Specific deletion of TGF-β receptor II in macrophages, CD11b+Gr1+, and dendritic cells inhibit tumor growth by increasing efficiency of the immune system.
Keywords: TGF-beta receptor II, LysM-Cre mice, tumor progression
Abstract
By crossing LysM-Cre and TGF-β type II receptor (Tgfbr2) floxed mice we achieved specific deletion of Tgfbr2 in myeloid cells (Tgfbr2MyeKO mice). S.c.-injected (LLC, EL4-OVA) and implanted (MMTV-PyMT) carcinoma cells grow slower in Tgfbr2MyeKO mice. The number of CD45+ cells in the tumor tissue was the same in both genotypes of mice, but upon analysis, the percentage of T cells (CD45+CD3+) in the KO mice was increased. By flow cytometry analysis, we did not detect any differences in the number and phenotype of TAMs, CD11b+Gr1+, and DCs in Tgfbr2MyeKO compared with Tgfbr2MyeWT mice. ELISA and qRT-PCR data showed differences in myeloid cell functions. In Tgfbr2MyeKO TAMs, TNF-α secretion was increased, basal IL-6 secretion was down-regulated, TGF-β did not induce any VEGF response, and there was decreased MMP9 and increased MMP2 and iNOS expression. TGF-β did not have any effect on CD11b+Gr1+ cells isolated from Tgfbr2MyeKO mice in the regulation of Arg, iNOS, VEGF, and CXCR4, and moreover, these cells have decreased suppressive activity relative to T cell proliferation. Also, we found that DCs from tumor tissue of Tgfbr2MyeKO mice have increased antigen-presented properties and an enhanced ability to stimulate antigen-specific T cell proliferation. We conclude that Tgfbr2 in myeloid cells has a negative role in the regulation of anti-tumorigenic functions of these cells, and deletion of this receptor decreases the suppressive function of CD11b+Gr1+ cells and increases antigen-presenting properties of DCs and anti-tumorigenic properties of TAMs.
Introduction
The tumor microenvironment is formed through the interactions of different types of cells, including malignant, immune, fibroblasts/stromal, and endothelial cells. Many investigators have focused on TGF-β signaling in specific cell populations during tumor progression, as this protein has numerous effects on cell functions. Most of them vary depending on the cell type, tumor age, and stage of tumor progression. One approach to investigating the roles of TGF-β signaling is through deletion of the Tgfbr2 in specific cell types during tumorigenesis. Today, the dominant conclusion is that loss of the TGF-β response in tumor cells and in fibroblasts results in tumor initiation, progression, and metastasis [1]. In our laboratory, we have shown that conditional deletion of Tgfbr2 in mammary epithelial cells resulted in shortened tumor latency and a fivefold increase in lung metastases compared with tumors with intact TGF-β signaling [2]. Loss of TGF-β signaling on carcinoma cells increased CXCL1 and CXCL5 secretion [3], which increased migration of CD11b+Gr1+ cells to tumor tissue [4], and played indirect roles in increasing the number of Th17 cells that have a protumorigenic effect in mammary carcinoma [5]. In the pancreas, Tgfbr2 deletion specific to the epithelium in the context of Kras activation results in the development of aggressive pancreatic ductal adenocarcinoma and regulates tumor-stromal interactions via a Cxcr2-dependent chemokine and connective tissue growth factor [6]. Recently, Borczuk et al. [7] showed that loss of Tgfbr2 induced a highly invasive phenotype of lung cancer and that tumor-associated stromal cells displayed an immunosuppressive profile marked by increased numbers of B and T cells. In fibroblasts, deletion of Tgfbr2 promotes metastasis in MMTV-PyMT mammary tumor progression, and the mechanism is dependent on CXCL12 and CCL2 chemokines [8].
Deletion of TGF-β signaling in immune cells has different effects, depending on the type of cell from which it is deleted. Loss of TGF-β signaling in T cells results in a fatal autoimmune pathology, with the animals dying by 3 weeks of age [9]. Selective loss of Smad4-dependent signaling in T cells leads to spontaneous gastrointestinal cancers, whereas epithelial-specific deletion of the Smad4 gene does not [10]. Deletion of Tgfbr1 in DCs using Langerin-Cre mice results in Langerhans cell deficiency and reduced contact hypersensitivity [11]. TGF-β can promote the generation of DCs with an immature phenotype [12] and regulate antigen-presenting functions by regulation expression of MHC-II and secretion of IL-12 [13]. In monocytes, TGF-β induces MMPs, IL-1, and IL-6, and it acts as a chemoattractant [14, 15]. Once monocytes differentiate into macrophages, TGF-β functions mostly as an inhibitory molecule [16]. CD36 and SR-A, two scavenger receptors involved in phagocytosis, are down-regulated by TGF-β [17, 18]. In vitro, TGF-β inhibits the expression of TNF-α and MMP12 as well as chemokines, including MIP-1α and MIP-2 [19, 20]. Moreover, TGF-β down-regulates the production of NO and superoxide ion and inhibits the expression of iNOS in activated macrophages [21, 22]. TGF-β regulation of PMN activation and effector functions, such as phagocytosis, degranulation, and respiratory burst, is controversial and requires further investigation.
In the current study, we examined the role of impaired TGF-β signaling in myeloid cells during tumor progression. With the use of Lys M-Cre mice crossed with Tgfbr2 floxed mice, we generated mice with Tgfbr2 deleted in myeloid cells. In these mice, we analyzed normal hematopoiesis, dynamics of tumor growth in different models, as well as phenotype and functions of subpopulations of myeloid cells: CD11b+Gr1−F4/80+ (TAM), CD11b+Gr1+, and CD11b+CD11c+ (DCs).
MATERIALS AND METHODS
Cell lines and mice
LLC cell line (CRL-1642) and EL4-OVA (CRL-2113) were obtained from American Type Culture Collection (Manassas, VA, USA) and maintained following the manufacturer's protocols. MMTV-PyMT carcinoma cell lines were derived from primary tumors of MMTV-PyMT/Tgfbr2flox/flox mice, established and cultured in DMEM/F12 with 5% adult bovine serum, as described previously [3, 4]. LLC cells (5×105 cells) and EL4-OVA cells (5×105 cells) were injected s.c. into the right flank of mice. MMTV-PyMT carcinoma cells were implanted to mammary fat pad of #4 mammary gland via collagen plugs (5×105 cells/plug). The size of LLC tumors was determined by direct measurement of tumor dimensions at 2- to 3-day intervals using calipers. The equation volume = length × (width)2 × 0.5 was used to calculate tumor volume [23]. All studies were performed on Tgfbr2MyeKO and Tgfbr2MyeWT mice. To generate these mice, we crossed LysM-Cre mice, ordered from The Jackson Laboratory (Bar Harbor, ME, USA), with Tgfbr2 floxed mice, which were established and maintained as described [24]. These mice are on a pure C57BL/6 background. For additional experiments, 8- to 10-week-old OT-I (003831) mice were purchased from The Jackson Laboratory. The studies were approved by IACUC at Vanderbilt University Medical Center (Nashville, TN, USA).
