Ubiquitination, the attachment of a 76 a.a. protein ubiquitin (Ub) or of a polyubiquitin (polyUb) chain to a protein target, is involved in a wide range of cellular processes in eukaryotes, ranging from progression through the cell cycle, to transcriptional activation, antigen processing, and vesicular trafficking of proteins[1]. Ub monomers are usually linked to each other via an isopeptide bond between the ε-amino group of a lysine residue in the proximal Ub and the C-terminal Gly76 of the successive Ub also known as the distal Ub. Thanks to the seven lysines in the Ub sequence, polyUb chains can be formed of various linkages and lengths (e.g.,[2]). A central question in Ub biology concerns the molecular mechanisms underlying the ability of different polyUb chains to act as distinct molecular signals for diverse cellular events. For example, K48-linked polyUbs act as a universal signal for proteasomal degradation, while K63-linked chains serve as regulatory rather than proteolytic signals. The biological role and the structural and recognition properties of polyUb chains assembled through the other five lysines (6, 11, 27, 29, and 33) or head-to-tail are currently the focus of extensive research (e.g., [2, 3]). It is believed that the specificity in the recognition signal carried by each polyUb chain depends on the linkage type that defines the unique conformation(s) that the chain can adopt.[2] Uncovering the mechanisms that allow various polyUb chains to serve as distinct molecular signals requires the ability to make these chains with the native connectivity, defined linkage and composition, controlled length, and in sufficient quantities for structural/biochemical studies.
Unfortunately, current enzyme-based methods could not provide a general solution to controlled polyUb chain assembly and production of sufficient amounts of most of these chains, because linkage-specific Ub-conjugating enzymes (E2) are not available for all lysines. Moreover, the commonly used methods[4, 5] require the introduction of some mutations, in order to control the enzymatic chain assembly, which inevitably results in polyUb having unnatural linkages and/or some lysines permanently replaced with other residues (e.g., Arg or Cys). The latter issue can be addressed by using lysines with removable protecting groups (incorporated into Ub as genetically encoded unnatural amino acids), which result in fully native polyUb chains,[6] however, the lack of linkage-specific E2s remains a significant bottleneck. These limitations have motivated several research groups to adopt non-enzymatic ubiquitination methods to form the isopeptide bond.[7] These accomplishments have been an important milestone in chemical biology of Ub systems. Notably, our group reported[8] the first total chemical synthesis of all seven variants of isopeptide-bond linked di-Ub (Ub2) chains in milligram quantities, as well as K48-linked tetra-Ub,[9] thus paving the way for various biochemical and structural studies, opening new, previously unavailable opportunities to characterize the properties of these chains.
Unravelling the mechanisms underlying linkage-specific recognition of polyUb signals requires the ability to obtain monomer-specific information on their interactions with receptors under physiological conditions and at atomic-level resolution. NMR is particularly suited for such studies. However, the homo-polymeric nature of polyUb presents a major challenge for NMR, because all Ub monomers in the chain are virtually identical both chemically and spectroscopically (except for the linkage-related modifications),[5] which makes it almost impossible to resolve the signals from each individual Ub unit. An obvious solution to this problem lies in segmental isotopic labelling,[5, 10] to provide polyUb chains with the individual Ub units differentially labelled and hence separately “observed” by NMR. This however could be formidably expensive if polyUb chains are fully synthesized chemically. On the other hand, the accessible and cost-effective isotope enrichment of proteins using bacterial expression could serve as a means to incorporate isotope-labelled recombinant Ub monomers into the chemically synthesized chains.
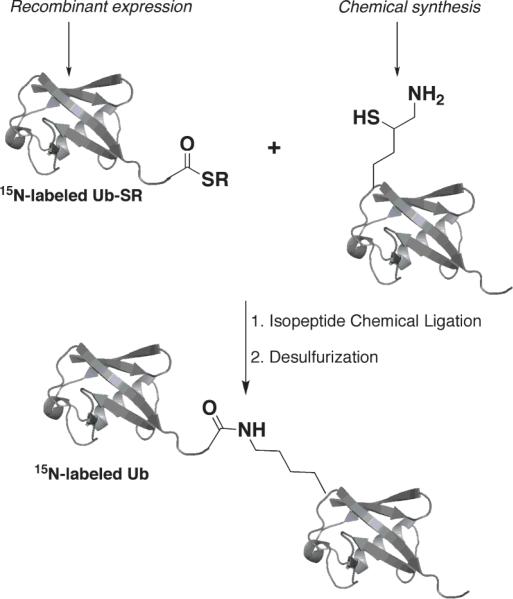
Herein we demonstrate the semi-synthetic assembly of segmentally isotopically labelled Ub2 chains. This method is based on chemical synthesis[8] of the proximal Ub carrying a δ-mercaptolysine at the desired position (e.g., 33 or 48 in this study) to allow isopeptide formation with the isotope-labelled recombinant distal Ub species (Scheme 1, see the details in Supporting Information). Using this method, we successfully assembled Ub2 in milligram quantities and with very high purity (Fig.1) linked via a natural isopeptide bond through K48 (as control) or K33 and having uniformly 15N-enriched wild type Ub as the distal unit. This allowed us to unravel, for the first time, the structural, conformational, and ligand-binding properties of K33-linked di-Ub.
