upon activation of the Hippo pathway by cell-to-cell contacts, YAP becomes phosphorylated at various S residues, including S127, by a concerted action of two upstream kinases, MST and LATS [2]. The pS127 and the flanking residues in turn serve as a docking site for 14-3-3 proteins and the resulting interaction is primarily responsible for the cytoplasmic localization of YAP in response to activation of Hippo pathway [3]. Within the cytoplasm, YAP mediates pro-apoptotic signals. However, phosphorylation of S residues other than S127 is believed to lead to ubiquitination and proteosomal degradation of YAP, thereby down-regulating pro-apoptotic signaling through YAP [2]. Remarkably, the absence of phosphorylation of YAP at S127 serves as a signal for its translocation to the cell nucleus where it forms a complex with members of the TEAD family of transcription factors that drive transcription of growth-promoting and anti-apoptotic genes [1, 4].
Importantly, YAP is a bona fide oncogene. The amplification or over-expression of the YAP gene was demonstrated in human cancers of various organs, and YAP over-expression in mammalian cells was shown to elicit a plethora of oncogenic parameters [5, 6]. The nuclear localization of YAP in tumor biopsies correlates with poor prognosis for cancer patients [1, 2].
As mentioned in previous chapters of this issue, the Hippo tumor suppressor pathway was originally delineated in Drosophila by genetic screening approaches [7]. YAP is the mammalian ortholog of Drosophila Yki, and MST kinase is the mammalian ortholog of the Drosophila Hippo kinase from which the name of the pathway was derived.
Several structural studies have provided valuable insight into the details of YAP signaling via complexes with partner proteins. In particular, the crystal structures of YAP in complex with TEAD4 and the WW domain of YAP with its PPxY sequence-containing ligands were solved and will be discussed in detail [8, 9]. We will shed light on how the cardiac glycoside digitoxin [10] may serve as a lead compound for the development of drugs that could antagonize the oncogenic activity of YAP in cells and animal models, and ultimately be of use in managing cancer in clinics. We will also briefly address PDZ domains that recognize the tail sequence of YAP protein and act as mediators of important regulatory complexes with YAP oncogene, representing potential targets for developing anti-cancer drugs [11].
1. Modular structure of YAP1 and YAP2 isoforms
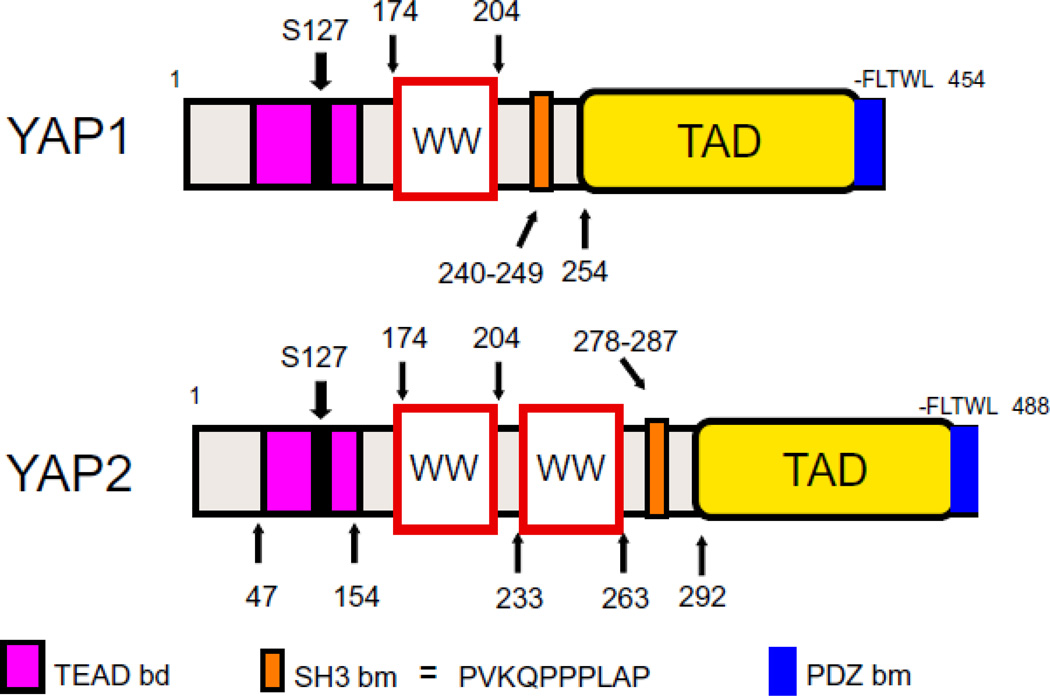
There are two major isoforms of YAP that are derived by differential splicing [12–14]. These are YAP1, containing one WW domain and YAP2, containing two WW domains (FIGURE 1). Actually, this very difference between the two YAP isoforms observed in the process of characterization of various cDNA clones of YAP led to identification of the WW domain as a signaling module [13–15]. Today, we know that there are more than 2 isoforms of YAP (at least 8), which are generated by differential splicing of short exons located within the transcriptional activation domain of YAP.
Figure 1.
Modular structures of the two major isoforms of YAP protein. TEAD binding domain (bd), WW domains, SH3 domain-binding motif (bm) transcriptional activation domain (TAD), and PDZ domain-binding motif (bm) are demarcated on the scheme of structures. See text for more details.
