Abstract
OBJECTIVE
Epidemiological and clinical studies suggest that rates of antisocial behavior, depression, and impulsive substance use are increased among individuals diagnosed with alcohol dependence relative to those who are not. Thus, the present study conducted genome-wide linkage scans of antisocial behavior, depression, and impulsive substance use in the University of California at San Francisco Family Alcoholism Study.
METHODS
Antisocial behavior, depressive symptoms, and impulsive substance use were assessed using three scales from the MMPI-2, the Antisocial Practices content scale (ASP), the Depression content scale (DEP), and the revised MacAndrew Alcoholism scale (MAC-R). Linkage analyses were conducted using a variance components approach.
RESULTS
Suggestive evidence of linkage to three genomic regions independent of alcohol and cannabis dependence diagnostic status was observed: the ASP scale showed evidence of linkage to chromosome 13 at 11 cM, the MAC-R scale showed evidence of linkage to chromosome 15 at 47 cM, and all 3 scales showed evidence of linkage to chromosome 17 at 57–58 cM.
CONCLUSIONS
Each of these regions has shown prior evidence of linkage and association to substance dependence as well as other psychiatric disorders such as mood and anxiety disorders, ADHD, and schizophrenia thus suggesting potentially broad relations between these regions and psychopathology.
Keywords: Genetic Linkage, Depressive Symptoms, Antisocial Practices, Substance Use Disorders, MMPI-2
Epidemiological (e.g., Grant and Harford, 1995; Grant et al., 2004; Kessler et al., 1997; Regier et al., 1990; Stinson et al., 2005) and clinical studies (e.g., Babor et al., 1992; Cloninger, 1987b; Hesselbrock et al., 1985; Morey and Blashfield, 1981; Ross et al., 1988) suggest that rates of depression, antisocial personality disorder, and illicit substance use are elevated among individuals diagnosed with alcohol dependence relative to individuals without this diagnosis. Given these high rates of co-occurrence, samples selected for alcohol dependence can be expected to be enriched for depression, antisocial behavior, and other substance use disorders. Further, each of these associated phenotypes show substantial genetic influences in their etiology as demonstrated by numerous twin studies (depression - Cadoret et al., 1985; Kendler et al., 1994, antisocial behavior - Lyons et al., 1995; Slutske et al., 1997, substance misuse - Kendler et al., 2003a; Tsuang et al., 1996). Thus, it is reasonable to utilize populations selected for alcohol dependence to identify genetic loci that contribute to the development of depression, antisocial behavior, and substance use phenotypes.
Such an approach is not without risks, however. Twin studies suggest that a shared genetic etiology is, in part, responsible for the phenotypic correlation between antisocial behavior and alcohol dependence (Hicks et al., 2004; Kendler et al., 2003b). Similarly, shared genetic influences have been reported to underlie the phenotypic correlations between mood and anxiety disorders and alcohol dependence (Kendler et al., 1993; Khan et al., 2005; Wender et al., 1986), and shared genetic influences have also been reported to underlie the phenotypic correlations between illicit substance use and alcohol dependence (Han et al., 1999; Young et al., 2006). Although such studies suggest that significant overlap exists in the genetic risk associated with these phenotypes, they also demonstrate that unique genetic influences contribute to their development. As a result, studies investigating the genetic contributions to these phenotypes must be careful to evaluate the extent to which linked genomic loci are due to the unique genetic etiologies of each phenotype and the extent to which they are the result of a shared genetic etiology.
The present study builds upon our previous efforts using scales from the Minnesota Multiphasic Personality Inventory - 2nd edition (MMPI-2; Butcher et al., 1989) to identify genetic influences that contribute to the development of depressive symptoms, antisocial behavior, and substance misuse (Gizer et al., 2010) and alcohol dependence (Gizer et al., 2011) in the University of California at San Francisco Family Alcoholism Study (SFFAM). Results from the former study suggested that the Depression (DEP), Antisocial Practices (ASP), and MacAndrew Alcoholism-Revised (MAC-R) scales from the MMPI-2 yielded quantitative measures of depression, antisocial behavior, and impulsive substance use, respectively, that were elevated among individuals diagnosed with alcohol dependence relative to those without the diagnosis in the SFFAM. Further, each scale showed evidence of familial transmission suggesting their utility in molecular genetic studies.
The ASP scale of the MMPI-2 was designed to assess antisocial attitudes and behaviors associated with psychopathy (Butcher et al., 1990), and has shown moderate correlations with similar measures (Lilienfeld, 1996). The DEP scale is primarily composed of items assessing an individual's feelings of inadequacy and low self-worth (Butcher et al., 1990). It has shown moderate correlations with other depression measures such as the Beck Depression Inventory (Ben-Porath et al., 1993; Boone, 1994). The MAC-R scale was originally constructed to differentiate alcoholic outpatients from outpatients with other psychiatric conditions (MacAndrew, 1965). Subsequent research has demonstrated that the MAC-R assesses general substance misuse rather than alcoholism specifically and may be more accurately interpreted as a measure of impulsive drug use and reward seeking (Allen, 1991; Greene and Garvin, 1988; MacAndrew, 1979). Reward seeking behavior, which refers to an array of approach and consumption behaviors, is influenced by dopamine neurons in the mesolimbic pathway, which includes the nucleus accumbens and ventral tegmental area (Ikemoto and Panksepp, 1999). Several drugs including cocaine and methamphetamine directly stimulate this pathway, though research has shown that activities such as eating, sexual behavior, and even aggression can stimulate this pathway as well (Berridge, 2004). Thus, high reward-seeking behavior has been related to substance use disorders and antisocial behavior, which are thought to result from dysregulation of the mesolimbic pathway (Cloninger, 1987a).
To further explore the conclusion that the ASP, DEP, and MAC-R scales may represent useful phenotypes for molecular genetic studies of antisocial behavior, depression, and impulsive substance use, respectively, we conducted genome-wide linkage scans for each of these scales in the present study. Because the SFFAM sample was selected for alcohol dependence and previous studies of the SFFAM have also shown high rates of cannabis use among the participants (Ehlers et al., 2010a; Ehlers et al., 2010b; Vieten et al., 2004), a second set of linkage scans using alcohol and then cannabis dependence diagnoses as covariates were conducted to determine the extent to which genetic loci identified in the initial analyses were unique or shared with the etiology of alcohol or cannabis dependence. The results of these analyses and the implications of these findings for future molecular genetic studies of substance dependence are discussed.
Materials and Methods
The protocol for the study was approved by the Institutional Review Board at the University of California at San Francisco. Each recruited individual was fully briefed on the nature of the study and provided written informed consent prior to participation. Ongoing management and analysis of study data has been approved by the Institutional Review Board at the University of North Carolina at Chapel Hill.
Participants
Participants were recruited as part of the UCSF Family Alcoholism Study, a nationwide study on the genetics of alcoholism and other substance dependence. In brief, probands were sampled from the community through semi-targeted direct mail, a web site, press releases, advertisements and from alumni of treatment centers across the nation. Probands were invited to participate if they met screening criteria for alcohol dependence at some point in their lifetime and had at least one sibling or both parents available to participate in the study. With the permission of the proband, relatives were invited by mail to participate.
