Abstract
The occurrence of the noncomplementary G-U base pair at the end of a helix is found to be governed by stacking interactions. As a rule, a G-U pair with G on the 5'-side of a Watson-Crick base pair exhibits strikingly greater stacking overlap with the Watson-Crick base pair than a G-U pair on the 3'-side of a Watson-Crick base pair. The former arrangement is expected to be more stable and indeed is observed 29 times out of 32 in the known transfer RNA molecules. In accordance with this rule, the major wobble base pairs G-U or I-U in codon-anticodon interactions have G or I on the 5'-side of the anticodon. Similarly, in initiator tRNAs, this rule is obeyed where now the G is the first letter of the codon (5'-side). In the situation where U is in the wobble position of the anticodon, it is usually substituted at C(5) andmay also have a 2-thio group and it can read one to four codons depending on its modifications. A G at the wobble position of the anticodon can recognize the two codons ending with U or C and modification of G (unless it is I) does not change its reading properties.
Full text
PDF
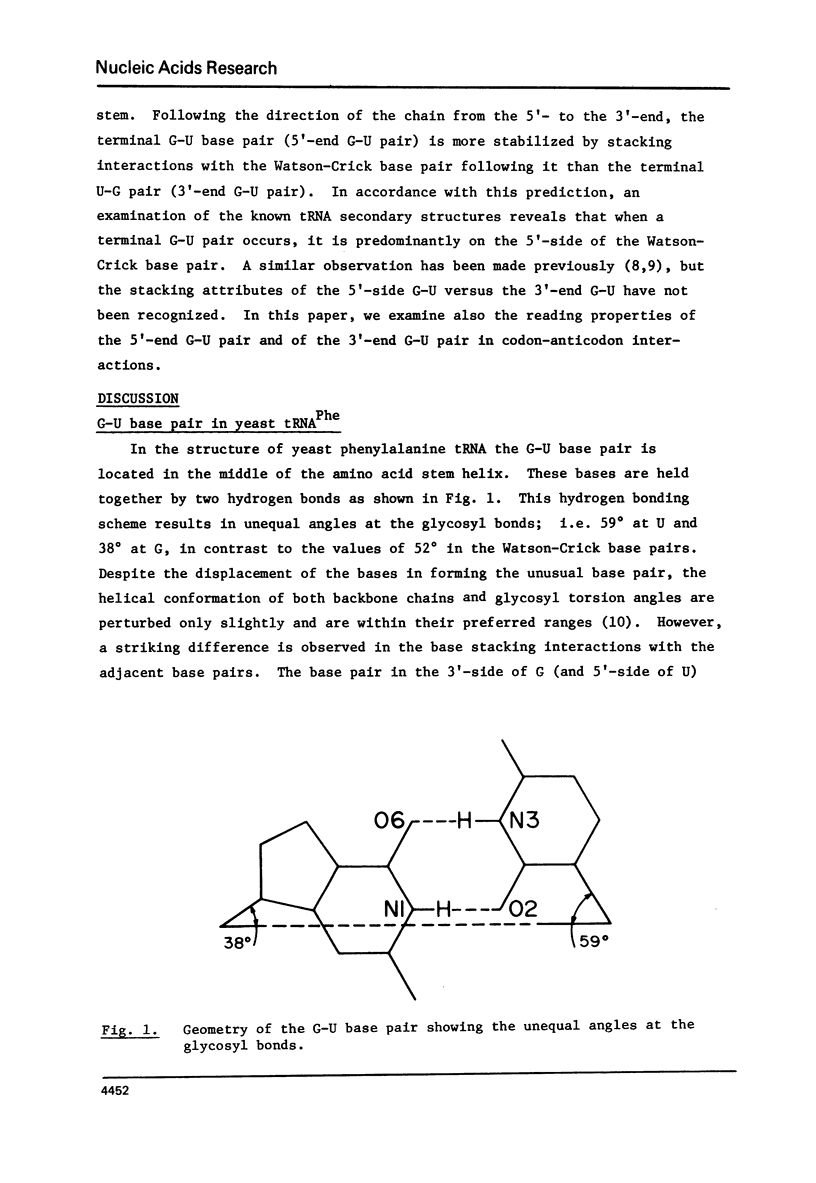
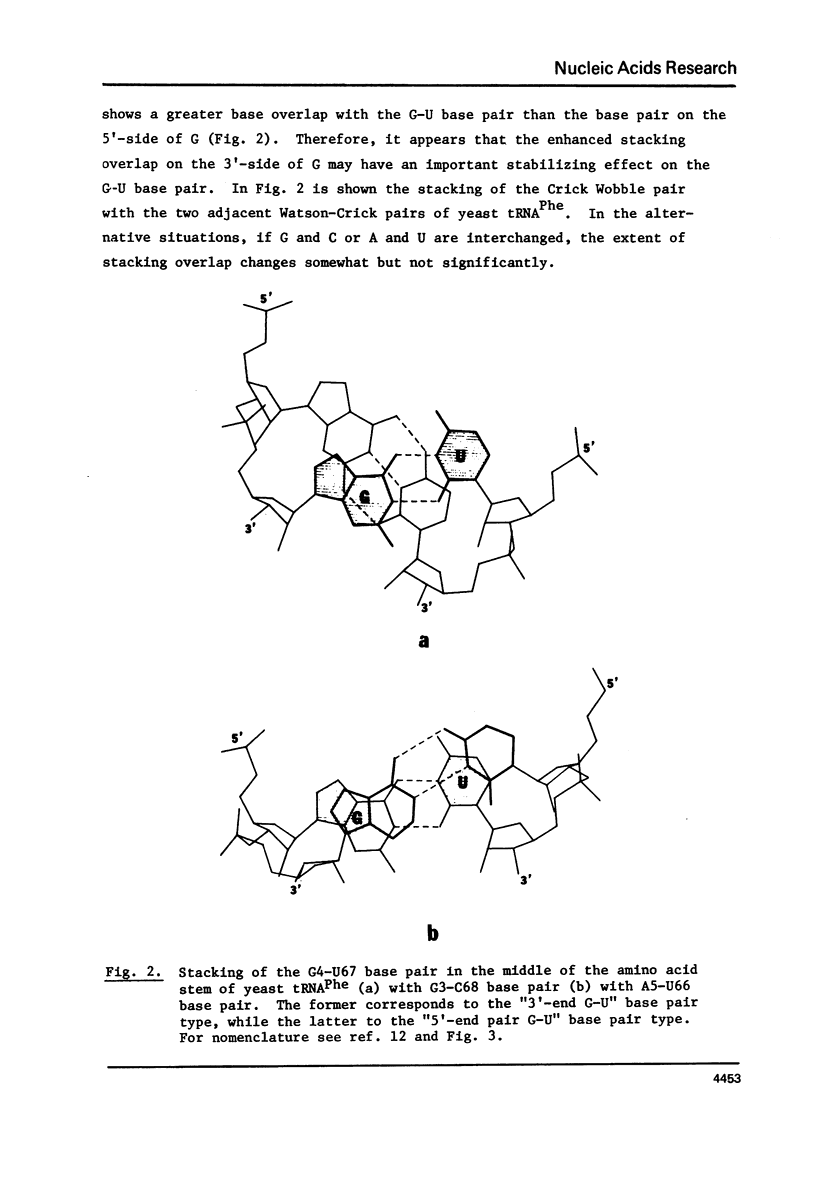




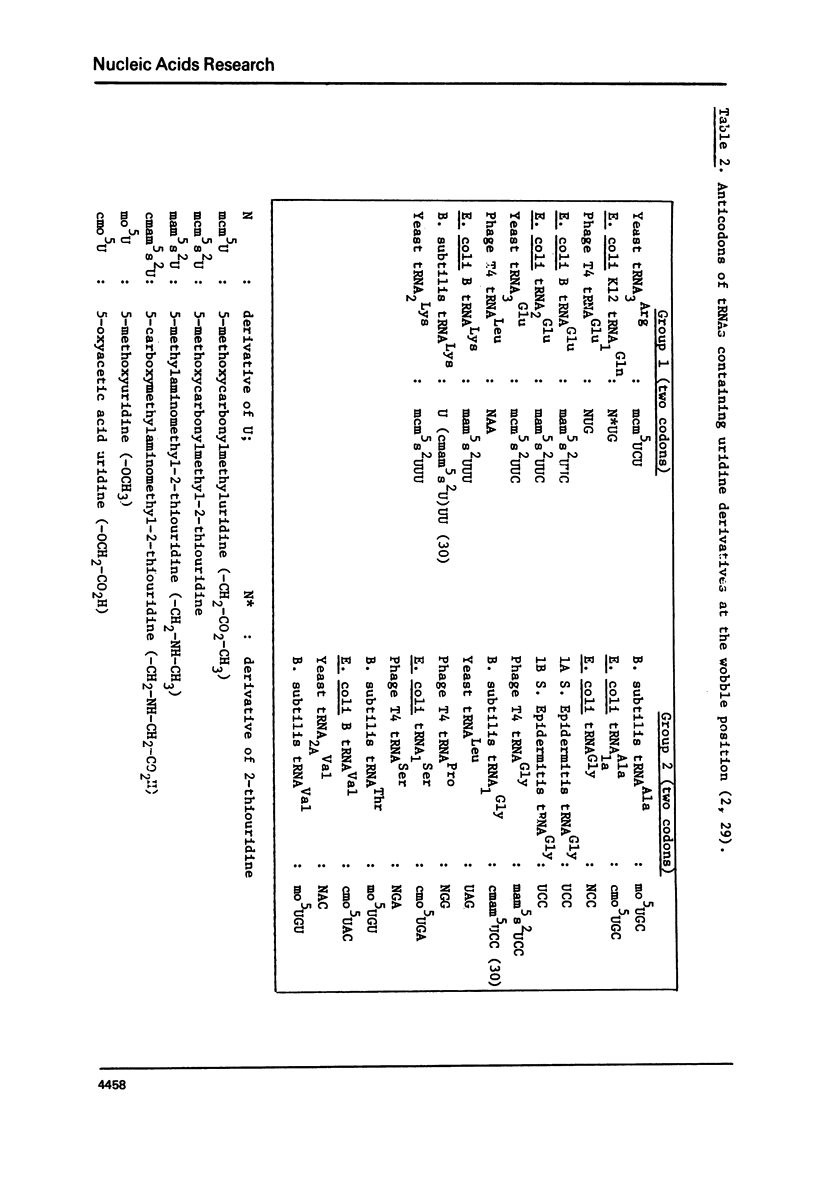



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barrell B. G., Seidman J. G., Guthrie C., McClain W. H. Transfer RNA biosynthesis: the nucleotide sequence of a precursor to serine and proline transfer RNAs. Proc Natl Acad Sci U S A. 1974 Feb;71(2):413–416. doi: 10.1073/pnas.71.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H. M., Marcu D., Narayanan P. Modified bases in tRNA: the structures of 5-carbamoylmethyl- and 5-carboxymethyl uridine. Nucleic Acids Res. 1978 Mar;5(3):893–903. doi: 10.1093/nar/5.3.