The modular structure of both YAP1 and YAP2 contains at the amino terminal region a TEAD factor-binding domain that is located between amino acids 47 and 154 [4]. The first WW domain is located between amino acids 174 and 204, and the second WW domain that is present only in YAP2 is located between amino acids 233 and 263. Both YAP1 and YAP2 also contain an SH3-binding motif [12], and a transcriptional activation domain, the latter located at the carboxy-terminal half of the protein [14]. A PDZ-binding motif composed of 5 terminal amino acids, –FLTWL, critical for nuclear translocation and binding to PDZ domains of ZO-1 and ZO-2 proteins, is located on the very carboxy-terminal end of YAP1 and YAP2 proteins [11].
2. YAP-TEAD complex as a primary target of drugs
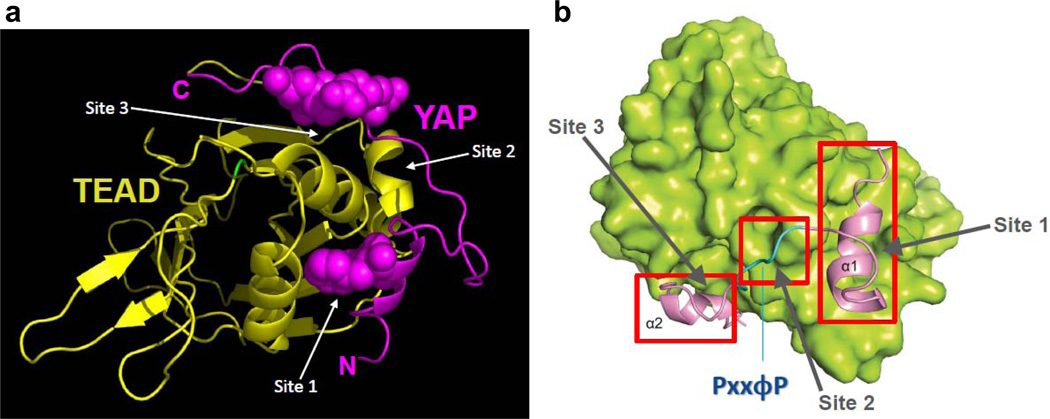
The complex between YAP and TEAD proteins is necessary for growth promoting activity of YAP oncogene [12], therefore it is a subject of intense structure-function analyses. Two groups have solved crystal structures of YAP and TEAD complexes at ~3Å resolution, revealing interesting molecular details of the binding interface [8, 9]. Two α-helices (α1 and α2) of YAP, along with the connecting hydrophobic linker, sequester TEAD in a manner akin to a pair of forceps (FIGURE 2). Indeed, three major interaction sites that accommodate the clip are clearly discernible in the TEAD protein in this complex: site 1, where the α1 helix fits in a rather large groove of TEAD surface, site 2 that accommodates the linker sequence, and site 3 into which the α2 helix of YAP fits snugly [8]. The relatively high resolution of both structures allowed for unequivocal identification of specific residues that make hydrogen bonds, van der Waals contacts and hydrophobic interactions between TEAD and YAP. Notably, the structures provided the rationale for the Y421H disease-causative mutation present in TEAD1, responsible for Sveinsson’s chorioretinal atrophy syndrome [8, 16]. The Y421 residue is located in a specific portion of the carboxyterminal-region of TEAD1, which was mapped as the region required for physical and functional interaction with YAP. From the structure, one could deduce that the replacement of Y421 with H eliminated a hydrogen bond with a neighboring S and affected a hydrophobic contact with closely located F, both of these neighboring residues present in YAP. From this rather significant change in molecular contacts, one could predict that the complex between YAP and TEAD1 (one of the four members of the TEAD family of transcription factors) would be affected. Indeed, the prediction was well confirmed by earlier biochemical results showing that the Sveinsson’s Y421H mutation abrogated the TEAD1 complex with YAP [17].
Figure 2.
In A is the ribbon structure of the amino-terminal region of YAP (in magenta) in complex with the carboxy-terminal region of TEAD transcription factor (matte gold). Note that YAP clips TEAD structure like a pair of forceps. In B, three major sites on TEAD (shown in green), which accommodate YAP, are demarcated in red. The first site binds α1 helix, the second side accommodates the linker sequence, and the third site binds the α2 helix. See text for more details. Both figures are a gift from Drs. Wan Jin Hong and Hai Wei Song (8). The permission to reproduce Figure 2B (from reference 8) was granted by the authors and the Cold Spring Harbor Laboratory Press.
As is evident from the structure itself, but also from the biochemical interrogations of various point mutants of the interface residues in terms of the stability of the complex, it seems that all three sites of interaction act in concert to mediate the YAP-TEAD complex [8, 9]. It is important to stress that the deletion of the short linker sequence in YAP results in a diminished interaction between YAP and TEAD [9]. This conformation closely resembles that of TAZ, the ortholog of YAP, whose amino-terminal TEAD-binding region harbors α1 and α2 helices, but lacks the linker sequence [9, 18]. Since TAZ and TEAD were shown to affect the transcriptional program of cell proliferation and EMT (epithelial-to-mesenchymal transition) [2], it is curious to know how TAZ and TEAD interact. Rather than proposing that TAZ interacts more weakly with TEAD when compared to YAP, we would like to suggest that two molecules of TAZ interact with one molecule of TEAD to form a transcriptional complex. The α1 helix of one TAZ molecule and the α2 helix of the other TAZ molecule could dock into TEAD’s grooves without any constrains from the lack of linker sequence. This scenario would better explain the function of TAZ, whose oncogenic and stemness activities are not weaker compared to those of YAP, but are rather qualitatively different. As two YAP-TEAD complexes form a structure of dimerized heterodimer, perhaps the higher order structure of the TAZ-TEAD complex holds secrets to the differences in signaling between TAZ and YAP.