Flow cytometry analysis
Single-cell suspensions were made from the spleens and bone marow of normal and tumor-bearing mice [23, 25] and tumor tissues [26]. These cells were labeled with fluorescence-conjugated antibodies (BioLegend, eBioscience, Becton Dicksinson, all San Diego, CA, USA) and isotype-matched IgG controls. The cells were analyzed on a LSRII flow cytometer (Becton Dickinson), and the data were analyzed with FlowJo software. For intracellular staining, cells were fixed and permeabilized using BD Cytofix/Cytoperm Fixation/Permeabilization Solution Kit, following the manufacturer's protocol. T cells were stimulated with leukocyte activation cocktail, with BD GolgiPlug (Becton Dickinson), following the manufacturer's protocols.
Magnetic cell separation
CD11b+Gr1+ cells, T cells (CD3+), and B cells (CD19+) from spleen were magnetically separated by Gr1-, CD3-, or CD19-biotin, respectively, with Streptavidin-magnetic microbeads following application protocols of the manufacturer (Miltenyi Biotec, Auburn, CA, USA). Lin− cells from bone marrow were isolated by negative magnetic separation following application protocols of the manufacturer (Miltenyi Biotec).
Single-cell sorting
Single-cell suspensions of tumor tissue were stained with fluorescence-labeled antibodies and sorted with a FACSAria flow cytometer (Becton Dickinson). CD11b+Gr1+F/80- and CD11b+F4/80+Gr1− cells were collected for gene expression and cytokine secretion analysis.
Functional assays
T cell proliferation was measured using a MLR. CD11b+Gr1+ cells were isolated from spleen of LLC tumor-bearing mice and mixed with splenocytes from OT-I mice in a different ratio, started from 1:1. DCs from EL4-OVA tumor-bearing mice were sorted by flow cytometry and mixed with splenocytes from OT-I mice in a different ratio, started from 1:10. Also, MLR was performed by mixing splenocytes from EL4-OVA tumor-bearing mice with splenocytes from OT-I mice in different ratios, starting from 1:1. Cells were incubated for 72 h, and 3[H]-thymidine was then added at 1 μCi/200 μl cells/well for an additional 18 h, followed by cell harvesting and radioactivity count using a liquid scintillation counter.
qRT-PCR
Total RNA was extracted from isolated spleen CD11b+Gr1+ cells and also from sorted CD11b+Gr1+ and CD11b+Gr1−F4/80+ cells, as described, using an RNeasy Mini Kit (Qiagen, Valencia, CA, USA). cDNA was synthesized using Invitrogen Superscript First-Strand synthesis system for RT-PCR (Invitrogen, Carlsbad, CA, USA). Primers specific for Tgfbr2, Arg, iNOS, VEGF, CXCR4, S100A8, S100A9, MMP2, MMP9, and TNF-α were used, and relative gene expression was determined using the ABI PRISM 7900HT sequence detection system (PE Applied Biosystems, Foster City, CA, USA). The CT cycle method was used to calculate gene expression normalized to β-actin as a gene reference. Primer sequences are available upon request.
ELISA
Cytokine levels in macrophage supernatants were measured using the mouse VEGF and IL-6 ELISA kits (R&D Systems, Minneapolis, MN, USA), following the manufacturer's protocol.
Statistical analysis
Data were analyzed using GraphPad Prism 4.0 software (GraphPad Software, San Diego, CA, USA) and presented as mean ± sem. Comparisons among several treatment groups were performed using one-way ANOVA, followed by appropriate post-tests. Comparisons between two groups were performed using two-tailed unpaired t tests. A P value < 0.05 was considered significant.
RESULTS
Generation of Tgfbr2MyeKO mice
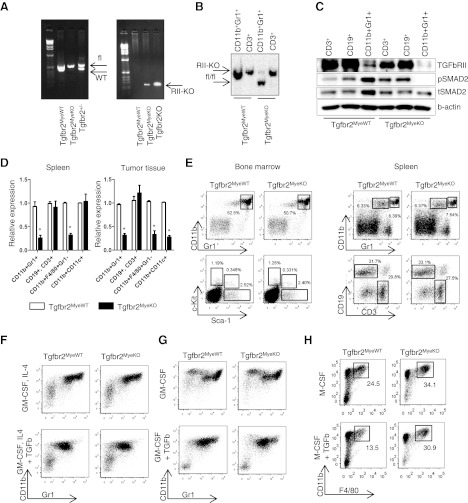
To delete Tgfb2 in myeloid cells, we used LysM-Cre mice crossed with Tgfb2 floxed mice (Supplemental Fig. 1). Tgfbr2MyeKO mice, LysM Cre+/−Tgfbr2fl/fl, were the experimental group, and LysM Cre−/−Tgfbr2fl/fl mice were the control group (Tgfbr2MyeWT). Mice were maintained in a pure C57bl/6 background. To prove specific deletion of Tgfbr2 in myeloid cells, we isolated CD11b+Gr1+ cells from the spleen of naïve mice and performed PCR analysis of genomic DNA as in previously published methods [24]. In parallel with PCR, we performed Southern blot and Western blot analysis on isolated CD11b+Gr1+ cells from tumor tissue to show the degree of Tgfbr2 recombination in Tgfbr2MyeKO mice. In these assays, we found complete recombination of Tgfbr2 in CD11b+Gr1+ cells isolated from Tgfbr2MyeKO mice (Fig. 1A–C). LysM is expressed in the myeloid cell lineage, including monocytes, mature macrophages, and granulocytes [27]. To check the expression of Tgfbr2 in different cell populations, we isolated CD11b+Gr1+ cells, T cells, B cells, monocyes/macrophages, and DCs and performed qRT-PCR analysis for expression of Tgfbr2 (Fig. 1D). In cells derived from the spleen, no differences were observed in T and B cells and DCs (CD11b+CD11c+), but expression of Tgfbr2 was much less in CD11b+Gr1+ cells and in monocytes/macrophages (CD11b+F4/80+Gr1−). In tumor tissue, in addition to CD11b+Gr1+ cells and macrophages, DCs have decreased expression of Tgfbr2. In the original manuscript about creating the LysM-Cre mice [27], the authors mentioned that partial deletion could be detected in CD11c+ splenic DCs, which are closely related to the monocyte/macrophage lineage. We found similar results in spleens of the mice that we generated, but in the injected or implanted tumor tissue, all DCs are negative for Tgfbr2. The DCs are heterogeneous, and CD11c is common marker for different types of DCs. We analyzed various subsets of DCs in the spleen and in tumor tissue and found that expression of LysM is high in myeloid DCs, and it was associated with deletion of Tgfbr2 (Supplemental Figs. 2 and 3). Moreover, tumor tissue contains only the myeloid type of DCs, and because of this, we found just a partial deletion of Tgfbr2 in the spleen but a compete deletion of Tgfbr2 on CD11c+ cells in tumor tissue.
Figure 1. Tgfbr2MyeKO mice and normal hematopoiesis.