Diagnostic and Statistical Manual of Mental Disorders (DSM-IV) diagnoses of alcohol and other substance (e.g., nicotine, cannabis, and stimulant) dependence were made using a modified version of the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA; Bucholz et al., 1994). Participants also completed the MMPI-2 (Butcher et al., 1989). Scores from three scales from the DEP, ASP, and MAC-R scales were evaluated. Probands with serious drug addictions (e.g., stimulants, cocaine, or opiates) and those who reported any history of intravenous substance use were excluded given that the primary aim of the UCSF Family Alcoholism Study was to identify genetic loci that conferred risk for alcohol dependence. Probands were excluded if, upon screening, they reported a current or past diagnosis of schizophrenia, bipolar disorder, or other psychiatric illness involving psychotic symptoms (those with depressive and anxiety disorders were accepted); a life-threatening illness; or an inability to speak and read English.
Two thousand five hundred and twenty-four individuals were enrolled in the UCSF Family Alcoholism Study of which 1647 were included in the present study. Participants' mean age was 49.9 ± 12.8 years. Racial distribution was 93% Caucasian, 3% Hispanic American, 2% African-American, and 1% each Native American and other. No attempt was made to exclude or over sample minorities. Probands were 62% female. A total of 700 participants met criteria only for DSM-IV Alcohol Dependence, 46 participants met criteria only for DSM-IV Cannabis Dependence, and 169 participants met criteria for both disorders. Sample means for the three MMPI scales were as follows: DEP - M = 51.19 ± 11.70, ASP - M = 49.48 ± 9.78, MAC-R - M = 52.48 ± 10.73. Correlations between these scales and Alcohol and Cannabis Dependence are presented in Table 1.
Table 1.
Pearson correlations among DSM-IV dependence diagnoses and MMPI-2 scales.
| Scale | Alcohol Dependence |
Cannabis Dependence |
ASP | DEP |
|---|---|---|---|---|
| Cannabis Dependence | 0.201 | |||
| ASP | 0.197 | 0.085a | ||
| DEP | 0.193 | 0.067b | 0.418 | |
| MAC-R | 0.312 | 0.095 | 0.465 | 0.215 |
Notes: p-values for all bivariate correlations were <0.001 except for
p = 0.001 and
p = 0.010.
DNA Collection and Genotyping
The DNA extraction procedure and genotyping protocol have been previously described (Wilhelmsen et al., 2003). Briefly, DNA was isolated from whole blood using a commercial kit (Gentra, Minneapolis, MN), and genotypes for a panel of microsatellite polymorphisms were generated using fluorescently labeled polymerase chain reaction (PCR) primers (HD5, version 2.0; Applied Biosystems, Foster City, CA). The HD5 panel set consisted of 811 markers with an average marker-to-marker distance of 4.6 cM (maximum, 14 cM) and an average heterozygosity of greater than 77%. A small subset of markers was omitted from the panel because of null alleles, irregular allele spacing or other problems with reproducibility. None of the omitted markers were adjacent to other omitted markers.
The sizes of marker amplimers were determined (blinded to pedigree structure and subject characteristics) from electropherograms produced with an ABI 3700 (Applied Biosystems, Foster City, CA) using the Genotyper software package (ABI). All electropherograms were visually inspected and exported from Genotyper in base pair sizes relative to the standard measured to one hundredth of a base pair. Allele frequencies observed in the founders were used for all analysis. The sex-averaged marker map order obtained from the manufacturer was used and verified with the family data from the current sample.
Genotypes for autosomal markers were analyzed using the Pedigree Relationship Statistical Test (PREST; McPeek and Sun, 2000) software to detect sample and pedigree structure errors. Genotyping was repeated for any individual with a probable error, and problematic genotypes were treated as missing if the error persisted. Fifteen families were identified with pedigree structure errors, and five were resolved following re-genotyping. The program Pedcheck was used to detect non-Mendelian inheritance (O'Connell and Weeks, 1998). When isolated Mendelian errors were observed, the genotypes for the entire family were excluded for the marker yielding the error. Markers exhibiting a high rate of Mendelian errors across families were excluded from subsequent analysis. Pedcheck identified 3104 Mendelian errors resulting in 7714 lost genotypes and the exclusion of one marker. To further reduce errors, the error-checking algorithm implemented in MERLIN (Abecasis et al., 2002) was used to assess the probability that each genotype was correctly called. A total of 1867 genotypes with probabilities of less than 0.025 of being correct were removed from further analysis. A total of 1867 problematic genotypes were identified and removed by MERLIN. Following these quality control procedures, a total of 1,269,708 genotypes were accepted with a success rate of 99.6%.
Analysis
Both genotype and phenotype data were available for 1647 individuals, and phenotype but not genotype data were available for an additional 875 individuals (2 additional individuals were lacking complete phenotype data). Seven hundred and thirteen families were considered genetically informative for linkage analysis. Families that contained sibling, half-sibling, avuncular or cousin pairs were included as being potentially genetically informative. When considering all participants, these families ranged in size from 3 to 20 subjects (average 2.90±2.44). The data includes: 1085 sibling, 40 half sibling, 17 grandparent-grandchild, 238 avuncular, and 32 cousin genetically-informative relative pairs. An additional 177 families contained only a single individual with phenotype data. These individuals were included within some analyses to the extent that they contributed information about trait means and variance and the impact of covariates.
All linkage analyses were conducted using the variance components method implemented in SOLAR (Almasy and Blangero, 1998). SOLAR is able to incorporate information from all individuals in an extended pedigree whether it is a quantitative phenotype or a dichotomous phenotype indicating affected and unaffected individuals. Though analyses of dichotomous traits have lower statistical power than analyses of quantitative traits, this reduction in power is less for dichotomous traits with high prevalence rates such as that observed for alcohol dependence in the UCSF Family Alcoholism Study (Duggirala et al., 1997). Multipoint LOD scores were calculated across the genome at 1 cM intervals. Because the UCSF Family Alcoholism Study sample was selected for alcohol dependence, higher levels of antisocial behavior, depressive symptoms, and impulsive substance use were anticipated (see Table 1 for correlations between dependence diagnoses and MMPI-2 scales). While only small deviations of the MMPI-2 standardized scores from the expected mean of 50 were observed (maximum deviation = +2.54 for the MAC-R scale), means for each scale were constrained to 50 in the variance components models to correct for any bias due to ascertainment.
Initial linkage scans were conducted separately for the ASP, DEP, and MAC-R MMPI-2 scales. A second set of linkage scans were then conducted for each of these three scales using DSM-IV alcohol and then cannabis dependence diagnostic status as covariates and then as additional phenotypes in bivariate analyses. Alcohol and cannabis dependence were separately analyzed as covariates for two reasons: (1) to provide parallel analyses to the bivariate analyses that were conducted between each MMPI-2 scale and each diagnostic variable and (2) because an evaluation of the linkage model suggested that cannabis dependence diagnostic status was not a significant covariate after controlling for alcohol dependence diagnostic status for any of the three MMPI scales, likely a result of the high rates of alcohol dependence (79%) among individuals who met criteria for cannabis dependence. The latter multivariate linkage analyses are conducted in SOLAR by estimating the proportion of variance in each phenotype that can be explained by a genetic locus and the extent to which the locus can explain the genetic correlation between phenotypes. Linkage peaks exceeding a LOD score of 3.6 were reported as achieving genome-wide significance and peaks exceeding a LOD score of 2.2 were reported as yielding suggestive evidence for linkage as described by Lander and Kruglyak (1995). In addition, linkage peaks exceeding a LOD score of 1.5 were reported as regions of interest. While these latter peaks represent weaker evidence for linkage, reporting such findings may aid future studies and meta-analytic reviews. To further aid in the interpretation of the results, empirical p-values were calculated for reported linkage peaks by generating allele-sharing probabilities for a simulated locus under the null hypothesis of no linkage across 100,000 trials (Duggirala et al., 2001), an approach shown to yield appropriate Type I error rates (Jung et al., 2006). The distribution of simulated LOD scores was used to calculate point-wise estimates of significance. Support intervals for reported peaks were defined as the region surrounding a linkage peak yielding a LOD score that was greater than the maximum LOD – 1 in each direction.