YAP-TEAD complex is a formidable target for developing new cancer therapeutics. The YAP binding surface should be druggable through the fragment-based strategy approach and one could envision a battery of diverse compounds that could dock into one or any combination of the three binding sites to disrupt or weaken the YAP-TEAD complex. By targeting the function of deregulated Hippo pathway at the level of the nuclear effector of the pathway, one should diminish the potential side effects expected from targeting any of the upstream proteins of the pathway, which are more interconnected with other signaling networks. Because a significant percentage of patients affected by certain cancers, such as cancer of the liver, breast or pharynx, harbor causative amplification or over-expression of the YAP gene [1], the use of YAP-TEAD inhibiting drugs could be tailored to selected patients for optimal effects.
3. YAP WW domains and their roles in YAP signaling
In addition to YAP and its paralog TAZ, many other signaling proteins in the Hippo network contain WW domains [19, 20]. Moreover, there are a substantial number of Hippo pathway proteins that contain PPxY consensus sequences (where P is proline, Y is tyrosine and x is any amino acid), which represent a required core motif for ligands of WW domains [19].
The WW domain is one of the smallest among currently known modular protein domains [15, 21]. The domain is composed of ~35 amino acids that fold into an anti-parallel triple-stranded β-sheet forming a binding pocket for P-rich or P-containing ligands [22–24]. There are several classes of WW domains based on the ligand preference, but the largest class binds ligands that contain PPxY motif [25]. Curiously, this class of WW domains is represented in the Hippo network [20]. The Y in the PPxY motif must be in the non-phospho state for binding to occur, and when the Y is phosphorylated, it negatively regulates WW domain binding [13, 25, 26]. Therefore, the WW domain shares mechanisms of complex formation used by SH2 (Src homology 2) and SH3 (Src homology 3) domains in terms of regulation of binding by Y phosphorylation and the requirement of consecutive Ps in the ligand, respectively [25]. There are ~100 WW domains in the human proteome and almost 2,000 PPxY motifs scattered within human proteins [20, 27]. Several WW domains, including WW domains of Hippo pathway proteins, show a propensity for dimerization [20]. This molecular feature adds to the functional plasticity of WW domains and the Hippo pathway takes full advantage of these molecular attributes.
Interestingly, over-expression of the human YAP gene with mutated WW domains promoted transformation and migration of breast epithelial cells more efficiently than the wild type controls, suggesting that WW domains of YAP and the complexes they form with cognate proteins convey inhibitory signals [28]. To support this observation we, and others, have documented the existence of functional complexes between YAP WW domain and PPxY motif-containing LATS1 kinase, as well as between YAP WW domain and PPxY motif-containing AMOTL1 protein [29–33]. In agreement with the mutational analysis of YAP WW domains published by the team of Kieran Harvey [28], both LATS1 kinase and one of the cell-to-cell junction proteins, AMOTL1, anchor YAP protein in the cytoplasm, not dissimilar to the action of 14-3-3 protein [3], therefore inhibiting the proliferative activity of YAP. Based on these observations we speculate that the development of small molecule adaptors or modifiers that would stabilize YAP WW domain complexes could act in synergy with YAP-TEAD inhibitors to check the oncogenic activity of YAP. However, we need to consider more complex scenarios in which YAP WW domains also exert positive effects. YAP and its Drosophila ortholog Yki were shown to form WW domain-mediated complexes with proteins that positively regulate cell proliferation. For example, WBP2 (WW domain binding protein 2) [34] was shown to promote tissue growth and cell transformation via YAP and Yki [28, 35–38]. It is likely that the function of YAP WW domains may vary depending on the cell and tissue context.
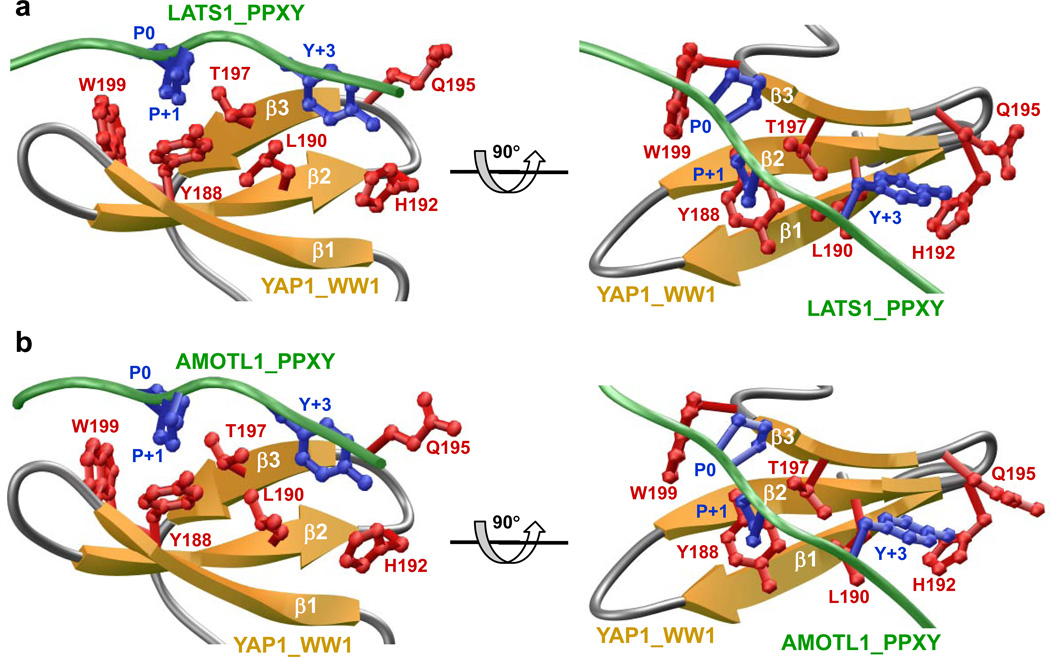
