Abstract
The gastrointestinal tract microbiota contributes to the development and differentiation of the mammalian immune system. The composition of the microbiota affects immune responses and affects susceptibility to infection by intestinal pathogens and development of allergic and inflammatory bowel diseases. Antibiotic administration, while facilitating clearance of targeted infections, also perturbs commensal microbial communities and decreases host resistance to antibiotic-resistant microbes. Here, we review recent advances that begin to define the interactions between complex intestinal microbial populations and the mammalian immune system and how this relation is perturbed by antibiotic administration. We further discuss how antibiotic-induced disruption of the microbiota and immune homeostasis can lead to disease and we review strategies to restore immune defenses during antibiotic administration.
Keywords: antibiotics, microbiota, immunity, infection, allergy
Commensal microbes inhabiting the intestinal tract
The gastrointestinal tract is inhabited by complex microbial populations, which, in aggregate, are referred to as the microbiota. The relation between the microbiota and the host is symbiotic, with the host providing a physical niche and nutrition to intestinal bacteria that in turn benefit the host by enhancing resistance to infection and facilitating the absorption of ingested food [1,2]. The intestinal microbiota contributes to the development and differentiation of the host immune system [3], and changes in the microbiota have been linked to infections, atopy, inflammatory bowel disease, diabetes and arthritis [4–8]. Massively parallel DNA sequencing platforms have revealed the complexity of bacterial populations inhabiting the gut, with thousands of different bacterial phylotypes belonging predominantly to the Firmicutes and Bacteroidetes phyla, and smaller populations belonging to the Proteobacteria, Actinobacteria, Verrucomicrobia or Fusobacteria phyla [9]. The microbiota of the large intestine is more dense and diverse than the microbiota of the small intestine [10,11] and the bacterial taxa in these two sites differ [11]. The microbial populations associated with the mucus layer differ from those found in the intestinal lumen [9,12]. Antibiotic administration perturbs the intestinal microbiota and affects immune defense against pathogens [4]. Deep 16S rDNA sequencing following antibiotic reatment has revealed dramatic and long-term changes to the intestinal microbiota that have implications for immune defense. Here, we review recent advances that begin to define the complex relation between the intestinal microbiota and the mammalian immune system and how this relation is perturbed by antibiotic administration.
Impact of the microbiota on the immune system
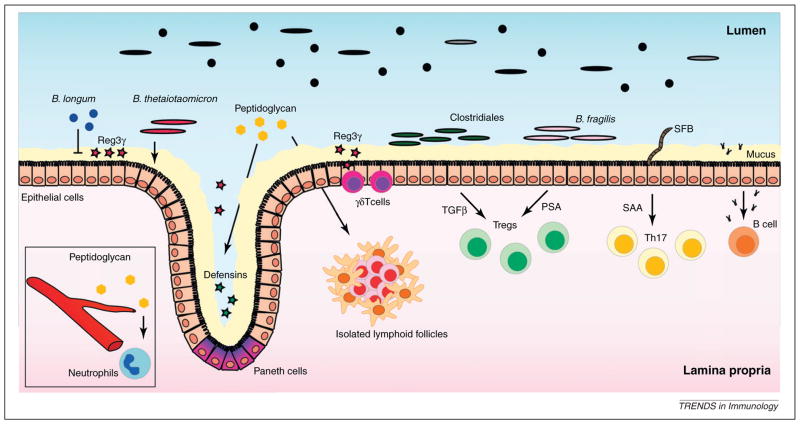
Colonization of the gastrointestinal tract by commensal microbes after birth has a profound effect on the immune system [3] (Figure 1), which influences the host’s ability to combat infections. Intestinal epithelial cells (IECs) are in close contact with commensal microbes and physically restrict commensal and pathogenic microorganisms to the gut lumen. In addition to this barrier function, IECs and Paneth cells, a specialized epithelial cell population at the base of intestinal crypts, secrete antimicrobial peptides (AMPs). Commensal bacteria in the intestinal lumen can stimulate the expression of AMPs, and IECs and Paneth cells from germ-free (GF) mice express lower levels of regenerating islet-derived protein 3 gamma (Reg3γ), a C-type lectin with bactericidal activity against Gram-positive bacteria [13]. The microbiota induces Reg3γ expression by activating Toll-like receptors (TLRs) in IECs and Paneth cells [14,15]. Mice deficient in myeloid differentiation primary response gene 88 (MyD88), an adaptor protein that mediates TLR signaling, have reduced expression of Reg3γ in the gut, rendering them more susceptible to intestinal Listeria monocytogenes infection [14]. Interestingly, different commensal microbes induce AMPs differently in IECs. Reconstitution of GF mice with the Gram-negative bacterium Bacteroides thetaiotaomicron induces whereas the Gram-positive bacterium Bifidobacterium longum inhibits Reg3γ expression [16]. Microbiota-mediated induction of Reg3γ reduces bacterial density in the ~50-μm mucus layer immediately adjacent to the intestinal epithelium [17]. Thus, by stimulating TLRs and inducing Reg3γ expression, the microbiota promotes its own segregation from the intestinal epithelium. In addition to TLRs, Paneth cells express NOD2, a cytosolic protein containing a nucleotide binding oligomerization domain (NOD) that facilitates innate immune signaling in response to cytosolic muramyl dipeptide (MDP), a peptidoglycan fragment [18]. NOD2 activation by microbiota-derived MDP induces Paneth cell expression of defensins, a subset of AMPs. Deletion of NOD2 diminishes expression of defensin related cryptdin (Defcr4) and Defcrrs10 and increases susceptibility to intestinal L. monocytogenes infection [19]. Activation of NOD1 receptor by peptidoglycan fragments derived from Gram-negative bacteria contributes to the development of isolated lymphoid follicles (ILFs), a tertiary lymphoid tissue in the small and large intestine [20]. NOD1 activation by microbiota-derived peptidoglycan absorbed from the gut also primes neutrophils, enhancing their ability to kill pathogenic bacteria [21]. Translocation of TLR ligands derived from the microbiota into the bloodstream contributes to the pathogenesis of some viral infections. For example, elevation of serum lipopolysaccharide (LPS) levels in HIV-infected patients correlates with innate and adaptive immune activation [22]. Low concentrations of LPS in serum induce emigration of monocytes from the bone marrow into the bloodstream [23]. It is unclear whether differences in the composition of the intestinal microbiota under homeostatic conditions correlate with differences in circulating LPS levels.