Scheme 1.
Semi-synthesis of a di-Ub chain 15N-labelled on the distal Ub.
Figure 1.
Characterization of semi-synthetic di-Ub using (a) HPLC and mass spectrometry analysis (for the K33 case) showing the correct molecular weight of 17195 Da (Calcd. 17194 Da) and (b) SDS-PAGE of the K33 and K48 chains.
Initially, we examined by NMR the semi-synthesized K48-linked Ub2, to verify that the chain had the same spectral and conformational properties as the enzymatically assembled chain. NMR is extremely sensitive to the local chemical environment of a nucleus under observation and therefore can detect subtle changes in the structure or conformation that are undetectable by other methods. We used the K48-linked chain as a control because this is the most well studied polyUb by NMR, and we have significant experimental data to compare with.[5, 10, 11] Indeed, the NMR spectrum of the distal Ub in this chain is very similar to that of monomeric Ub (monoUb) (Fig.S2). As expected, the strongest signal perturbations are observed in the C-terminal residues (especially G76), and reflect changes in the local chemical environment due to formation of the isopeptide bond with K48 of the proximal Ub. Importantly, highly specific chemical shift perturbations (CSPs) are present in and around the so-called “hydrophobic-patch” residues L8, I44, and V70, which form the Ub/Ub interface in K48-linked Ub2 and Ub4.[10, 12] The CSPs observed in these residues are the hallmark of the closed conformation of K48-linked chains (Fig.S2), which is predominantly (~85%) populated at neutral pH[10]. As we showed earlier[10], the conformation of K48-linked Ub2 is pH-dependent and becomes predominantly open as the pH is lowered to 4.5. Moreover, the Ub2's ability to open is critical for allowing Ub-receptors access to the hydrophobic patch surface on the Ub units in the chain [13]. To verify that this functionally essential feature is retained in the semi-synthesized Ub2, we performed similar NMR measurements at pH 4.5. Indeed, our data (Fig.S2) clearly show absence of non-covalent contacts between Ub units in the semi-synthesized K48-Ub2 at acidic pH, consistent with an open conformation of the chain. To summarize these control studies, our NMR data demonstrate that the K48-linked Ub2 obtained using the semi-synthetic method is both structurally and conformationally virtually identical to the enzyme-assembled chain.
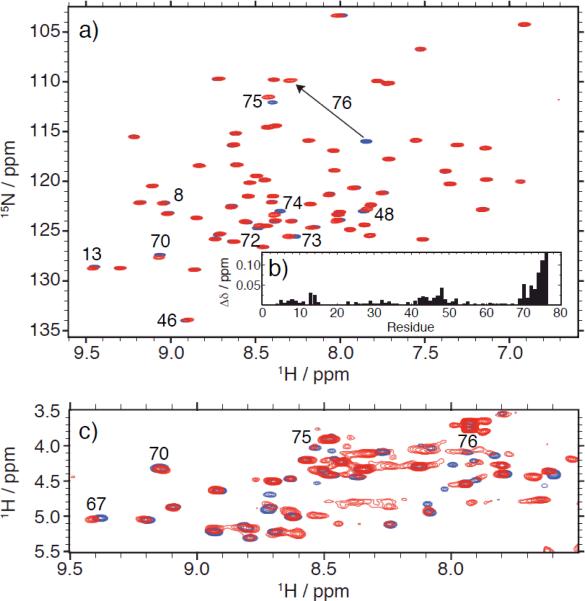
We then focused on K33-linked chains, which have recently been found to play a non-proteolytic, regulatory role in T-cell signalling.[14] Despite this finding, the precise biological function and the corresponding receptors for these chains are not known, and neither structural nor biochemical data are currently available. As shown in Figure 2 (see also Fig.S2 for K48-linked Ub2), using isotope editing/filtering NMR techniques, we recorded separately NMR spectra of the distal and the proximal Ubs in the same K33-linked Ub2. The excellent signal dispersion in the 1H-15N NMR spectrum of the distal Ub (Fig.2a) confirms that this unit is well folded. Moreover, the strong similarity of this spectrum with that of monoUb indicates that the structure of the distal Ub is intact. Furthermore, the signal dispersion in the 1H-1H TOCSY spectrum of the proximal Ub in the same chain (Fig.2c) and its overlay with that of monoUb indicate that this unit, too, is structurally intact.