Results
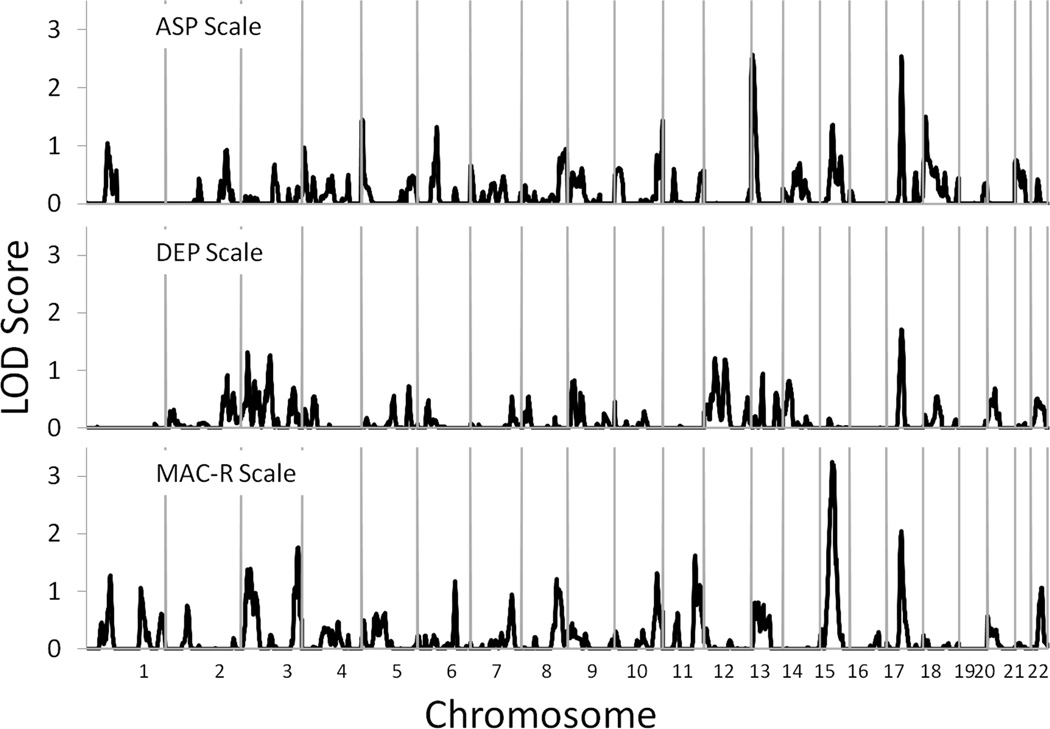
Analyses of the individual MMPI-2 scales yielded several loci that were suggestive of linkage at the genome-wide level, though none achieved genome-wide significance (see Table 2 for complete results). For the ASP scale, suggestive linkage peaks were observed on chromosomes 13 at 11 cM (LOD=2.57, point-wise empirical P=0.00032) nearest marker D13S1304 with a support interval extending from 5–22 cM and chromosome 17 at 58 cM (LOD=2.55, point-wise empirical P=0.00033) nearest marker D17S927 with a support interval extending from 54–63 cM. For the DEP scale, a single locus of interest was observed on chromosome 17 at 58 cM (LOD=1.71, point-wise empirical P=0.00200) nearest marker D17S927 with a support interval extending from 47–67 cM. For the MAC-R scale, a locus on chromosome 15 at 47 cM (LOD=3.25, point-wise empirical P=0.00004) nearest markers D15S117 and D15S1033 with a support interval extending from 37–58 cM yielded suggestive evidence for linkage. In addition, loci of interest were observed on chromosomes 3 at 218 cM (LOD=1.76, point-wise empirical P=0.00334) nearest markers D3S1265 with a support interval extending from 202–225, 11 at 123 cM (LOD=1.62, point-wise empirical P=0.00459) nearest markers D11S4094 with a support interval extending from 117–149, and chromosome 17 at 57 cM (LOD=2.05, point-wise empirical P=0.00164) nearest markers D17S927 with a support interval extending from 49–64.
Table 2.
Chromosomal Regions with Significant or Suggestive Evidence of Linkage to MMPI-2 Scales.
| MMPI-2 Scale | Chromosome | cM | LOD (p-value) |
Nearest Markers |
Previous evidence for linkage to related phenotypes |
|---|---|---|---|---|---|
| ASP | 13 | 11 | 2.57 (0.00032) | D13S1304 | Ehlers and Wilhelmsen, 20061; Ehlers et al., 20082; Saccone et al., 20031 |
| 17 | 58 | 2.55 (0.00033) | D17S927 | Gelernter et al., 20063; Hill et al., 20044; Holmans et al., 20075; Middledorp et al., 20095; Stallings et al., 20036 and 20056 | |
| DEP | 17 | 58 | 1.71 (0.00200) | D17S927 | Gelernter et al., 20063; Hill et al., 20044; Holmans et al., 20075; Middledorp et al., 20095; Stallings et al., 20036 and 20056 |
| MAC-R | 3 | 218 | 1.76 (0.00334) | D3S1265 | |
| 11 | 123 | 1.62 (0.00459) | D11S4094 | ||
| 15 | 47 | 3.25 (0.00004) | D15S117/D15S1033 | Beirut et al., 20041; Dick et al., 20027; Holmans et al., 20075; McGuffin et al., 20055; Middledorp et al., 20095; Zubenko et al., 20025 | |
| 17 | 57 | 2.05 (0.00164) | D17S927 | Gelernter et al., 20063; Hill et al., 20044; Holmans et al., 20075; Middledorp et al., 20095; Stallings et al., 20036 and 20056 |
Notes: cM - centimorgans,
tobacco use,
externalizing behavior,
substance dependence,
alcohol dependence,
depression,
conduct disorder,
anxiety,
bold text indicates regions that yielded suggestive evidence of linkage to the MMPI scales (i.e., LOD>2.50) prior to controlling for the presence of alcohol or cannabis dependence.
To follow up these results, we repeated each genome scan with alcohol and then cannabis dependence included as covariates to evaluate whether the observed findings could be attributed to the MMPI-2 scales independent of alcohol or cannabis dependence diagnostic status. The results of these analyses for the regions described above are shown in Table 3 alongside the corresponding results of linkage scans for alcohol and cannabis dependence with and without the MMPI-2 scales included as covariates. The results examining the overlap with alcohol dependence are described below. An inspection of Table 3 reveals fluctuations of similar magnitudes were observed when examining the overlap with cannabis dependence.
Table 3.