(A) Tgfbr2-specific PCR on genomic DNA from CD11b+Gr1+ cells from mice carrying the genotype indicated (left); Tgfbr2 recombination-specific PCR on genomic DNA from CD11b+Gr1+ cells from the same mice (right). (B) Southern blot analysis of genomic DNA from CD11b+Gr1+ cells FACS-sorted from LLC tumor tissue showing degree of Tgfbr2 recombination in Tgfbr2MyeKO mice. (C) Cells were isolated from spleen of LLC tumor-bearing mice by magnetic microbeads (Miltenyi Biotec) and incubated with TGF-β for 2 h at 1 ng/ml. Protein lysates were isolated through cell homogenization in cOmplete-M lysis buffer (Roche, Indianapolis, IN, USA) and quantified by Bradford DC assay (Bio-Rad, Hercules, CA, USA). Phospho-Smad2 (pSMAD2) was detected by using rabbit mAb (Cat. #04-1029; Millipore, Bedford, MA, USA), total-Smad2 (tSmad2) by using rabbit polyclonal antibody (Cat. #AB3849; Millipore), and TGFbRII by rabbit polyclonal antibody (Cat. #sc-400; Santa Cruz Biotechnology, Santa Cruz, CA, USA). (D) qRT-PCR for Tgfbr2 in different population of myeloid cells sorted by FACSAria from tumor tissue and spleen of tumor-bearing mice. (E) Representative FACS plots of basic populations of cells in spleen and bone marrow in naïve Tgfbr2MyeWT and Tgfbr2MyeKO mice. (F, G, H) Differentiation assay with Lin− cells isolated from bone marrow by negative magnetic separation. Shown are representative flow cytometry plots. *P < 0.05; data correspond with the mean ± sem of three individual mice from three experiments.
Before analysis of tumor progression in Tgfbr2MyeKO mice, we performed experiments to answer the question: can deletion of Tgfbr2 in LysM+ cells affect normal hematopoiesis? First, by flow cytometry, we analyzed the bone marrow and spleen for the number of basic cell populations. No differences were observed in the number of CD11b+Gr1+ cells, progenitor cells (Sca-1, c-Kit), and T and B cells in spleen and bone marrow (Fig. 1E). Second, we checked the ability of progenitor cells to differentiate into myeloid cells in the presence of TGF-β. We used isolated Lin− cells from bone marrow of naïve mice and incubated them with GM-CSF or GM-CSF + IL-4 for 5 days and then analyzed the phenotype of the cells. We found that TGF-β has the same effect on differentiation in Tgfbr2MyeKO and Tgfbr2MyeWT mice—down-regulation of Gr1 on the cell surface (Fig. 1F and G). The number of CD11b+Gr1+ cells (Fig. 1F and G), F4/80+ (data not shown), and CD11c (data not shown) was the same in Tgfbr2MyeKO mice compared with Tgfbr2MyeWT mice. It has been shown by many investigators that there are two types of MDSCs—G-MDSC and M-MDSC [28–30]. M-MDSCs migrated to tumor tissue can differentiate to macrophages compared with G-MDSCs, which don't have the ability for additional differentiation. Because of that, we isolated CD11b+Gr1+ cells from the spleen of tumor-bearing mice and differentiated them to macrophages in the presence of M-CSF, with and without TGF-β. We found that TGF-β decreased the number of macrophages (CD11b+F4/80+) in Tgfbr2MyeWT mice, and this effect was eliminated in Tgfbr2MyeKO mice (Fig. 1H). However, in additional experiments, we did not find significant differences in the number of macrophages in tumor tissue in Tgfbr2MyeKO mice versus control mice, and we suppose that it is because there is a different mechanism involved in the differentiation of myeloid cells besides TGF-β.
Deletion of Tgfbr2 in LysM+ cells decreases tumor growth
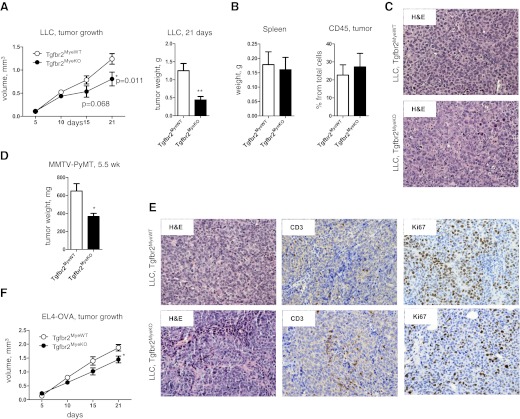
To analyze the effects of Tgrbr2 deletion on tumor growth, we implanted different tumor cells into their respective congenic background. First, we injected LLCs s.c. and analyze tumor growth (Fig. 2A–C). We found that for the first 2 weeks, tumor growth was the same in Tgfbr2MyeWT and Tgfbr2MyeKO mice, but between the 2nd and 3rd week, the rate of tumor growth decreased in Tgfbr2MyeKO mice compared with control mice (Fig. 2A). Tumor weight on the 21st day of tumor progression was less in Tgfbr2MyeKO mice compared with Tgfbr2MyeWT mice (0.44±0.09 g and 1.25±0.20 g, respectively; Fig. 2A). The weight of the spleen and the number of CD45+ cells in tumor tissue were the same in control and experimental groups (Fig. 2B). In a second model, we used MMTV-PyMT mammary epithelial cells, which were established by our lab [31], and we implanted these cells to the mammary gland and also observed decreased tumor weight at Tgfbr2MyeKO mice relative to Tgfbr2MyeWT mice (366.3±34.45 mg vs. 648.8±83.67 mg; Fig. 2D and E). By using IHC staining, we found an increased number of T cells (CD3+) and a decreased proliferative activity (Ki-67) in tumors from Tgfbr2MeyKO mice (Fig. 2E). For a third model, we used lymphoma cells s.c.-injected into the mice, and we observed the same effect as with LLC cells—decreased tumor size on the 3rd week of tumor progression at Tgfbr2MyeKO mice relative to controls (Fig. 2F).
Figure 2. Tumor growth.
(A) LLC growth after s.c. injection (left) and LLC tumor weight on Day 21 after s.c. injection (right). (B) Weight of spleen (left) and number of CD45+ cells in tumor tissue (right) in LLC tumor-bearing mice upon sacrifice on Day 21 of tumor progression. Data correspond with the mean ± sem of five individual mice from two experiments. (C) H&E staining indicated morphological changes in LLC tumors in Tgfbr2MyeWT and Tgfbr2MyeKO mice. (D) Tumor weight of MMTV-PyMT carcinoma cells implanted into the #4 mammary fat pad. Data correspond with the mean ± sem of five individual mice from two experiments. (E) H&E staining indicated morphological changes in MMTV-PyMT tumors in Tgfbr2MyeWT and Tgfbr2MyeKO mice and IHC staining for CD3 and Ki67 in same type of tumor. (F) Dynamics of tumor growth of EL4-OVA cells injected s.c. Data correspond with the mean ± sem of three individual mice from two experiments. *P < 0.05; **P < 0.01; data correspond with the mean ± sem.