Figure 1.
Microbiota effects on immunity. Products derived from commensal microbes stimulate the production of the antimicrobial peptide Reg3γ by intestinal epithelial cells (IECs), Paneth cells and γδ T cells. Different microbial species have different effects on Reg3γ expression. Although Bifidobacterium longum diminishes intestinal Reg3γ levels, Bacteroides thetaiotaomicron increases Reg3γ expression. Peptidoglycan induces expression of defensins by Paneth cells and upon translocation into the circulation primes neutrophils, enhancing their ability to kill pathogenic bacteria. In addition, peptidoglycan is essential for the development of isolated lymphoid follicles. Mucus production is also induced by the intestinal microbiota. Intestinal T regulatory cell differentiation is induced by polysaccharide A of Bacteroides fragilis and by induction of transforming growth factor β secretion by Clostridiales spp. Segmented filamentous bacteria associate with IECs and induces serum amyloid A expression, which, in conjunction with dendritic cells, induces differentiation of T helper 17 cells. Commensal microbes induce the production of highly specific IgA by B cells.
Inflammasomes are cytoplasmic protein complexes that induce innate immune responses and inflammation by responding to pathogen-derived molecules and also host-derived products released in response to a range of tissue perturbations [24]. Upon recognition of an inflammatory stimulus, inflammasomes assemble and activate caspase-1, which cleaves and activates the proinflammatory cytokines pro-interleukin (IL)-1β and pro-IL-18 [24]. Different inflammasomes are composed of one of several NOD-like receptor proteins including, NLRP1, NLRP2, NLRP3, NLRP6 or NLRC4. Specific inflammasomes are activated by different ligands. NLRC4 inflammasomes are activated by bacterial flagellin and PhoP repressed gene J (PrgJ) in a process that is enabled by NLR family, apoptosis inhibitory protein (NAIP) accessory proteins. NAIP2 is required for NLRC4 inflammasome activation by PrgJ, and NAIP5 and NAIP6 activate NLRC4 in response to flagellin [25]. Inflammasome deficiency induces changes in the microbiota that promote intestinal inflammation [26]. Nlrp6−/− mice develop intestinal hyperplasia, inflammatory cell recruitment and exacerbation of dextran sodium sulfate (DSS)-induced colitis. This colitogenic phenotype is not directly the result of defective inflammasome activation but results from changes in the microbiota. Deep sequencing of 16S rDNA has demonstrated increased representation of bacteria belonging to the family Prevotellaceae and the phylum TM7. Wild-type mice exposed to the altered microbiota of Nlrp6−/− mice develop enhanced DSS-induced colitis. Mice deficient in apoptosis-associated speck-like protein containing a caspase-recruitment domain (ASC), caspase-1 and IL-18, proteins involved in the NLRP6 inflammasome signaling pathway, also developed a colitogenic microbiota, suggesting that loss of NLRP6 inflammasomes and diminished IL18 production drive gut commensal bacterial communities towards a more inflammation-inducing phenotype. Indeed, the severity of nonalcoholic steatohepatitis (NASH) is increased in mice lacking caspase-1, ASC, NLRP3, NLRP6 or IL-18, and also results from the establishment of an inflammation-inducing microbiota [27].
Innate lymphoid cells (ILCs), including natural killer (NK)-like NKp46+ and lymphoid tissue inducer (LTi) cells, contribute to gut immune defense by producing IL22 [28–30], a cytokine that promotes IEC growth and the expression of Reg3γ [31]. ILCs depend on aryl hydrocarbon (Ah) receptor signaling for expansion, maintenance and functionality [32–34]. Ahr−/− mice have decreased ILC numbers and markedly reduced IL22 production [32–34]. Ah receptor ligands are present in some vegetables and can be produced by intestinal microbes, but the role of diet and microbiota in the establishment and maintenance of ILC populations remains controversial. For example, similar numbers of NK-like NKp46+ cells are present in GF mice and mice housed in a conventional barrier facility [32], and mice receiving an Ah-receptor-deficient diet have reduced intestinal ILC numbers, suggesting that diet plays a greater role than the microbiota in the generation of ILCs [34]. Other studies, however, have shown reduced frequencies and IL-22 production by NK-like NKp46+ cells in GF mice [28,30], and it has been demonstrated that intestinal bacteria produce Ah receptor ligands by metabolizing tryptophan [35].