To understand the physical basis of the binding of WW domain of YAP to PPxY motifs within LATS1 and AMOTL1, we built appropriate structural models (FIGURES 3a and 3b). Our models show that in each case, the PPxY peptides adopt the polyP type II (PPII) helical conformation and bind within the hydrophobic groove of the anti-parallel triple-stranded β-sheet fold of the WW domain in a canonical manner [22–24, 39]. In agreement with our previous study [40], only the consensus residues within the PPxY motifs appear to be engaged in key intermolecular contacts with specific residues lining the hydrophobic groove of the WW domain. Notably, the pyrrolidine moiety of P0, the first P within each PPxY motif [according to our earlier nomenclature [40], stacks against the indole side chain of W199 in WW domain. The side chains of Y188/T197 within the WW domain sandwich the pyrrolidine moiety of P+1 within each PPxY motif. The phenyl moiety of Y+3, the terminal Y within the PPxY motif, buries deep into the hydrophobic groove and is escorted by side chains of L190/H192/Q195 in the WW domain. The various interactions between specific side chains in the WW domain and the PPxY motifs appear to be stabilized by an extensive network of van der Waals contacts and hydrogen bonding. In particular, the Hη phenolic hydrogen of Y+3 appears to hydrogen bond with the Nδ1 imidazole nitrogen of H192 in the WW domain. We note that non-consensus residues within and flanking the PPxY motifs make no discernable contacts with any residues within the WW domain, but may be important for stabilizing the PPII conformation of the PPxY peptides.
Figure 3.
Structural models of the WW domain of YAP in complex with PPxY peptides derived from LATS1 (a) and AMOTL1 (b). In each case, the β-strands in the WW domain are shown in yellow with loops depicted in gray and the ligand colored green. The side chain moieties of residues within WW domain engaged in key intermolecular contacts with the PPxY peptides are shown in red. The side chain moieties of counteracting residues within the peptides colored blue correspond to the PPxY motif. All structural models were built using the MODELLER software based on homology modeling [55]. For the structural models of WW domain of YAP1 in complex with PPxY peptides derived from LATS1 and AMOTL1, the NMR structure of WW domain of YAP1 bound to a peptide containing the PPxY motif was used as a template (PDB# 1JMQ). In each case, a total of 100 atomic models were calculated and the structure with the lowest energy, as judged by the MODELLER Objective Function, was selected for further analysis. The atomic models were rendered using RIBBONS [56].
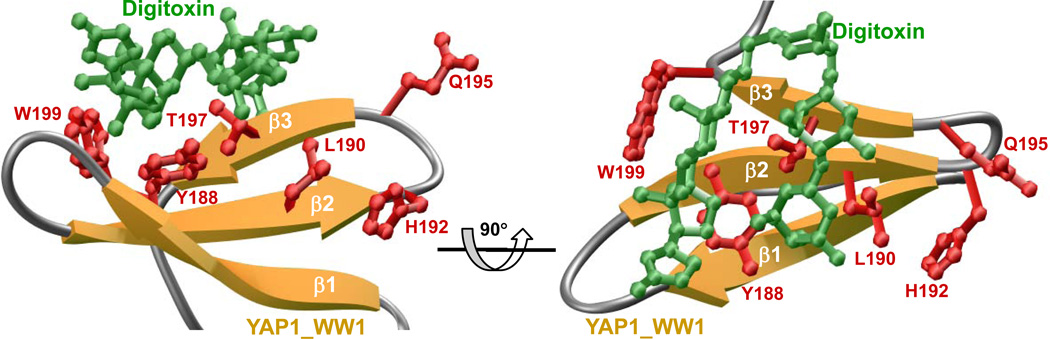
Through in silico analysis, we recently predicted that the cardiac glycoside digitoxin may have affinity for the WW domain of dystrophin with important implications on therapeutic design of small molecule modulators of WW domains [10]. In an effort to test the extent to which WW domains of other proteins may also be targeted by digitoxin, we built a structural model of the first WW domain of YAP bound to digitoxin (FIGURE 4). Our analysis reveals that digitoxin gravitates to the WW domain of YAP in a manner akin to the binding of canonical PPxY ligands. Thus, digitoxin docks to the canonical hydrophobic groove within the WW domain that is also shared by PPxY ligands. Importantly, digitoxin appears to engage in an extensive network of intermolecular van der Waals and hydrogen bonding contacts with an array of residues, such as Y188, L190, T197 and W199 lining the hydrophobic groove within the WW domain. These residues are also critical for the binding of PPxY ligands. However, other residues such as H192 and Q195 within the WW domain, which also play a key role in the binding of PPxY ligands, do not appear to be important for the binding of digitoxin. That this is so, suggests that digitoxin is unlikely to target all WW domains indiscriminately and that potential opportunities exist for the chemical modification of digitoxin so as to enhance its specificity toward a small group of WW domains involved in regulating a specific signaling cascade such as the Hippo pathway. We believe that digitoxin and its chemical analogs bear the potential to compete with PPxY ligands for binding to the WW1 domain of YAP in a mutually exclusive manner, and thus could be exploited as inhibitors for therapeutic intervention, especially in tissues where YAP WW complexes convey proliferative signals [34–36]. Since there are some examples of human cancers where YAP was shown to act as a tumor suppressor [41, 42], enhancing the activity of YAP by inhibiting WW domain complexes could have therapeutic ramifications.