Phenotype and function of macrophages
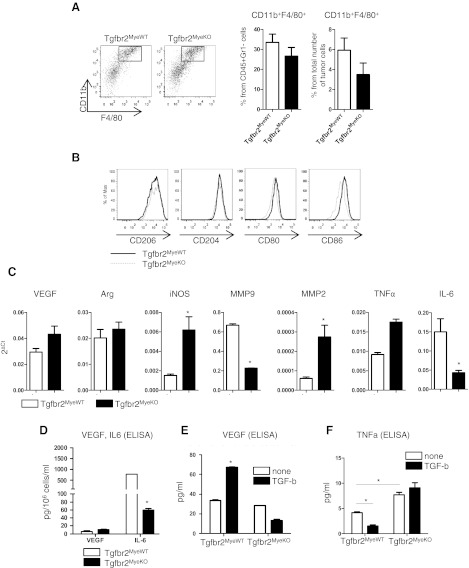
For the following experiments, we used the established LLC model of tumorigenesis and performed phonotypic and functional analysis of cells on the 14th day of tumor progression. The reason for this was because in the 2nd week, we found a marginal significant effect (Fig. 2A) in tumor growth, but in the 3rd week, there was a significant difference, but tumor tissue contained areas of necrosis that can affect cell functions. First, we analyzed the number of TAMs (CD11b+Gr1−F4/80+) in tumor tissue (Fig. 3A). No differences were observed in the percentage of these cells from CD45+Gr1− and from the total number of cells in Tgfbr2MyeKO mice compared with Tgfbr2MyeWT mice. As TAMs can be divided into two populations—M1 macrophages, which promote activity of killer T cells (anti-tumor), and M2 macrophages, which promote inflammatory responses (pro-tumor or alternatively activated) [32, 33]—we next analyzed the phenotype of CD11b+Gr1−F4/80+ cells in tumor tissue. We found no significant differences in expression of markers, which correspond with M2 TAM (CD206, CD204) and M1 TAM (CD80, CD86) [33–35] in Tgfbr2MyeKO and Tgfbr2MyeWT mice (Fig. 3B). We further analyzed the function of TAMs by sorting CD11b+Gr1−F4/80+ cells from tumor tissue on Day 14 of tumor progression and performing qRT-PCR analysis. We found no significant differences in VEGF, Arg, and TNF-α expression in cells with deleted Tgfbr2 and cells with intact signaling. However, in cells isolated from tumor tissue of Tgfbr2MyeKO mice, expression of iNOS was increased four times (P=0.02) and MMP2, 4.5 times (P=0.01), and expression of MMP9 decreased three times (P=0.051) and IL-6, three times (P=0.021) compared with Tgfbr2MyeWT (Fig. 3C). It is known that TGF-β is a regulator of VEGF, IL-6, and TNF-α secretion [16], and because of this, we examined the secretion of these proteins in TAMs. We used two approaches to address this question. First, we isolated i.p. macrophages after 4% thioglycolate i.p. injection, and second, we differentiated monocytes from Lin− cells isolated from bone marrow in the presence of M-CSF. Results were consistent in i.p. macrophages and differentiated monocytes. The basal level of VEGF was the same in cells from Tgfbr2MyeKO and Tgfbr2MyeWT mice, but the basal level of IL-6 was decreased by tenfold (P=0.001) in Tgfbr2MyeKO mice compared with Tgfbr2MyeWT mice. Adding TGF-β to macrophages up-regulates secretion of VEGF in control cells, but this effect is absent in cells with impaired TGF-β signaling (Fig. 3E). The basal level of TNF-α was increased in cells without Tgfbr2 (P=0.035) and adding TGF-β, decreased by 2.5-fold secretion of this protein in control cells but has no effect in KO cells (Fig. 3F).
Figure 3. Phenotype and functions of macrophages.
(A) Number of macrophages (CD11b+F4/80+) in tumor tissue. Cells were gated as CD45+Gr1−. Representative FACS plots (left) and quantatative data (right). (B) Flow cytometry data for phenotype of CD11b+F4/80+Gr1− cells in tumor tissue. (C) qRT-PCR data for CD11b+F4/80+Gr1− cells isolated by FACS sorting from LLC tumor tissue. (D) VEGF and IL-6 secretion by macrophages. Cells were isolated from peritoneal area on the 4th day after 4% thioglycolate injection. (E) VEGF and (F) TNF-α secretion stimulated by TGF-β in peritoneal macrophages. *P < 0.05; data correspond with the mean ± sem of two to three individual mice from three experiments.
Phenotype and function of CD11b+Gr1+ cells
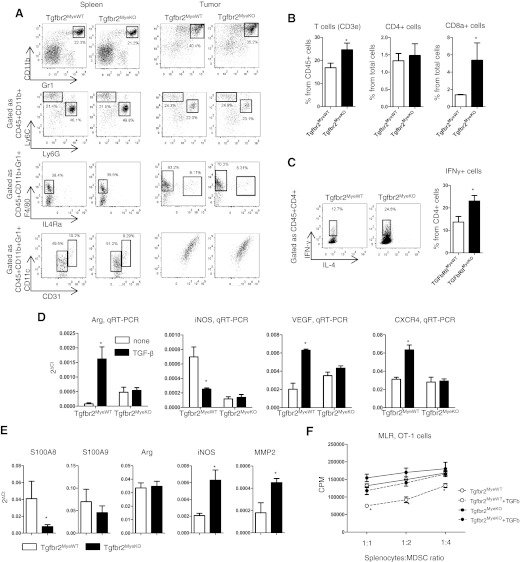
Analysis of phenotype and function of CD11b+Gr1+ cells in spleen and tumor tissue of Tgfbr2MyeKO and Tgfbr2MyeWT mice were performed after the 14th day of tumor progression. The number of these cells in the spleen and tumor tissue was the same in both groups of mice, and it is ∼20% in spleen and ∼40% in tumor tissue of all CD45+ cells (Fig. 4A). It has been shown that this population of cells can be monocytic or granulocytic and is dependent on Ly6C and Ly6G expression [36]. We performed analysis of a subpopulation of these cells, and also, no differences were observed in the number of CD11b+Ly6ChighLy6Glow and CD11b+Ly6GhighLy6Clow cells in the spleen and tumor tissue (Fig. 4A). CD11b+Gr1+ cells can be neutrophils in early stages of tumor progression, but usually, these cells are MDSCs, which are characterized by the presence of markers of other types of cells (CD115, CD244, CD31, CD11c, etc.) and can suppress T cell proliferation [28, 37]. All CD11b+Ly6GhighLy6Clow cells (G-MDSC) in tumor tissue were CD244+ [37], which means that they are not neutrophils (data not shown). We examined the phenotype of CD11b+Gr1+ cells for the presence of F4/80, IL-4Ra (CD124), CD11c, and CD31. No differences were observed in expression of these markers on CD11b+Gr1+ cells in both groups of mice, except for a slightly decreased number of CD11b+Gr1+IL-4Ra+F4/80+ cells in Tgfbr2MyeKOmice (Fig. 4A). This is interesting, as Marigo et al. [38] have shown that the human IL-4Ra+ populations of MDSC have a strong, suppressive effect on T cell proliferation.
Figure 4. Phenotype and functions of CD11b+Gr1+ cells.