Intestinal T cell numbers are also influenced by the intestinal microbiota. T cell receptor (TCR)γδ T cells are present in the intraepithelial compartment of the gut and can limit bacterial invasion of the intestinal mucosa [36]. γδ T cells depend on dietary Ah receptor ligands for maintenance and not commensal microbes [36,37]. However, members of the epithelium-associated microbiota such as Escherichia coli are necessary for γδ T cell-mediated induction of AMP expression [36] and restriction of bacterial penetration.
CD4+ TCRαβ T helper (Th) cells, including Th1, Th2 and Th17 cells, are also influenced by the microbiota. A single bacterial species, Bacteroides fragilis, can restore the im-balanced Th1/Th2 cell ratio in GF mice [38]. Th17 cells contribute to defense against extracellular pathogens by producing cytokines IL-17 and IL-22, which induce antibody class switching in B cells [39] and Reg3γ production by epithelial cells [31], respectively. Th17 cells contribute to the development of inflammatory disorders such as rheumatoid arthritis, multiple sclerosis and inflammatory bowel disease [40]. The development of Th17 cells depends on commensal microbes, and the frequency of Th17 cells is markedly reduced in GF mice [41,42]. Colonization by a single bacterial species, segmented filamentous bacteria (SFB), is sufficient for reconstitution of Th17 populations in the small intestine [43]. SFBs closely associate with IECs and induce serum amyloid A expression, which, in conjunction with dendritic cells (DCs), induce differentiation of Th17 cells [43]. Intestinal bacteria can also induce Th17 differentiation by producing high levels of ATP [41]. Certain intestinal bacteria induce anti-inflammatory T regulatory (Treg) cells. Although polysaccharide A from B. fragilis can direct the differentiation of CD4+ T cells into Foxp3+ Treg cells [44], recent studies have shown that bacteria belonging to Clostridiales clusters IV and XIVa induce accumulation of Treg cells in the colonic lamina propria (LP) by increasing production of transforming growth factor (TGF)β, a critical regulator of Treg cell development [45]. Some colonic Treg cells are specific to microbiota-derived antigens. Lathrop et al. have demonstrated that the TCR repertoire of colonic Foxp3+ Treg cells is distinct from that of effector/memory and naïve Foxp3− T cells or from Foxp3+ T cells found in other organs, suggesting that colonic Treg cell populations are shaped by intestine-derived antigens [46]. To determine if colonic Treg cells are specific for antigens derived from the microbiota, colonic Treg TCRs were expressed in hybridomas. Five out of eight hybridomas were reactive with intestinal contents of conventionally housed but not GF mice, and two reacted exclusively with specific members of the intestinal microbiota. In contrast to Treg TCRs found at other peripheral locations, transgenic expression of cloned colonic Treg TCRs did not induce Foxp3+ thymocytes, suggesting that colonic Treg cells develop extrathymically in response to bacterial antigens.
The microbiota also induces antibody responses. Hapfelmeier et al. have studied IgA responses induced by commensal bacteria using reversible colonization of GF mice with mutant E. coli that cannot divide or persist in vivo. Using this approach, they demonstrated that live but not dead E. coli induced highly specific IgA responses that persisted beyond the period of colonization [47]. Persistence of IgA responses to specific bacteria was limited, however, when mice were colonized with other commensal bacterial species, suggesting that the mucosal immune system modifies IgA responses to commensals in response to microbial shifts.
Antibiotics alter the composition of the microbiota
Intestinal commensal microbes influence the immune system and changes in the microbiota composition can affect immune responses. In modern societies, widespread antibiotic administration is probably a major factor contributing to changes in the mucosal microbiota. A rigorous study of healthy human volunteers before, during and after a 5-day course of oral ciprofloxacin demonstrated decreased diversity, richness and evenness of the fecal microbiota during antibiotic administration [48]. The abundance of roughly one third of the bacterial taxa changed during ciprofloxacin administration. Although the microbiota largely returned to pretreatment community composition at 4 weeks following ciprofloxacin treatment, several bacterial taxa failed to recover, indicating that changes to the microbiota can persist following a short course of oral antibiotics. Another study of the human fecal microbiota demonstrated that a 7-day course of clindamycin reduced diversity within the Bacteroides genus and led to increased frequencies of highly antibiotic-resistant clones. The Bacteroides population did not return to its original composition for up to 2 years after clindamycin treatment [49]. Along similar lines, mouse models have shown that treatment with clindamycin, ampicillin, cefoperazone or vancomycin induces long-lasting changes in the microbiota that persist after cessation of antibiotic treatment [11,50,51]. The microbial populations of the intestine are interdependent, therefore, treatment with antibiotics specific for one class of bacteria may indirectly lead to the depletion of antibiotic-resistant bacteria. For example, administration of vancomycin, an antibiotic that kills Gram-positive bacteria, also depletes the Gram-negative phylum Bacteroidetes [11,52] from the gut of treated mice. In this circumstance, however, it is possible that nonabsorbable antibiotics such as vancomycin achieve high enough concentrations in the gut to inhibit bacteria belonging to the Bacteroidetes phylum. Interestingly, 2 weeks after ceasing vancomycin treatment, mice recovered an intestinal microbiota with higher abundances of Enterococcus spp., Clostridium spp. and Enterobacteriaceae [11]. Similarly, Enterococcus spp. and Enterobacteriaceae also increase in newborn humans treated with oral cefalexin, a broad-spectrum antibiotic [53]. Increases in these bacterial populations is of potential clinical importance because they are major causes of hospital-acquired infections.