LOD Scores for Linkage Analysis of MMPI-2 Scales, Alcohol Dependence, and Cannabis Dependence.
| MMPI-2 Scale | Alcohol Dependence | Cannabis Dependence | Bivariate | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Chrom | cM | Univariate | Alc. Dep. as covariate |
Can. Dep. as covariate |
Univariate | MMPI-2 Scale as covariate |
Univariate | MMPI-2 Scale as covariate |
MMPI-2 Scale & Alc. Dep. |
MMPI-2 Scale & Can. Dep. |
| ASP | ||||||||||
| 13 | 11 | 2.57 | 3.03 | 2.79 | 0.00 | 0.00 | 0.00 | 0.00 | 2.55 | 2.56 |
| 17 | 58 | 2.55 | 2.08 | 2.52 | 0.64 | 0.12 | 0.49 | 0.44 | 1.96 | 1.94 |
| DEP | ||||||||||
| 17 | 58 | 1.71 | 1.11 | 1.65 | 0.64 | 0.13 | 0.49 | 0.49 | 1.18 | 1.81 |
| MAC-R | ||||||||||
| 3 | 218 | 1.76 | 2.12 | 1.78 | 0.00 | 0.00 | 0.04 | 0.01 | 1.20 | 1.34 |
| 11 | 123 | 1.62 | 1.82 | 1.10 | 0.00 | 0.00 | 0.00 | 0.04 | 1.53 | 1.29 |
| 15 | 47 | 3.25 | 2.80 | 3.45 | 0.89 | 0.94 | 0.00 | 0.17 | 2.46 | 2.36 |
| 17 | 57 | 2.05 | 1.97 | 1.73 | 0.64 | 0.14 | 0.49 | 0.38 | 1.36 | 2.05 |
Notes: Chrom - chromosome, cM - centimorgans, Alc. Dep. - Alcohol Dependence, Can. Dep - Cannabis Dependence, bold text indicates regions that yielded suggestive evidence of linkage to the MMPI scales (i.e., LOD>2.50) after controlling for the presence of alcohol or cannabis dependence.
For the ASP scale, the linkage peak on chromosome 13 at 11 cM was strengthened (LOD score increased from 2.57 to 3.03) when the presence of alcohol dependence was included as a covariate, but the peak on chromosome 17 at 58 cM decreased in strength (LOD score decreased from 2.55 to 2.08). For the DEP scale, the linkage peak on chromosome 17 at 58 cM decreased in strength (LOD score decreased from 1.71 to 1.11). For the MAC-R scale, the strongest linkage peak, which was observed on chromosome 15 at 47 cM, decreased in strength (LOD score decreased from 3.25 to 2.80) as did the peak observed on chromosome 17 at 57 cM (LOD score decreased from 2.05 to 1.97). In contrast, the peaks observed on chromosomes 3 at 218 cM (LOD score increased from 1.76 to 2.12) and 11 at 123 cM (LOD score increased 1.62 to 1.82) were both strengthened when the presence of alcohol dependence was included as a covariate. In addition to these analyses, bivariate analyses of each MMPI-2 scale and alcohol dependence diagnostic status were conducted to determine whether this would yield LOD scores stronger than those observed in the univariate analyses. These tended to yield results that were either weaker than or similar to the univariate analyses of the MMPI-2 scales without alcohol dependence diagnostic status included as a covariate (see Table 3). Further, there was no evidence to suggest linkage between alcohol dependence and any of these genomic regions.
In conducting the described analyses, it was observed that the locus on chromosome 17 at 57–58 cM was reported as a region of interest for each of the MMPI-2 scales. To further explore this result, a series of analyses spanning the support interval for this peak were conducted to determine which scales were contributing to the evidence for linkage at this locus. First, the evidence for linkage of each scale to this region was re-evaluated when the two remaining scales were included as covariates in the analysis. For example in the analysis of the ASP scale, the DEP and MAC-R scales were included as covariates. Second, a set of bivariate analyses were run for each pair of scales with the remaining scale included as a covariate in the analysis. Finally, a multivariate analysis was conducted including all three scales. The results of these analyses are presented in Table 4. It was notable that including the remaining two scales as covariates in the univariate analyses substantially reduced the LOD scores for each MMPI-2 scale, and further, the strongest result was observed for the trivariate analysis including all 3 MMPI-2 scales (LOD=3.16), though this result was weakened when alcohol or cannabis dependence diagnostic status was included in the analysis as a covariate (LOD=2.78 and 2.96, respectively).
Table 4.
Univariate and Multivariate Linkage Analysis of MMPI-2 Scales to Chromosome 17 at 57–58 Centimorgans.
| Univariate Analysisa |
Univariate Analysis w/ added Covariatesb |
Bivariate Analysisc | Trivariate Analysis | ||||
|---|---|---|---|---|---|---|---|
| Scale | LOD Score | Scale | LOD Score | Scales | LOD Score | Scales | LOD Score |
| ASP | 2.55 | ASP | 0.37 | ASP-DEP | 1.20 | ASP-DEP-MAC-R | 3.16 |
| DEP | 1.71 | DEP | 1.02 | ASP-MAC-R | 1.88 | ||
| MAC-R | 2.05 | MAC-R | 0.94 | DEP-MAC-R | 1.04 | ||
Notes:
No MMPI-2 scale scores were included as covariates,
The remaining two MMPI-2 scales scores were included as covariates,
The remaining MMPI-2 scale was included as a covariate.
Discussion
The present study sought to identify genomic loci that confer risk for antisocial behavior, depression, and impulsive substance use by conducting linkage scans of three scales from the MMPI-2 that were constructed to measure these behaviors, the Antisocial Practices content scale (ASP), the Depression content scale (DEP), and the revised MacAndrew Alcoholism scale (MAC-R), respectively. Suggestive evidence of linkage was observed to three genomic regions that proved independent of alcohol and cannabis dependence diagnostic status. Specifically, the ASP scale showed evidence of linkage to chromosome 13 at 11 cM, the MAC-R scale showed evidence of linkage to chromosome 15 at 47 cM, and all 3 scales showed evidence of linkage to chromosome 17 at 57–58 cM. Of note, previous linkage scans of alcohol and cannabis dependence conducted in the SFFAM failed to detect evidence of linkage to any of these regions (Gizer et al., 2011 and Ehlers et al., 2010b, respectively). This lack of overlap in findings is somewhat surprising given the evidence from twin studies suggesting that common genetic influences underlie to some extent the phenotypic correlations between the dependence diagnoses and antisocial behavior, impulsive behavior, and even depression (e.g., Hicks et al., 2004; Kendler et al., 1993; Young et al., 2006). It is possible that that the lack of overlap in findings between the diagnoses and MMPI-2 scales could be due to the reduced power of variance components linkage analysis for dichotomous traits, especially those with lower prevalence rates such as cannabis dependence, relative to quantitative traits (Duggirala et al., 1997). Nonetheless, the uniformly low LOD scores for the diagnostic variables in those linkage regions identified for the MMPI-2 scales (all LODs <1.0) suggest that the discrepant results are not simply the result of low power. Further, the relatively modest correlations between the dependence diagnoses and the examined MMPI-2 scales in the UCSF sample (see Table 1) suggest that these traits are largely distinct, thus providing a compelling explanation for the lack of overlapping findings.
