Review on how the integration of bioenergetics and metabolism controls inflammation progression.
Keywords: AMP, epigenetics, fatty acid oxidation, glycolysis, HIF-1α, NAD+, Nampt, obesity, PGC-1, sepsis
Abstract
We review the emerging concept that changes in cellular bioenergetics concomitantly reprogram inflammatory and metabolic responses. The molecular pathways of this integrative process modify innate and adaptive immune reactions associated with inflammation, as well as influencing the physiology of adjacent tissue and organs. The initiating proinflammatory phase of inflammation is anabolic and requires glucose as the primary fuel, whereas the opposing adaptation phase is catabolic and requires fatty acid oxidation. The fuel switch to fatty acid oxidation depends on the sensing of AMP and NAD+ by AMPK and the SirT family of deacetylases (e.g., SirT1, -6, and -3), respectively, which couple inflammation and metabolism by chromatin and protein reprogramming. The AMP-AMPK/NAD+-SirT axis proceeds sequentially during acute systemic inflammation associated with sepsis but ceases during chronic inflammation associated with diabetes, obesity, and atherosclerosis. Rebalancing bioenergetics resolves inflammation. Manipulating cellular bioenergetics is identifying new ways to treat inflammatory and immune diseases.
Introduction
Inflammation is an evolutionarily conserved, coordinated response to harmful stimuli, with a goal of returning to homeostasis [1]. Thus, the inflammatory response has a unifying purpose encoded in the germline: protection and restoration. The cellular and soluble components of innate and adaptive immunity fulfill this purpose, during which a typical inflammatory response progresses from a proinflammatory to an adaptive phase and eventually restores homeostasis.
Emerging data support that switches in bioenergy inextricably link metabolism with inflammation and immunity to protect cells and organisms and to restore homeostasis [2]. As a result, metabolic polarity exists between the anabolic proinflammatory phase, which requires glycolysis to meet the rapid demands for high energy during the early response to a threat, and the catabolic adaptation phase, which depends on fatty acid oxidation to heal and restore homeostasis [3, 4]. The adaptation phase has been variously called a compensatory anti-inflammatory response or endotoxin tolerance, but the term “adaptation phase” better reflects this complex reprogramming state for inflammatory and metabolic signaling pathways and genes. During acute systemic inflammatory diseases, such as sepsis, the polarity is sequential and predictable, shifting from the proinflammatory phase to the adaptation phase, whereas in chronic inflammatory diseases, such as diabetes, obesity with metabolic syndrome, and atherosclerosis, the proinflammatory phase dominates and persists, unless there are external changes in nutrition and bioenergy requirements [5]. Importantly, the adaptation phase of acute systemic inflammation from sepsis is associated with immunosuppression of innate and antigen-specific acquired immunity.
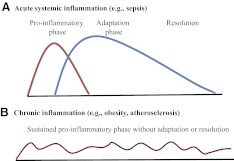
The interplay of cellular bioenergetics, metabolism, and inflammation occurs in innate and adaptive immune responses associated with inflammatory diseases, and similar changes may occur in organ-specific tissues. Prime examples of this integrated relationship and its distinctions occur during the acute systemic inflammatory response associated with sepsis and during chronic inflammatory responses characteristic of obesity with metabolic syndrome and atherosclerosis. Fig. 1 illustrates the distinctions between acute systemic inflammation and chronic inflammation in two graphs. Fig. 1A depicts the shift of the acute systemic inflammatory response from a proinflammatory phase to an adaptation phase, and Fig. 1B depicts the inflexibility of a sustained proinflammatory phase of chronic inflammatory diseases. Here, we review how bioenergy sensing sequentially coordinates shifts of metabolism with the conversion of the proinflammatory phase to the adaptation phase during acute systemic inflammation and in contrast, how this transition is unsuccessful in chronic inflammation, where the process remains a proinflammatory phase, unless there is an intervention. We divide our discussion into the following components: 1) reprogramming inflammatory genes; 2) reprogramming glycolysis and fatty acid oxidation metabolic pathways during inflammation; 3) connecting cellular bioenergetics with energy sensors; 4) illustrating how bioenergy sensing integrates switches in metabolism and inflammation to generate clinical phenotypes; and 5) identifying challenges, opportunities, and questions derived from this unified concept.
Figure 1. Inflammation processes.
(A) Acute systemic inflammation switches from the proinflammatory phase to the adaptation phase and eventually progresses to resolution. (B) Chronic inflammation sustains a proinflammatory phase.
REPROGRAMMING INFLAMMATORY GENES
Acute inflammation
As described in ancient writings, acute inflammation starts abruptly and usually resolves [6]. During acute inflammation, mild or strong stimulation of TLR4 by severe trauma in humans reprograms ∼80% of the protein-encoding genome of human blood leukocytes [7]; expression of ∼50% of these genes increased, and 50% decreased. Remarkably, this broad response varied only in duration, which was determined by the magnitude of the environmental stress. The same 80% of genes were reprogrammed even after a small dose of endotoxin administered i.v. to humans. This important study shows the breadth of gene programming after an acute inflammatory response and emphasizes that inflammation reprogramming is determined epigenetically.
The proinflammatory phase of reprogramming induces gene products such as TNF-α and IL-1β and then represses genes that ignited the proinflammatory phase and reprograms many other sets of genes to support the adaptation phase [8], which lasts much longer than the initiating proinflammatory-phase response before inflammation resolves to homeostasis [9]. The switch from the proinflammatory phase to the adaptation phase cannot be identified clinically, which provides one reason why early sepsis trials using a variety of anti-inflammatory approaches failed [10].