Antibiotic-induced changes in the microbiota impact immunity
Depletion of commensal microbes and changing the microbiota composition by antibiotic administration affects intestinal immune defenses (Table 1). Amoxicillin, a frequently used antibiotic, affects gene expression in the proximal and distal small intestine and colon of suckling rats [54]. Amoxicillin treatment essentially eradicates Lactobacillus spp. in the small intestine and reduces the density of colonic anaerobic and aerobic bacteria. Amoxicillin-induced microbiota changes reduce expression of MHC class I and II genes in the small and large intestine, reduce AMP expression in the proximal intestine, and increase mast cell protease expression in the distal small intestine. Treatment of mice with metronidazole, neomycin and vancomycin diminishes expression of the Reg3γ in the distal ileum [4]. Treatment with metronidazole, an antibiotic that kills anaerobic bacteria, increases Reg3γ and IL-25 expression in the murine colon [55]. Metronidazole treatment results in higher numbers of macrophages and NK cells in the colonic LP and diminishes intestinal expression of Muc2, the major component of the mucin layer [55]. Thinning of the mucus layer may increase contact between epithelial cells and the microbiota, thereby enhancing innate immune stimulation and elevating the inflammatory tone of the intestine. Metronidazole treatment does not diminish bacterial density in the intestine but does result in depletion of obligate anaerobic Bacteroidales and Clostridium coccoides bacteria and expansion of aerotolerant bacteria populations, including lactobacilli. How changes in these microbial communities affect Muc2 expression remains to be elucidated.
Table 1.
Antibiotic-induced changes in the microbiota impact immunity.
| Antibiotic | Effect on the microbiota | Effect on immunity | References |
|---|---|---|---|
| Amoxicillin |
Lactobacillus spp. depletion in SI ↓aerobic and anaerobic bacterial numbers in the colon |
↓ MHC I and MHC II expression in SI and LI ↓AMPs expression in SI ↑mast cell proteases expression in SI |
[54] |
| Metronidazole, neomycin and vancomycin | ↓ bacterial numbers in SI and LI Multiple effects on composition, including: ↓ Bacteroidetes ↑ Enterobacteriaceae |
↓ Reg3γ expression in SI | [4,11] |
| Metronidazole | Bacteroidales and Clostridium coccoides depletion ↑ Lactobacilli |
↑ Reg3γ and IL-25 expression in colon ↑ numbers of macrophages and NK cells in colon ↓ mucus |
[55] |
| Colistin | ND (Gram-negative spectrum)a | ↓ numbers of ILFs | [20] |
| Ampicillin, neomycin, metronidazole, vancomycin | Microbiota depletion ↓ peptidoglycan levels in serum |
↓neutrophil-mediated killing of pathogenic bacteria ↓Reg3γ expression by γδ T cells ↓pro-IL1β, pro-IL18, NLRP3 |
[21] [36] [65] |
| Amoxicillin/clavulanate | ND | ↓ IgG serum levels | [56] |
| Ampicillin, gentamicin, metronidazole, neomycin, vancomycin | ↓ bacterial numbers in LI Multiple effects on composition, including: ↓ luminal Firmicutes in LI ↓ mucosal associate Lactobacillus in LI |
↓ IFNγ and IL-17 production by CD4+ T cells in SI ↑ IgE serum levels ↑ basophils in blood |
[5] [12] |
| Vancomycin | ↓ Gram-positive bacteria ↑ Enterobacteriaceae |
↓ Treg cells in colon ↓ Th17 in SI ↓ ILFs to a lesser extent than colistin |
[20,42,45] |
LI, large intestine; ND, not determined; SI, small intestine.
ND (Not determined but only Gram-negative bacteria are sensitive to colistin).
Development of intestinal lymphoid tissues is affected by antibiotic treatment. Mice treated from birth with colistin or vancomycin, which kill Gram-negative or Gram-positive bacteria, respectively, have reduced numbers of ILFs [20]. Consistent with the role of Gram-negative peptidoglycan in the development of ILFs by NOD1 stimulation, colistin has a greater effect on ILF numbers than vancomycin has [20]. Depletion of the microbiota with a cocktail of antibiotics containing ampicillin, neomycin, metronidazole and vancomycin diminishes circulating peptidoglycan levels, thereby reducing neutrophil-mediated killing of Streptococcus pneumoniae [21]. Intraperitoneal administration of the NOD1 ligand MurNAcTriDAP to antibiotic-treated mice enhanced neutrophil killing capacity [21]. In another study, administration of amoxicillin/clavulanate diminished IgG serum levels in humans without affecting IgA or IgM levels [56].