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan T., Sundaralingam M. Structlre of transfer RNA molecules containing the long variable loop. Nucleic Acids Res. 1976 Nov;3(11):3235–3250. doi: 10.1093/nar/3.11.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F. H., Brenner S., Klug A., Pieczenik G. A speculation on the origin of protein synthesis. Orig Life. 1976 Dec;7(4):389–397. doi: 10.1007/BF00927934. [DOI] [PubMed] [Google Scholar]
- Crick F. H. Codon--anticodon pairing: the wobble hypothesis. J Mol Biol. 1966 Aug;19(2):548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- Fuller W., Hodgson A. Conformation of the anticodon loop intRNA. Nature. 1967 Aug 19;215(5103):817–821. doi: 10.1038/215817a0. [DOI] [PubMed] [Google Scholar]
- Grosjean H. J., de Henau S., Crothers D. M. On the physical basis for ambiguity in genetic coding interactions. Proc Natl Acad Sci U S A. 1978 Feb;75(2):610–614. doi: 10.1073/pnas.75.2.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith G., Dirheimer G. The primary structure of rabbit, calf and bovine liver tRNAPhe. Biochim Biophys Acta. 1978 Jan 26;517(1):133–149. doi: 10.1016/0005-2787(78)90041-2. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Irie T., Yoshida M., Takeishi K., Ukita T. The primary structure of yeast glutamic acid tRNA specific to the GAA codon. Biochim Biophys Acta. 1974 Oct 11;366(2):168–181. doi: 10.1016/0005-2787(74)90331-1. [DOI] [PubMed] [Google Scholar]
- Ladner J. E., Jack A., Robertus J. D., Brown R. S., Rhodes D., Clark B. F., Klug A. Atomic co-ordinates for yeast phenylalanine tRNA. Nucleic Acids Res. 1975 Sep;2(9):1629–1637. doi: 10.1093/nar/2.9.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladner J. E., Jack A., Robertus J. D., Brown R. S., Rhodes D., Clark B. F., Klug A. Atomic co-ordinates for yeast phenylalanine tRNA. Nucleic Acids Res. 1975 Sep;2(9):1629–1637. doi: 10.1093/nar/2.9.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumdar S. K., Saenger W. Molecular structure of poly-2-thiouridylic acid, a double helix with non-equivalent polynucleotide chains. J Mol Biol. 1974 May 15;85(2):213–219. doi: 10.1016/0022-2836(74)90361-1. [DOI] [PubMed] [Google Scholar]