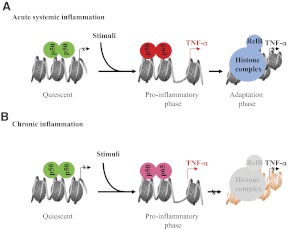
Acute inflammation ignites after TLRs and other sensors, such as complement or antibody receptors, react to danger [11]. Until then, the rapid-response proinflammatory genes maintain a poised chromatin state, wherein RNA polymerase II is terminated prematurely, and the proximal promoters remain epigenetically silenced [12]. Sensing a threat rapidly removes the repressor complex to prompt the broad response of innate myeloid-derived monocytes and neutrophils—as well as Teffs—whose signaling paths converge on chromatin to initiate inflammation's epigenetic reprogramming [8]. The rate-limiting signals for igniting the acute systemic inflammatory phase include p65, stress kinases (e.g., ERK, JNK, p38), and protein methylases and acetyl transferases. During acute systemic inflammation— and likely, acute local inflammation—the proinflammatory-phase response switches within hours to the adaptation phase, which opposes many acute cellular responses. This shift is determined epigenetically. First, p65 promoter binding at proinflammatory-phase genes (e.g., TNF-α and IL-1β) is deactivated by deacetylation and degradation [13, 14]. A second feature of the switch requires de novo synthesis NF-κB member RelB, which binds to NF-κB p50 dimers and replaces p65 [15–18]. Continued RelB expression sustains the adaptation phase by directly assembling and stabilizing facultative (reversible) heterochromatin to silence proinflammatory-phase genes (e.g., TNF-α and IL-1β) but activate euchromatin at adaptation-phase genes [19] (e.g., IκBα). Importantly, chromatin silencing can be reversed by removing RelB or other members of the facultative heterochromatin repressor complex (e.g., histone and methyltransferases and structural proteins) [20]. These modifications in chromatin structure are not isolated, as many other repressor proteins in the cytosol and nucleus assure the adaptation phase [21]. Fig. 2A depicts a simplified scheme of the epigenetic switch between the pro- and adaptation phase in acute systemic inflammation.
Figure 2. Epigenetic reprogramming of inflammatory genes.
(A) Proinflammatory phase requires p65, and adaptation phase requires RelB epigenetic silencing during acute systemic inflammation. (B) Chronic inflammation does not deactivate p65 nor develop an appropriate opposing adaptation phase. Green, Quiescent state; red, proinflammatory phase; blue, adaptation phase.
Chronic inflammation
Fig. 1A and B highlights a distinction between acute and chronic inflammation, where acute systemic inflammation in survivors can eventually subside after the adaptation phase, but chronic inflammation sustains a waxing and waning proinflammatory phase. This does not mean that phase polarity is totally absent in chronic inflammation, as mixtures of dominating proinflammatory-phase M1 macrophages and activated Teffs coexist with low levels of adaptation-phase M2 macrophages and suppressor Tregs in adipose tissue of obese diabetics [22–26] and in atherosclerosis lesions [27]. However, the two phases occur in cells located in different anatomical regions of the pathologic tissue. In contrast, others and our studies of a THP1 cell promonocyte model of TLR4-dependent polarity and changes in sepsis blood leukocytes support a linear transformation from proinflammatory phase to adaptation phase during acute systemic inflammation [5, 8]. That linear changes in phases can occur sequentially within the same cell is also supported by recent reports showing that a single quiescent cell can be the progenitor for polarity in phenotypes [28, 29]. Still, the polarity between the proinflammatory phase and the adaptation phase associated with sepsis may differ in vivo and distinct phenotypes of proinflammatory and adaptation phase, where monocytes, neutrophils, and T cells may coexist in blood during sepsis. One explanation for in vivo heterogeneity of circulating phenotypes is that bone marrow or spleen cells exist in distinct phases that enter the circulation and target tissues. In any event, the unrestrained and persistent proinflammatory phase of chronic inflammation is distinct from acute inflammation, and the mechanisms responsible for this are unknown.
Another difference between acute and chronic inflammation is “flame” intensity. What makes chronic inflammation simmer, and why has evolution not adapted to simmering by switching to an adaptation phase to restore homeostasis? Growing reports clarify that proximal sensing and signaling mechanisms for inducing chronic inflammation may differ from those igniting acute inflammation. For example, extracellular sensors of fatty acids or very low doses of circulating gut-derived endotoxin may generate distinct signaling paths to chromatin reprogramming. A reported example is that macrophages, sensing very low concentrations of endotoxin, bypass NF-κB pathways, using an alternative route that accentuates subsequent TLR4 responses [30]. This distinct path for priming macrophages with low-dose endotoxin requires that IL-1R-activating kinase selectively displaces repressor nuclear receptors and activates C/EBPδ and IKKϵ.
REPROGRAMMING GLYCOLYSIS AND FATTY ACID OXIDATION METABOLIC PATHWAYS DURING INFLAMMATION
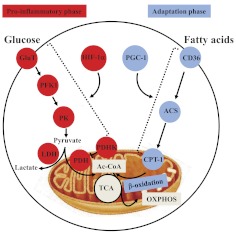
Many studies of acute or chronic systemic inflammatory diseases now support that the proinflammatory phase and the innate and adaptive immune effector responses depend on glucose as fuel and that the adaptation-phase responses of both arms of immunity depend on fatty acid oxidation [3, 31–33]. The principal metabolic pathways of glucose and fatty acid oxidation that fuel the polarity of the proinflammation phase and anti-inflammation phase are shown in Fig. 3. Early glycolytic reprogramming provides a surge of ATP for anabolic energy, which is generated under normoxia conditions, thereby simulating the Warburg effect of glycolysis and reduced mitochondrial glucose oxidation typical of many cancer cells [34]. It also occurs during hypoxia. Glycolysis also activates the pentose shunt to kill bacteria by NADPH oxidase and to provide amino acids and fats for anabolism. The regulatory components of amplified glucose fueling include increased expression of Glut1, elevated expression of the series of glycolysis regulatory genes, and disrupted mitochondrial glucose oxidation by PDHK, which deactivates mitochondrial-located PDH. This on and off switching, which limits mitochondrial glucose oxidation, generates increased intracellular and extracellular pyruvate and lactate [35]. The glycolysis surge and reduced glucose mitochondrial oxidation depend on HIF-1α, which is stabilized by inactivating prolyl hydroxylase activity and transactivated by the NF-κB and other signaling events [36]. Thus, the pivotal HIF-1α and p65 pathways provide a bridge for regulating inflammation and modifying glucose metabolism. Increased glucose flux is essential to support transactivation of DNA of host defense genes, such as TNF-α and IL-1β, perhaps by increasing levels of acetate and acetyl CoA and acetylation of nuclear regulators [37–39]. Importantly, increased glucose flux is absolutely required for immunocompetent effector responses. This effective host defensive and proinflammatory-phase path generates ROS to kill microbes, but ROS also injure proteins and DNA. Among essential organelles damaged are mitochondria, which enter autophagic vacuoles (mitophagy), leading to pronounced decreases in ATP and mitochondrial mass [40, 41].
Figure 3. Metabolic regulation of inflammation.
The proinflammatory phase requires HIF-1α-dependent glycolysis, and adaptation phase requires PGC-1-dependent fatty acid oxidation and mitochondrial biogenesis. Red, Proinflammatory phase; blue, adaptation phase. PFK1, Phosphofructokinase-1; PK, pyruvate kinase; Ac-CoA, acetyl CoA; TCA, tricarboxylic acid; ACS, acyl-CoA synthetase; CPT-1, carnitine palmitoyltransferase 1.