T cell function is also affected by antibiotic therapy. γδ T cells in mice that have been treated with ampicillin, vancomycin, neomycin and metronidazole express less Reg3γ [36]. A similar cocktail of antibiotics containing ampicillin, gentamycin, metronidazole, neomycin and vancomycin reduces the number of bacteria in the large intestine by 10-fold and reduces luminal bacteria of the Firmicutes phylum and mucosa-associated Lactobacillus species [57]. Antibiotic administration also reduced production of interferon (IFN)γ and IL-17 cytokines by CD4+ T lymphocytes in the small intestine [57]. Consistent with the role of Gram-positive Clostridiales spp. in driving colonic Treg differentiation, vancomycin treatment but not polymyxin B, which kills Gram-negative bacteria, reduces the numbers of Treg cells in the colon LP [45]. Along similar lines, induction of Th17 cells by Gram-positive SFB is impaired by vancomycin but not by neomycin plus metronidazole treatment [42].
Antibiotics, immunity and disease
Antibiotics, by changing the composition of the microbiota and altering the development of the immune system, can predispose the host to infections. Administration of streptomycin alters the gut microbiota and renders mice susceptible to infection with Salmonella spp. [52,58,59]. Recovery of the complex microbiota eliminates Salmonella from the murine intestinal tract by undefined mechanisms that are independent of IgA production [58]. It is possible that the microbiota stimulates the host innate immune system to eliminate Salmonella. Alternatively, as suggested by older studies, the microbiota may suppress Salmonella infection directly by producing inhibitory molecules such as short chain fatty acids [60].
Disruption of the microbiota with metronidazole, neomycin and vancomycin enhances vancomycin-resistant Enterococcus (VRE) colonization of the intestinal tract by diminishing the expression of Reg3γ in IECs [4]. The diminished innate immune tone during antibiotic treatment promotes VRE growth in the small and large intestine, leading to a state of intestinal domination in which > 97% of the microbiota is VRE [11].
Clostridium difficile is the principal cause of hospital-acquired diarrhea in the United States (reviewed in [61]). C. difficile produces spores that can persist and contaminate hospital environments, and, upon ingestion, infect the gut when the intestinal microbiota is altered or absent. Consequently, C. difficile infection is almost always associated with administration of antibiotics such as clindamycin, cephalosporins or fluoroquinolones, and the microbiota of patients with C. difficile infection has decreased diversity [62]. A mouse model has been developed to investigate how alterations in the microbiota promote C. difficile infection. Administration of clindamycin to mice followed by C. difficile administration results in a state of chronic infection and shedding of C. difficile spores [63]. A single dose of clindamycin produces long-lasting changes in the small and large intestinal microbiota, with a loss of approximately 90% of the cecal microbial taxa [51]. Clindamycin-induced changes in the microbiota enable C. difficile to infect the gut and induce diarrhea and colitis, and mice remain susceptible to C. difficile infection for up to 10 days following clindamycin treatment. Mice surviving acute infection remain highly colonized with C. difficile, with persistent colitis for up to 28 days. In other studies, however, mice only remain highly infected with C. difficile during clindamycin treatment [63] and, with recovery of the microbiota upon cessation of clindamycin, C. difficile is almost completely cleared from the gastrointestinal tract. It is likely that different results with respect to C. difficile persistence can be accounted for by inter institutional differences in animal husbandry and microbiota composition. Microbiota recovery after ampicillin treatment can differ dramatically in mice that are separately housed, and it is possible that such differences can influence susceptibility to infection [11]. Although it is likely that the microbiota directly suppresses C. difficile growth in the gut, recent studies have demonstrated that innate immune defenses also provide protection against C. difficile infection. MyD88-deficient mice, for example, have more pronounced intestinal disease, including increased hyperplasia, cellular infiltrate and edema, and higher mortality following C. difficile infection [63]. NOD1-deficient mice are also more susceptible to C. difficile infection, with impaired neutrophil recruitment to the colon and increased bacterial translocation across the epithelium [64].
Changes in the microbiota following antibiotic treatment also affect immune responses against viral infection. The microbiota induces expression of pro-IL-1β and pro-IL-18, two pro-cytokines that are processed by inflammasomes and that contribute to defense against influenza virus [65]. Antibiotic treatment diminishes expression of the two pro-cytokines, impairing responses against influenza virus. Neomycin, an antibiotic that predominantly kills Gram-negative bacteria, is most effective at impairing anti-influenza responses. Another study has shown that antibiotic treatment enhances immune responses against the mouse mammary tumor virus (MMTV) [66], which is transmitted from the mother to offspring by ingestion of milk. Under normal circumstances, viral particles bind microbiota-derived LPS and trigger TLR4 in the spleen, inducing anti-inflammatory IL-10 in an IL-6-dependent manner, thereby evade immune responses. Antibiotic-treated mothers do not transmit the virus to their offspring because of diminished viral LPS binding.
