Abstract
The development of tools for patterning cocultures of cells is a fundamental interest among cell biologists and tissue engineers. Although a variety of systems exist for micropatterning cells, the methods used to generate cell micropatterns are often cumbersome and difficult to adapt for tissue engineering purposes. This study combines acoustic droplet ejection and aqueous two-phase system exclusion patterning to introduce a method for patterning cocultures of cells in multiplexed arrays. This new method uses focused acoustic radiation pressure to eject discrete droplets of uniform size from the surface of a dextran solution containing cells. The size of droplets is controlled by adjusting ultrasound parameters, such as pulse, duration, and amplitude. The ejected dextran droplets are captured on a cell culture substrate that is manipulated by a computer-controlled 3D positioning system according to predesigned patterns. Polyethylene glycol solution containing an additional cell type is then added to the culture dish to produce a two-phase system capable of depositing different types of cells around the initial pattern of cells. We demonstrate that our method can produce patterns of islands or lines with two or more cell types. Further, we demonstrate that patterns can be multiplexed for studies involving combinations of multiple cell types. This method offers a tool to transfer cell-containing samples in a contact-free, nozzle-less manner, avoiding sample cross-contamination. It can be used to pattern cell cocultures without complicated fabrication of culture substrates. These capabilities were used to examine the response of cancer cells to the presence of a ligand (CXCL12) secreted from surrounding cocultured cells.
Introduction
Cell patterning and organization are essential to the development of most tissues and organs.1–3 These processes also factor into many diseases, including cancer, where transformed cells form tumors that can subsequently invade the surrounding tissue and metastasize to distant sites.4 To better understand tissue development and disease it is necessary to advance experimental systems that either preserve native tissue structure or promote interactions among populations of cells.
In vivo, the interactions that occur among cells and their environments are so numerous and complex that detailed mechanistic studies of cell behavior and cell–cell interactions are difficult to undertake and interpret. Conversely, in vitro systems offer simplified experimental setups, but often lack the ability to organize cells into patterned structures that resemble tissue. Organotypic cultures represent an intermediate between in vivo systems and cell culture that allow native tissue structure to be preserved to some extent. However, preparation of organotypic cultures from animals requires a high level of expertise, and results can be difficult to interpret due to the presence of poorly defined molecules and proteins in native tissues. As a result, development of tools for patterning cocultures of cells has become an important theme in cell culture research, particularly, in the field of tissue engineering where information about cell–cell interactions may factor into the choice of material or cell type that will be used for tissue reconstruction and where development of physiological drug testing platforms are essential for testing new therapies.
Micropatterned cell culture systems can facilitate high-throughput formats and multiplexed data collection. However, most cell micropatterning methods are expensive and complicated (e.g., photolithography and dielectrophoresis),5,6 or rely on specialized conditions that limit the types of experiments that can be performed (e.g., inkjet printing, polydopamine-based cell patterning on polydimethylsiloxane [PDMS] substrates, PDMS stamp patterning, and parafilm patterning).7–10 Thus, development of techniques that are both low cost and flexible is highly desirable in order to facilitate cell micropatterning.
Micropatterning strategies fall broadly into two categories: (i) those that rely on substrate features, such as biochemical or topographic patterns to position cells,11–13 and (ii) those that actively dispense cells.14,15 Techniques that rely on substrate patterning are usually limited to applications that involve just one cell type growing on a single type of material. This limitation arises because it is difficult to control cell cross-reactivity between different physical or biochemical features and because it is difficult to fabricate features in composite materials and to manipulate materials with nonplanar geometries. Techniques that dispense cells circumvent these issues and allow patterning on a wide range of materials, but can be limited by their ability to reliably dispense high-viability cell preparations (cells can be damaged during printing or by drying, and orifices can become clogged by cells or medium). Dispensing techniques are also limited by the long length of time required to print large areas.
Another strategy for dispensing cells involves aqueous two-phase systems (ATPSs) that can be used for exclusion patterning of cells.16 ATPS exclusion patterning is performed by dispensing a droplet of dextran onto a substrate of interest to form an exclusion dome. A solution of polyethylene glycol (PEG) containing cells is then added to cover the dextran. Phase separation of the two polymer solutions causes the cells in the PEG phase to attach outside of the dextran droplet, producing a zone of cell exclusion. In contrast to other techniques, ATPS exclusion patterning is simple, low cost, and flexible.
Methods for liquid handling are often required for cell patterning, including low-cost and flexible patterning techniques, such as ATPS exclusion patterning. Acoustic droplet ejection (ADE) has been exploited as a method for liquid handling. ADE uses acoustic radiation force associated with an ultrasound field to transfer momentum into the ejection of a liquid droplet from an air–liquid interface.17–19 High intensity can be derived from a focused ultrasound beam. When the focus of the ultrasound is placed on the air–liquid interface, it forms a small cross-sectional area with the highest energy in the dimension of the ultrasound beam width. When the acoustic pressure of the applied ultrasound field is higher than the interface-restoring surface energy of the liquid, a discrete volume of liquid is ejected from the surface of the liquid. The liquid droplet travels upward and can be collected by a destination plate placed on the path of the upward-traveling liquid droplet. In a previous report ADE was used to eject single cells in picoliter droplets to study single-cell epitaxy20; however, the study used a microfabicated device for droplet ejection and cell culture, which limited its potential applications.
In this study, we combined ADE and ATPS exclusion patterning to provide a contact-free, nozzle-less liquid transfer system for micropatterning cells. We improved the exclusion pattering technique by incorporating an additional cell type in the ejected dextran droplet, allowing islands or lines of one cell type to be surrounded by a second cell type. We used an automated 3D positioning system and multiplexed liquid reservoir to produce a variety of coculture patterns. We then performed an in vitro cancer cell colony growth assay that allowed us to monitor the effects of CXCL12/CXCR4 signaling (a signaling axis with known importance in cell proliferation, cell trafficking, and metastatic cancer) to demonstrate the feasibility of the ADE-ATPS method.
Methods
ADE setup
For ADE, a low-cost, customizable source plate was fabricated using the following procedure. First, a PDMS membrane with a 100-μm thickness (10:1 polymer:curing agent) (GE) was created by spin coating the uncured polymer onto a silanized glass slide. The PDMS membrane was then cured at 120°C. A PDMS slab (5-mm thickness), in which a cylindrical well (6-mm diameter) was punched to house the source solution, was bonded to the PDMS membrane bottom (to allow ultrasound transmission) (Fig. 1). Finally, a custom-cut 50 mL conical tube was affixed to the PDMS slab to prevent water from entering the device during immersion. The complete device was then removed from the silanized glass.