Metabolism also links with the adaptation phase of acute systemic inflammation, but it is distinctly different, wherein the critical HIF 1α-dependent glycolytic phase switches to increased fatty acid oxidation and increased expression of PGC-1α and -β [42, 43], and PGC-1α and/or -β support expression of NRF1 and -2), which transactivate multiple mitochondrial structural and OXPHOS genes used in mitochondrial biogenesis and respirator physiology [44]. This coupled sequence occurs during animal and human sepsis [45, 46] and when M2 alternative adaptation-phase-like macrophages are generated in vitro by IL-4 [47, 48]. Importantly, PGC-1α and -β also control expression of the genes used for fatty acid oxidation and mitochondrial biogenesis [49]. Thus, the resulting adaptation phase in macrophages exists in a catabolic state of low-energy requirements [50] with fatty acids as the predominant fuel. Recent data support that metabolic polarity during acute and chronic inflammation also characterizes the shift from glycolytic Teffs, such as T17 cells, to fatty acid oxidative repressor T cells, such as Tregs and antigen-tolerant Th1 T cells [51, 52]. Together, emerging data define a new paradigm, in which a unifying mechanism regulates innate and acquired immunity, inflammatory and immune polarity to balance effector and adaptation responses.
The concept for the M2 alternative macrophage, which has adaptation-phase characteristics, was identified in chronic inflammation associated with obesity, where M1 proinflammatory-phase and M2 alternative adaptation-phase-like macrophages coexist in inflamed adipose tissue [24, 53]. Proinflammatory-phase M1 cells and Teffs aggregate in hypoxic and necrotic areas containing large adipocytes, whereas adaptation-phase cells are located near normal-appearing fat cells and connective tissue. Apparently, local environmental differences inform the two phenotypes. It is not known what proportion of the M1 and M2 phenotypes and Teffs and Tregs comes from blood and/or is generated within the site of chronic inflammation. However, it is clear that the proinflammatory phase of diabetic or atherosclerosis states persist for months or years, as compared with days to weeks, for acute inflammation. It is also known that the proinflammatory phase of innate and immune T cells requires the HIF-1α-dependent glycolytic path, which with transactivated p65, promotes proinflammation of diabetes and atherosclerosis [54, 55]. Glucose intolerance and liver and cardiac steatosis in mice can be reduced by increasing expression of PGC-1α, which switches the glycolysis-dependent proinflammatory phase to the fatty acid-dependent adaptation phase [56, 57]. Thus, growing evidence indicates that the proinflammatory-phase immunocytes of chronic inflammation are “suspended” in a glycolytic and low mitochondrial glucose oxidation state, accompanied by elevated ROS and dysregulated mitochondrial biogenesis [58].
CONNECTING CELLULAR BIOENERGETICS WITH ENERGY SENSORS
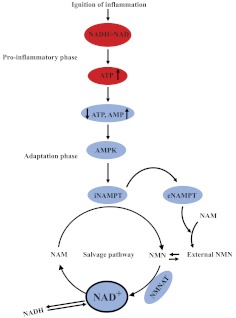
Bioenergy balance between AMP and ATP and oxidized NAD+ and reduced NADH inform many cellular functions associated with inflammation and metabolism, including intracellular signaling pathways, nuclear transcription factors, and chromatin structure [59]. Increases in ATP production and NADH formation decrease the ratios of AMP/ATP, and NAD+/NADH occurs during the early proinflammatory-phase response of innate and adaptive immunity effector responses. During the switch from the proinflammatory and proimmune phase to the adaptation phase, elevated ratios of AMP/ATP and NAD+/NADH and/or the de novo generation of NAD+ become pivotal regulators of cellular metabolism. In animals, NAD+ production is controlled primarily by the rate-limiting enzyme, Nampt, which is expressed as iNampt and eNampt [60], induced by AMP sensor AMPK [61]. Surprisingly eNampt has proinflammatory-phase properties and provides an extracellular source of NAD+ [62]. Changes in iNampt provide the rate-limiting intracellular step for producing intracellular NAD+. The Nampt pathway for NAD+ synthesis during acute systemic inflammation is depicted in Fig. 4. Important features of NAD+ biosynthesis, in addition to generating NAD+, include NMN production and activation of NMNAT, which occurs in three forms that translocate in distinct regions within cells (nucleus, Golgi complex, and mitochondria) to provide compartment-specific production of NAD+ [59]. NAD+, generated by iNampt and eNampt, is critically important for regulating the metabolic requirements of inflammation, as well as reprogramming expression of genes encoding mediators of inflammation.
Figure 4. NAD+ generation by rate-limiting NAMPT.
Red, proinflammatory phase; blue, adaptation phase.
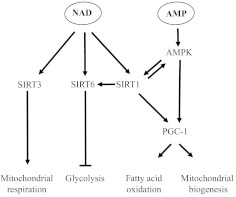
The importance of NAD+ in cell biology markedly expanded after the discovery of the Sir2 family [63]. By sensing NAD+, these proteins function as deacetylases that reverse the transactivating acetylation of proinflammatory-phase genes and metabolic enzymes used in glycolysis [64]. Sir2, the founding SirT member of Class III deacetylases, was identified in yeast at heterochromatin loci. There are seven members of the Sir2 family in mammals. Three of these, SirT1, SirT3, and SirT6, by NAD+ bioenergy sensing are emerging as providing a critically important, proximal axis for regulating metabolism and inflammation [65]. SirT1 and -6 are most prominent in the nucleus and influence gene programming by modifying transcription factor competence and silencing genes by promoting heterochromatin formation. Examples of nuclear regulation of genes controlling inflammation and/or metabolism include the direct deacetylation and deactivation of p65, histone H4 deacetylation, and increased expression and promoter loading of RelB dual chromatin modifier of SirT1 [66], which also partners with AMPK, preceding SirT deactivation of proinflammatory-phase genes. SirT6 deacetylates histone H3 lysine 9 to promote silent chromatin at NF-κB-dependent genes [67]. Thus, SirT1 and SirT6 bridge bioenergy shifts with reprogramming of inflammatory genes. Importantly, SirT 6 also is an essential regulator of glucose metabolism, and SirT6 knockout mice die from hypoglycemia [68]. To control glycolysis, SirT6 deacetylates histone H3K9 and silences multiple genes supporting glycolysis, thereby opposing HIF-1α induction of glycolysis [69]. SirT3, by contrast, is primarily located in mitochondria, where it senses compartment-specific increases in NAD+ and deacetylates multiple mitochondrial proteins linked to metabolism and inflammation [70, 71]. Among these are LCADH, which is required for fatty acid β oxidation; mnSOD, which counters ROS; isocitrate dehydrogenase, which supports the citric acid cycle; and members of the electron transport Complexes I and III. Not surprisingly, SirT-3, and -6 are tightly regulated by transcription, microRNA-directed mRNA degradation, and translational and post-translational degradation [4]. PGC-1α supports increased expression of SirT1 and of SirT3, and SirT1 regulates SirT6 [72, 73]. AMPK and NAD+ as a cofactor for SirTs meet all of the preliminary requirements for a cell physiology coordinating system. Fig. 5 depicts how AMPK, SirT1, -3, and -6 bioenergy sensors can modify cell function.