As described, the locus on chromosome 17 at 57–58 cM appeared to be linked to scores on all three MMPI-2 scales that were evaluated. Further, the evidence for linkage was strongest when the 3 scales were included in a multivariate analysis. It should be noted that these scales do exhibit modest correlations in clinic-referred samples despite minimal item overlap (Greene, 2000; see Table 1 for correlations in the UCSF Family Sample). Such correlations clearly demonstrate overlap in what each scale is measuring, but they also demonstrate that each scale is capturing unique information. Given the observed increase in LOD scores when all 3 scales were considered in the analysis, it appears that the unique information provided by each scale is contributing to the observed linkage signal. This would suggest that the identified region on chromosome 17 is influencing psychopathology in some general fashion rather than substance dependence, antisocial behavior, or depression specifically. As an example, twin studies suggest the presence of genetic influences that jointly predispose an individual to externalizing (e.g., antisocial behavior, substance dependence) and internalizing disorders (e.g., major depressive disorder) (Cosgrove et al., 2011; O'Conner et al., 1998). Low effortful control, which can manifest as high levels of reward seeking behavior as well as high levels of negative emotionality, has been suggested by some researchers as a broad risk factor for psychopathology that could account for the shared genetic influences common to externalizing and internalizing disorders (MacDonald, 2008; Nigg, 2006). Supporting the conclusion that this region might confer a broad risk for psychopathology, studies have reported evidence of linkage to this region with a range of psychiatric disorders including major depressive disorder (Holmans et al., 2007; Middeldorp et al., 2009), alcohol dependence (Hill et al., 2004), substance dependence (Gelernter et al., 2006), and conduct disorder (Stallings et al., 2005; Stallings et al., 2003).
This chromosome 17 locus is located in a relatively gene-rich region and includes several candidate genes that have been widely studied in relation to psychiatric disorders and related phenotypes. For example, the gene encoding for the serotonin transporter (SLC6A4) is one of the most widely studied genes in the psychiatric genetics literature and polymorphisms of this gene have shown replicable evidence of association with disorders such as depression (Lopez-Leon et al., 2008), alcohol dependence (McHugh et al., 2010), ADHD (Gizer et al., 2009), and schizophrenia (Allen et al., 2008) in meta-analytic reviews. Additional candidate genes in the region are involved in dopaminergic (PPP1R1B/DARPP32), noradrenergic (PNMT), and cholinergic (VAT1) neurotransmission, neuronal development (MAPT, NEUROD2), and the stress response hormone system (CRHR1). Given the diversity of psychiatric phenotypes that have been related to this genomic region as well as the diversity in neurobiological systems represented by potential candidate genes in the region, a substantial amount of work will be required to refine our understanding of the observed relations; however, preliminary findings such as those described in the present report as well as those reviewed above suggest that this chromosomal region may either harbor several genetic variants that each confer risk to a specific disorder, and/or genetic variants that confer risk to psychopathology more broadly.
In addition to the locus on chromosome 17, suggestive evidence for linkage for the ASP scale to chromosome 13 at 11 cM and for the MAC-R scale to chromosome 15 at 47 cM was identified and proved to be independent of alcohol and cannabis dependence diagnostic status. Both of these loci have shown evidence of linkage to phenotypically similar traits in previous studies. For example, the locus on chromosome 13, which was linked to antisocial behavior as measured by the ASP scale, has previously shown evidence of linkage to externalizing behavior in a Native American population (Ehlers et al., 2008) as well as tobacco use in the same Native American population (Ehlers and Wilhelmsen, 2006) and the COGA sample (Saccone et al., 2003). Additionally, recent genome-wide association studies have reported suggestive association signals in this region with several psychiatric disorders including mood disorders (Huang et al., 2010), attention-deficit hyperactivity disorder (ADHD) (Anney et al., 2008; Lasky-Su et al., 2010; Sonuga-Barke et al., 2008), and schizophrenia (Sullivan et al., 2008) suggesting a relation between this region and psychiatric phenotypes. Despite this convergence of findings to the described region, it is a relatively gene-poor region without any obvious candidate genes, and the top hits reported in several of the GWAS’s described above come from intergenic regions or genes that would appear to lack a direct relation with psychiatric disorders.
The locus on chromosome 15, which was linked to impulsive substance use as assessed by the MAC-R scale in the present study, has also shown previous evidence of linkage to psychiatric phenotypes. For example, COGA has reported evidence of linkage between this region and alcohol misuse related to heightened anxiety (Dick et al., 2002) and tobacco use (Bierut et al., 2004), and evidence of linkage to this region for alcohol withdrawal was reported in a Native American sample (Ehlers et al., 2004). Further, a locus approximately 50 cM telomeric has shown evidence of linkage to depression phenotypes in four previous studies (Holmans et al., 2007; McGuffin et al., 2005; Middeldorp et al., 2009; Zubenko et al., 2002). Notably, Terracciano and colleagues (2010) recently reported that a SNP in the RORA gene located within the linkage region reported in the present study represented the top hit in their GWAS of trait depression. This gene was also implicated in bipolar disorder using a functional genomics approach integrating data from human and animal studies (Le-Niculescu et al., 2009). Thus, it may be that the linkage signal reported for the MAC-R in the present study is related to cyclic changes in mood that co-occur with substance misuse. Additional candidate genes in the region include two genes that encode for the glutamate receptor, ionotropic, N-methyl D-aspartate-like 1A (GCOM1, GRINL1A) and CYP19A1, which encodes for the aromatase enzyme responsible for the conversion of androgen to estrogen.
Despite the potential importance of these findings, there are limitations of the present study that should be considered. First, families in the UCSF Family Study sample were selected to be enriched for the alcohol dependence diagnosis, and as a result, it is not clear how findings from the present study will generalize to other populations. Second, it should be noted that multiple phenotypes were evaluated in the present study, though corrections were not made for multiple testing due to the generally low power of linkage analysis for complex traits. Nonetheless, we attempted to limit spurious evidence for linkage by restricting follow-up analyses (i.e., those including covariates and the multivariate analyses) to those regions of interest identified in the initial univariate analyses. Third, the scales from the MMPI-2 included in the present study, while well-studied, have not been previously included in molecular genetic studies of psychiatric disorders. Thus, replication of the reported results is needed. Nonetheless, the convergence between the reported results and studies using alternative assessment methods is promising.
In summary, the present study attempted to identify genetic loci that confer risk to antisocial behavior, depressive symptoms, and impulsive substance use and reward-seeking behavior. Suggestive evidence for linkage that was independent of alcohol and cannabis dependence diagnostic status was observed between antisocial behavior and chromosome 13 at 11 cM and between impulsive substance use and chromosome 15 at 47 cM. In addition, a region on chromosome 17 at 57–58 cM showed evidence of linkage in a multivariate analysis to antisocial behavior, depressive symptoms, and impulsive substance use. These regions have shown prior evidence of linkage and association to substance dependence and externalizing behavior as well as other psychiatric disorders such as depression suggesting potentially broad relations with psychopathology.
Figure 1.
Multipoint linkage analysis for the Antisocial Practices (ASP), Depression (DEP), and MacAndrew Revised Alcoholism (MAC-R) MMPI-2 scales. Chromosome numbers are displayed on the x-axis, and LOD scores are displayed on the y-axis. Results for each chromosome are aligned end to end with the pterminus on the left. Vertical lines indicate the boundaries between chromosomes.