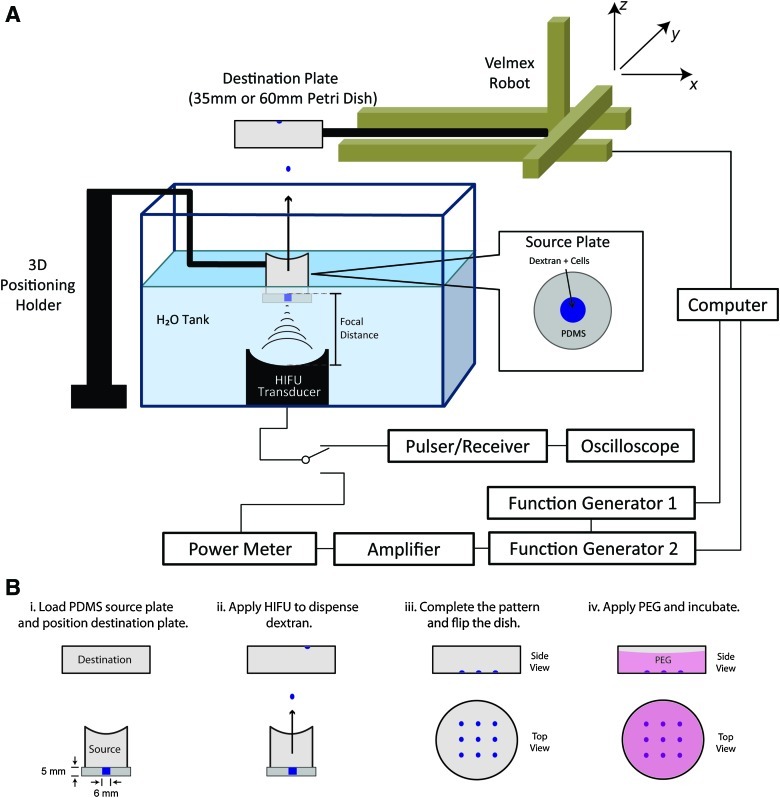
FIG. 1.
ADE and ATPS setup. (A) The source plate containing dextran is positioned above the ultrasound transducer, both immersed in degassed water. Movement of the destination plate is controlled by an automated positioning system synchronized with the ultrasound pulse. (B) Two-phase patterning is achieved in four steps. ADE, acoustic droplet ejection; ATPS, aqueous two-phase system; PDMS, polydimethylsiloxane; PEG, polyethylene glycol; HIFU, high intensity focused ultrasound. Color images available online at www.liebertonline.com/tec
The ADE procedure was carried out in a tank of filtered and degassed water (Fig. 1A). The focused ultrasound transducer (Blatek, Inc.) was an immersion transducer with a center frequency of 4 MHz, and a donut-shaped aperture (outer diameter of 6 cm, inner diameter of 40 mm, −3dB beam width of 0.5 mm, and a focal length of 63.3 mm) (Supplementary Fig. S1; Supplementary Data are available online at www.liebertonline.com/scd). The ultrasound transducer was calibrated before experiments in free field in water using a 40 μm calibrated needle hydrophone (HPM04/1; Precision Acoustics).
During experiments, the transducer faced upward in the water tank aiming toward the source plate. Function/arbitrary waveform generators (33220A; Agilent) and a power amplifier (model 75A250; Amplifier Research) were used to produce signals with desired pulse duration and amplitude to drive the ultrasound transducer for droplet ejection. A power meter (PM-1; JJ&A Instruments) was used to monitor the power during ultrasound application. A pulser/receiver (5052PR; Panametrics) was used to operate the transducer in pulse-echo mode and an oscilloscope (54603B; Hewlett Packard) was used to measure the distance between the transducer and the source plate, which was positioned using a 3D positioning system to align the surface of the source solution with the focal point of the transducer.
A holder for a destination plate (e.g., 35-mm or 60-mm Petri dish) was attached to a 3D computer-controlled system (Velmex, Inc.), which moved according to a designed pattern programmed in Matlab (MathWorks).
Dextran droplet ejection
An ATPS consisting of 12.8% wt/wt dextran T500 (Pharmacosmos) and 5.0% wt/wt PEG 35000 (Sigma), reconstituted as separate solutions in culture medium, was used for all experiments. Both phases were prepared 1 day prior to use and stored separately at 4°C. Dextran was loaded in the source plate well using a pipette.
To maximize droplet ejection efficiency, the distance between the transducer and the source plate (dts=df−dsol) was adjusted so that the transducer focus (df) coincided with the surface of the source solution based on the depth of the source solution (dsol). The values for speed of sound in water (1480 m/s) and in 12.8% dextran (1714 m/s) were used to determine the distance (d) based on the traveling time of the ultrasound pulse from the transducer to the bottom of the source plate (tts) and from the transducer to the surface of the source solution (tsol) as measured by the oscilloscope (dts=1480×tts and dsol=1714×tsol). After dts was fixed, the transducer was disconnected from the pulser/receiver and connected to the power amplifier for droplet ejection.
Pulsed ultrasound exposures were used to generate dextran droplets that were collected on the destination plate. The relationship of the ultrasound parameters to droplet properties (maximum traveling height and droplet volume) was characterized for pulse durations ranging from 250 μs to 1000 μs with 250 μs intervals, and pulse amplitudes ranging from 1.46 MPa to 2.47 MPa. Maximum height of droplet travel was determined by lowering the destination plate with respect to the source plate and recording the distance between source and destination plates at which ejected droplets were first captured. The droplet volume was determined from the density of the 12.8% dextran and the average droplet weight was obtained from measurement of 50 ejected droplets.
To produce the desired pattern of droplet deposition, the destination plate was moved horizontally in a two-dimensional (2D) plane after each ejected droplet. Afterward, the destination plate was flipped over and PEG was added to cover the droplets and form an ATPS (Fig. 1B).
Singleplex cell patterning
All cells were maintained at 37°C, 5% CO2 in a humidified incubator in Dulbecco's modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum and antibiotic solution. C2C12 myofibroblasts were grown to 70% confluence, trypsinized, and collected for labeling with either Cell Tracker Green or Cell Tracker Red (Molecular Probes). Separate populations of cell-tracker-labeled cells were pelleted, and resuspended in dextran at 2000 cells/μL and in PEG at 200 cells/μL. Dextran-containing cells labeled with Cell Tracker Red were loaded in the source plate well. Droplets were ejected to form spatial patterns of red-labeled cells. PEG-containing cells labeled with Cell Tracker Green were then applied to the Petri dish.
Propidium iodide (PI) (5 μg/mL; Molecular Probes) was used to assess the cell viability for each cell type patterned by ADE. Cell numbers and viability were determined by counting cells labeled with PI immediately after adding PEG. Therefore, our viability data account for the complete process of cell patterning including ADE and ATPS formation. Fluorescence and brightfield images of the cells were acquired using a Nikon TE300 fluorescent microscope (Nikon).