Figure 5. Bioenergy sensing by SirTs and AMPK regulate mitochondrial respiration, mitochondrial biogenesis, glycolysis, and fatty acid oxidation.
ILLUSTRATING HOW BIOENERGY SENSING INTEGRATES SWITCHES IN METABOLISM AND INFLAMMATION TO GENERATE CLINICAL PHENOTYPES
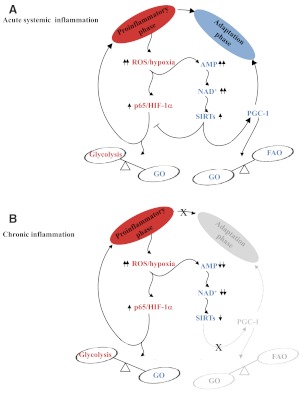
Accumulating data support that bioenergy sensors couple metabolism with inflammation to switch physiologic and clinical phenotypes. During acute systemic inflammation of sepsis, bioenergy shifts rapidly follow TLR stimulation. With the surge of glycolysis and induction of proinflammatory-phase genes, there are increases in glycolysis-induced ATP, reductions in ATP derived from glucose oxidation, and direct disruptions of the electron transport chain by NO [74–76]. An accompanying increase in AMP supports activation of AMPK, inducing expression of Nampt and increases NAD+ [77], which can prompt SirT1 to coordinate the switch from the proinflammatory phase to the adaptation phase [78]. We discovered that NAD+ informs chromatin to switch from the proinflammation phase to the adaptation phase in a THP-1 promonocyte sepsis cell model and in human sepsis leukocytes [66]. To initiate this process, NAD+ sensor (SirT1) deacetylates and deactivates p65 and deacetylates euchromatin histone H4. SirT1 then supports de novo RelB expression and promotes its binding to the TNF-α and IL-1β promoters, where RelB assembles a silencing complex of facultative heterochromatin [8], as discussed previously. To sustain the adaptation phase, RelB, SirT1, and Nampt levels increase. This NAD+-dependent pathway is gene-specific and does not modify expression of adaptation-phase gene IκBα. Importantly, depleting NAD+, SirT1, or RelB reverses the facultative heterochromatin state, which silences proinflammatory-phase TNF-α and IL-1β [15, 20]. SirT1 also induces SirT6, which limits expression of HIF-1α target glycolysis genes, and reduces glucose fueling. Interestingly, RelB may repress HIF-1α transcription, which is responsible for enhanced glycolysis [79], perhaps by the same epigenetic process that silences TNF-α and IL-1β [8]. The switch from the proinflammatory-phase response to the adaptation phase also increases expression of PGC-1α and -β, which are directly activated by SirT1 [80]. PGC-1α and -β regulate fatty acid oxidation and promote mitochondrial biogenesis, which is needed to restore homeostasis. The duration of the adaptation phase, which is immunosuppressive during sepsis, depends on the magnitude of the danger and amount of energy needed to defend the host from the initial threat [7]. Ultimately, bioenergetics, glucose oxidation, and fatty acid oxidation are rebalanced and the flame quenched. SirT1 also regulates sepsis resolution [81]. However, evidence in rats supports that epigenetic marks may persist for months after sepsis is clinically resolved [82]. The integrative pathways and network of bioenergy, metabolism, and acute systemic inflammation are depicted in Fig. 6A.
Figure 6. Bioenergy coordinates metabolic and inflammatory responses.
SirT sensors of NAD+ provide an axis that integrates glycolysis and proinflammation with fatty acid oxidation and adaptation responses, which oppose the proinflammatory state and support restoration and resolution. GO, Glucose oxidation; FAO, fatty acid oxidation. Red, Proinflammatory phase; blue, Adaptation phase.
The phase shifts of acute systemic inflammation appear linear and involve innate and adaptive immunity. As discussed previously, this contrasts with chronic inflammation of diseases, such as obesity and atherosclerosis, which remain in a proinflammatory phase, where M1 proinflammatory macrophages and Teffs predominate over M2 adaptation-phase-like macrophages and T cell-derived suppressor cells. Under these conditions, HIF-1α induces increased glycolysis in M1 cells and Teffs, and PGC-1 increases in fatty acid oxidation in M2 adaptation phase and Tregs. Another important distinction between the proinflammatory-phase and adaptation-phase phenotypes is seen during chronic inflammation associated with obesity. M1 proinflammatory macrophages have repressed levels of AMPK, Nampt, NAD+, SirT1, and PCG-1, whereas the adaptation-phase M2 macrophages have enhanced levels of these gene products [58, 83, 84]. In other words, the critical bioenergy sensors that support the switch from the proinflammatory phase to the adaptation phase during acute inflammation apparently are unavailable to the M1 macrophage and Teffs of chronic inflammation, although they are available to generate M2 macrophages and Tregs in different areas of inflamed adipose tissue. Thus, the bioenergetics of chronic inflammation is complex and varies within different tissue regions. The arrested proinflammatory phase in M1 and Teffs associated with chronic inflammation is depicted in Fig. 6B. This figure does not show the minority population and M2 macrophages and Tregs, which reflect gene expression and metabolic patterns of the adaptation phase.
Recent data further support the bioenergy network concept in chronic inflammation, in that SirT1 expression in adipocytes regulates the polarity in obese mice [58]. Although the roles of SirT6 and SirT3 are poorly defined in acute or chronic inflammation, it is likely that both are critical for switching from the proinflammatory phase to the adaptation phase. In support of this are studies showing that liver-specific SirT6 knockout mice have chronic hepatic steatosis associated with chronic inflammation [85]. We speculate that SirT6-mediated balance of HIF-1α-induced glycolysis is compromised in chronic inflammation and thereby limits the switch to an adaptation phase [86].