Figure 2.
NMR characterization of both Ub units in K33-linked Ub2. (a) 1H-15N TROSY spectrum of the distal Ub (red) superimposed with a similar spectrum of monoUb (blue). (b) Amide chemical shift perturbations (CSPs) in the distal Ub of K33-Ub2 versus monoUb, quantified as a weighted average of the shifts in 1H (ΔδH) and 15N (ΔδN) resonances: Δδ=(ΔδH2+ΔδH2/25)½. Strong CSPs in the C-terminal residues (R72-G76) reflect chemical modification of the C-terminus as a result of its conjugation to the proximal Ub. Small but consistent site-specific CSPs are evident in and around the hydrophobic patch residues L8, I44, V70 and point to their involvement in transient contacts with the proximal Ub. (c) The HN-Hα region of the 15N-filtered 1H-1H TOCSY spectrum of the proximal Ub (red) overlaid with the similar spectrum of monoUb (blue).
As in the case of K48-Ub2 (Fig.S2), we observed strong CSPs in the C-terminal residues of the distal Ub in K33-linked Ub2 (Fig.2b), consistent with their involvement in the isopeptide linkage, this time to K33 of the proximal Ub. These data provide direct evidence that the 15N-labelled distal Ub unit is conjugated at its C-terminus, and the huge signal shift observed for G76 is a signature of this residue's involvement in the formation of a covalent bond [5]. Interestingly, our NMR data also revealed small but systematic signal perturbations (Fig.2b) clustered in and around the hydrophobic-patch residues L8, I44, and V70. As mentioned above, the same residues form the Ub/Ub interface in K48-linked Ub2 (compare with Fig.S2). The observation of these highly specific CSPs indicates the presence of interdomain contacts in K33-linked Ub2, mediated by the hydrophobic patch on the distal Ub. Judging by the magnitude of the CSPs, the interface is relatively weak (or transient) compared to that in K48-linked Ub2; nevertheless, its very existence is significant, because it provides important insights into the ability of the Ub monomers to form non-covalent contacts within the K33-linked chain. Noteworthy, no such contacts were observed in K63-linked Ub2[15] or in the open state of K48-linked Ub2 (Fig.S2). Interestingly, the CSPs observed in the distal Ub of K33-linked Ub2 (Fig.2b) agree with our in silico prediction[16] that this chain should not be able to form a strong hydrophobic patch-to-patch interface between the two Ub units, and instead a possible contact could involve the hydrophobic patch residues of the distal Ub and the β1/β2-loop of the proximal Ub.
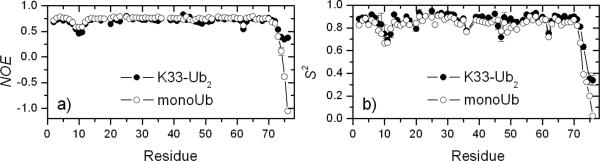
As the next step in the characterization of K33-linked Ub2, we measured 15N relaxation rates (R1, R2) and heteronuclear 15N{1H} NOEs for all observed amides in the distal Ub (Figs.3 and S3). With these data, we determined the overall rotational diffusion tensor of K33-linked Ub2 and characterized the backbone dynamics in the distal Ub (see Supporting Information). The overall tumbling of K33-Ub2 (“sensed” by the distal Ub) is anisotropic, in contrast to an almost isotropic rotational diffusion of monoUb[17]. Judging by the slower overall tumbling time (τc=8.1 ns versus 4.5 ns for monoUb) and strong rotational anisotropy (~1.52), the two Ub moieties in the chain tumble together as one entity (to a first approximation)[10]. The backbone dynamics are essentially same as in monoUb, except for the most C-terminal residues G75 and G76, which in the distal Ub experience significantly smaller amplitudes of motion, as evident from the noticeably higher hetero-NOEs and order parameters (Fig.3). This indicates that the distal Ub's C-terminus in K33-linked Ub2 behaves like an extended loop rather than a free C-terminus (as in monoUb), which is consistent with its attachment to the proximal Ub. A similar behavior was observed in K48-linked Ub2.[10] Thus, our 15N relaxation data independently confirm that the distal Ub is (i) tethered (in contrast to tumbling freely in solution) and (ii) its attachment is achieved via the C-terminus. These results further demonstrate the utility of the semi-synthesized chains for domain-specific characterization by NMR.