Acknowledgements
Funding for this study was provided by the State of California for medical research on alcohol and substance abuse through the University of California at San Francisco. Additional support was provided by the Ernest Gallo Clinic and Research Center (KCW), AA10201 and U54 RR02502024 (CLE), T32 AA007573 (IRG), and National Institute of Drug Abuse (NIDA) grant DA019333 (KCW, CLE, IRG). The authors wish to acknowledge the technical support of James Lee and Samantha Segal.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Allen JP. Personality correlates of the MacAndrew alcoholism scale: A review of the literature. Psychol Addict Behav. 1991;5:59–65. [Google Scholar]
- Allen NC, Bagade S, McQueen MB, Ioannidis JP, Kavvoura FK, Khoury MJ, et al. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat Genet. 2008;40:827–834. doi: 10.1038/ng.171. [DOI] [PubMed] [Google Scholar]
- Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. American J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anney RJ, Lasky-Su J, O'Dushlaine C, Kenny E, Neale BM, Mulligan A, et al. Conduct disorder and ADHD: evaluation of conduct problems as a categorical and quantitative trait in the international multicentre ADHD genetics study. Am J Med Genet B Neuropsychiatr Genet. 2008;147:1369–1378. doi: 10.1002/ajmg.b.30871. [DOI] [PubMed] [Google Scholar]
- Babor TF, Hofmann M, DelBoca FK, Hesselbrock VM, Meyer RE, Dolinsky ZS, et al. Types of alcoholics, I. Evidence for an empirically derived typology based on indicators of vulnerability and severity. Arch Gen Psychiatry. 1992;49:599–608. doi: 10.1001/archpsyc.1992.01820080007002. [DOI] [PubMed] [Google Scholar]
- Ben-Porath YS, McCully E, Almagor M. Incremental Validity of the MMPI-2 Content Scales in the Assessment of Personality and Psychopathology by Self-Report. J Pers Assess. 1993;61:557–575. doi: 10.1207/s15327752jpa6103_12. [DOI] [PubMed] [Google Scholar]
- Berridge KC. Motivation concepts in behavioral neuroscience. Physiol Behav. 2004;81:179–209. doi: 10.1016/j.physbeh.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Bierut LJ, Rice JP, Goate A, Hinrichs AL, Saccone NL, Foroud T, et al. A genomic scan for habitual smoking in families of alcoholics: common and specific genetic factors in substance dependence. Am J Med Genet. 2004;124:19–27. doi: 10.1002/ajmg.a.20329. [DOI] [PubMed] [Google Scholar]
- Boone DE. Validity of the MMPI-2 depression content scale with psychiatric inpatients. Psychol Rep. 1994;74:159–162. doi: 10.2466/pr0.1994.74.1.159. [DOI] [PubMed] [Google Scholar]
- Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI, Jr, et al. A new, semi-structured psychiatric interview for use in genetic linkage studies: A report on the reliability of the SSAGA. J Stud Alcohol. 1994;55:149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- Butcher JN, Dahlstrom WG, Graham JR, Tellegen A, Kaemmer B. MMPI-2: Manual for administration and scoring. Minneapolis, MN: University of Minnesota Press; 1989. [Google Scholar]
- Butcher JN, Graham JR, Williams CL, Ben-Porath YS. Development and use of the MMPI-2 Content scales. Minneapolis, MN: University of Minnesota Press; 1990. [Google Scholar]
- Cadoret RJ, O'Gorman TW, Heywood E, Troughton E. Genetic and environmental factors in major depression. J Affect Disord. 1985;9:155–164. doi: 10.1016/0165-0327(85)90095-3. [DOI] [PubMed] [Google Scholar]
- Cloninger CR. A systematic method for clinical description and classification of personality variants. A proposal. Arch Gen Psychiatry. 1987a;44:573–588. doi: 10.1001/archpsyc.1987.01800180093014. [DOI] [PubMed] [Google Scholar]
- Cloninger CR. Neurogenetic adaptive mechanisms in alcoholism. Science. 1987b;236:410–410. doi: 10.1126/science.2882604. [DOI] [PubMed] [Google Scholar]
- Cosgrove VE, Rhee SH, Gelhorn HL, Boeldt D, Corley RC, Ehringer MA, et al. Structure and etiology of co-occurring internalizing and externalizing disorders in adolescents. J Abnorm Child Psychol. 2011;39:109–123. doi: 10.1007/s10802-010-9444-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick DM, Nurnberger JI, Jr, Edenberg HJ, Goate AM, Crowe R, Rice J, et al. Suggestive linkage on chromosome 1 for a quantitative alcohol-related phenotype. Alcohol Clin Exp Res. 2002;26:1453–1460. doi: 10.1097/01.ALC.0000034037.10333.FD. [DOI] [PubMed] [Google Scholar]
- Duggirala R, Blangero J, Almasy L, Arya R, Dyer TD, Williams KL, et al. A major locus for fasting insulin concentrations and insulin resistance on chromosome 6q with strong pleiotropic effects on obesity-related phenotypes in nondiabetic Mexican Americans. Am J Hum Genet. 2001;68:1149–1164. doi: 10.1086/320100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggirala R, Williams JT, Williams-Blangero S, Blangero J. A variance component approach to dichotomous trait linkage analysis using a threshold model. Genet Epidemiol. 1997;14:987–992. doi: 10.1002/(SICI)1098-2272(1997)14:6<987::AID-GEPI71>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Gilder DA, Slutske WS, Lind PA, Wilhelmsen KC. Externalizing disorders in American Indians: Comorbidity and a genome wide linkage analysis. Am J Med Genet B Neuropsychiatr Genet. 2008;147:690–698. doi: 10.1002/ajmg.b.30666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers CL, Gilder DA, Wall TL, Phillips E, Feiler H, Wilhelmsen KC. Genomic screen for loci associated with alcohol dependence in Mission Indians. Am J Med Genet B Neuropsychiatr Genet. 2004;129:110–115. doi: 10.1002/ajmg.b.30057. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Gizer IR, Vieten C, Gilder DA, Stouffer GM, Lau P, et al. Cannabis dependence in the San Francisco Family Study: age of onset of use, DSM-IV symptoms, withdrawal, and heritability. Addict Behav. 2010a;35:102–110. doi: 10.1016/j.addbeh.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers CL, Gizer IR, Vieten C, Wilhelmsen KC. Linkage analyses of cannabis dependence, craving, and withdrawal in the San Francisco family study. Am J Med Genet B Neuropsychiatr Genet. 2010b;153:802–811. doi: 10.1002/ajmg.b.31050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers CL, Wilhelmsen KC. Genomic screen for loci associated with tobacco usage in Mission Indians. BMC Med Genet. 2006;7:1–9. doi: 10.1186/1471-2350-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Panhuysen C, Wilcox M, Hesselbrock V, Rounsaville B, Poling J, et al. Genomewide linkage scan for opioid dependence and related traits. Am J Hum Genet. 2006;78:759–769. doi: 10.1086/503631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gizer IR, Ehlers CL, Vieten C, Seaton-Smith KL, Feiler HS, Lee JV, et al. Linkage scan of alcohol dependence in the UCSF Family Alcoholism Study. Drug Alcohol Depend. 2011;113:125–132. doi: 10.1016/j.drugalcdep.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gizer IR, Ficks C, Waldman ID. Candidate gene studies of ADHD: a meta-analytic review. Hum Genet. 2009;126:51–90. doi: 10.1007/s00439-009-0694-x. [DOI] [PubMed] [Google Scholar]