- Mitra S. K., Lustig F., Akesson B., Lagerkvist U. Codon-acticodon recognition in the valine codon family. J Biol Chem. 1977 Jan 25;252(2):471–478. [PubMed] [Google Scholar]
- Ninio J. Recognition in nucleic acids and the anticodon families. Prog Nucleic Acid Res Mol Biol. 1973;13:301–337. doi: 10.1016/s0079-6603(08)60106-3. [DOI] [PubMed] [Google Scholar]
- Nishimura S. Minor components in transfer RNA: their characterization, location, and function. Prog Nucleic Acid Res Mol Biol. 1972;12:49–85. [PubMed] [Google Scholar]
- RajBhandary U. L., Chang S. H. Studies on polynucleotides. LXXXII. Yeast phenylalanine transfer ribonucleic acid: partial digestion with ribonuclease T-1 and derivation of the total primary structure. J Biol Chem. 1968 Feb 10;243(3):598–608. [PubMed] [Google Scholar]
- Randerath K., Chia L. S., Gupta R. C., Randerath E. Structural analysis of nonradioactive RNA by postlabeling: the primary structure of baker's yeast tRNA Leu/CUA. Biochem Biophys Res Commun. 1975 Mar 3;63(1):157–163. doi: 10.1016/s0006-291x(75)80024-6. [DOI] [PubMed] [Google Scholar]
- Roberts R. J. Staphylococcal transfer ribonucleic acids. II. Sequence analysis of isoaccepting glycine transfer ribonucleic acids IA and IB from Staphylococcus epidermidis Texas 26. J Biol Chem. 1974 Aug 10;249(15):4787–4796. [PubMed] [Google Scholar]
- Sekiya T., Takeishi K., Ukita T. Specificity of yeast glutamic acid transfer RNA for codon recognition. Biochim Biophys Acta. 1969 Jun 17;182(2):411–426. doi: 10.1016/0005-2787(69)90192-0. [DOI] [PubMed] [Google Scholar]