Figure 3.
Characterization of backbone motions in the distal Ub of K33-linked Ub2. (a) 15N{1H} NOEs and (b) order parameter (S2) as a function of residue number. Also shown are data for monoUb, for comparison. The rest of relaxation data and analysis are in figure S3.
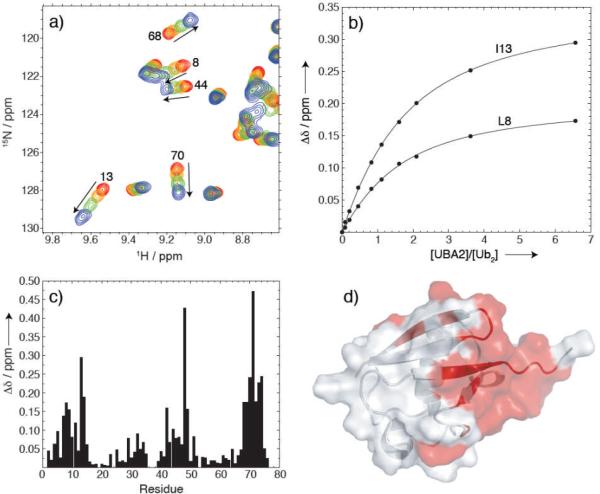
Finally, we used NMR to examine recognition of K33-linked Ub2 by hHR23a (the human homologue of protein Rad23), an extra-proteasomal shuttle that mediates the interaction of polyubiquitiquitinated substrates with the 26S proteasome. Specifically, the C-terminal Ub-associated domain (UBA2) of hHR23a serves as a polyUb receptor with strong binding preference for K48-linked chains versus K63-linked ones.[11, 15, 18] To monitor UBA2 binding, unlabelled UBA2 was titrated into a solution of K33-linked Ub2, and 1H-15N NMR spectra of the distal Ub were recorded at each titration step (Fig.4). This allowed us to map the UBA2-interaction surface on the distal Ub to the hydrophobic patch, which was in full agreement with similar findings for UBA2's recognition by monoUb and by K63-linked Ub2.[11, 15] In fact, the pattern of CSPs shown in Figure 4c is almost identical to those induced by UBA2 binding to monoUb or the distal Ub in K63-linked Ub2.[11, 15] To assess the stoichiometry of the UBA2/K33-Ub2 binding we measured the 15N T1 value at saturation ([UBA2]:[Ub2]=6.6). The result (T1=913 ± 58 ms) is close to that (~900 ms) expected for a 1:1 complex, thus suggesting that a single UBA2 molecule is bound per K33-Ub2 (either to the proximal or the distal Ub). Fitting the titration data to a 1:1 binding model (Fig.4b) gave the microscopic dissociation constant Kd = 216 ± 43 μM (averaged over 12 residues with CSP > 0.1 ppm), which is comparable to that reported for UBA2 binding to monoUb[15, 18, 19] or K63-Ub2[15], and approximately ten-fold higher than the Kd for UBA2 binding to K48-Ub2.[11] This is an important finding, because it indicates, for the first time, that the K33-linked chain is recognized by the proteasomal shuttle hHR23a differently from the K48-linked chain[11] but similarly to K63-linked Ub2.[15] These results suggest that, like the K63-linked chain, the K33-linked chain does not serve as a signal for proteasomal degradation.
Figure 4.
NMR titration studies of K33-Ub2 binding to the UBA2 domain of hHR23a. (a) Fragment of the overlay of 1H-15N SOFAST spectra of the distal Ub in K33-Ub2 recorded at four steps in its titration with the UBA2 domain of hHR23a. Residue numbers for selected NMR signals are indicated. In total, ten titration points were recorded, with the [UBA2]/[Ub2] molar ratio ranging from 0 to 6.6. Shown here are spectra for four titration points: at the start ([UBA2]=0, red contours), two intermediate points (orange and green), and the endpoint (blue). (b) Representative titration curves for L8 and I13, the lines represent the results of fitting to a 1:1 stoichiometry model (see text). (c) Amide CSPs at the endpoint of titration ([UBA2]/[Ub2]= 6.6) as a function of the residue number in Ub. (d) Mapping of the CSPs (residues with Δδ>0.1ppm in panel (c) colored red) onto the structure of Ub indicates that the binding interface involves the hydrophobic patch-surface of the distal Ub.