- Gizer IR, Seaton-Smith KL, Ehlers CL, Vieten C, Wilhelmsen KC. Heritability of MMPI-2 scales in the UCSF family alcoholism study. J Addict Dis. 2010;29:84–97. doi: 10.1080/10550880903436002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant BF, Harford TC. Comorbidity between DSM-IV alcohol use disorders and major depression: results of a national survey. Drug Alcohol Depend. 1995;39:197–206. doi: 10.1016/0376-8716(95)01160-4. [DOI] [PubMed] [Google Scholar]
- Grant BF, Stinson FS, Dawson DA, Chou SP, Dufour MC, Compton W, et al. Prevalence and co-occurrence of substance use disorders and independent mood and anxiety disorders: results from the National Epidemiologic Survey on Alcohol and Related Conditions. Arch Gen Psychiatry. 2004;61:807–816. doi: 10.1001/archpsyc.61.8.807. [DOI] [PubMed] [Google Scholar]
- Greene RL. The MMPI-2: An Interpretive Manual. Needham Heights, MA: Allyn and Bacon; 2000. [Google Scholar]
- Greene RL, Garvin RD. Substance abuse/dependence. In: Greene MA, editor. The MMPI: Use in specific populations. San Antonio, TX: Grune & Stratton; 1988. pp. 159–197. [Google Scholar]
- Han C, McGue MK, Iacono WG. Lifetime tobacco, alcohol and other substance use in adolescent Minnesota twins: univariate and multivariate behavioral genetic analyses. Addiction. 1999;94:981–993. doi: 10.1046/j.1360-0443.1999.9479814.x. [DOI] [PubMed] [Google Scholar]
- Hesselbrock MN, Meyer RE, Keener JJ. Psychopathology in hospitalized alcoholics. Arch Gen Psychiatry. 1985;42:1050–1055. doi: 10.1001/archpsyc.1985.01790340028004. [DOI] [PubMed] [Google Scholar]
- Hicks BM, Krueger RF, Iacono WG, McGue M, Patrick CJ. Family transmission and heritability of externalizing disorders: a twin-family study. Arch Gen Psychiatry. 2004;61:922–928. doi: 10.1001/archpsyc.61.9.922. [DOI] [PubMed] [Google Scholar]
- Hill SY, Shen S, Zezza N, Hoffman EK, Perlin M, Allan W. A genome wide search for alcoholism susceptibility genes. Am J Med Genet B Neuropsychiatr Genet. 2004;128:102–113. doi: 10.1002/ajmg.b.30013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmans P, Weissman MM, Zubenko GS, Scheftner WA, Crowe RR, Depaulo JR, Jr, et al. Genetics of recurrent early-onset major depression (GenRED): final genome scan report. Am J Psychiatry. 2007;164:248–258. doi: 10.1176/ajp.2007.164.2.248. [DOI] [PubMed] [Google Scholar]
- Huang J, Perlis RH, Lee PH, Rush AJ, Fava M, Sachs GS, et al. Cross-disorder genomewide analysis of schizophrenia, bipolar disorder, and depression. Am J Psychiatry. 2010;167:1254–1263. doi: 10.1176/appi.ajp.2010.09091335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemoto S, Panksepp J. The role of nucleus accumbens dopamine in motivated behavior: A unifying interpretation with special reference to reward-seeking. Brain Res Rev. 1999;31:6–41. doi: 10.1016/s0165-0173(99)00023-5. [DOI] [PubMed] [Google Scholar]
- Jung J, Weeks DE, Feingold E. Gene-dropping vs. empirical variance estimation for allele-sharing linkage statistics. Genet Epidemiol. 2006;30:652–665. doi: 10.1002/gepi.20177. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Heath AC, Neale MC, Kessler RC, Eaves LJ. Alcoholism and major depression in women. A twin study of the causes of comorbidity. Arch Gen Psychiatry. 1993;50:690–698. doi: 10.1001/archpsyc.1993.01820210024003. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Jacobson KC, Prescott CA, Neale MC. Specificity of genetic and environmental risk factors for use and abuse/dependence of cannabis, cocaine, hallucinogens, sedatives, stimulants, and opiates in male twins. Am J Psychiatry. 2003a;160:687–695. doi: 10.1176/appi.ajp.160.4.687. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Prescott CA, Myers J, Neale MC. The structure of genetic and environmental risk factors for common psychiatric and substance use disorders in men and women. Arch Gen Psychiatry. 2003b;60:929–937. doi: 10.1001/archpsyc.60.9.929. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Walters EE, Truett KR, Heath AC, Neale MC, Martin NG, et al. Sources of individual differences in depressive symptoms: analysis of two samples of twins and their families. Am J Psychiatry. 1994;151:1605–1614. doi: 10.1176/ajp.151.11.1605. [DOI] [PubMed] [Google Scholar]
- Kessler RC, Crum RM, Warner LA, Nelson CB, Schulenberg J, Anthony JC. Lifetime co-occurrence of DSM-III-R alcohol abuse and dependence with other psychiatric disorders in the National Comorbidity Survey. Arch Gen Psychiatry. 1997;54:313–321. doi: 10.1001/archpsyc.1997.01830160031005. [DOI] [PubMed] [Google Scholar]
- Khan AA, Jacobson KC, Gardner CO, Prescott CA, Kendler KS. Personality and comorbidity of common psychiatric disorders. Brit J Psychiatry. 2005;186:190–196. doi: 10.1192/bjp.186.3.190. [DOI] [PubMed] [Google Scholar]
- Lander ES, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Lasky-Su J, Won S, Mick E, Anney RJ, Franke B, Neale B, et al. On genome-wide association studies for family-based designs: an integrative analysis approach combining ascertained family samples with unselected controls. Am J Hum Genet. 2010;86:573–580. doi: 10.1016/j.ajhg.2010.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le-Niculescu H, Patel SD, Bhat M, Kuczenski R, Faraone SV, Tsuang MT, et al. Convergent functional genomics of genome-wide association data for bipolar disorder: comprehensive identification of candidate genes, pathways and mechanisms. Am J Med Genet B Neuropsychiatr Genet. 2009;150:155–181. doi: 10.1002/ajmg.b.30887. [DOI] [PubMed] [Google Scholar]
- Lilienfeld SO. The MMPI-2 Antisocial Practices Content scale: Construct validity and comparison with the Psychopathic Deviate scale. Psychol Assess. 1996;8:281–293. [Google Scholar]
- Lopez-Leon S, Janssens AC, Gonzalez-Zuloeta Ladd AM, Del-Favero J, Claes SJ, Oostra BA, et al. Meta-analyses of genetic studies on major depressive disorder. Mol Psychiatry. 2008;13:772–785. doi: 10.1038/sj.mp.4002088. [DOI] [PubMed] [Google Scholar]
- Lyons MJ, True WR, Eisen SA, Goldberg J, Meyer JM, Faraone SV, et al. Differential heritability of adult and juvenile antisocial traits. Arch Gen Psychiatry. 1995;52:906–915. doi: 10.1001/archpsyc.1995.03950230020005. [DOI] [PubMed] [Google Scholar]