- Sprinzl M., Grüter F., Gauss D. H. Collection of published tRNA sequences. Nucleic Acids Res. 1978 May;5(5):r15–r27. [PMC free article] [PubMed] [Google Scholar]
- Stahl S., Paddock G. V., Abelson J. Nucleotide sequence determination of bacteriophage T4 glycine transfer ribonucleic acid. Nucleic Acids Res. 1974 Oct;1(10):1287–1304. doi: 10.1093/nar/1.10.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout C. D., Mizuno H., Rubin J., Brennan T., Rao S. T., Sundaralingam M. Atomic coordinates and molecular conformation of yeast phenylalanyl tRNA. An independent investigation. Nucleic Acids Res. 1976 Apr;3(4):1111–1123. doi: 10.1093/nar/3.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman J. L., Kim S. H. Idealized atomic coordinates of yeast phenylalanine transfer RNA. Biochem Biophys Res Commun. 1976 Jan 12;68(1):89–96. doi: 10.1016/0006-291x(76)90014-0. [DOI] [PubMed] [Google Scholar]
- Weissenbach J., Dirheimer G., Falcoff R., Sanceau J., Falcoff E. Yeast tRNALeu (anticodon U--A--G) translates all six leucine codons in extracts from interferon treated cells. FEBS Lett. 1977 Oct 1;82(1):71–76. doi: 10.1016/0014-5793(77)80888-0. [DOI] [PubMed] [Google Scholar]
- Weissenbach J., Dirheimer G. Pairing properties of the methylester of 5-carboxymethyl uridine in the wobble position of yeast tRNA3Arg. Biochim Biophys Acta. 1978 May 23;518(3):530–534. doi: 10.1016/0005-2787(78)90171-5. [DOI] [PubMed] [Google Scholar]
- Williams R. J., Nagel W., Roe B., Dudock B. Primary structure of E. coli alanine transfer RNA: relation to the yeast phenylalanyl tRNA synthetase recognition site. Biochem Biophys Res Commun. 1974 Oct 23;60(4):1215–1221. doi: 10.1016/0006-291x(74)90328-3. [DOI] [PubMed] [Google Scholar]
- Woese C. Molecular mechanics of translation: a reciprocating ratchet mechanism. Nature. 1970 May 30;226(5248):817–820. doi: 10.1038/226817a0. [DOI] [PubMed] [Google Scholar]


