Abstract
The liver is a primary target of growth hormone (GH). GH signals are mediated by the transcription factor signal transducer and activator of transcription 5 (STAT5). Here, we focus on recent discoveries about the role of GH-STAT5 signaling in hepatic physiology and pathophysiology. We discuss roles of the GH-STAT5 axis in body growth, lipid metabolism, and the cell cycle pertaining to hepatosteatosis, fibrosis, and hepatocellular carcinoma. Finally, we discuss recent discoveries about the role of GH-STAT5 in sex-specific gene expression and bile acid, steroid, and drug metabolism.
Keywords: growth hormone, STAT5, liver function
Introduction
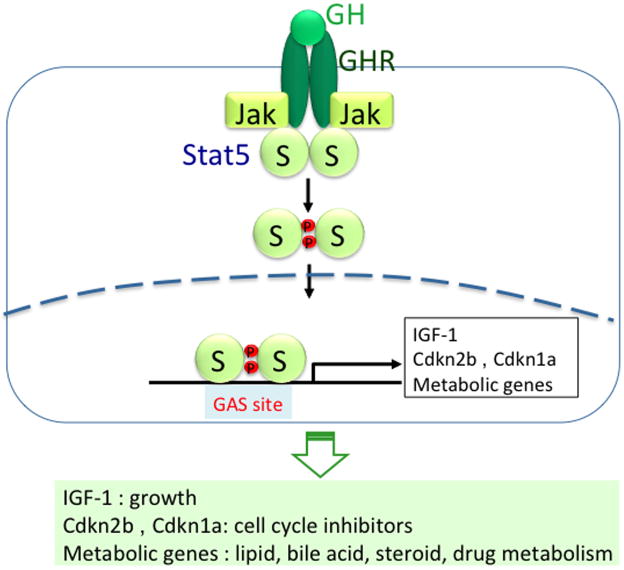
In the liver, growth hormone (GH) binds its cognate receptor (GHR), which results in the activation of JAK2, which in turn phosphorylates the signal transducer and activator of transcription 5 (STAT5) (Fig. 1A). Phosphorylated STAT5 translocates to the nucleus, where it binds to GAS motifs to regulate target genes. In the liver, GH-STAT5 signaling regulates expression of target genes associated with several physiological processes, including body growth, cell cycle, lipid, bile acid (BA), steroid, and drug metabolism (Fig. 1; Table 1). Disruption of STAT5 signaling is associated with liver disease, including fatty liver, fibrosis, and hepatocellular carcinoma (HCC). Here, we review current status of GH-STAT5 signaling on these topics.
Figure 1.
GH-STAT5 signaling pathway. GH binds its receptor and then activates Jak2, which in turn activates STAT5 by phosphorylation. Phosphorylated STAT5 goes to the nucleus, where it binds to gamma interferon–activated sequence (GAS) site of target gene's promoters. It activates transcription of Igf-1 gene.3,4 It activates cell cycle inhibitors Cdkn2b (cyclin-dependent kinase inhibitor 2B; p15INK4B) and Cdkn1a (p21CIP1).13 It also activates metabolic genes associated with lipid, bile acid, and steroid/drug metabolism. STAT5.
Table 1. Genes regulated by STAT5 and sex in the liver.
| Gene | STAT5 deletion effect in males | STAT5 regulation | References |
|---|---|---|---|
| Growth | |||
| IGF-1, IGFALS, IGFBP3, EGFR | ↓ | STATA/B | 3,4 |
| Lipid metabolism | |||
| PPARγ, CD36 | ↑ | STATA/B | 3 |
| Cell cycle inhibitor | |||
| Cdkn1a, Cdkn2b | ↓ | STATA | 13 |
| Cell cycle regulator | |||
| Cyclin D1 | ↑ | STATA/B | 3,4,13 |
| JAK-STAT pathway associated genes | |||
| Socs2, Cish | ↓ | STATA/B | 3,4 |
| Socs3, STAT1, STAT3 | ↑ | STATA/B | 3,4 |
| Male-specific genes | |||
| Cyp4a12, Gstπ, Slp, Elovl3, Mup3, Cyp7b1, Slco1a1(Oatp1), Hsd3b5, Moxdl1 | ↓ | STAT5B | 22-34 |
| B-cell leukemia/lymphoma 6 (Bcl6) | - | STAT5A/B | 40,44 |
| Female-specific genes | |||
| Cyp2a4, Cyp39a1, Sult1e1, Nnmt | ↑ | STAT5B | 22, 35-37 |
| Cyp2b9, Cyp2b13 | ↑ | - | 22,38,39 |
| Cyp2b10 | ↓ | STAT5B | 17 |
Arrows indicate changes in gene expression (up, down, no change).