Figure 4.
Structural models of the WW domain of YAP in complex with digitoxin. For the model shown in the figure, the WW domain of dystrophin in complex with digitoxin was used as a template [10]. For more details see legend to Figure 3 and the text.
4. WW domain and fine analysis of protein folding
When the first structure of YAP WW domain was revealed by the collaborative efforts between the group led by Hartmut Oschkinat at EMBL in Heidelberg and the Sudol team at The Rockefeller University in New York, it was amazing to see the compact meander of three β strands formed by less than 30 amino acids and folding without the aid of cofactors or disulfide bonds [22]. In a way, the WW domain defied past predictions stipulating that a stable, autonomous protein fold must be at least 40 amino acids long [21].
Several laboratories promptly zoomed in on the WW domain as an attractive subject of protein folding studies [43–47]. They considered that the detailed biophysical analysis of the WW domain would shed light on the mechanism of β-strand formation. Two of our colleagues, Jeff Kelly from the Scripps Research Institute in California and Rama Ranganathan from the University of Texas, Southwestern Medical Center provided fine insight into the dynamics of WW domain folding by identifying several residues that are critical for the stability and the dynamics of the fold [43–45]. The identification of the hydrophobic core of four amino acids, which lie beneath the binding pocket of the domain and form a stabilizing foundation of the WW domain fold, has direct ramifications for explaining the Golabi-Ito-Hall syndrome that is caused by a point mutation in the WW domain of PQBP1 [27, 44]. However, a quantum leap in the study of the WW domain fold came from the cutting edge analysis of the dynamic of WW protein domain folding, which was completed by the team of David E. Shaw, a recognized academician at the Columbia University in New York City [46, 47]. He and his team employed equilibrium simulations of a WW domain and captured eight folding and seven unfolding events that follow the same and well–defined folding pathway [41]. The specialized machine was developed by the Shaw laboratory for elucidating the atomic-level behavior of proteins on a biologically critical time scale. With such a fine understanding of the WW domain fold, it should be possible to predict and test, at first in silico, a large repertoire of WW domain variants that would display attenuated or enhanced folding and binding activities of the domains, and therefore their host proteins. If so, such WW domain variants could be used either by themselves or as parts of modified YAP genes in gene therapy approaches to modulate the Hippo tumor suppressor network for anti-proliferative outputs. The ultimate aim of such an exercise would be to develop modified genes for managing cancers caused by dysregulation of the Hippo pathway.
The occurrence of a single class of WW domains and its cognate PPxY ligand sequences in the Hippo pathway is remarkable and unmatched by any other known signaling pathway [20]. Because of this unusual concentration of WW domain in the Hippo network, we anticipate a more robust cross-talk via WW domains and PPxY ligand cores among the members of the Hippo network then what was already deciphered. We suggest that the potential for new multi-component complexes and feedback regulatory loops that rely on reiterated use of WW domains in the Hippo pathway is very high.
5. Nuclear localization of YAP is controlled by its PDZ-binding motif
We have shown that the PDZ-binding motif of YAP, which is composed of 5 terminal amino acids, –FLTWL, is critical for its nuclear translocation [11]. YAP delta C mutant, missing these 5 amino acids, tends to stay in the cytoplasm [11]. Since this complex regulates a critical step in YAP oncogenic signaling, it deserves further functional and structural analyses. Small molecule inhibitors that mimic the -FLTWL or even -TWL peptide could be of therapeutic value. Targeting the PDZ domain complexes of YAP could be quite successful because the -TWL sequence is present only in three human proteins. Apart from YAP and its oncogenic paralog TAZ, the –TWL carboxy-terminal sequence is found only in the mitochondrial ribosomal protein L43, suggesting that mimetics of this sequence could be specific as drugs because they would not interfere with many other PDZ domain complexes. However, there are quite a few PDZ domains that are likely to bind the –TWL motif, based on comprehensive mapping data with PDZ domains [48]. Nevertheless, a –TWL mimetic could be considered as an inhibitor of proliferative signaling of the Hippo pathway, possibly with moderate or minimal side effects.
An interesting study from the laboratory of Yutaka Hata at the Tokyo Medical and Dental University revealed that dobutamine, a sympathomimetic drug frequently used in the treatment of heart failure and cardiogenic shock, was able to stimulate YAP translocation from the nucleus to the cytoplasm in an osteosarcoma cell line, and it significantly suppressed YAP-TEAD complex-mediated gene transcription [49]. Although the precise mechanism by which dobutamine affects YAP is not known, it is conceivable that this drug targets YAP-PDZ complexes. This study is a good example of a tailored, low-throughput screen of approved drugs to uncover desirable inhibitory effects on the oncogenic signaling of the Hippo pathway.