In summary, we have demonstrated the use of segmental isotopic labelling of Ub chains using semi-synthesis to characterize, for the first time, the structural, conformational, and ligand binding properties of K33-linked Ub2. This should also enable similar studies of the other currently uncharacterized chains (e.g. K6-, K27-, or K29-linked) to dissect their monomer-specific inter and intra-molecular interactions in cellular processes. This strategy opens up numerous possibilities for using various isotope-incorporation schemes (ranging from uniform enrichment to residue- or group specific labelling[20]), to enable studies of polyUb chains by NMR, small-angle neutron scattering (SANS, e.g., using contrast variation), and other biophysical techniques.
Supplementary Material
Footnotes
Supported by NIH grant GM065334 to D.F., NSF Postdoctoral Fellowship DBI-0905967 to C.A.C., and by the Edmond J. Safra Foundation and Israel Science Foundation (A.B.)
Supporting information for this article is available on the WWW under http://www.angewandte.org.
Contributor Information
Carlos A. Castañeda, Department of Chemistry and Biochemistry, Center for Biomolecular Structure and Organization University of Maryland College Park, Maryland, 20742, USA
Liat Spasser, Department of Chemistry and National Institute for Biotechnology in the Negev Ben-Gurion University of the Negev Beer Sheva, 84105, Israel.
Sudhir N. Bavikar, Department of Chemistry and National Institute for Biotechnology in the Negev Ben-Gurion University of the Negev Beer Sheva, 84105, Israel
Ashraf Brik, Department of Chemistry and National Institute for Biotechnology in the Negev Ben-Gurion University of the Negev Beer Sheva, 84105, Israel.
David Fushman, Department of Chemistry and Biochemistry, Center for Biomolecular Structure and Organization University of Maryland College Park, Maryland, 20742, USA.
References
- 1.a Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425–480. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]; b Glickman MH, Ciechanover A. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]; c Ikeda f., Dikic I. EMBO Rep. 2008;9:536–42. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Wickliffe K, Williamson A, Jin L, Rape M. Chem Rev. 2009;109:1537–48. doi: 10.1021/cr800414e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pickart CM, Fushman D. Curr Opin Chem Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 3.Ye Y, Rape M. Nat Rev Mol Cell Biol. 2009;10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a Piotrowski J, Beal R, Hoffmann L, Wilkinson KD, Cohen RE, Pickart CM. J.Biol.Chem. 1997;272:23712. doi: 10.1074/jbc.272.38.23712. [DOI] [PubMed] [Google Scholar]; b Pickart CM, Raasi S. Methods Enzymol. 2005;399:21–36. doi: 10.1016/S0076-6879(05)99002-2. [DOI] [PubMed] [Google Scholar]
- 5.Varadan CR, Assfalg M, Fushman D. Methods Enzymol. 2005;399:177–192. doi: 10.1016/S0076-6879(05)99012-5. [DOI] [PubMed] [Google Scholar]
- 6.Castaneda CA, Liu J, Kashyap TR, Singh RK, Fushman D, Cropp TA. Chem Commun (Camb) 2011;47:2026–2028. doi: 10.1039/c0cc04868b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a Chatterjee C, McGinty RK, Pellois JP, Muir TW. Angew Chem Int Ed Engl. 2007;46:2814–2818. doi: 10.1002/anie.200605155. [DOI] [PubMed] [Google Scholar]; b Yang R, Pasunooti KK, Li F, Liu X-W, Liu C-F. J. Am. Chem. Soc. 2009;131:13592–13593. doi: 10.1021/ja905491p. [DOI] [PubMed] [Google Scholar]; c Kumar KSA, Haj-Yahya M, Olschewski D, Lashuel HA, Brik A. Angew. Chem. Int. Ed. 2009;48:8090–8094. doi: 10.1002/anie.200902936. [DOI] [PubMed] [Google Scholar]; d Virdee S, Ye Y, Nguyen D, Komander D, Chin J. Nat. Chem. Biol. 2010;6:750–757. doi: 10.1038/nchembio.426. [DOI] [PubMed] [Google Scholar]; e Kumar KSA, Spasser L, Ohayon S, Erlich LA, Brik A. Bioconjugate Chem. 2011;22:137–143. doi: 10.1021/bc1004735. [DOI] [PubMed] [Google Scholar]
- 8.Kumar KS, Spasser L, Erlich LA, Bavikar SN, Brik A. Angew Chem Int Ed Engl. 2010;49:9126–9131. doi: 10.1002/anie.201003763. For other studies related to the synthesis of di-Ub chains with the native isopeptide bond see Yang R, Pasunooti KK, Li F, Liu X-W, Liu C-F. Chem. Commun. 2010;46:7199–7201. doi: 10.1039/c0cc01382j.; El Oualid F, Merkx R, Ekkebus R, Hameed DS, Smit JJ, de Jong A, Hilkmann H, Sixma TK, Ovaa H. Angew. Chem. Int. Ed. 2010;49:10149–10153. doi: 10.1002/anie.201005995.