- MacAndrew C. The differentiation of male alcohol outpatients from nonalcoholic psychiatric outpatients by means of the MMPI. Q J Stud Alcohol. 1965;26:238–246. [PubMed] [Google Scholar]
- MacAndrew C. On the possibility of the psychometric detection of persons who are prone to the abuse of alcohol and other substances. Addict Behav. 1979;4:11–20. doi: 10.1016/0306-4603(79)90016-9. [DOI] [PubMed] [Google Scholar]
- MacDonald KB. Effortful control, explicit processing, and the regulation of human evolved predispositions. Psychol Rev. 2008;115:1012–1031. doi: 10.1037/a0013327. [DOI] [PubMed] [Google Scholar]
- McGuffin P, Knight J, Breen G, Brewster S, Boyd PR, Craddock N, et al. Whole genome linkage scan of recurrent depressive disorder from the depression network study. Hum Mol Genet. 2005;14:3337–3345. doi: 10.1093/hmg/ddi363. [DOI] [PubMed] [Google Scholar]
- McHugh RK, Hofmann SG, Asnaani A, Sawyer AT, Otto MW. The serotonin transporter gene and risk for alcohol dependence: a meta-analytic review. Drug Alcohol Depend. 2010;108:1–6. doi: 10.1016/j.drugalcdep.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPeek MS, Sun L. Statistical tests for detection of misspecified relationships by use of genome-screen data. Am J Hum Genet. 2000;66:1076–1094. doi: 10.1086/302800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middeldorp CM, Sullivan PF, Wray NR, Hottenga JJ, de Geus EJ, van den Berg M, et al. Suggestive linkage on chromosome 2, 8, and 17 for lifetime major depression. Am J Med Genet B Neuropsychiatr Genet. 2009;150:352–358. doi: 10.1002/ajmg.b.30817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey LC, Blashfield RK. Empirical classifications of alcoholism: a review. J Stud Alcohol. 1981;42:925–937. doi: 10.15288/jsa.1981.42.925. [DOI] [PubMed] [Google Scholar]
- Nigg JT. Temperament and developmental psychopathology. J Child Psychol Psychiatry. 2006;47:395–422. doi: 10.1111/j.1469-7610.2006.01612.x. [DOI] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor TG, McGuire S, Reiss D, Hetherington EM, Plomin R. Co-occurrence of depressive symptoms and antisocial behavior in adolescence: a common genetic liability. J Abnorm Psychol. 1998;107:27–37. doi: 10.1037//0021-843x.107.1.27. [DOI] [PubMed] [Google Scholar]
- Regier DA, Farmer ME, Rae DS, Locke BZ, Keith SJ, Judd LL, et al. Comorbidity of mental disorders with alcohol and other drug abuse. Results from the Epidemiologic Catchment Area (ECA) Study. JAMA. 1990;264:2511–2518. [PubMed] [Google Scholar]
- Ross HE, Glaser FB, Germanson T. The prevalence of psychiatric disorders in patients with alcohol and other drug problems. Arch Gen Psychiatry. 1988;45:1023–1031. doi: 10.1001/archpsyc.1988.01800350057008. [DOI] [PubMed] [Google Scholar]
- Saccone NL, Neuman RJ, Saccone SF, Rice JP. Genetic analysis of maximum cigarette-use phenotypes. BMC Genet. 2003;4:S105. doi: 10.1186/1471-2156-4-S1-S105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slutske WS, Heath AC, Dinwiddie SH, Madden PA, Bucholz KK, Dunne MP, et al. Modeling genetic and environmental influences in the etiology of conduct disorder: a study of 2,682 adult twin pairs. J Abnorm Psychol. 1997;106:266–279. doi: 10.1037//0021-843x.106.2.266. [DOI] [PubMed] [Google Scholar]
- Sonuga-Barke EJ, Lasky-Su J, Neale BM, Oades R, Chen W, Franke B, et al. Does parental expressed emotion moderate genetic effects in ADHD? An exploration using a genome wide association scan. Am J Med Genet B Neuropsychiatr Genet. 2008;147:1359–1368. doi: 10.1002/ajmg.b.30860. [DOI] [PubMed] [Google Scholar]
- Stallings MC, Corley RP, Dennehey B, Hewitt JK, Krauter KS, Lessem JM, et al. A genome-wide search for quantitative trait loci that influence antisocial drug dependence in adolescence. Arch Gen Psychiatry. 2005;62:1042–1051. doi: 10.1001/archpsyc.62.9.1042. [DOI] [PubMed] [Google Scholar]
- Stallings MC, Corley RP, Hewitt JK, Krauter KS, Lessem JM, Mikulich SK, et al. A genome-wide search for quantitative trait loci influencing substance dependence vulnerability in adolescence. Drug Alcohol Depend. 2003;70:295–307. doi: 10.1016/s0376-8716(03)00031-0. [DOI] [PubMed] [Google Scholar]
- Stinson FS, Grant BF, Dawson DA, Ruan WJ, Huang B, Saha T. Comorbidity between DSM-IV alcohol and specific drug use disorders in the United States: results from the National Epidemiologic Survey on Alcohol and Related Conditions. Drug Alcohol Depend. 2005;80:105–116. doi: 10.1016/j.drugalcdep.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Lin D, Tzeng JY, van den Oord E, Perkins D, Stroup TS, et al. Genomewide association for schizophrenia in the CATIE study: results of stage 1. Mol Psychiatry. 2008;13:570–584. doi: 10.1038/mp.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terracciano A, Tanaka T, Sutin AR, Sanna S, Deiana B, Lai S, et al. Genome-wide association scan of trait depression. Biol Psychiatry. 2010;68:811–817. doi: 10.1016/j.biopsych.2010.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuang MT, Lyons MJ, Eisen SA, Goldberg J, True W, Lin N, et al. Genetic influences on DSM-III-R drug abuse and dependence: a study of 3,372 twin pairs. Am J Med Genet. 1996;67:473–477. doi: 10.1002/(SICI)1096-8628(19960920)67:5<473::AID-AJMG6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Vieten C, Seaton KL, Feiler HS, Wilhelmsen KC. The University of California, San Francisco Family Alcoholism Study. I. Design, methods, and demographics. Alcohol Clin Exp Res. 2004;28:1509–1516. doi: 10.1097/01.alc.0000142261.32980.64. [DOI] [PubMed] [Google Scholar]
- Wender PH, Kety SS, Rosenthal D, Schulsinger F, Ortmann J, Lunde I. Psychiatric disorders in the biological and adoptive families of adopted individuals with affective disorders. Arch Gen Psychiatry. 1986;43:923–929. doi: 10.1001/archpsyc.1986.01800100013003. [DOI] [PubMed] [Google Scholar]
- Wilhelmsen KC, Schuckit M, Smith TL, Lee JV, Segall SK, Feiler HS, et al. The search for genes related to a low-level response to alcohol determined by alcohol challenges. Alcohol Clin Exp Res. 2003;27:1041–1047. doi: 10.1097/01.ALC.0000075551.02714.63. [DOI] [PubMed] [Google Scholar]
- Young SE, Rhee SH, Stallings MC, Corley RP, Hewitt JK. Genetic and environmental vulnerabilities underlying adolescent substance use and problem use: general or specific? Behav Genet. 2006;36:603–615. doi: 10.1007/s10519-006-9066-7. [DOI] [PubMed] [Google Scholar]
- Zubenko GS, Hughes HB, Stiffler JS, Zubenko WN, Kaplan BB. Genome survey for susceptibility loci for recurrent, early-onset major depression: results at 10cM resolution. Am J Medical Genetics. 2002;114:413–422. doi: 10.1002/ajmg.10381. [DOI] [PubMed] [Google Scholar]