Multiplexed cell patterning
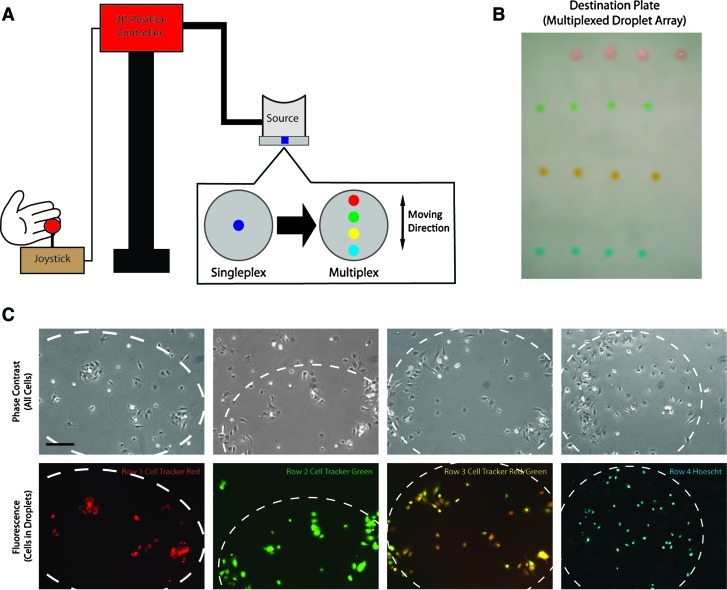
For the purpose of patterning multiple types of cells in the same dish, some modifications were made to the singleplex setup. First, multiple wells were punched in the PDMS slab of the source plate to house each different cell type. Second, instead of moving the ultrasound transducer to eject droplets from each well, an additional 3D position controller that could be manipulated by a user-controlled joystick was mounted on the source plate holder to move the source plate to align each well to the same location as the previous well with respect to the ultrasound transducer. After multiple cell types were transferred to the destination plate, an additional cell type was placed around them in the PEG phase through ATPS exclusion.
To demonstrate multiplexed cell patterning, we used the same cell types but labeled them with different trackers to distinguish the different pools of cells. C2C12 cells were labeled with four different fluorescent trackers (Cell Tracker Red, Cell Tracker Green, a combination of Cell Tracker Red and Green, and Hoescht 33342) (Molecular Probes). C2C12 cells in the PEG phase were not labeled. Fluorescent images were acquired after cell attachment and replacement of the ATPS medium with culture medium.
Cancer cell cocultures
Cocultures of MDA MB 231 breast cancer cells with a surrounding feeder population of HEK 293 cells were generated using the ADE-ATPS technique. A pattern of 2 rows of 4 droplets of MDA MB 231 cells was generated. To examine the effects of CXCL12 on the growth of MDA MB 231, we used two populations of HEK 293 cells, one overexpressing the cell-secreted CXCL12 chemokine and the other expressing minimal levels of CXCL12. These HEK 293 cells were combined with patterns of MDA MB 231 cells that either expressed baseline levels of CXCR4 (the receptor for CXCL12) or overexpressed CXCR4. All cells used in these experiments have previously been characterized by us and by others in terms of their biological responses to CXCL12.21–23 Colonies of patterned MDA MB 231 cells were cocultured with HEK 293 cells for 7 days over which time phase-contrast images were collected to monitor colony growth characteristics. Cell culture medium was replaced every other day by removing half of the conditioned medium and replacing it with fresh medium. After 7 days the cells were fixed with 4.0% paraformaldehyde and stained with Hoescht 33342. Samples were quantified in terms of total colony area (main colony+satellites), number of satellite colonies, and mean satellite area for each of the four possible combinations of cell types.
Data analysis
One-way analysis of variance and Student's t-test were used as statistical tests of significance where appropriate. Image analysis, quantifications, and data analyses were carried out using Image J (NIH), SigmaStat, and SigmaPlot.
Results
Droplet characterization
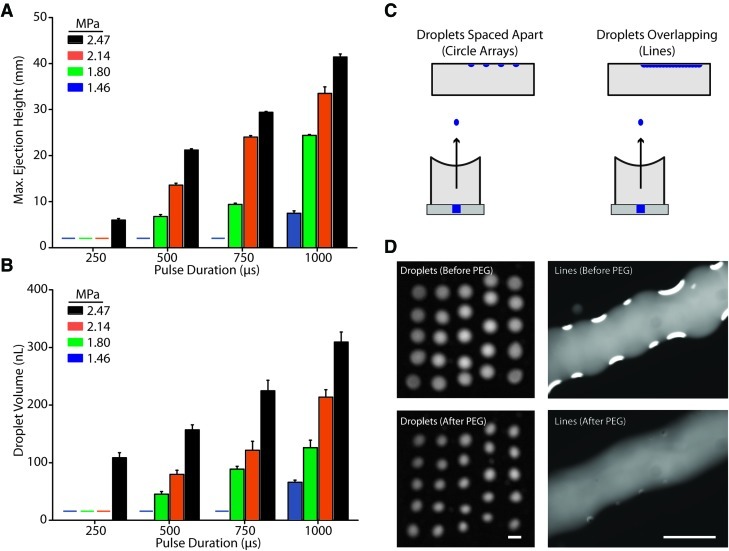
For each pulse duration, there were thresholds of pulse acoustic amplitude, below which no droplets were ejected. The threshold amplitude for discrete droplet ejection was inversely correlated with the pulse duration. The 250 μs duration required the highest pressure amplitude for droplet ejection to occur (2.47 MPa); however, at 1000 μs pulse duration, droplets could be ejected at pressures as low as 1.46 MPa. The maximum traveling height of the dextran droplets ranged from 60 mm (2.47 MPa, 250 μs pulse duration) to 414 mm (2.47 MPa, 1000 μs pulse duration). The maximum height of travel for an ejected droplet increased linearly with increasing acoustic pressure amplitude and pulse duration of the applied ultrasound (Fig. 2A). For a fixed pressure, the ejection height could be described by y=ax+b, where y was the ejection height and x was the pulse width, with a and b as constants. The constant a increased with higher pressure amplitude. For fixed pulse width, the ejection height also followed a linear relationship with the pressure (in MPa), where y was the ejection height and x was the pressure amplitude. The constant a also increased with longer pulse width. Droplet volume increased linearly with increasing acoustic pressure and pulse duration, following a similar trend to what was observed for ejection height. The dextran droplet volume ranged from 44.9 nL (1.80 MPa, 500 μs pulse width) to 309.2 nL (2.47 MPa, at 1000 μs pulse width) (Fig. 2B).
FIG. 2.