Fig. 6 emphasizes how important mitochondria are for linking the processes of inflammation and metabolism. Mitochondrial glucose oxidation is reduced during the proinflammatory-phase response of acute systemic inflammation and during chronic inflammation. Mitochondria apparently cannot switch to fatty acid oxidation during chronic inflammation. Little is known about the molecular features of the SirT bioenergy axis in mitochondria during acute or chronic inflammation, although mitochondrial physiology links to both states, and mitochondrial biogenesis plays a critical role in restoring homeostasis in acute systemic inflammation from sepsis. Mitochondrial-specific SirT3 likely coordinates metabolism and inflammation concomitant with other SirT, as SirT3 protects against ROS by activating mnSOD, enhances the citrate cycle, and activates LCADH, which is required for fatty acid oxidation by mitochondria [70]. We expect that the network of SirT1, -6, and -3 as bioenergy sensors in inflammation critically modifies mitochondria and that SirT3 and perhaps other mitochondrial located SirTs, SirT4 and -5, form a mitochondrial regulatory network that aligns with inflammation resolution [87].
HIGHLIGHTS
HIF-1α and p65 pathways converge to coordinate cellular defenses and ignite inflammation by glucose fueling.
PGC-1-induced fatty acid oxidation and mitochondrial biogenesis quench inflammation.
NAD+ sensors SirT1, -3, and -6 coordinate the switch between glucose and fatty acid oxidation during inflammation.
The NAD+ sensing network is dysregulated during chronic inflammation.
IDENTIFYING CHALLENGES, OPPORTUNITIES, AND QUESTIONS DERIVED FROM THIS UNIFIED CONCEPT
Bioenergy coupling to metabolism and inflammation is emerging as a unifying concept for stress responses that should boost designing new therapeutic agents. Clinical diseases that could benefit from this unified concept include acute systemic inflammation from sepsis, blunt or burn trauma, and systemic autoimmune diseases such as lupus erythematosis. Equal promise exists for better understanding and treating chronic inflammatory diseases, such as obesity with glucose intolerance or diabetes, atherosclerosis, Alzheimer's dementia, and accelerated muscular disability during aging. However challenges in applying this concept to new therapies will occur, as bioenergetics and cellular nutrition and metabolism couple with proinflammatory- and adaptation-phase phenotypes, and these often coexist in the same host. To address this coexisting polarity, clinical phenotypic markers in blood and/or tissue are needed, as clinical signs to identify phase shifts are insensitive. Another challenge is that tissue specificities for bioenergy regulation are likely (e.g., heart, liver, and skeletal muscle), and managing the double-edged sword of compromising the proinflammatory phase is treacherous.
There are also important unanswered questions: Do linear shifts from the proinflammatory- to the adaptation-phase phenotype occur in the circulation and tissue? Does the presence of coexisting proinflammatory and hypoinflammatory phagocytes and T cells indicate that they originate in bone marrow and spleen? Why are proinflammatory-phase immunocytes of chronic inflammation suspended in the glycolytic anabolic state? Is mitochondrial biogenesis responsible for resolving the adaptation phase? What are the bioenergy-driven changes in mitochondrial physiology during inflammation, and what roles do specific SirTs play? Will reversing the hypoinflammatory state during sepsis by altering the bioenergy axis improve or worsen outcomes? There is much to be learned about this new paradigm of inflammation and metabolism reprogramming.
ACKNOWLEDGMENTS
This work was supported by U.S. National Institutes of Health (NIH) RO1 grants AI-065791 (C.E.M.), AI-079144 (C.E.M.), K08 grants GM086470 (V.T.V.) and MO-1RR 007,122.
Footnotes
- eNampt
- extracellular nicotinamide phosphoribosyltransferase
- HIF-1α
- hypoxia-inducing factor 1α
- iNampt
- intracellular nicotinamide phosphoribosyltransferase
- LCADH
- long-chain acyl-CoA dehydrogenase
- mnSOD
- manganese SOD
- Nam
- nicotinamide
- Nampt
- nicotinamide phosphoribosyltransferase
- NMN
- nicotinamide mononucleotide
- NMNAT
- nicotinamide mononucleotide adenyltransferase
- NRF
- nuclear respiratory factor
- OXPHOS
- oxidative phosphorylation
- p65
- NF-κB p65
- PDH
- pyruvate dehydrogenase
- PDHK
- pyruvate dehydrogenase kinase
- PGC-1
- peroxisome proliferator-activated receptor-γ coactivator 1
- Sir2
- silence information regulatory 2
- SirT
- sirtuin
- Teff
- T effector cell
- Treg
- regulatory T cell
AUTHORSHIP
T.F.L. and C.E.M. prepared the manuscript. C.M.B., M.E.G., L.M., P.M., A.R., V.T.V., and B.K.Y. closely discussed the design of this review and read and provided critical comments about this review.