(A) Representative FACS plots of number and phenotype of CD11b+Gr1+ cells in spleen and tumor tissue. (B) Number of T cells in tumor tissue. (C) Intracellular staining of T cells in tumor tissue. Before flow cytometry analysis, single-cell suspension of tumor tissue was stimulated for 6 h by PMA/ionomycin. Data correspond with the mean ± sem of three individual mice from three experiments. (D) Regulation of genes by TGF-β in CD11b+Gr1+ cells. Gr1+ cells were isolated by magnetic microbeads from spleen of tumor-bearing mice and incubated for 24 h with 5 ng/ml TGF-β. After incubation, cells were harvested, and total RNA was isolated. (E) qRT-PCR analysis of CD11b+Gr1+ cells isolated by FACS sorting from tumor tissue. (F) MLR. Gr1+ cells were isolated from spleen by magnetic sorting of tumor-bearing mice and incubated overnight with 5 ng/ml TGF-β, washed, and then mixed with splenocytes from OT-I transgenic mice in the presence of SIINFEKL. Data correspond with the mean ± sem of three individual mice from two experiments. *P < 0.05; data correspond with the mean ± sem.
Because of the decreased IL-4Ra+ population in CD11b+Gr1+ cells and because one of the most basic of the many functions of MDSCs is to inhibit the proliferation of T cells, we analyzed the number and phenotype of T cells in tumor tissue. The number of all T cells (CD3+) was increased in tumor tissue of Tgfbr2MyeKO mice (24.65±2.8 vs. 16.83±1.97 in control mice; Fig. 4B), which confirmed previous findings by IHC (Fig. 2E). The number of Th cells (CD3+CD4+) was the same in both groups of mice; however, the number of cytotoxic T cells (CD3+CD8+) was increased fourfold in Tgfbr2MyeKO mice (5.38±1.99 vs. 1.375±0.04 in control mice; P=0.032; Fig. 4B). By intracellular staining, we analyzed the phenotype of CD4+ cells to know what proportion of Th cells was found in the tumors (Th1 or Th2). We found an increased number of IFN-γ+ cells (Th1) in Tgfbr2MyeKO mice and no IL-4+ cells (Th2) in both groups of mice (Fig. 4C).
We next wanted to know what role TGF-β plays in regulating the functions of CD11b+Gr1+ cells. From the spleen of tumor-bearing mice, we isolated CD11b+Gr1+ cells and incubated them with TGF-β for 24 h, followed by qRT-PCR analysis. We found that Arg, VEGF, and CXCR4 were up-regulated by TGF-β in Tgfbr2MyeWT mice, but as expected, TGF-β does not have any effect on these genes in Tgfbr2MyeKO mice. However, TGF-β has the opposite effect in regulation of iNOS expression (Fig. 4D). In CD11b+Gr1+ cells isolated from tumor tissue, we did not find any differences in Arg expression, but we found increased iNOS and MMP2 and decreased S100A8 in Tgfbr2MyeKO mice (Fig. 4E). As molecules, such as Arg and iNOS, are the most important effectors in the suppressive function of MDSCs [29, 36], we asked whether the ability to suppress T cell proliferation by CD11b+Gr1+ cells is dependent on TGF-β signaling. We isolated CD11b+Gr1+ cells from the spleen of LLC tumor-bearing mice on the 14th day of tumor progression, incubated them overnight with TGF-β, washed them, and cocultured at different ratios with splenocytes from OT-I transgenic mice in the presence of OT-I peptide. CD11b+Gr1+ cells from Tgfbr2MyeWT andTgfbr2MyeKO mice without incubation with TGF-β did not have any differences in their ability to suppress T cell proliferation. However, TGF-β increased the suppressive activity of these cells isolated from Tgfbr2MyeWT mice in all ratios with splenocytes. In cells that do not have Tgfbr2, we observed no differences in suppressive activity compared with untreated cells (Fig. 4F).
Antigen-presenting function of DCs
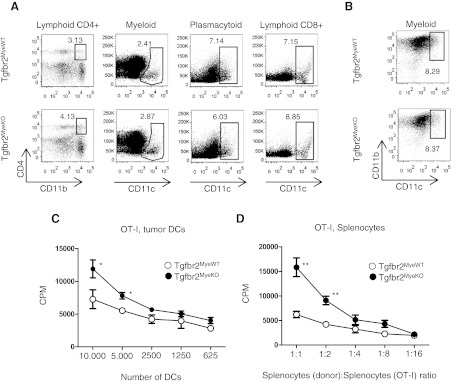
DCs are APCs that play a critical role in specific immune responses against tumors, and as we have shown above, myeloid DCs as well as TAMs and CD11b+Gr1+ cells are negative for Tgfbr2 in tumor tissue of Tgfbr2MyeKO mice. We performed phenotypic and functional analysis of different subpopulations of DCs in spleen and tumor tissue. By flow cytometry, we found no differences in the number of lymphoid, myeloid, or plasmacytoid DCs in the spleen and myeloid DCs in tumor tissue at 14 days of tumor progression at Tgfbr2MyeWT and Tgfbr2MyeKO mice (Fig. 5A and B). Next, we analyzed the antigen-presenting function of these cells and the ability to stimulate T cell proliferation in the presence of MDSCs. First, we s.c. injected tumorigenic EL4 cells transfected with OVA and 14 days later, FACS-sorted DCs from tumor tissue and mixed them with splenocytes from transgenic OT-I mice to stimulate a specific response of CD8+ cells in a MLR. Second, in same tumor model and in the same time frame, we took splenocytes from tumor-bearing mice, which included DCs and CD11b+Gr1+, and mixed them with splenocytes from OT-I mice to check the effect on proliferation of CD8+ cells, stimulated by DCs in parallel with inhibition by MDSCs. Results are shown in Fig. 5C and D. DCs with deleted Tgfbr2 have a significantly increased (P<0.05) ability to stimulate specific T cell proliferation (in 10,000 and 5000 cells), which shows an important role of TGF-β signaling to regulate antigen presentation in DCs during tumorigenesis. Decreased antigen presentation function of DCs will lower the specific stimulation of T cell proliferation against the tumor. A stronger effect (P<0.01 in 1:1 and 1:2 ratios) was observed when we mixed splenocytes from two types of mice. In this type of experiment, increased T cell proliferation was driven by increased AP function of DCs, in parallel with a decreased ability of MDSCs to inhibit T cell proliferation. In summary, the effect of Tgfbr2 deletion in regulation of immune response clearly showed a protumorigenic role of TGF-β signaling in myeloid cells by decreasing AP function of DCs and increasing of suppressive function of MDSCs.
Figure 5. Phenotype and functions of DCs.
(A) Representative FACS plots of a number of subpopulations of DCs in spleen. Flow cytometry strategy is shown in Supplemental Fig. 2A. Lymphoid CD4+ DCs are gated for CD11c+CD8−B220−, myeloid DCs as CD11c+B220−CD8−CD4−, plasmacytoid DCs B220+CD11b−, and lymphoid CD8+ DCs as CD11b−B220−CD4−CD8+. (B) Number of myeloid DCs in tumor tissue; cells are gated as CD45+CD4−CD8−B220−. Data correspond with the mean ± sem of three individual mice from three experiments. (C and D) MLR; DCs were isolated by FACS sorting from EL4-OVA tumor tissue and then mixed in different ratios with splenocytes from OT-I mice (C). MLR; splenocytes from tumor-bearing mice (EL4-OVA) were mixed in different ratios with splenocytes from OT-I mice (D). Data correspond with the mean ± sem of three individual mice from two experiments. *P < 0.05; **P < 0.01; data correspond with the mean ± sem.