- 9.Kumar KS, Bavikar SN, Spasser L, Moyal T, Ohayon S, Brik A. Angew Chem. 2011;50:6137–6141. doi: 10.1002/anie.201101920. [DOI] [PubMed] [Google Scholar]
- 10.Varadan R, Walker O, Pickart C, Fushman D. J Mol Biol. 2002;324:637–647. doi: 10.1016/s0022-2836(02)01198-1.; Fushman D, Varadan R, Assfalg M, Walker O. Progr. NMR Spectr. 2004;44:189–214.; Ryabov YE, Fushman D. Proteins. 2006;63:787–796. doi: 10.1002/prot.20917.; Ryabov YE, Fushman D. J. Am. Chem. Soc. 2007;129:3315–27. doi: 10.1021/ja067667r. For the early example of segmental isotope labelling of protein domains see Xu R, Ayers B, Cowburn D, Muir TW. Proc.Natl.Acad. Sci. USA. 1999;96:388–393. doi: 10.1073/pnas.96.2.388.
- 11.Varadan R, Assfalg M, Raasi S, Pickart C, Fushman D. Mol Cell. 2005;18:687–698. doi: 10.1016/j.molcel.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 12.a Cook WJ, Jeffrey LC, Carson M, Zhijian C, Pickart CM. J Biol Chem. 1992;267:16467–16471. doi: 10.2210/pdb1aar/pdb. [DOI] [PubMed] [Google Scholar]; b Eddins MJ, Varadan R, Fushman D, Pickart CM, Wolberger C. J Mol Biol. 2007;367:204–211. doi: 10.1016/j.jmb.2006.12.065. [DOI] [PubMed] [Google Scholar]
- 13.Dickinson BC, Varadan R, Fushman D. Protein Sci. 2007;16:369–378. doi: 10.1110/ps.062508007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang H, Jeon MS, Liao L, Yang C, Elly C, Yates JR, 3rd, Liu YC. Immunity. 2010;33:60–70. doi: 10.1016/j.immuni.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Varadan R, Assfalg M, Haririnia A, Raasi S, Pickart C, Fushman D. J Biol Chem. 2004;279:7055–7063. doi: 10.1074/jbc.M309184200. [DOI] [PubMed] [Google Scholar]
- 16.Fushman D, Walker O. J Mol Biol. 2010;395:803–814. doi: 10.1016/j.jmb.2009.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tjandra N, Feller SE, Pastor RW, Bax A. J. Am. Chem. Soc. 1995;117:12562–12566. [Google Scholar]
- 18.a Raasi S, Orlov I, Fleming KG, Pickart CM. J Mol Biol. 2004;341:1367–1379. doi: 10.1016/j.jmb.2004.06.057. [DOI] [PubMed] [Google Scholar]; b Raasi S, Varadan R, Fushman D, Pickart CM. Nat Struct Mol Biol. 2005;12:708–714. doi: 10.1038/nsmb962. [DOI] [PubMed] [Google Scholar]
- 19.a Ryu KS, Lee KJ, Bae SH, Kim BK, Kim KA, Choi BS. J Biol Chem. 2003;278:36621. doi: 10.1074/jbc.M304628200. [DOI] [PubMed] [Google Scholar]; b Mueller TD, Kamionka M, Feigon J. J Biol Chem. 2004;279:11926–36. doi: 10.1074/jbc.M312865200. [DOI] [PubMed] [Google Scholar]
- 20.Tugarinov V, Hwang PM, Kay LE. Annu Rev Biochem. 2004;73:107–146. doi: 10.1146/annurev.biochem.73.011303.074004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.