REFERENCES
- 1. Medzhitov R. (2010) Inflammation 2010: new adventures of an old flame. Cell 140, 771–776 [DOI] [PubMed] [Google Scholar]
- 2. Gillum M. P., Erion D. M., Shulman G. I. (2011) Sirtuin-1 regulation of mammalian metabolism. Trends Mol. Med. 170, 8–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhong L., Mostoslavsky R. (2010) SIRT6: a master epigenetic gatekeeper of glucose metabolism. Transcription 1, 17–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Verdin E., Hirschey M. D., Finley L. W., Haigis M. C. (2010) Sirtuin regulation of mitochondria: energy production, apoptosis, and signaling. Trends Biochem. Sci. 35, 669–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McCall C. E., El Gazzar M., Liu T., Vachharajani V., Yoza B. (2011) Epigenetics, bioenergetics, and microRNA coordinate gene-specific reprogramming during acute systemic inflammation. J. Leukoc. Biol. 90, 439–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mayno G. (1975) The Healing Hand: Man and Wound in the Ancient World, Harvard University Press, Cambridge, MA, USA [Google Scholar]
- 7. Xiao W., Mindrinos M. N., Seok J., Cuschieri J., Cuenca A. G., Gao H., Hayden D. L., Hennessy L., Moore E. E., Minei J. P., Bankey P. E., Johnson J. L., Sperry J., Nathens A. B., Billiar T. R., West M. A., Brownstein B. H., Mason P. H., Baker H. V., Finnerty C. C., Jeschke M. G., López M. C., Klein M. B., Gamelli R. L., Gibran N. S., Arnoldo B., Xu W., Zhang Y., Calvano S. E., McDonald-Smith G. P., Schoenfeld D. A., Storey J. D., Cobb J. P., Warren H. S., Moldawer L. L., Herndon D. N., Lowry S. F., Maier R. V., Davis R. W., Tompkins R. G. (2011) Inflammation and host response to injury large-scale collaborative research program A genomic storm in critically injured humans. J. Exp. Med. 208, 2581–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McCall C. E., Yoza B., Liu T., El Gazzar M. (2010) Gene-specific epigenetic regulation in serious infections with systemic inflammation. J. Innate Immun. 2, 395–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McCall C. E., Yoza B. K. (2007) Gene silencing in severe systemic inflammation. Am. J. Respir. Crit. Care Med. 175, 763–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hotchkiss R. S., Opal S. (2010) Immunotherapy for sepsis—a new approach against an ancient foe. N. Engl. J. Med. 363, 87–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Riedemann N. C., Guo R. F., Ward P. A. (2003) The enigma of sepsis. J. Clin. Invest. 112, 460–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Glass C. K., Saijo K. (2010) Nuclear receptor transrepression pathways that regulate inflammation in macrophages and T cells. Nat. Rev. Immunol. 10, 365–376 [DOI] [PubMed] [Google Scholar]
- 13. Natoli G. (2009) Control of NF-κB-dependent transcriptional responses by chromatin organization. Cold Spring Harb. Perspect. Biol. 1, a000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hoffmann A., Natoli G., Ghosh G. (2006) Transcriptional regulation via the NF-κB signaling module. Oncogene 25, 6706–6716 [DOI] [PubMed] [Google Scholar]
- 15. Chen X., El Gazzar M., Yoza B. K., McCall C. E. (2009) The NF-κB factor RelB and histone H3 lysine methyltransferase G9a directly interact to generate epigenetic silencing in endotoxin tolerance. J. Biol. Chem. 284, 27857–27865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. El Gazzar M., Yoza B. K., Hu J. Y., Cousart S. L., McCall C. E. (2007) Epigenetic silencing of tumor necrosis factor α during endotoxin tolerance. J. Biol. Chem. 282, 26857–26864 [DOI] [PubMed] [Google Scholar]
- 17. Yoza B. K., Hu J. Y., Cousart S. L., Forrest L. M., McCall C. E. (2006) Induction of RelB participates in endotoxin tolerance. J. Immunol. 177, 4080–4085 [DOI] [PubMed] [Google Scholar]
- 18. Chan C., Li L., McCall C. E., Yoza B. K. (2005) Endotoxin tolerance disrupts chromatin remodeling and NF-κB transactivation at the IL-1β promoter. J. Immunol. 175, 461–468 [DOI] [PubMed] [Google Scholar]
- 19. Chen X., Yoza B. K., El Gazzar M., Hu J. Y., Cousart S. L., McCall C. E. (2009) RelB sustains IκBα expression during endotoxin tolerance. Clin. Vaccine Immunol. 16, 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yoza B. K., McCall C. E. (2011) Facultative heterochromatin formation at the IL-1 β promoter in LPS tolerance and sepsis. Cytokine 53, 145–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liew F. Y., Xu D., Brint E. K., O'Neill L. A. (2005) Negative regulation of Toll-like receptor-mediated immune responses. Nat. Rev. Immunol. 5, 446–458 [DOI] [PubMed] [Google Scholar]
- 22. Hirschey M. D., Shimazu T., Jing E., Grueter C. A., Collins A. M., Aouizerat B., Stancakova A., Goetzman E., Lam M. M., Schwer B., Stevens R. D., Muehlbauer M. J., Kakar S., Bass N. M., Kuusisto J., Laakso M., Alt F. W., Newgard C. B., Farese R. V., Jr., Kahn C. R., Verdin E. (2011) SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol. Cell 44, 177–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chawla A., Nguyen K. D., Goh Y. P. (2011) Macrophage-mediated inflammation in metabolic disease. Nat. Rev. Immunol. 11, 738–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Olefsky J. M., Glass C. K. (2010) Macrophages, inflammation, and insulin resistance. Annu. Rev. Physiol. 72, 219–246 [DOI] [PubMed] [Google Scholar]
- 25. Lumeng C. N., Saltiel A. R. (2011) Inflammatory links between obesity and metabolic disease. J. Clin. Invest. 121, 2111–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lumeng C. N., Maillard I., Saltiel A. R. (2009) T-ing up inflammation in fat. Nat. Med. 15, 846–847 [DOI] [PubMed] [Google Scholar]
- 27. Shibata N., Glass C. K. (2009) Regulation of macrophage function in inflammation and atherosclerosis. J. Lipid Res. 50 (Suppl.), S277–S281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Littman D. R., Singh H. (2007) Immunology. Asymmetry and immune memory. Science 315, 1673–1674 [DOI] [PubMed] [Google Scholar]
- 29. Littman D. R., Rudensky A. Y. (2010) Th17 and regulatory T cells in mediating and restraining inflammation. Cell 140, 845–858 [DOI] [PubMed] [Google Scholar]
- 30. Maitra U., Gan L., Chang S., Li L. (2011) Low-dose endotoxin induces inflammation by selectively removing nuclear receptors and activating CCAAT/enhancer-binding protein δ. J. Immunol. 186, 4467–4473 [DOI] [PubMed] [Google Scholar]