Characterization of droplet ejection and patterning. (A) Maximum ejection height increases linearly with ultrasound pulse width and peak pressure amplitude. (B) Droplet volume increases linearly with ultrasound pulse width and peak pressure amplitude. Bars represent mean±SEM. (C–D) Droplet ejection can produce a diverse range of pattern types, including arrays of circles and lines that were stable enough to permit cell attachment. Scale=1 mm; n≥3 for all experiments. Horizontal lines in (A) and (B) represent conditions that were not capable of ejecting droplets. Color images available online at www.liebertonline.com/tec
Based on these data, pulse durations of 750 μs with pressure amplitudes of either 2.14 MPa or 2.47 MPa were chosen for cell patterning. Two simple pattern types, circle arrays and lines, were created to demonstrate the principal of spatial patterning. The triggering of each ultrasound pulse and the movement of the destination plate were synchronized. For a pulse repetition frequency of 0.5 Hz used for the pulsed ultrasound exposures, the time interval between each movement of the source plate was set to be 2 s to ensure that there was only one droplet at each step along the path. If step size of the velmex was set to be <0.6 mm, droplets could form consecutive lines as sequentially generated droplets overlapped in space (Fig. 2C, D). Fluorescein isothiocyanate (FITC)-dextran was used to monitor pattern quality before and after adding PEG.
Circle/droplet arrays and lines of FITC-dextran droplets were generated (Fig. 2C, D). Arrays of discrete circles (5×5) were patterned within a 10 mm by 10 mm area and continuous lines (1 mm in width) were generated by adjusting the interdroplet spacing such that neighboring droplets merged together on the destination plate. The automated procedure permitted generation of precisely reproducible patterns on the order of minutes (depending on pattern complexity), without dehydration of the droplets before the addition of PEG. Droplet arrays and lines of dextran were stable after PEG was added to form the ATPS with only slight shrinkage of the dextran patterns after addition of PEG (due to equilibration between the two phases).
Singleplex cell patterning
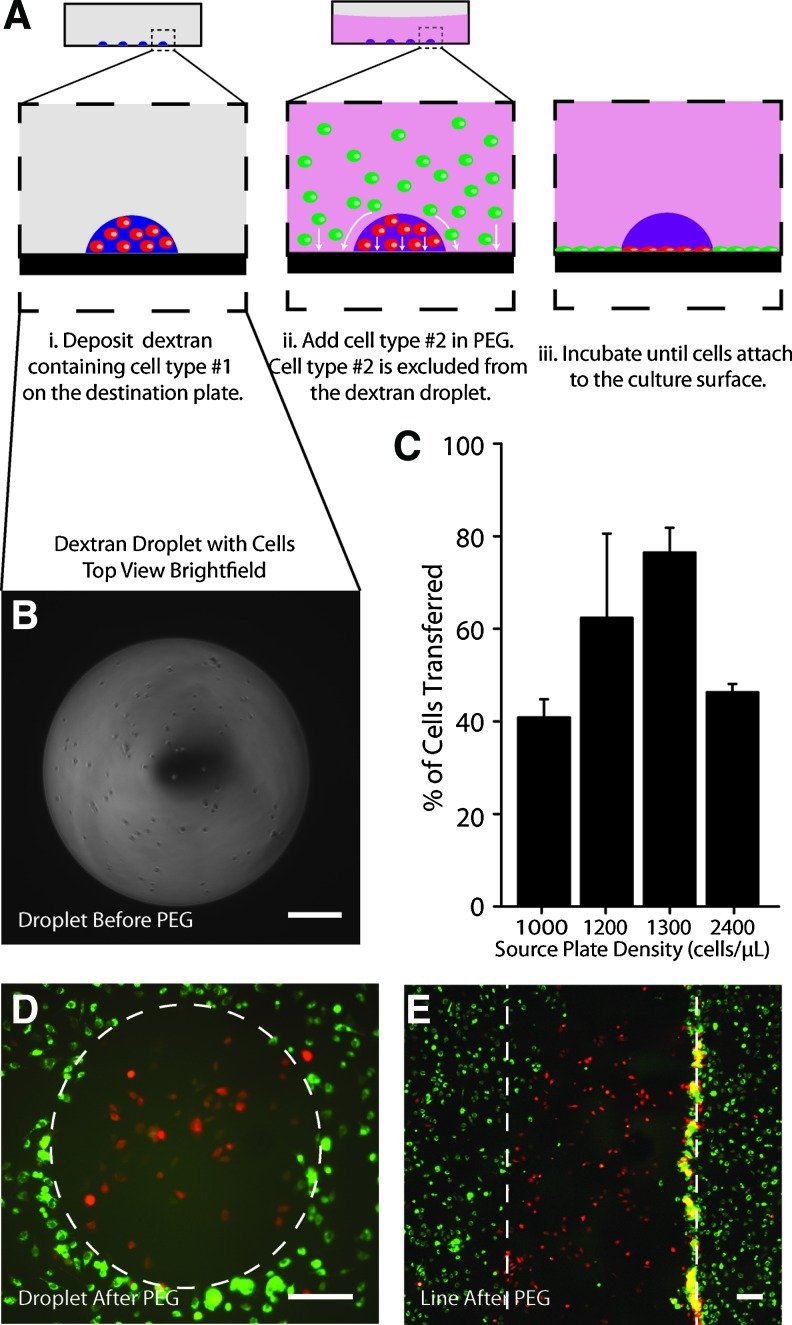
Patterned cocultures were generated by performing ATPS exclusion on dextran droplets patterned by ADE-ATPS (Fig. 3A). An ATPS was required to produce this type of patterned coculture (Supplementary Fig. S2). Pattern fidelity was lost after adding PEG when cells were dispensed in droplets containing only culture medium. Cells dispensed in culture medium alone were also subjected to rapid dehydration. Before adding PEG, the deposited dextran droplets were of the shape of a spherical cap with cells distributed evenly throughout (Fig. 3B). Generally ∼40% to 80% of the total cells added into the source well were transferred to the destination plate because of cell settling in the source well. This could be improved by stirring the solution in the source plate periodically to better suspend the cells. The percentage of cells transferred did not exhibit consistent dependence on cell density (Fig. 3C). The viability of cells in dextran droplets dispensed by ADE was compared with cells dispensed using a micropipetter. PI staining revealed that greater than 90% of cells remained viable after ADE (Fig. S3).
FIG. 3.
Arrays of cocultured cells can be generated using ADE with ATPS exclusion patterning. (A) Cell exclusion patterning is achieved by depositing one cell population within dextran, then surrounding the dextran pattern with a second cell type contained in PEG. (B) Ejected droplets are spherical with cells distributed evenly in the X-Y dimensions. (C) 20%–60% of cells are lost during the ADE process. Bars represent mean±SEM. (D–E) Exclusion patterned C2C12 cells, patterned as either droplets or lines. Scale=200 μm. Images were acquired ∼4 h after the cocultures were generated. Color images available online at www.liebertonline.com/tec
After exclusion patterning and cell attachment, red-labeled cells were well-retained within the dextran patterns with very few green-labeled cells (excluded cells) infiltrating the patterned regions (Fig. 3D, E). The shape of the cell patterns was true to the original patterns, even after 24 h of incubation. Some cells were captured by the PEG/Dex interface during exclusion patterning, manifesting as either a local increase in the density of attached cells near the interface or a loss of clusters of cells that were trapped by the interface and could not attach to the culture substrate. After several days of culturing, areas of high local cell density became less evident, as cell proliferation and migration resulted in a more even distribution of cells within the patterned regions. For example, in the cancer cell cocultures presented in Figure 5, it is clear that after 3 days or longer in culture the cells completely and uniformly fill the interior of the patterned region.