6. Concluding remarks
The Hippo tumor suppressor pathway has exploded in the past several years as a novel signaling pathway that is directly relevant to human cancer and the biology of cancer stem cells [48]. New discoveries that are relevant to this pathway are reported at a very fast pace. For example, one of the emerging but still unexplored frontiers for the development of new strategies to control oncogenic function of YAP is the competitive relationship of YAP with the family of WW domain-containing ubiquitin ligases [51–53]. New animal transgenic and knock-out models of Hippo pathway genes have been constructed and are being used now to analyze this pathway at the level of the organism. These will be useful for drug validation efforts. Detailed structural studies of Hippo pathway proteins, as briefly reviewed here, should complement biological studies and impact the search for effective therapies. One of the more elegant approaches, which resulted in promising drug leads for controlling activity of other oncogenes, is the genetic screen of synthetic lethality [54]. Several existing cell culture models of the Hippo pathway are amenable to this approach and may reveal cooperative signals from naturally-occurring biochemical rather than synthetic molecules to bind to the folds of protein domains discussed in this review.
ACKNOWLEDGMENTS
We thank our colleagues Wan Jin Hong and Hai Wei Song for their permission to use in Figure 2 the pictures of YAP-TEAD complexes that they published previously, and the Cold Spring Harbor Laboratory Press for giving us a formal permission to reproduce the data from the Genes and Development journal. We thank all the participants of Hippo I and Hippo II workshops in Rome for stimulating discussions that provided seeds for some of the ideas presented here. We appreciate kind words from David E. Shaw approving the proposal of WW domain repertoires with partial folding and binding activities as modulators of the Hippo pathway. And we also thank our colleagues Andrea Uetrecht, Virginia Mazack, Gary Bader, Dev Sidhu, Bob Varelas and Henning Wackerhage for valuable comments on the first version of the manuscript.
This work was supported by the National Institutes of Health Grants R01-GM083897 and funds from the USylvester Braman Family Breast Cancer Institute (to AF), by Science Foundation Ireland (#08/IN1/B1864) to (DCS), and by PA Breast Cancer Coalition Grants (#60707 an #920093) plus the Geisinger Clinic (to MS)
Abbreviations used
- A
Alanine
- AMOTL1
Angiomotin-Like 1
- EMT
epithelial-to-mesenchymal transition
- LATS
large tumor suppressor
- MST
mammalian ste20-like protein kinase
- PDZ domain
Psd-95 (Post Synaptic Density Protein), DlgA (Drosophila Disc Large Tumor Suppressor) and ZO1 (Zonula Occludens-1 Protein)
- P
Proline
- S
Serine
- TAZ
transcriptional co-activator with PDZ-binding motif
- also known as WWTR1
WW-domain-containing transcription regulator 1
- TEAD factor
TEA domain-containing transcription factor
- WW domain
Tryptophan-Tryptophan domain
- Y
Tyrosine
- YAP
Yes kinase-associated protein
- ZO
zona occludens
References
- 1.Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao B, Li L, Lei Q, Guan KL. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24:862–874. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basu S, Totty NF, Irwin MS, Sudol M, Downward J. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol Cell. 2003;11:11–23. doi: 10.1016/s1097-2765(02)00776-1. [DOI] [PubMed] [Google Scholar]
- 4.Vassilev A, Kaneko KJ, Shu H, Zhao Y, DePamphilis ML. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 2001;15:1229–1241. doi: 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, et al. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci U S A. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harvey K, Tapon N. The Salvador-Warts-Hippo pathway - an emerging tumour-suppressor network. Nat Rev Cancer. 2007;7:182–191. doi: 10.1038/nrc2070. [DOI] [PubMed] [Google Scholar]
- 8.Chen L, Chan SW, Zhang X, Walsh M, Lim CJ, Hong W, et al. Structural basis of YAP recognition by TEAD4 in the hippo pathway. Genes Dev. 2010;24:290–300. doi: 10.1101/gad.1865310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z, Zhao B, Wang P, Chen F, Dong Z, Yang H, et al. Structural insights into the YAP and TEAD complex. Genes Dev. 2010;24:235–240. doi: 10.1101/gad.1865810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casey FP, Pihan E, Shields DC. Discovery of small molecule inhibitors of protein-protein interactions using combined ligand and target score normalization. J Chem Inf Model. 2009;49:2708–2717. doi: 10.1021/ci900294x. [DOI] [PubMed] [Google Scholar]
- 11.Oka T, Remue E, Meerschaert K, Vanloo B, Boucherie C, Gfeller D, et al. Functional complexes between YAP2 and ZO-2 are PDZ domain-dependent, and regulate YAP2 nuclear localization and signalling. Biochem J. 2010;432:461–472. doi: 10.1042/BJ20100870. [DOI] [PubMed] [Google Scholar]
- 12.Sudol M. Yes-associated protein (YAP65) is a proline-rich phosphoprotein that binds to the SH3 domain of the Yes proto-oncogene product. Oncogene. 1994;9:2145–2152. [PubMed] [Google Scholar]