An increase of allergic disorders in humans, including allergic asthma, has been observed in recent decades [67]. The ‘hygiene hypothesis’ proposes that cleaner lifestyles with more limited exposure to environmental microbes alter immune maturation and predispose to allergy and asthma [68]. The prevalence of Helicobacter pylori, a bacterial species that is commonly found in the stomach in humans, has decreased in developed countries as a result of antibiotic usage. The presence of H. pylori is inversely associated with the development of childhood asthma [69]. H. pylori colonization induces accumulation of lymphoid cells in the gastric LP, including Treg and Th cells, which are essentially absent in H. pylori-negative persons, suggesting that H. pylori activates the immune system in a way that prevents immune disorders such as asthma. Additional research is necessary to determine if this hypothesis is correct and to identify mechanisms by which H. pylori infection may prevent asthma. Several studies using mice have shown that antibiotic-mediated alterations of the microbiota can enhance allergic disorders. Administration of kanamycin to NC/Nga mice, a mouse model of atopic dermatitis, increased IgE levels and exacerbated allergic symptoms 8 weeks after antibiotic treatment [70]. Antibiotic-treated mice can also be more susceptible to anaphylactic shock following oral administration of a food allergen. TLR4-deficient mice that have not received antibiotics also develop anaphylaxis that is abrogated by CpG-mediated TLR9 activation, suggesting that TLR stimulation by the microbiota can suppress the development of food allergies [71]. Administration of vancomycin, but not streptomycin, to neonatal mice increases the severity of allergic asthma. Vancomycin treatment diminishes intestinal microbial diversity and changes the microbiota composition to a greater extent than streptomycin, suggesting that disruption of the intestinal microbiota by specific antibiotics correlates with increased susceptibility to allergic disorders [72]. Another recent study has demonstrated that antibiotic-induced changes in the microbiota predispose to allergic disorders by affecting the development of basophils [5]. In this study, Hill and coworkers have demonstrated that administration of an antibiotic cocktail containing ampicillin, gentamicin, metronidazole, neomycin and vancomycin increases basophil-mediated Th2 cell responses and allergic inflammation in mice. Antibiotic-induced alterations in the microbiota increase serum IgE levels, which drive an increase in the frequency and number of basophils in the blood. IgE increases expression of CD123 in bone-marrow-resident basophil precursors (BaPs), thereby enhancing basophil development. BaPs from antibiotic-treated mice express higher levels of CD123 and, in the presence of IL-3, generate higher numbers of mature basophils than BaPs from untreated mice. MyD88 signaling in B cells limits serum IgE levels and circulating basophil populations, suggesting that products of the microbiota, by inhibiting IgE production by B cells, can prevent allergic disorders.
Antibiotic-induced changes in the microbiota may also have an impact on cancer. H. pylori infection increases the risk of noncardia gastric cancer [73]. By contrast, a recent study suggests that the risk of developing esophageal adenocarcinoma inversely correlates with the presence of H. pylori, especially cagA+ strains [74].
Counteracting the negative effect of antibiotics on immunity
As described above, antibiotics can have a negative impact on the microbiota, immunity and health. Avoidance of antibiotics, however, is often not feasible, because many infections can only be survived with antibiotic treatment. For this reason, several strategies have been tested to counteract the negative effect of antibiotics on the microbiota and immune responses. One strategy is to provide probiotics, usually live bacteria, to supplement antibiotic-induced deficits in the microbiota. In studies done with streptomycin-treated mice, oral administration of anaerobic microbiota cultures partially prevents Salmonella infection [59]. Several studies in humans have investigated probiotics to prevent and treat infectious disease (for review, see [75]). Administration of a mixture of Streptococcus thermophilus, Lactobacillus casei and Lactobacillus bulgaricos prevents intestinal disease caused by C. difficile [76]. An alternative to live probiotics is the administration of bacterial ligands to boost the immune tone during antibiotic therapy. Impaired innate immune defense against the parasite Encephalitozoon cuniculi following antibiotic administration is corrected by oral administration of DNA extracted from commensal bacteria [77]. Administration of flagellin or oral LPS to antibiotic-treated mice restricts VRE intestinal colonization by inducing the expression of Reg3γ, which can kill VRE [4,78]. Flagellin administration induces production of IL-23 by CD103+CD11b+ LP DCs, which drives IL-22-dependent Reg3γ production [79]. Flagellin administration also diminishes C. difficile intestinal colonization after clindamycin treatment [80]. Although, these strategies correct some of the negative effects of antibiotic administration, ongoing studies will probably provide a more detailed understanding of the bacterial species that communicate with the immune system and the molecular triggers that enhance intestinal resistance to infection. Identifying the microbes and their products that sustain optimal immune defense on mucosal surfaces will provide exciting new opportunities to limit infections and enhance clearance of microbial pathogens.
Concluding remarks
Commensal microbial species that inhabit the intestinal tract contribute to the development, maintenance and function of the immune system. Microbiota effects on immunity range from enhancement of AMP expression by IECs to induction of intestinal ILFs, antibody production, and T cell differentiation. Components of the microbiota that translocate into the circulation can also affect immune cell populations located in sites beyond the intestine. The plethora of microbial species and components has a wide range of effects on the immune system. Although some commensal bacteria are anti-inflammatory, other bacterial species promote the differentiation of proinflammatory immune cells or induce the expression of proinflammatory cytokines. Antibiotic administration can alter intestinal commensal bacterial populations and, as a consequence, modify immune defenses leading, in some cases, to detrimental effects on health. Viral and bacterial infections or allergies can be promoted by antibiotic-induced changes in the microbiota. Some of the alterations in the microbiota and immunity produced by antibiotic usage can be restored with the administration of probiotic bacteria or bacterial ligands of innate immune receptors. A better understanding of how specific commensal microbes shape the immune system will lead to new approaches to counteract the negative effects of antibiotic treatment.
Acknowledgments
C.U. is funded by the grant SAF2011-29458 from the Spanish MICINN and the Marie-Curie Career Integration Grant PCIG09-GA-2011-293894. E.G.P. is funded by the NIH grants RO1-AI042135 and PO1-CA023766.