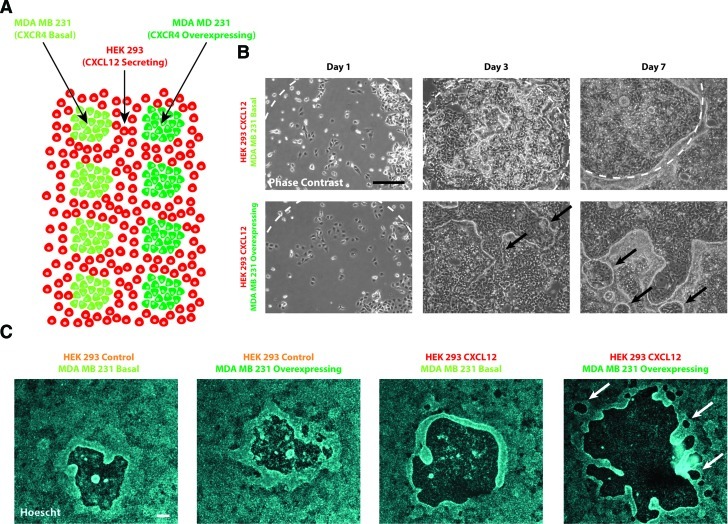
FIG. 5.
Multiplexed cocultures were used to assess cancer cell migration in response to cell-secreted factors. (A) Schematic of coculture setup. (B) Phase-contrast images were acquired over 7 days to monitor growth of the cocultured cells. (C) Hoechst 33342 staining revealed differences in colony growth characteristics in response to the presence of CXCL12 and its receptor CXCR4. White dashed lines in (B) indicate the location of the main colony when a circular colony could be traced. Arrows indicate the location of satellite colonies. Scale=200 μm. Color images available online at www.liebertonline.com/tec
Multiplexed cell patterning
Multiplexed cell patterning was achieved using a source plate with multiple wells controlled by a separate 3D controller (Fig. 4A). Food dyes without cells were placed in the dextran wells (red, green, yellow, and blue) to illustrate the formation of multiplexed patterns (Fig. 4B). Arrays of droplets containing C2C12 cells labeled with different cell-tracking dyes (as described in the Methods section) were also generated. Comparison of phase-contrast images and the fluorescent images showed that the cells in the dextran phase (with fluorescent trackers) were confined within the dextran boundaries (Fig. 4C).
FIG. 4.
The ADE setup can be adapted for multiplexed patterning. (A) Multiplexing is facilitated by adding additional source wells and manipulating the source plate using an additional 3D controller. (B) Multiplexed dextran droplets containing different food dyes. (C) Multiplexed patterning of labeled C2C12 cells surrounded by unlabeled cells. All images were acquired from the same culture plate ∼4 h after the cultures were generated. Scale=200 μm. Color images available online at www.liebertonline.com/tec
To demonstrate an application, multiplexed arrays of cocultured cells were used to study the response of breast cancer cells to the cell-secreted factor CXCL12. Two populations of breast cancer cells (one population overexpressing CXCR4 [the receptor for CXCL12] and the other expressing basal levels of CXCR4) were patterned within a feeder population of HEK 293 cells (either CXCL12 secreting or control) and monitored over the course of 7 days (Fig. 5A, B). On day 1, the two populations of MDA MB 231 breast cancer cells were similar in appearance. On day 3, the MDA MB 231 cells that overexpressed CXCR4 started to proliferate and migrate away from the original colony, while MDA MB 231 cells that expressed baseline levels of CXCR4 tended to proliferate and stay within the original pattern. By day 7, MDA MB 231 cells that overexpressed CXCR4 began to form satellite colonies (black arrows). At 7 days, the MDA MB 231 cells with baseline levels of CXCR4 were mostly confined to the original colony pattern.
The cocultured colonies were fixed and stained with Hoechst 33342 to allow for a more detailed comparison (Fig. 5C). The overall sizes of MDA MB 231 colonies cultured with the HEK 293 control feeder were much smaller than colonies cultured with HEK 293 CXCL12 feeder cells (normalized areas of 7.78±1.06 for MDA MB 231 control and 7.39±0.49 for MDA MB 231 CXCR4 as compared with 13.31±1.19 and 16.164±1.55, respectively) (N≥4 colonies for each condition). As expected, MDA cells overexpressing CXCR4 formed larger colonies with satellite colonies that were greater both in number (6.7±1.0 satellites on average vs. 1.4±0.5) and total area (normalized average satellite area of 0.58±0.19 vs. 0.23±0.07) than basal MDA MB 231 colonies when cultured with feeder cells secreting CXCL12. These observations are quantified in Supplementary Figure S4 for each of the 4 cell conditions tested. This demonstrates that cell-secreted CXCL12 can drive differential responses in cancer cells expressing different levels of the CXCR4 receptor. The results from the exclusion patterned system were consistent with results obtained from conventional experimental methods.24–26
Discussion
In the first instance of nozzle-less ADE demonstrated by Elrod et al., acoustic frequencies ranging from 5 to 300 MHz generated discrete droplets of water with diameters of 5–300 μm.17 ADE techniques that use specially fabricated self-focusing ultrasound transducers based on Fresnel rings or sleeves/nozzles to control droplet diameter have also been reported.27–29 A commercially available ADE device has since become available from Labcyte, Inc., which was validated by Grant et al.30 ADE techniques have also been used to deposit photoresist31,32 and other materials,33,34 as well as hydrogels containing cells.35
In this study, we developed a new approach combing ADE and ATPS to provide a versatile strategy for patterning cells and demonstrated an application by patterning cocultures of cells and examining their interactions. ATPSs have been used since the 1950s for separation of biological materials, including cells.36 This owes to one of the most well-known properties of ATPS, partitioning, in which materials distribute preferentially to one phase or the other.36,37 More recently, ATPSs have been included in microfluidic separation methods and for patterning cells in standard culture setups.38–42 A recent report from our lab focused on using the phase boundary formed between dextran and PEG to exclusion pattern a single cancer cell type.16 Over time, cancer cells migrated into the open exclusion zones. In the present report we build upon this system to generate cocultures of cells. By combining ATPS exclusion patterning with ADE, we introduce a method for multiplexed coculture cell patterning. This method is flexible in terms of pattern design, and also allows the use of a variety of culture substrates, providing benefits over a number of other patterning techniques.5–10,20 ADE provides an additional benefit in that it allows nozzle-less transfer of solutions, thus avoiding sample cross-contamination.
A specific discussion regarding our results and the system configuration is included below.