DISCUSSION
Myeloid cells (neutrophils/MDSCs, monocytes/macrophages, and DCs) are involved in tumorigenesis with one primary goal: to decrease tumor growth and help cytotoxic T cells to eliminate malignant cells. However, different soluble factors, which comprise a portion of the tumor microenvironment, can change their properties from anti- to protumorigenic, and TGF-β is one of them. The role of TGF-β in the function of immune cells was summarized very well in several reviews [16, 39–41]. However, these reviews focused on the role of TGF-β in lymphoid cells or in myeloid cells during the specific responses to wound healing, infection, or normal physiological conditions, and it is not clear what the exact functions are of TGF-β signaling on myeloid cells during tumor progression.
By using LysM-Cre mice and Tgfbr2 floxed mice, we generated mice with deleted Tgfbr2 in LysM+ cells, which are myeloid cells [27]. This deletion did not affect normal hematopoiesis parameters measured (Fig. 1), but we found that tumors grew more slowly in Tgfbr2MyeKO mice (Fig. 2). An important detail is that tumor growth slowed in Tgfbr2MyeKO mice after 2 weeks following tumor injection, not from the 1st day. We suggest that this time frame is necessary to format the specific immune response against tumor tissue, and this process is more efficient in mice with deleted Tgfbr2 in myeloid cells. Also, during tumor growth, the level of TGF-β is increasing in the tumor microenvironment, which makes the differences of loss of Tgfbr2 more visible.
It has been shown that TGF-β plays a direct and indirect role as a chemoattractant for macrophages [14, 42] and acts initially to stimulate an immune response by recruitment of PMN neutrophils, but when inflammation is ongoing, TGF-β limits the influx through indirect mechanisms [43, 44]. We found no differences in the number of myeloid cells on the 14th day of tumor progression. This can be explained in two ways. First, we did not analyze the number of these cells at early or late stages of tumor progression, where differences could occur, as we wanted to check the functions of these cells when a difference in tumor growth had appeared—the 14th day after injection. Second, many other factors play a primary, important role in the migration of myeloid cells, such as CXCL1, CXCL5, or MCP1, and these chemokines can eliminate the effect of TGF-β as a chemoattractant.
TGF-β can inhibit macrophage phagocytosis [17, 18], inhibit production of TNF-α [19], down-regulate the production of NO, and inhibit iNOS [21]. We observed that in macrophages with intact TGF-β signaling, isolated from tumor tissue, expression of iNOS, MMP2, and TNF-α is increased in parallel with decreased MMP9. In vitro data showed decreased basal secretion of IL-6 and no response in VEGF and TNF-α secretion when stimulated with TGF-β (Fig. 3). It has been shown that high levels of iNOS correspond with anti-tumorigenic, classically activated M1 macrophages [33, 45]. The activated inflammatory macrophage plays a crucial role in matrix remodeling by producing MMPs. MMP2/9 efficiently degrades collagen IV and laminin-5, thereby assisting the metastatic cancerous cells to pass through the basement membrane. TAMs have been reported to correlate with the metastatic potential of a variety of human cancers, and they have also been shown to be a major source of MMP9 [46, 47]. We propose that higher MMP2 will help in migration of macrophages to tumor tissue by remodeling the ECM and in parallel, with higher iNOS and TNF-α and the absence of VEGF response increases the anti-tumorigenic properties of these cells.
In the analysis of the second major population of myeloid cells, CD11b+Gr1+ cells, we found no differences in the number of this cell type and their phenotype, except a slightly decreased number of subpopulations of these cells—IL-4Ra+ in tumors (Fig. 4). It has been reported that most CD11b+Gr1+ cells in tumor tissue are MDSCs [28, 29, 48, 49], and they suppress T cell proliferation. The arginase mechanism is common for this effect, and we found that TGF-β signaling is involved in increasing Arg expression. In Tgfbr2MyeKO mice, MDSCs have a decrease in their suppressive function, and in parallel, we found an increased number of CD8a+ cytotoxic T cells in tumor tissue. Also, we found decreased expression of S100A8, a protein that correlates highly with the suppressive function of MDSC [50, 51]. These results mean that development of the suppressive function of MDSCs is dependent on TGF-β signaling. Additionally, we detected that TGF-β increased CXCR4 expression in MDSCs, and we showed previously that this receptor plays an important role in aggressive breast cancer development [31]. As in macrophages, we found increased iNOS and MMP2 in MDSCs with deleted Tgfbr2, but because it was cells isolated from tumor tissue, we cannot say that it is a direct or indirect effect of TGF-β, but we suggest that these genes increased anti-tumorigenic properties in Tgfbr2MyeKO cells.
The proper function of DCs during tumorigenesis is a key factor in having a sufficiently specific immune response. Deletion of Tgfbr2 does not affect the number or phenotype of these cells (Fig. 5A), but the antigen presentation of DCs in tumor tissue and their ability to stimulate specific T cell proliferation are increased in cells without Tgfbr2. Moreover, DCs mixed with MDSCs (both from Tgfbr2MyeKO mice) can simulate specific T cell proliferation more sufficiently than cells with intact TGF-β signaling (Fig. 5B). Taken together, this clearly shows us that TGF-β on myeloid cells has a negative effect in the regulation of the immune system during tumorigenesis. In summary, our results demonstrated that deletion of Tgfbr2 in myeloid cells inhibits tumor growth by increasing efficiency of the immune system. The mechanism behind this is modifying the function of TAMs to an anti-tumorigenic phenotype, decreased suppressive function of CD11b+Gr1+ cells, and increased antigen-presenting function of DCs.
Supplementary Material
ACKNOWLEDGMENTS
This work is supported by U.S. National Institutes of Health grants CA085492, A102162, and CA126505 and the T.J. Martell Foundation to H.L.M. and CA068485 for core laboratory support.
The online version of this paper, found at www.jleukbio.org, includes supplemental information.
- Arg
- arginine
- CT
- comparative threshold
- G-MDSC
- granulocyte myeloid-derived suppressor cell
- IHC
- immunohistochemistry
- IL-4ra
- IL-4R-α
- KO
- knockout
- LLC
- Lewis lung carcinoma
- LysM
- lysozyme M
- M-MDSC
- monocyte myeloid-derived suppressor cell
- MDSC
- myeloid-derived suppressor cell
- MLR
- mixed leukocyte reaction
- MMP
- matrix metalloproteinase
- MMTV-PyMT
- mouse mammary tumor virus-polyoma middle T antigen
- qRT-PCR
- quantitative RT-PCR
- TAM
- tumor-associated macrophage
- Tgfbr2
- TGF-β type II receptor
- Tgfbr2MyeKO
- deletion of TGF-β type II receptor in myeloid cells
AUTHORSHIP
S.V.N. and H.L.M. performed research, analyzed and interpreted data, and wrote the manuscript. M.W.P., A.C., D.P., and P.O. performed research.