References
- 1.Hooper LV, et al. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- 2.Vollaard EJ, Clasener HA. Colonization resistance. Antimicrob Agents Chemother. 1994;38:409–414. doi: 10.1128/aac.38.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hill DA, Artis D. Intestinal bacteria and the regulation of immune cell homeostasis. Annu Rev Immunol. 2010;28:623–667. doi: 10.1146/annurev-immunol-030409-101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brandl K, et al. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455:804–807. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hill DA, et al. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat Med. 2012;18:538–546. doi: 10.1038/nm.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009;361:2066–2078. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wen L, et al. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu HJ, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eckburg PB. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garner CD, et al. Perturbation of the small intestine microbial ecology by streptomycin alters pathology in a Salmonella enterica serovar Typhimurium murine model of infection. Infect Immun. 2009;77:2691–2702. doi: 10.1128/IAI.01570-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ubeda C, et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest. 2010;120:4332–4341. doi: 10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hill DA, et al. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 2009;3:148–158. doi: 10.1038/mi.2009.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cash HL. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brandl K, et al. MyD88-mediated signals induce the bactericidal lectin RegIII gamma and protect mice against intestinal Listeria monocytogenes infection. J Exp Med. 2007;204:1891–1900. doi: 10.1084/jem.20070563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vaishnava S, et al. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci USA. 2008;105:20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sonnenburg JL, et al. Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol. 2006;4:e413. doi: 10.1371/journal.pbio.0040413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaishnava S, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Artis D. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat Rev Immunol. 2008;8:411–420. doi: 10.1038/nri2316. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi KS, et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 20.Bouskra D, et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 2008;456:507–510. doi: 10.1038/nature07450. [DOI] [PubMed] [Google Scholar]
- 21.Clarke TB, et al. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010;16:228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brenchley JM, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 23.Shi C, et al. Bone marrow mesenchymal stem and progenitor cells induce monocyte emigration in response to circulating toll-like receptor ligands. Immunity. 2011;34:590–601. doi: 10.1016/j.immuni.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strowig T, et al. Inflammasomes in health and disease. Nature. 2012;481:278–286. doi: 10.1038/nature10759. [DOI] [PubMed] [Google Scholar]
- 25.Kofoed EM, Vance RE. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature. 2011;477:592–595. doi: 10.1038/nature10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elinav E, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henao-Mejia J, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–185. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanos SL, et al. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat Immunol. 2009;10:83–91. doi: 10.1038/ni.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luci C, et al. Influence of the transcription factor RORgammat on the development of NKp46+ cell populations in gut and skin. Nat Immunol. 2009;10:75–82. doi: 10.1038/ni.1681. [DOI] [PubMed] [Google Scholar]
- 30.Satoh-Takayama N, et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008;29:958–970. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 31.Bird L. Host response: antimicrobial function for IL-22. Nat Rev Microbiol. 2008;6:259. [Google Scholar]
- 32.Lee JS, et al. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat Immunol. 2011;13:144–151. doi: 10.1038/ni.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiu J, et al. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity. 2012;36:92–104. doi: 10.1016/j.immuni.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kiss EA, et al. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science. 2011;334:1561–1565. doi: 10.1126/science.1214914. [DOI] [PubMed] [Google Scholar]
- 35.Perdew GH, Babbs CF. Production of Ah receptor ligands in rat fecal suspensions containing tryptophan or indole-3-carbinol. Nutr cancer. 1991;16:209–218. doi: 10.1080/01635589109514159. [DOI] [PubMed] [Google Scholar]
- 36.Ismail AS, et al. Gammadelta intraepithelial lymphocytes are essential mediators of host-microbial homeostasis at the intestinal mucosal surface. Proc Natl Acad Sci USA. 2011;108:8743–8748. doi: 10.1073/pnas.1019574108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y, et al. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell. 2011;147:629–640. doi: 10.1016/j.cell.2011.09.025. [DOI] [PubMed] [Google Scholar]
- 38.Mazmanian S, et al. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 39.Mitsdoerffer M, et al. Proinflammatory T helper type 17 cells are effective B-cell helpers. Proc Natl Acad Sci USA. 2010;107:14292–14297. doi: 10.1073/pnas.1009234107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tesmer LA, et al. Th17 cells in human disease. Immunol Rev. 2008;223:87–113. doi: 10.1111/j.1600-065X.2008.00628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Atarashi K, et al. ATP drives lamina propria TH17 cell differentiation. Nature. 2008;455:808–812. doi: 10.1038/nature07240. [DOI] [PubMed] [Google Scholar]
- 42.Ivanov II, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lathrop SK, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hapfelmeier S, et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science. 2010;328:1705–1709. doi: 10.1126/science.1188454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dethlefsen L, et al. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jernberg C, et al. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 2007;1:56–66. doi: 10.1038/ismej.2007.3. [DOI] [PubMed] [Google Scholar]