Droplet size and ultrasound properties
For droplets generated in nozzle-less systems, the droplet diameter is inversely proportional to the frequency of the focused ultrasound beam as the droplet size is related to the acoustic wavelength.33,43 More precisely, Elrod et al. reported the relationship as d ∼ f− 0.9± 0.1, where f is the ultrasound frequency, resulting in droplets of water with diameters of 5–300 μm using acoustic frequencies ranging from 5 to 300 MHz.17
In our study, the center frequency of the transducer was fixed at 4 MHz. Assuming the droplets to be spherical, the droplet diameter ranged from 400 μm to 800 μm with variable ultrasound pulse duration and pulse pressure amplitude, which was in accordance with the trend described in Elrod's report. It is worth noting that the focused beam shape also affects the formation of droplets. Demirci proposed that the relationship of droplet size is ddroplet=1.77λF4/3,31 where λ is the wavelength (=sound speed/frequency) and F is the f-number (the ratio of focal length over transducer diameter). Following this equation, the theoretical diameter of droplets generated from our setup should be around 700 μm, which is in the range of our experimental data. The discrepancies between our results and theoretical models may be accounted for by differences in pressure amplitude, pulse width, type of solution (affects the surface tension, resistance, and inertial force, etc.), and others factors.
Maximum height of travel for dextran droplets was linearly correlated with ultrasound pressure amplitude as well as pulse duration, so these factors should be optimized for specific applications. The droplet volume was also linearly dependent on ultrasound pulse width and pressure amplitude that are both related to the amount of energy expended to the surface to overcome the surface tension. The spatial peak time average intensity (Ispta) we used for characterizing the droplets ranged from 147.7 W/cm2 to 448.3 W/cm2. The phenomenon that droplet size increases when the distributed energy increases at longer pulse widths was also mentioned in previous reports.29 It can be similarly concluded that when ultrasound pressure amplitude increases, the droplet size increases as the applied energy increases. Similar to droplet diameter, the maximum ejection height increases as the pulse energy increases. This also agrees with the relationship of hrise ∼ E2f4 mentioned by Elrod et al.17
To contain sufficient number of cells in each droplet, but still provide colonies small enough for detailed microscopic evaluation of cell migration, we wanted droplet volumes between 100 nL and 200 nL. Based on our droplet characterization results, we chose pulse durations of 750 μs with pressure amplitudes of either 2.14 MPa or 2.47 MPa for cell patterning. Other droplet volumes, if desired, could be generated by adjusting the ultrasound parameters.
System configuration
Although an automated positioning system is not required to generate coculture arrays using ADE-ATPS, it is helpful for rapid generation of well-defined patterns, and can offer more precise control than can be achieved using a micropipetter. The following conditions should be considered for system configuration.
As solution volume in the source well decreases with each ejected droplet, the surface level of the dextran in the reservoir must be adjusted to be always aligned with the focal length of the ultrasound transducer, as shown in Figure 1, to allow optimized ejection conditions. To generate multiplexed arrays we found it helpful to use a joystick to move between consecutive source plate wells and fine tune this alignment. However, it should be noted that this can be easily accomplished with a computer-controlled source plate alignment system.
It is also important to note that other materials and configurations can be used for the source plate (e.g., it is possible to use a modified 96-well plate). We found that PDMS could be used to construct a low-cost, reusable source plate that could be easily maneuvered and aligned with respect to the ultrasound setup and the water tank. Further, the thickness of the PDMS slab used in the source plate (5 mm) and the diameter of the opening (6 mm) provided a relatively small well (smaller than a 96-well plate well) that allowed us to use smaller amounts of solutions containing fewer cells in our experiments. This becomes important when working with cells that are not available in large quantities, such as stem cells, patient-derived cells, or primary cell preparations. Our source plate could also withstand autoclaving to ensure that surfaces were free of microbial contaminants. Although limitations might exist on the size of the source plate well and choice of material, in terms of the maximum heights and droplet volumes that could be obtained, few deleterious effects on the cells were observed and the range of ejection heights and volumes was well suited to our application.
During droplet ejection and patterning, cells were lost due to settling to the bottom of the source well. Cells were also lost by capture at the PEG/dextran interface when they did not have the opportunity to attach. In our experience, it was unavoidable to capture some cells at the interface, but this effect was abolished at later time points in culture as cells tended to migrate and proliferate to fill any voids within the patterned regions. Because of these two factors, it was necessary to use a 10-fold higher seeding density in the dextran phase to achieve a homogeneous seeding density between the PEG region and the dextran pattern. The viability of the remaining cells was maintained at a very high level (>90%).
Examination of cell–cell interactions
Singleplexed and multiplexed coculture cell patterning were readily achieved using our method. Although we only demonstrated patterning of rectangular arrays of circular droplets and straight lines, any desired patterns could be produced with modification of our Matlab control program, including circles of various sizes, letters, or other desired shapes. One can envisage a number of tissue engineering applications that could utilize this type of cell patterning, for example, liver/kidney cell culture to recapitulate sinusoid or nephron organization, as well as feeder culture support for stem cells or other cell types.
We used cocultures of breast cancer cells that express basal or excess chemokine receptor CXCR4 with HEK 293 cells that secrete or do not secrete the CXCR4 ligand (CXCL12) to demonstrate an application of our method. CXCR4 and CXCL12 mediate cancer metastasis in vivo and cell proliferation and migration in vitro.24,26,44,45 During breast cancer metastasis, CXCR4-expressing cancer cells respond to gradients of CXCL12 that are present within the surrounding tissue and other target tissues.46 We used the ADE-ATPS method to study cell migration and proliferation of cancer cells in response to the presence of CXCL12 and obtained results that were consistent with those reported using conventional cell culture methods. Compared with conventional methods, however, multiplexed patterning with ATPS and ADE can pattern multiple types of cells on the same feeder layer, thus reducing the number of cells required and providing more consistent comparisons (all cancer cells are subjected to the same set of experimental conditions). It is important to note that in these studies we used cells that had been previously characterized in terms of their biological responses21–23; however, for future studies that explore new biological signaling pathways it will be important to incorporate additional controls, such as blocking antibodies or small molecules capable of disrupting specific signaling pathways.
Our study used immortalized cell lines to drive the expression of CXCL12 and CXCR4 such that the interaction between cell types mediated by these molecules could be examined in a well-controlled system. However, CXCL12/CXCR4 signaling also has been appreciated in the context of stem cell homing and chemotaxis. In several reports, mesenchymal stem cells were observed to migrate in response to CXCL12 as well as to home from bone marrow to target tissues that express CXCL12.47–50 Taking these studies into consideration, it may be possible in future studies to use our system to explore these signaling mechanisms in other cell types (such as stem cells), highlighting the relevance of our system for tissue engineering. It can also be predicted that apart from surface receptor/ligand interactions between neighboring cells, this patterning method has other potential applications related to scaling down of conventional biochemical assays (e.g., immunoprecipitations or examining bacterial interactions within a microbiome).