REFERENCES
- 1. Mueller M. M., Fusenig N. E. (2011) Tumor-Associated Fibroblasts and Their Matrix. Springer, Dordrecht, Germany [Google Scholar]
- 2. Forrester E., Chytil A., Bierie B., Aakre M., Gorska A. E., Sharif-Afshar A. R., Muller W. J., Moses H. L. (2005) Effect of conditional knockout of the type II TGF-β receptor gene in mammary epithelia on mammary gland development and polyomavirus middle T antigen induced tumor formation and metastasis. Cancer Res. 65, 2296–2302 [DOI] [PubMed] [Google Scholar]
- 3. Bierie B., Chung C. H., Parker J. S., Stover D. G., Cheng N., Chytil A., Aakre M., Shyr Y., Moses H. L. (2009) Abrogation of TGF-β signaling enhances chemokine production and correlates with prognosis in human breast cancer J. Clin. Invest. 119, 1571–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yang L., Huang J., Ren X., Gorska A. E., Chytil A., Aakre M., Carbone D. P., Matrisian L. M., Richmond A., Lin P. C., Moses H. L. (2008) Abrogation of TGF β signaling in mammary carcinomas recruits Gr-1+CD11b+ myeloid cells that promote metastasis. Cancer Cell 13, 23–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Novitskiy S. V. (2011) TGF-β receptor II loss promotes mammary carcinoma progression by Th17 dependent mechanisms. Cancer Discov. 1, 430–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ijichi H., Chytil A., Gorska A. E., Aakre M. E., Bierie B., Tada M., Mohri D., Miyabayashi K., Asaoka Y., Maeda S., Ikenoue T., Tateishi K., Wright C. V., Koike K., Omata M., Moses H. L. (2011) Inhibiting Cxcr2 disrupts tumor-stromal interactions and improves survival in a mouse model of pancreatic ductal adenocarcinoma. J. Clin. Invest. 121, 4106–4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Borczuk A. C., Sole M., Lu P., Chen J., Wilgus M. L., Friedman R. A., Albelda S. M., Powell C. A. (2011) Progression of human bronchioloalveolar carcinoma to invasive adenocarcinoma is modeled in a transgenic mouse model of K-ras-induced lung cancer by loss of the TGF-β type II receptor. Cancer Res. 71, 6665–6675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fang W. B., Jokar I., Chytil A., Moses H. L., Abel T., Cheng N. (2011) Loss of one Tgfbr2 allele in fibroblasts promotes metastasis in MMTV: polyoma middle T transgenic and transplant mouse models of mammary tumor progression. Clin. Exp. Metastasis 28, 351–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marie J. C., Liggitt D., Rudensky A. Y. (2006) Cellular mechanisms of fatal early-onset autoimmunity in mice with the T cell-specific targeting of transforming growth factor-β receptor. Immunity 25, 441–454 [DOI] [PubMed] [Google Scholar]
- 10. Kim B. G., Li C., Qiao W., Mamura M., Kasprzak B., Anver M., Wolfraim L., Hong S., Mushinski E., Potter M., Kim S. J., Fu X. Y., Deng C., Letterio J. J. (2006) Smad4 signalling in T cells is required for suppression of gastrointestinal cancer. Nature 441, 1015–1019 [DOI] [PubMed] [Google Scholar]
- 11. Zahner S. P., Kel J. M., Martina C. A., Brouwers-Haspels I., van Roon M. A., Clausen B. E. (2011) Conditional deletion of TGF-βR1 using Langerin-Cre mice results in Langerhans cell deficiency and reduced contact hypersensitivity. J. Immunol. 187, 5069–5076 [DOI] [PubMed] [Google Scholar]
- 12. Yamaguchi Y., Tsumura H., Miwa M., Inaba K. (1997) Contrasting effects of TGF-β 1 and TNF-α on the development of dendritic cells from progenitors in mouse bone marrow. Stem Cells 15, 144–153 [DOI] [PubMed] [Google Scholar]
- 13. Geissmann F., Revy P., Regnault A., Lepelletier Y., Dy M., Brousse N., Amigorena S., Hermine O., Durandy A. (1999) TGF-β 1 prevents the noncognate maturation of human dendritic Langerhans cells. J. Immunol. 162, 4567–4575 [PubMed] [Google Scholar]
- 14. Wahl S. M., Allen J. B., Weeks B. S., Wong H. L., Klotman P. E. (1993) Transforming growth factor β enhances integrin expression and type IV collagenase secretion in human monocytes. Proc. Natl. Acad. Sci. USA 90, 4577–4581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wahl S. M., Hunt D. A., Wakefield L. M., McCartney-Francis N., Wahl L. M., Roberts A. B., Sporn M. B. (1987) Transforming growth factor type β induces monocyte chemotaxis and growth factor production. Proc. Natl. Acad. Sci. USA 84, 5788–5792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li M. O., Wan Y. Y., Sanjabi S., Robertson A. K., Flavell R. A. (2006) Transforming growth factor-β regulation of immune responses. Annu. Rev. Immunol. 24, 99–146 [DOI] [PubMed] [Google Scholar]
- 17. Bottalico L. A., Wager R. E., Agellon L. B., Assoian R. K., Tabas I. (1991) Transforming growth factor-β 1 inhibits scavenger receptor activity in THP-1 human macrophages. J. Biol. Chem. 266, 22866–22871 [PubMed] [Google Scholar]
- 18. Han J., Hajjar D. P., Tauras J. M., Feng J., Gotto A. M., Jr., Nicholson A. C. (2000) Transforming growth factor-β1 (TGF-β1) and TGF-β2 decrease expression of CD36, the type B scavenger receptor, through mitogen-activated protein kinase phosphorylation of peroxisome proliferator-activated receptor-γ. J. Biol. Chem. 275, 1241–1246 [DOI] [PubMed] [Google Scholar]
- 19. Bogdan C., Paik J., Vodovotz Y., Nathan C. (1992) Contrasting mechanisms for suppression of macrophage cytokine release by transforming growth factor-β and interleukin-10. J. Biol. Chem. 267, 23301–23308 [PubMed] [Google Scholar]
- 20. Werner F., Jain M. K., Feinberg M. W., Sibinga N. E., Pellacani A., Wiesel P., Chin M. T., Topper J. N., Perrella M. A., Lee M. E. (2000) Transforming growth factor-β 1 inhibition of macrophage activation is mediated via Smad3. J. Biol. Chem. 275, 36653–36658 [DOI] [PubMed] [Google Scholar]
- 21. Vodovotz Y., Bogdan C., Paik J., Xie Q. W., Nathan C. (1993) Mechanisms of suppression of macrophage nitric oxide release by transforming growth factor β. J. Exp. Med. 178, 605–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tsunawaki S., Sporn M., Ding A., Nathan C. (1988) Deactivation of macrophages by transforming growth factor-β. Nature 334, 260–262 [DOI] [PubMed] [Google Scholar]
- 23. Yang L., DeBusk L. M., Fukuda K., Fingleton B., Green-Jarvis B., Shyr Y., Matrisian L. M., Carbone D. P., Lin P. C. (2004) Expansion of myeloid immune suppressor Gr+CD11b+ cells in tumor-bearing host directly promotes tumor angiogenesis. Cancer Cell 6, 409–421 [DOI] [PubMed] [Google Scholar]