- 50.Antonopoulos DA, et al. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun. 2009;77:2367–2375. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buffie CG, et al. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect Immun. 2012;80:62–73. doi: 10.1128/IAI.05496-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sekirov I, et al. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect Immun. 2008;76:4726–4736. doi: 10.1128/IAI.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanaka S, et al. Influence of antibiotic exposure in the early postnatal period on the development of intestinal microbiota. FEMS Immunol Med Microbiol. 2009;56:80–87. doi: 10.1111/j.1574-695X.2009.00553.x. [DOI] [PubMed] [Google Scholar]
- 54.Schumann A. Neonatal antibiotic treatment alters gastrointestinal tract developmental gene expression and intestinal barrier transcriptome. Physiol Genomics. 2005;23:235–245. doi: 10.1152/physiolgenomics.00057.2005. [DOI] [PubMed] [Google Scholar]
- 55.Wlodarska M, et al. Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated Citrobacter rodentium-induced colitis. Infect Immun. 2011;79:1536–1545. doi: 10.1128/IAI.01104-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dufour V, et al. Effects of a short-course of amoxicillin/clavulanic acid on systemic and mucosal immunity in healthy adult humans. Int Immunopharmacol. 2005;5:917–928. doi: 10.1016/j.intimp.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 57.Hill DA, et al. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 2010;3:148–158. doi: 10.1038/mi.2009.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Endt K, et al. The microbiota mediates pathogen clearance from the gut lumen after non-typhoidal Salmonella diarrhea. PLoS Pathog. 2010;6:e1001097. doi: 10.1371/journal.ppat.1001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miller C, Bohnhoff M. Changes in the mouse’s enteric microflora associated with enhanced susceptibility to Salmonella infection following streptomycin treatment. J Infect Dis. 1963;113:59–66. doi: 10.1093/infdis/113.1.59. [DOI] [PubMed] [Google Scholar]
- 60.Bohnhoff M, Miller C. Resistance of the mouse’s intestinal tract to experimental Salmonella infection. J Exp Med. 1964;120:805–816. doi: 10.1084/jem.120.5.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rupnik M, et al. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol. 2009;7:526–536. doi: 10.1038/nrmicro2164. [DOI] [PubMed] [Google Scholar]
- 62.Chang JY, et al. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis. 2008;197:435–438. doi: 10.1086/525047. [DOI] [PubMed] [Google Scholar]
- 63.Lawley TD, et al. Antibiotic treatment of Clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect Immun. 2009;77:3661–3669. doi: 10.1128/IAI.00558-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hasegawa M, et al. Nucleotide-binding oligomerization domain 1 mediates recognition of Clostridium difficile and induces neutrophil recruitment and protection against the pathogen. J Immunol. 2011;186:4872–4880. doi: 10.4049/jimmunol.1003761. [DOI] [PubMed] [Google Scholar]
- 65.Ichinohe T, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci USA. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kane M, et al. Successful transmission of a retrovirus depends on the commensal microbiota. Science. 2011;334:245–249. doi: 10.1126/science.1210718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eder W, et al. The asthma epidemic. N Engl J Med. 2006;355:2226–2235. doi: 10.1056/NEJMra054308. [DOI] [PubMed] [Google Scholar]
- 68.Strachan DP. Hay fever, hygiene, and household size. BMJ. 1989;299:1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen Y, Blaser MJ. Helicobacter pylori colonization is inversely associated with childhood asthma. J Infect Dis. 2008;198:553–560. doi: 10.1086/590158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Watanabe J, et al. Administration of antibiotics during infancy promoted the development of atopic dermatitis-like skin lesions in NC/Nga mice. Biosci Biotechnol Biochem. 2010;74:358–363. doi: 10.1271/bbb.90709. [DOI] [PubMed] [Google Scholar]
- 71.Bashir MEH, et al. Toll-like receptor 4 signaling by intestinal microbes influences susceptibility to food allergy. J Immunol. 2004;172:6978–6987. doi: 10.4049/jimmunol.172.11.6978. [DOI] [PubMed] [Google Scholar]
- 72.Russell SL, et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 2012;13:440–447. doi: 10.1038/embor.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cover TL, Blaser MJ. Helicobacter pylori in health and disease. Gastroenterology. 2009;136:1863–1873. doi: 10.1053/j.gastro.2009.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Islami F, Kamangar F. Helicobacter pylori and esophageal cancer risk: a meta-analysis. Cancer Prev Res (Phila) 2008;1:329–338. doi: 10.1158/1940-6207.CAPR-08-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wolvers D, et al. Guidance for substantiating the evidence for beneficial effects of probiotics: prevention and management of infections by probiotics. J Nutr. 2010;140:698S–712S. doi: 10.3945/jn.109.113753. [DOI] [PubMed] [Google Scholar]
- 76.Hickson M, et al. Use of probiotic Lactobacillus preparation to prevent diarrhoea associated with antibiotics: randomised double blind placebo controlled trial. BMJ. 2007;335:80. doi: 10.1136/bmj.39231.599815.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hall JA, et al. Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity. 2008;29:637–649. doi: 10.1016/j.immuni.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kinnebrew MA, et al. Bacterial flagellin stimulates Toll-like receptor 5-dependent defense against vancomycin-resistant Enterococcus infection. J Infect Dis. 2010;201:534–543. doi: 10.1086/650203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kinnebrew MA, et al. Interleukin 23 production by intestinal CD103(+)CD11b(+) dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity. 2012;36:276–287. doi: 10.1016/j.immuni.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jarchum I, et al. Toll-like receptor 5 stimulation protects mice from acute Clostridium difficile colitis. Infect Immun. 2011;79:1498–1503. doi: 10.1128/IAI.01196-10. [DOI] [PMC free article] [PubMed] [Google Scholar]