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (R01CA136553, R01CA136829, R01CA142750, R01CA116592, and P50CA093990), the Coulter Foundation Translational Research Grant, and a generous gift from the Beyster Foundation.
Disclosure Statement
The authors declare no competing financial interests.
References
- 1.Borello U. Pierani A. Patterning the cerebral cortex: traveling with morphogens. Curr Opin Genet Dev. 2010;20:408. doi: 10.1016/j.gde.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Costantini F. Kopan R. Patterning a complex organ: branching morphogenesis and nephron segmentation in kidney development. Dev Cell. 2010;18:698. doi: 10.1016/j.devcel.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zaret K.S. Regulatory phases of early liver development: paradigms of organogenesis. Nat Rev Genet. 2002;3:499. doi: 10.1038/nrg837. [DOI] [PubMed] [Google Scholar]
- 4.Chambers A.F. Groom A.C. MacDonald I.C. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 5.Chen Y.P. Zhao Y. Qiu K.Q. Chu J. Lu R. Sun M. Liu X.W. Sheng G.P. Yu H.Q. Chen J. Li W.J. Liu G. Tian Y.C. Xiong Y. An innovative miniature microbial fuel cell fabricated using photolithography. Biosens Bioelectron. 2010;26:2841. doi: 10.1016/j.bios.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 6.Ho C.T. Lin R.Z. Chang W.Y. Chang H.Y. Liu C.H. Rapid heterogeneous liver-cell on-chip patterning via the enhanced field-induced dielectrophoresis trap. Lab on a Chip. 2006;6:724. doi: 10.1039/b602036d. [DOI] [PubMed] [Google Scholar]
- 7.Javaherian S. O'Donnell K.A. McGuigan A.P. A Fast and Accessible Methodology for Micro-Patterning Cells on Standard Culture Substrates Using Parafilm (TM) Inserts. Plos One. 2011;6:e20909. doi: 10.1371/journal.pone.0020909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ku S.H. Lee J.S. Park C.B. Spatial control of cell adhesion and patterning through mussel-inspired surface modification by polydopamine. Langmuir. 2010;26:15104. doi: 10.1021/la102825p. [DOI] [PubMed] [Google Scholar]
- 9.Roth E.A. Xu T. Das M. Gregory C. Hickman J.J. Boland T. Inkjet printing for high-throughput cell patterning. Biomaterials. 2004;25:3707. doi: 10.1016/j.biomaterials.2003.10.052. [DOI] [PubMed] [Google Scholar]
- 10.Yang I.H. Co C.C. Ho C.C. Spatially controlled co-culture of neurons and glial cells. J Biomed Mater Res A. 2005;75:976. doi: 10.1002/jbm.a.30509. [DOI] [PubMed] [Google Scholar]
- 11.Folch A. Toner M. Microengineering of cellular interactions. Annu Rev Biomed Eng. 2000;2:227. doi: 10.1146/annurev.bioeng.2.1.227. [DOI] [PubMed] [Google Scholar]
- 12.Jung D.R. Kapur R. Adams T. Giuliano K.A. Mrksich M. Craighead H.G. Taylor D.L. Topographical and physicochemical modification of material surface to enable patterning of living cells. Crit Rev Biotechnol. 2001;21:111. doi: 10.1080/20013891081700. [DOI] [PubMed] [Google Scholar]
- 13.Khademhosseini A. Langer R. Borenstein J. Vacanti J.P. Microscale technologies for tissue engineering and biology. Proc Natl Acad Sci U S A. 2006;103:2480. doi: 10.1073/pnas.0507681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boland T. Xu T. Damon B. Cui X. Application of inkjet printing to tissue engineering. Biotechnol J. 2006;1:910. doi: 10.1002/biot.200600081. [DOI] [PubMed] [Google Scholar]
- 15.Ringeisen B.R. Othon C.M. Barron J.A. Young D. Spargo B.J. Jet-based methods to print living cells. Biotechnol J. 2006;1:930. doi: 10.1002/biot.200600058. [DOI] [PubMed] [Google Scholar]
- 16.Tavana H. Kaylan K., et al. Rehydration of Polymeric, Aqueous, Biphasic System Facilitates High Throughput Cell Exclusion Patterning for Cell Migration Studies. Adv Mater. 2011;21:2920. doi: 10.1002/adfm.201002559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elrod S.A. Hadimioglu B. Khuriyakub B.T. Rawson E.G. Richley E. Quate C.F. Mansour N.N. Lundgren T.S. Nozzleless Droplet Formation with Focused Acoustic Beams. J Appl Phys. 1989;65:3441. [Google Scholar]
- 18.Torr G.R. The Acoustic Radiation Force. Am J Phys. 1984;52:402. [Google Scholar]
- 19.Wood R.W. Loomis A.L. The physical and biological effects of high-frequency sound-waves of great intensity. Philos Mag. 1927;4:417. [Google Scholar]
- 20.Demirci U. Montesano G. Single cell epitaxy by acoustic picolitre droplets. Lab Chip. 2007;7:1139. doi: 10.1039/b704965j. [DOI] [PubMed] [Google Scholar]
- 21.Luker K. Gupta M. Luker G. Bioluminescent CXCL12 fusion protein for cellular studies of CXCR4 and CXCR7. Biotechniques. 2009;47:625. doi: 10.2144/000113126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luker K.E. Steele J.M. Mihalko L.A. Ray P. Luker G.D. Constitutive and chemokine-dependent internalization and recycling of CXCR7 in breast cancer cells to degrade chemokine ligands. Oncogene. 2010;29:4599. doi: 10.1038/onc.2010.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Torisawa Y.S. Mosadegh B. Bersano-Begey T. Steele J.M. Luker K.E. Luker G.D. Takayama S. Microfluidic platform for chemotaxis in gradients formed by CXCL12 source-sink cells. Integr Biol (Camb) 2010;2:680. doi: 10.1039/c0ib00041h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandis A.Z. Prasad A. Band H. Klosel R. Ganju R.K. Regulation of CXCR4-mediated chemotaxis and chemoinvasion of breast cancer cells. Oncogene. 2004;23:157. doi: 10.1038/sj.onc.1206910. [DOI] [PubMed] [Google Scholar]
- 25.Rubin J.B. Kung A.L. Klein R.S. Chan J.A. Sun Y. Schmidt K. Kieran M.W. Luster A.D. Segal R.A. A small-molecule antagonist of CXCR4 inhibits intracranial growth of primary brain tumors. Proc Natl Acad Sci U S A. 2003;100:13513. doi: 10.1073/pnas.2235846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh S. Singh U.P. Grizzle W.E. Lillard J.W., Jr. CXCL12-CXCR4 interactions modulate prostate cancer cell migration, metalloproteinase expression and invasion. Lab Invest. 2004;84:1666. doi: 10.1038/labinvest.3700181. [DOI] [PubMed] [Google Scholar]