- 13.Sudol M, Bork P, Einbond A, Kastury K, Druck T, Negrini M, et al. Characterization of the mammalian YAP (Yes-associated protein) gene and its role in defining a novel protein module, the WW domain. J Biol Chem. 1995;270:14733–14741. doi: 10.1074/jbc.270.24.14733. [DOI] [PubMed] [Google Scholar]
- 14.Komuro A, Nagai M, Navin NE, Sudol M. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J Biol Chem. 2003;278:33334–33341. doi: 10.1074/jbc.M305597200. [DOI] [PubMed] [Google Scholar]
- 15.Bork P, Sudol M. The WW domain: a signalling site in dystrophin? Trends Biochem Sci. 1994;19:531–533. doi: 10.1016/0968-0004(94)90053-1. [DOI] [PubMed] [Google Scholar]
- 16.Fossdal R, Jonasson F, Kristjansdottir GT, Kong A, Stefansson H, Gosh S, et al. A novel TEAD1 mutation is the causative allele in Sveinsson's chorioretinal atrophy (helicoid peripapillary chorioretinal degeneration) Hum Mol Genet. 2004;13:975–981. doi: 10.1093/hmg/ddh106. [DOI] [PubMed] [Google Scholar]
- 17.Kitagawa M. A Sveinsson's chorioretinal atrophy-associated missense mutation in mouse Tead1 affects its interaction with the co-factors YAP and TAZ. Biochem Biophys Res Commun. 2007;361:1022–1026. doi: 10.1016/j.bbrc.2007.07.129. [DOI] [PubMed] [Google Scholar]
- 18.Kanai F, Marignani PA, Sarbassova D, Yagi R, Hall RA, Donowitz M, et al. TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J. 2000;19:6778–6791. doi: 10.1093/emboj/19.24.6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sudol M, Harvey KF. Modularity in the Hippo signaling pathway. Trends Biochem Sci. 2010;35:627–633. doi: 10.1016/j.tibs.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 20.Sudol M. Newcomers to the WW Domain-Mediated Network of the Hippo Tumor Suppressor Pathway. Genes Cancer. 2010;1:1115–1118. doi: 10.1177/1947601911401911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sudol M. Structure and function of the WW domain. Prog Biophys Mol Biol. 1996;65:113–132. doi: 10.1016/s0079-6107(96)00008-9. [DOI] [PubMed] [Google Scholar]
- 22.Macias MJ, Hyvonen M, Baraldi E, Schultz J, Sudol M, Saraste M, et al. Structure of the WW domain of a kinase-associated protein complexed with a proline-rich peptide. Nature. 1996;382:646–649. doi: 10.1038/382646a0. [DOI] [PubMed] [Google Scholar]
- 23.Huang X, Poy F, Zhang R, Joachimiak A, Sudol M, Eck MJ. Structure of a WW domain containing fragment of dystrophin in complex with beta-dystroglycan. Nat Struct Biol. 2000;7:634–638. doi: 10.1038/77923. [DOI] [PubMed] [Google Scholar]
- 24.Kanelis V, Rotin D, Forman-Kay JD. Solution structure of a Nedd4 WW domain-ENaC peptide complex. Nat Struct Biol. 2001;8:407–412. doi: 10.1038/87562. [DOI] [PubMed] [Google Scholar]
- 25.Sudol M, Hunter T. NeW Wrinkles for an old domain. Cell. 2000;103:1001–1004. doi: 10.1016/s0092-8674(00)00203-8. [DOI] [PubMed] [Google Scholar]
- 26.Chen HI, Sudol M. The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc Natl Acad Sci U S A. 1995;92:7819–7823. doi: 10.1073/pnas.92.17.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tapia VE, Nicolaescu E, McDonald CB, Musi V, Oka T, Inayoshi Y, et al. Y65C missense mutation in the WW domain of the Golabi-Ito-Hall syndrome protein PQBP1 affects its binding activity and deregulates pre-mRNA splicing. J Biol Chem. 2010;285:19391–19401. doi: 10.1074/jbc.M109.084525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X, Milton CC, Humbert PO, Harvey KF. Transcriptional output of the Salvador/warts/hippo pathway is controlled in distinct fashions in Drosophila melanogaster and mammalian cell lines. Cancer Res. 2009;69:6033–6041. doi: 10.1158/0008-5472.CAN-08-4592. [DOI] [PubMed] [Google Scholar]
- 29.Oka T, Mazack V, Sudol M. Mst2 and Lats kinases regulate apoptotic function of Yes kinase-associated protein (YAP) J Biol Chem. 2008;283:27534–27546. doi: 10.1074/jbc.M804380200. [DOI] [PubMed] [Google Scholar]
- 30.Oka T, Schmitt AP, Sudol M. Opposing roles of angiomotin-like-1 and zona occludens-2 on pro-apoptotic function of YAP. Oncogene. 2012;31:128–134. doi: 10.1038/onc.2011.216. [DOI] [PubMed] [Google Scholar]
- 31.Chan SW, Lim CJ, Chong YF, Pobbati AV, Huang C, Hong W. Hippo pathway-independent restriction of TAZ and YAP by angiomotin. J Biol Chem. 2011;286:7018–7026. doi: 10.1074/jbc.C110.212621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao B, Li L, Lu Q, Wang LH, Liu CY, Lei Q, et al. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 2011;25:51–63. doi: 10.1101/gad.2000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paramasivam M, Sarkeshik A, Yates JR, 3rd, Fernandes MJ, McCollum D. Angiomotin family proteins are novel activators of the LATS2 kinase tumor suppressor. Mol Biol Cell. 2011;22:3725–3733. doi: 10.1091/mbc.E11-04-0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen HI, Einbond A, Kwak SJ, Linn H, Koepf E, Peterson S, et al. Characterization of the WW domain of human yes-associated protein and its polyproline-containing ligands. 1997;272:17070–17077. doi: 10.1074/jbc.272.27.17070. [DOI] [PubMed] [Google Scholar]