- 24. Chytil A., Magnuson M. A., Wright C. V., Moses H. L. (2002) Conditional inactivation of the TGF-β type II receptor using Cre: Lox. Genesis 32, 73–75 [DOI] [PubMed] [Google Scholar]
- 25. Novitskiy S. V., Ryzhov S., Zaynagetdinov R., Goldstein A. E., Huang Y., Tikhomirov O. Y., Blackburn M. R., Biaggioni I., Carbone D. P., Feoktistov I., Dikov M. M. (2008) Adenosine receptors in regulation of dendritic cell differentiation and function. Blood 112, 1822–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ljung B. M., Mayall B., Lottich C., Boyer C., Sylvester S. S., Leight G. S., Siegler H. F., Smith H. S. (1989) Cell dissociation techniques in human breast cancer—variations in tumor cell viability and DNA ploidy. Breast Cancer Res. Treat. 13, 153–159 [DOI] [PubMed] [Google Scholar]
- 27. Clausen B. E., Burkhardt C., Reith W., Renkawitz R., Forster I. (1999) Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Trans. Res. 8, 265–277 [DOI] [PubMed] [Google Scholar]
- 28. Youn J. I., Gabrilovich D. I. (2010) The biology of myeloid-derived suppressor cells: the blessing and the curse of morphological and functional heterogeneity. Eur. J. Immunol. 40, 2969–2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gabrilovich D. I., Nagaraj S. (2009) Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunology. 9, 162–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nagaraj S., Gabrilovich D. I. (2007) Myeloid-derived suppressor cells. Adv. Exp. Med. Biol. 601, 213–223 [DOI] [PubMed] [Google Scholar]
- 31. Yang L., Moses H. L. (2008) Transforming growth factor β: tumor suppressor or promoter? Are host immune cells the answer? Cancer Res. 68, 9107–9111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Marx J. (2008) Cancer immunology. Cancer's bulwark against immune attack: MDS cells. Science 319, 154–156 [DOI] [PubMed] [Google Scholar]
- 33. Mantovani A., Sozzani S., Locati M., Allavena P., Sica A. (2002) Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 23, 549–555 [DOI] [PubMed] [Google Scholar]
- 34. Qian B. Z., Pollard J. W. (2010) Macrophage diversity enhances tumor progression and metastasis. Cell 141, 39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zaynagetdinov R., Sherrill T. P., Polosukhin V. V., Han W., Ausborn J. A., McLoed A. G., McMahon F. B., Gleaves L. A., Degryse A. L., Stathopoulos G. T., Yull F. E., Blackwell T. S. (2011) A critical role for macrophages in promotion of urethane-induced lung carcinogenesis. J. Immunol. 187, 5703–5711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Youn J. I., Nagaraj S., Collazo M., Gabrilovich D. I. (2008) Subsets of myeloid-derived suppressor cells in tumor-bearing mice. J. Immunol. 181, 5791–5802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Youn J. I., Collazo M., Shalova I. N., Biswas S. K., Gabrilovich D. I. (2012) Characterization of the nature of granulocytic myeloid-derived suppressor cells in tumor-bearing mice. J. Leukoc. Biol. 91, 167–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Marigo I., Dolcetti L., Serafini P., Zanovello P., Bronte V. (2008) Tumor-induced tolerance and immune suppression by myeloid derived suppressor cells. Immunol. Rev. 222, 162–179 [DOI] [PubMed] [Google Scholar]
- 39. Yoshimura A., Muto G. (2011) TGF-β function in immune suppression. Curr. Top. Microbiol. Immunol. 350, 127–147 [DOI] [PubMed] [Google Scholar]
- 40. Yang L., Pang Y., Moses H. L. (2010) TGF-β and immune cells: an important regulatory axis in the tumor microenvironment and progression. Trends Immunol. 31, 220–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Flavell R. A., Sanjabi S., Wrzesinski S. H., Licona-Limon P. (2010) The polarization of immune cells in the tumour environment by TGFβ. Nat. Rev. Immunol. 10, 554–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Allen J. B., Manthey C. L., Hand A. R., Ohura K., Ellingsworth L., Wahl S. M. (1990) Rapid onset synovial inflammation and hyperplasia induced by transforming growth factor β. J. Exp. Med. 171, 231–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Reibman J., Meixler S., Lee T. C., Gold L. I., Cronstein B. N., Haines K. A., Kolasinski S. L., Weissmann G. (1991) Transforming growth factor β 1, a potent chemoattractant for human neutrophils, bypasses classic signal-transduction pathways. Proc. Natl. Acad. Sci. USA 88, 6805–6809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brandes M. E., Mai U. E., Ohura K., Wahl S. M. (1991) Type I transforming growth factor-β receptors on neutrophils mediate chemotaxis to transforming growth factor-β. J. Immunol. 147, 1600–1606 [PubMed] [Google Scholar]
- 45. Sindrilaru A., Peters T., Wieschalka S., Baican C., Baican A., Peter H., Hainzl A., Schatz S., Qi Y., Schlecht A., Weiss J. M., Wlaschek M., Sunderkotter C., Scharffetter-Kochanek K. (2011) An unrestrained proinflammatory M1 macrophage population induced by iron impairs wound healing in humans and mice. J. Clin. Invest. 121, 985–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Illemann M., Bird N., Majeed A., Sehested M., Laerum O. D., Lund L. R., Dano K., Nielsen B. S. (2006) MMP-9 is differentially expressed in primary human colorectal adenocarcinomas and their metastases. Mol. Cancer Res. 4, 293–302 [DOI] [PubMed] [Google Scholar]
- 47. John A., Tuszynski G. (2001) The role of matrix metalloproteinases in tumor angiogenesis and tumor metastasis. Pathol. Oncol. Res. 7, 14–23 [DOI] [PubMed] [Google Scholar]
- 48. Lu T., Ramakrishnan R., Altiok S., Youn J. I., Cheng P., Celis E., Pisarev V., Sherman S., Sporn M. B., Gabrilovich D. (2011) Tumor-infiltrating myeloid cells induce tumor cell resistance to cytotoxic T cells in mice. J. Clin. Invest. 121, 4015–4029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Condamine T., Gabrilovich D. I. (2011) Molecular mechanisms regulating myeloid-derived suppressor cell differentiation and function. Trends Immunol. 32, 19–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ichikawa M., Williams R., Wang L., Vogl T., Srikrishna G. (2011) S100A8/A9 activate key genes and pathways in colon tumor progression. Mol. Cancer Res. 9, 133–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sinha P., Okoro C., Foell D., Freeze H. H., Ostrand-Rosenberg S., Srikrishna G. (2008) Proinflammatory S100 proteins regulate the accumulation of myeloid-derived suppressor cells. J. Immunol. 181, 4666–4675 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.