- 27.Kwon J.W. Zou Q. Kim E.S. Directional ejection of liquid droplets through sectoring half-wave-band sources of self-focusing acoustic transducer. Fifteenth IEEE International Conference on Micro Electro Mechanical Systems; Las Vegas, NV. Technical Digest; 2002. [Google Scholar]
- 28.Meacham J.M. Varady M.J. Degertekin F.L. Fedorov A.G. Droplet formation and ejection from a micromachined ultrasonic droplet generator: visualization and scaling. Phys Fluids. 2005;17 [Google Scholar]
- 29.Yu H. Kwon J.W. Kim E.S. Chembio extraction on a chip by nanoliter droplet ejection. Lab Chip. 2005;5:344. doi: 10.1039/b413697g. [DOI] [PubMed] [Google Scholar]
- 30.Grant R.J. Roberts K. Pointon C. Hodgson C. Womersley L. Jones D.C. Tang E. Achieving accurate compound concentration in cell-based screening: validation of acoustic droplet ejection technology. J Biomol Screen. 2009;14:452. doi: 10.1177/1087057109336588. [DOI] [PubMed] [Google Scholar]
- 31.Demirci U. Droplet-based photoresist deposition. Appl Phys Lett. 2006;88:144104. [Google Scholar]
- 32.Demirci U. Yaralioglu G.G. Haeggstrom E. Khuri-Yakub B.T. Femtoliter to picoliter droplet generation for organic polymer deposition using single reservoir ejector arrays. IEEE Trans Semiconductor Manufacturing. 2005;18:709. [Google Scholar]
- 33.Demirci U. Acoustic picoliter droplets for emerging applications in semiconductor industry and biotechnology. J Microelectromechanical Syst. 2006;15:957. [Google Scholar]
- 34.Demirci U. Ozcan A. Picolitre acoustic droplet ejection by ferntosecond laser micromachined multiple-orifice membrane-based 2D ejector arrays. Electron Lett. 2005;41:1219. [Google Scholar]
- 35.Xu F. Wu J.H. Wang S.Q. Durmus N.G. Gurkan U.A. Demirci U. Microengineering methods for cell-based microarrays and high-throughput drug-screening applications. Biofabrication. 2011;3:034101. doi: 10.1088/1758-5082/3/3/034101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Albertsson P.A. Partition of proteins in liquid polymer-polymer two-phase systems. Nature. 1958;182:709. doi: 10.1038/182709a0. [DOI] [PubMed] [Google Scholar]
- 37.Rosa P.A. Azevedo A.M. Sommerfeld S. Mutter A. Aires-Barros M.R. Backer W. Application of aqueous two-phase systems to antibody purification: a multi-stage approach. J Biotechnol. 2009;139:306. doi: 10.1016/j.jbiotec.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 38.Frampton J.P. Lai D. Sriram H. Takayama S. Precisely targeted delivery of cells and biomolecules within microchannels using aqueous two-phase systems. Biomed Microdevices. 2011;13:1043. doi: 10.1007/s10544-011-9574-y. [DOI] [PubMed] [Google Scholar]
- 39.Soohoo J.R. Walker G.M. Microfluidic aqueous two phase system for leukocyte concentration from whole blood. Biomed Microdevices. 2009;11:323. doi: 10.1007/s10544-008-9238-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tavana H. Jovic A. Mosadegh B. Lee Q.Y. Liu X. Luker K.E. Luker G.D. Weiss S.J. Takayama S. Nanolitre liquid patterning in aqueous environments for spatially defined reagent delivery to mammalian cells. Nat Mater. 2009;8:736. doi: 10.1038/nmat2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamada M. Kasim V. Nakashima M. Edahiro J. Seki M. Continuous cell partitioning using an aqueous two-phase flow system in microfluidic devices. Biotechnol Bioeng. 2004;88:489. doi: 10.1002/bit.20276. [DOI] [PubMed] [Google Scholar]
- 42.Tavana H. Mosadegh B. Takayama S. Polymeric aqueous biphasic systems for non-contact cell printing on cells: engineering heterocellular embryonic stem cell niches. Adv Mater. 2010;22:2628. doi: 10.1002/adma.200904271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee C.Y. Pang W. Hill S.C. Yu H.Y. Kim E.S. Airborne particle generation through acoustic ejection of particles-in-droplets. Aerosol Sci Technol. 2008;42:832. [Google Scholar]
- 44.Engl T. Relja B. Marian D. Blumenberg C. Muller I. Beecken W.D. Jones J. Ringel E.M. Bereiter-Hahn J. Jonas D. Blaheta R.A. CXCR4 chemokine receptor mediates prostate tumor cell adhesion through alpha5 and beta3 integrins. Neoplasia. 2006;8:290. doi: 10.1593/neo.05694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith M.C. Luker K.E. Garbow J.R. Prior J.L. Jackson E. Piwnica-Worms D. Luker G.D. CXCR4 regulates growth of both primary and metastatic breast cancer. Cancer Res. 2004;64:8604. doi: 10.1158/0008-5472.CAN-04-1844. [DOI] [PubMed] [Google Scholar]
- 46.Hsu E.L. Chen N. Westbrook A. Wang F. Zhang R. Taylor R.T. Hankinson O. Modulation of CXCR4, CXCL12, and tumor cell invasion potential in vitro by phytochemicals. J Oncol. 2009;2009:491985. doi: 10.1155/2009/491985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ryser M.F. Ugarte F. Thieme S. Bornhauser M. Roesen-Wolff A. Brenner S. mRNA transfection of CXCR4-GFP fusion—simply generated by PCR-results in efficient migration of primary human mesenchymal stem cells. Tissue Eng Part C Methods. 2008;14:179. doi: 10.1089/ten.tec.2007.0359. [DOI] [PubMed] [Google Scholar]
- 48.Shi M. Li J. Liao L. Chen B. Li B. Chen L. Jia H. Zhao R.C. Regulation of CXCR4 expression in human mesenchymal stem cells by cytokine treatment: role in homing efficiency in NOD/SCID mice. Haematologica. 2007;92:897. doi: 10.3324/haematol.10669. [DOI] [PubMed] [Google Scholar]
- 49.Sordi V. Malosio M.L. Marchesi F. Mercalli A. Melzi R. Giordano T. Belmonte N. Ferrari G. Leone B.E. Bertuzzi F. Zerbini G. Allavena P. Bonifacio E. Piemonti L. Bone marrow mesenchymal stem cells express a restricted set of functionally active chemokine receptors capable of promoting migration to pancreatic islets. Blood. 2005;106:419. doi: 10.1182/blood-2004-09-3507. [DOI] [PubMed] [Google Scholar]
- 50.Stich S. Haag M. Haupl T. Sezer O. Notter M. Kaps C. Sittinger M. Ringe J. Gene expression profiling of human mesenchymal stem cells chemotactically induced with CXCL12. Cell Tissue Res. 2009;336:225. doi: 10.1007/s00441-009-0768-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.