- 35.Zhang X, Milton CC, Poon CL, Hong W, Harvey KF. Wbp2 cooperates with Yorkie to drive tissue growth downstream of the Salvador-Warts-Hippo pathway. Cell Death Differ. 2011;18:1346–1355. doi: 10.1038/cdd.2011.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan SW, Lim CJ, Huang C, Chong YF, Gunaratne HJ, Hogue KA, et al. WW domain-mediated interaction with Wbp2 is important for the oncogenic property of TAZ. Oncogene. 2011;30:600–610. doi: 10.1038/onc.2010.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oh H, Irvine KD. In vivo analysis of Yorkie phosphorylation sites. Oncogene. 2009;28:1916–1927. doi: 10.1038/onc.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao B, Kim J, Ye X, Lai ZC, Guan KL. Both TEAD-binding and WW domains are required for the growth stimulation and oncogenic transformation activity of yes-associated protein. Cancer Res. 2009;69:1089–1098. doi: 10.1158/0008-5472.CAN-08-2997. [DOI] [PubMed] [Google Scholar]
- 39.Pires JR, Taha-Nejad F, Toepert F, Ast T, Hoffmuller U, Schneider-Mergener J, et al. Solution structures of the YAP65 WW domain and the variant L30 K in complex with the peptides GTPPPPYTVG, N-(n-octyl)-GPPPY and PLPPY and the application of peptide libraries reveal a minimal binding epitope. J Mol Biol. 2001;314:1147–1156. doi: 10.1006/jmbi.2000.5199. [DOI] [PubMed] [Google Scholar]
- 40.McDonald CB, McIntosh SK, Mikles DC, Bhat V, Deegan BJ, Seldeen KL, et al. Biophysical analysis of binding of WW domains of the YAP2 transcriptional regulator to PPXY motifs within WBP1 and WBP2 adaptors. Biochemistry. 2011;50:9616–9627. doi: 10.1021/bi201286p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan M, Tomlinson V, Lara R, Holliday D, Chelala C, Harada T, et al. Yes-associated protein (YAP) functions as a tumor suppressor in breast. Cell Death Differ. 2008;15:1752–1759. doi: 10.1038/cdd.2008.108. [DOI] [PubMed] [Google Scholar]
- 42.Nishikawa E, Osada H, Okazaki Y, Arima C, Tomida S, Tatematsu Y, et al. miR-375 is activated by ASH1 and inhibits YAP1 in a lineage-dependent manner in lung cancer. Cancer Res. 2011;71:6165–6173. doi: 10.1158/0008-5472.CAN-11-1020. [DOI] [PubMed] [Google Scholar]
- 43.Jager M, Nguyen H, Crane JC, Kelly JW, Gruebele M. The folding mechanism of a beta-sheet: the WW domain. J Mol Biol. 2001;311:373–393. doi: 10.1006/jmbi.2001.4873. [DOI] [PubMed] [Google Scholar]
- 44.Jager M, Dendle M, Kelly JW. Sequence determinants of thermodynamic stability in a WW domain – an all-beta-sheet protein. Protein Sci. 2009;18:1806–1813. doi: 10.1002/pro.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Russ WP, Lowery DM, Mishra P, Yaffe MB, Ranganathan R. Natural-like function in artificial WW domains. Nature. 2005;437:579–583. doi: 10.1038/nature03990. [DOI] [PubMed] [Google Scholar]
- 46.Shaw DE, Maragakis P, Lindorff-Larsen K, Piana S, Dror RO, Eastwood MP, et al. Atomic-level characterization of the structural dynamics of proteins. Science. 2010;330:341–346. doi: 10.1126/science.1187409. [DOI] [PubMed] [Google Scholar]
- 47.Piana S, Sarkar K, Lindorff-Larsen K, Guo M, Gruebele M, Shaw DE. Computational design and experimental testing of the fastest-folding beta-sheet protein. J Mol Biol. 2011;405:43–48. doi: 10.1016/j.jmb.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 48.Tonikian R, Zhang Y, Sazinsky SL, Currell B, Yeh JH, Reva B, et al. A specificity map for the PDZ domain family. PLoS Biol. 2008;6:e239. doi: 10.1371/journal.pbio.0060239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bao Y, Nakagawa K, Yang Z, Ikeda M, Withanage K, Ishigami-Yuasa M, et al. A cell-based assay to screen stimulators of the Hippo pathway reveals the inhibitory effect of dobutamine on the YAP-dependent gene transcription. J Biochem. 2011;150:199–208. doi: 10.1093/jb/mvr063. [DOI] [PubMed] [Google Scholar]
- 50.Mauviel A, Nallet-Staub F, Varelas X. Integrating developmental signals: a Hippo in the (path)way. Oncogene. 2011 doi: 10.1038/onc.2011.363. Epub. [DOI] [PubMed] [Google Scholar]
- 51.Levy D, Adamovich Y, Reuven N, Shaul Y. The Yes-associated protein 1 stabilizes p73 by preventing Itch-mediated ubiquitination of p73. Cell Death Differ. 2007;14:743–751. doi: 10.1038/sj.cdd.4402063. [DOI] [PubMed] [Google Scholar]
- 52.Aragon E, Goerner N, Zaromytiou AI, Xi Q, Escobedo A, Massague J, et al. A Smad action turnover switch operated by WW domain readers of a phosphoserine code. Genes Dev. 2011;25:1275–1288. doi: 10.1101/gad.2060811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang C, An J, Zhang P, Xu C, Gao K, Wu D, et al. The Nedd4-like Ubiquitin E3 Ligases Target Angimotin/P130 to Ubiquitin-Dependent Degradation. Biochm J. 2012 Mar 2; doi: 10.1042/BJ20111983. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 54.Kaelin WG., Jr The concept of synthetic lethality in the context of anti-cancer therapy. Nat Rev Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 55.Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 56.Carson M. Ribbons. Methods Enzymol. 1997;277:493–505. [PubMed] [Google Scholar]