- 31. Pearce E. L. (2010) Metabolism in T cell activation and differentiation. Curr. Opin. Immunol. 22, 314–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wellen K. E., Thompson C. B. (2010) Cellular metabolic stress: considering how cells respond to nutrient excess. Mol. Cell 40, 323–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jones R. G., Thompson C. B. (2007) Revving the engine: signal transduction fuels T cell activation. Immunity 27, 173–178 [DOI] [PubMed] [Google Scholar]
- 34. Vander Heiden M. G., Cantley L. C., Thompson C. B. (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Semenza G. L. (2011) Oxygen sensing, homeostasis, and disease. N. Engl. J. Med. 365, 537–547 [DOI] [PubMed] [Google Scholar]
- 36. Semenza G. L. (2011) Regulation of metabolism by hypoxia-inducible factor 1. Cold Spring Harb. Symp. Quant. Biol., Epub ahead of print [DOI] [PubMed] [Google Scholar]
- 37. Peyssonnaux C., Datta V., Cramer T., Doedens A., Theodorakis E. A., Gallo R. L., Hurtado-Ziola N., Nizet V., Johnson R.S. (2005) HIF-1α expression regulates the bactericidal capacity of phagocytes. J. Clin. Invest. 115, 1806–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cramer T., Yamanishi Y., Clausen B. E., Forster I., Pawlinski R., Mackman N., Haase V. H., Jaenisch R., Corr M., Nizet V., Firestein G. S., Gerber H. P., Ferrara N., Johnson R. S. (2003) HIF-1α is essential for myeloid cell-mediated inflammation. Cell 112, 645–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rius J., Guma M., Schachtrup C., Akassoglou K., Zinkernagel A. S., Nizet V., Johnson R. S., Haddad G. G., Karin M. (2008) NF-κB links innate immunity to the hypoxic response through transcriptional regulation of HIF-1α. Nature 453, 807–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim I., Rodriguez-Enriquez S., Lemasters J. J. (2007) Selective degradation of mitochondria by mitophagy. Arch. Biochem. Biophys. 462, 245–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Twig G., Elorza A., Molina A. J., Mohamed H., Wikstrom J. D., Walzer G., Stiles L., Haigh S. E., Katz S., Las G., Alroy J., Wu M., Py B. F., Yuan J., Deeney J. T., Corkey B. E., Shirihai O. S. (2008) Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 27, 433–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fernandez-Marcos P. J., Auwerx J. (2011) Regulation of PGC-1α, a nodal regulator of mitochondrial biogenesis. Am. J. Clin. Nutr. 93, 884S–8890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rodgers J. T., Lerin C., Gerhart-Hines Z., Puigserver P. (2008) Metabolic adaptations through the PGC-1 α and SIRT1 pathways. FEBS Lett. 582, 46–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vercauteren K., Gleyzer N., Scarpulla R. C. (2008) PGC-1-related coactivator complexes with HCF-1 and NRF-2β in mediating NRF-2(GABP)-dependent respiratory gene expression. J. Biol. Chem. 283, 12102–12111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Haden D. W., Suliman H. B., Carraway M. S., Welty-Wolf K. E., Ali A. S., Shitara H., Yonekawa H., Piantadosi C. A. (2007) Mitochondrial biogenesis restores oxidative metabolism during Staphylococcus aureus sepsis. Am. J. Respir. Crit. Care Med. 176, 768–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Suliman H. B., Carraway M. S., Welty-Wolf K. E., Whorton A. R., Piantadosi C. A. (2003) Lipopolysaccharide stimulates mitochondrial biogenesis via activation of nuclear respiratory factor-1. J. Biol. Chem. 278, 41510–41518 [DOI] [PubMed] [Google Scholar]
- 47. Chawla A. (2010) Control of macrophage activation and function by PPARs. Circ. Res. 106, 1559–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vats D., Mukundan L., Odegaard J. I., Zhang L., Smith K. L., Morel C. R., Wagner R. A., Greaves D. R., Murray P. J., Chawla A. (2006) Oxidative metabolism and PGC-1β attenuate macrophage-mediated inflammation. Cell Metab. 4, 13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gerhart-Hines Z., Rodgers J. T., Bare O., Lerin C., Kim S. H., Mostoslavsky R., Alt F. W., Wu Z., Puigserver P. (2007) Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1α. EMBO J. 26, 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Singer M. (2008) Cellular dysfunction in sepsis. Clin. Chest Med. 29, 655–656ix [DOI] [PubMed] [Google Scholar]
- 51. Rathmell J. C. (2011) T cell Myc-tabolism. Immunity 35, 845–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Michalek R. D., Gerriets V. A., Jacobs S. R., Macintyre A. N., Maciver N. J., Mason E. F., Sullivan S. A., Nichols A. G., Rathmell J. C. (2011) Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J. Immunol. 186, 3299–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Osborn O., Sears D. D., Olefsky J. M. (2010) Fat-induced inflammation unchecked. Cell Metab. 12, 553–554 [DOI] [PubMed] [Google Scholar]
- 54. Van U. P., Kenneth N. S., Webster R., Muller H. A., Mudie S., Rocha S. (2011) Evolutionary conserved regulation of HIF-1β by NF-κB. PLoS Genet. 7, e1001285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Van U. P., Kenneth N. S., Rocha S. (2008) Regulation of hypoxia-inducible factor-1α by NF-κB. Biochem. J. 412, 477–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schilling J., Kelly D. P. (2011) The PGC-1 cascade as a therapeutic target for heart failure. J. Mol. Cell. Cardiol. 51, 578–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Finck B. N., Kelly D. P. (2006) PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J. Clin. Invest. 116, 615–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gillum M. P., Kotas M. E., Erion D. M., Kursawe R., Chatterjee P., Nead K. T., Muise E. S., Hsiao J. J., Frederick D. W., Yonemitsu S., Banks A. S., Qiang L., Bhanot S., Olefsky J. M., Sears D. D., Caprio S., Shulman G. I. (2011) SirT1 regulates adipose tissue inflammation. Diabetes 60, 3235–3245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Imai S., Guarente L. (2010) Ten years of NAD-dependent SIR2 family deacetylases: implications for metabolic diseases. Trends Pharmacol. Sci. 31, 212–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Galli M., Van G. F., Rongvaux A., Andris F., Leo O. (2010) The nicotinamide phosphoribosyltransferase: a molecular link between metabolism, inflammation, and cancer. Cancer Res. 70, 8–11 [DOI] [PubMed] [Google Scholar]
- 61. Rodgers J. T., Lerin C., Haas W., Gygi S. P., Spiegelman B. M., Puigserver P. (2005) Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature 434, 113–118 [DOI] [PubMed] [Google Scholar]
- 62. Yang H., Lavu S., Sinclair D. A. (2006) Nampt/PBEF/Visfatin: a regulator of mammalian health and longevity? Exp. Gerontol. 41, 718–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Johnson F. B., Sinclair D. A., Guarente L. (1999) Molecular biology of aging. Cell 96, 291–302 [DOI] [PubMed] [Google Scholar]
- 64. Wang Q., Zhang Y., Yang C., Xiong H., Lin Y., Yao J., Li H., Xie L., Zhao W., Yao Y., Ning Z. B., Zeng R., Xiong Y., Guan K. L., Zhao S., Zhao G. P. (2010) Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 327, 1004–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Haigis M. C., Sinclair D. A. (2010) Mammalian sirtuins: biological insights and disease relevance. Annu. Rev. Pathol. 5, 253–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Liu T. F., Yoza B. K., El Gazzar M., Vachharajani V. T., McCall C. E. (2011) NAD+-dependent SIRT1 deacetylase participates in epigenetic reprogramming during endotoxin tolerance. J. Biol. Chem. 286, 9856–9864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kawahara T. L., Michishita E., Adler A. S., Damian M., Berber E., Lin M., McCord R. A., Ongaigui K. C., Boxer L. D., Chang H. Y., Chua K. F. (2009) SIRT6 links histone H3 lysine 9 deacetylation to NF-κB-dependent gene expression and organismal life span. Cell 136, 62–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Xiao C., Kim H. S., Lahusen T., Wang R. H., Xu X., Gavrilova O., Jou W., Gius D., Deng C. X. (2010) SIRT6 deficiency results in severe hypoglycemia by enhancing both basal and insulin-stimulated glucose uptake in mice. J. Biol. Chem. 285, 36776–36784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhong L., D'Urso A., Toiber D., Sebastian C., Henry R. E., Vadysirisack D. D., Guimaraes A., Marinelli B., Wikstrom J. D., Nir T., Clish C. B., Vaitheesvaran B., Iliopoulos O., Kurland I., Dor Y., Weissleder R., Shirihai O. S., Ellisen L. W., Espinosa J. M., Mostoslavsky R. (2010) The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1α. Cell 140, 280–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bell E. L., Guarente L. (2011) The SirT3 divining rod points to oxidative stress. Mol. Cell 42, 561–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lombard D. B., Alt F. W., Cheng H. L., Bunkenborg J., Streeper R. S., Mostoslavsky R., Kim J., Yancopoulos G., Valenzuela D., Murphy A., Yang Y., Chen Y., Hirschey M. D., Bronson R. T., Haigis M., Guarente L. P., Farese R. V., Jr., Weissman S., Verdin E., Schwer B. (2007) Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol. Cell. Biol. 27, 8807–8814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Schirmer H., Pereira T. C., Rico E. P., Rosemberg D. B., Bonan C. D., Bogo M. R., Souto A. A. (2012) Modulatory effect of resveratrol on SIRT1, SIRT3, SIRT4, PGC1α and NAMPT gene expression profiles in wild-type adult zebrafish liver. Mol. Biol. Rep. 39, 3281–3289 [DOI] [PubMed] [Google Scholar]
- 73. Kong X., Wang R., Xue Y., Liu X., Zhang H., Chen Y., Fang F., Chang Y. (2010) Sirtuin 3, a new target of PGC-1α, plays an important role in the suppression of ROS and mitochondrial biogenesis. PLoS One 5, e11707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Carre J. E., Orban J. C., Re L., Felsmann K., Iffert W., Bauer M., Suliman H. B., Piantadosi C. A., Mayhew T. M., Breen P., Stotz M., Singer M. (2010) Survival in critical illness is associated with early activation of mitochondrial biogenesis. Am. J. Respir. Crit. Care Med. 182, 745–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Suliman H. B., Sweeney T. E., Withers C. M., Piantadosi C. A. (2010) Co-regulation of nuclear respiratory factor-1 by NFκB and CREB links LPS-induced inflammation to mitochondrial biogenesis. J. Cell Sci. 123, 2565–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Suliman H. B., Welty-Wolf K. E., Carraway M. S., Schwartz D. A., Hollingsworth J. W., Piantadosi C. A. (2005) Toll-like receptor 4 mediates mitochondrial DNA damage and biogenic responses after heat-inactivated E. coli. FASEB J. 19, 1531–1533 [DOI] [PubMed] [Google Scholar]
- 77. Canto C., Gerhart-Hines Z., Feige J. N., Lagouge M., Noriega L., Milne J. C., Elliott P. J., Puigserver P., Auwerx J. (2009) AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458, 1056–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Yang Z., Kahn B. B., Shi H., Xue B. Z. (2010) Macrophage α1 AMP-activated protein kinase (α1AMPK) antagonizes fatty acid-induced inflammation through SIRT1. J. Biol. Chem. 285, 19051–19059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Frede S., Stockmann C., Winning S., Freitag P., Fandrey J. (2009) Hypoxia-inducible factor (HIF) 1α accumulation and HIF target gene expression are impaired after induction of endotoxin tolerance. J. Immunol. 182, 6470–6476 [DOI] [PubMed] [Google Scholar]
- 80. Canto C., Auwerx J. (2009) PGC-1α, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr. Opin. Lipidol. 20, 98–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zhang Z., Lowry S. F., Guarente L., Haimovich B. (2010) Roles of SIRT1 in the acute and restorative phases following induction of inflammation. J. Biol. Chem. 285, 41391–41401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Carson W. F., Cavassani K. A., Dou Y., Kunkel S. L. (2011) Epigenetic regulation of immune cell functions during post-septic immunosuppression. Epigenetics 6, 273–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. De Kreutzenberg S. V., Ceolotto G., Papparella I., Bortoluzzi A., Semplicini A., Dalla M. C., Cobelli C., Fadini G. P., Avogaro A. (2010) Downregulation of the longevity-associated protein sirtuin 1 in insulin resistance and metabolic syndrome: potential biochemical mechanisms. Diabetes 59, 1006–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Dahl T. B., Haukeland J. W., Yndestad A., Ranheim T., Gladhaug I. P., Damas J. K., Haaland T., Loberg E. M., Arntsen B., Birkeland K., Bjøro K., Ulven S. M., Konopski Z., Nebb H. I., Aukrust P., Halvorsen B. (2010) Intracellular nicotinamide phosphoribosyltransferase protects against hepatocyte apoptosis and is down-regulated in nonalcoholic fatty liver disease. J. Clin. Endocrinol. Metab. 95, 3039–3047 [DOI] [PubMed] [Google Scholar]
- 85. Kim H. S., Xiao C., Wang R. H., Lahusen T., Xu X., Vassilopoulos A., Vazquez-Ortiz G., Jeong W. I., Park O., Ki S. H., Gao B., Deng C. X. (2010) Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell Metab. 12, 224–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Jia G., Su L., Singhal S., Liu X. (2012) Emerging roles of SIRT6 on telomere maintenance, DNA repair, metabolism and mammalian aging. Mol. Cell. Biochem. 364, 345–350 [DOI] [PubMed] [Google Scholar]
- 87. Chalkiadaki A., Guarente L. (2012) Sirtuins mediate mammalian metabolic responses to nutrient availability. Nat. Rev. Endocrinol. 8, 287–296 [DOI] [PubMed] [Google Scholar]