STAT5, growth, lipid metabolism, cell cycle, and HCC
STAT5 and body growth
GH-STAT5 signaling regulates post-natal body growth by inducing Igf-1 gene expression.1 Kofoed et al. described a patient with a homozygous mutation in the Stat5b gene, which resulted in IGF-1 deficiency and growth hormone insensitivity with impairment of body growth.2 The Igf-1 gene is a direct target of STAT5, and our studies revealed that IGF-1 concentrations were reduced in STAT5-deficient mice.3,4 Two distinct GH-inducible STAT5B binding motifs have been identified in human Igf-1 and rat Igf-1 loci.5 Other studies have suggested the potential existence of multiple STAT5B-binding sites in the Igf-1 gene.6 Recently, it has been demonstrated that upon GH stimulation, STAT5B is recruited to a least seven distinct chromosomal domains throughout the Igf-1 locus, some of them with the potential of long-range enhancers.7
The six-exon Igf-1 gene in rats contains two promoters with distinct tissue-limited profiles of expression.8 GH-mediated signaling caused acute alterations in hepatic chromatin architecture at the Igf-1 locus.9 At promoter 1, GH caused RNA Pol II to be released from a previously recruited paused preinitiation complex. In contrast, at promoter 2, hormone treatment facilitates recruitment followed by the activation of RNA Pol II to initiate transcription. It appears that both Igf-1 promoters reside in open chromatin, despite minimal transcription in the absence of GH, as inferred from relatively high levels of core histone acetylation and enhanced histone H3K4 trimethylation. It has been suggested that this landscape of open chromatin may reflect the impact of previous long-term exposure to GH.
STAT5 and lipid metabolism
Several studies have suggested that GH-STAT5 signaling plays an important role in controlling hepatic lipid metabolism. Mice with a liver-specific STAT5 ablation developed hepatosteatosis, glucose intolerance, insulin resistance, late-onset obesity, and impaired liver regeneration.3 Notably, expression of genes associated with adipogenesis (PPARγ) and fatty acid uptake (CD36) was up-regulated in Stat5a/b-deficient mice.3 These changes may partially explain hepatosteatosis induced by loss of STAT5. Liver-specific deletion of the growth hormone receptor also displayed marked hepatic steatosis, insulin resistance, glucose intolerance, and increased triglyceride synthesis and decreased efflux.10 Recent studies with STAT5-deficient mice suggests that elevated CD36, PPARγ, and PGC1α/β, along with increased fatty acid synthesis, lipoprotein lipase, and very low-density lipoprotein receptor, are responsible for hepatic steatosis in these mice.11 Further studies with hepatocyte-specific Stat5-null mice will contribute to an understanding of the underlying molecular mechanisms.
STAT5, cell cycle, and HCC
Loss of STAT5 in hepatocytes caused steatosis, liver fibrosis, and promoted chemically induced liver cancer,12 which was associated with altered expression of cell cycle regulators.13 Loss of STAT5 from mouse embryonic fibroblasts (MEFs) leads to enhanced proliferation and was linked to reduced levels of the cell cycle inhibitors Cdkn2b (cyclin-dependent kinase inhibitor 2B; p15INK4B) and Cdkn1a (p21CIP1). Growth hormone, through STAT5 binding to promoter-bound GAS motifs, enhanced expression of the Cdkn2b and Cdkn1a genes. In mouse liver STAT5, like in MEFs, activated expression of the Cdkn2b gene. This study demonstrated that cytokines, through STAT5, induce the expression of key cell cycle inhibitors. STAT5 might promote cell cycle arrest in chronically injured liver hepatocyte and HCC as a critical tumor suppressor. STAT5 also regulates tumor suppressor and antiproliferating activities in other cell types. In T cells, oncogenic tyrosine kinase NPM1-ALK induces epigenetic silencing of Stat5a gene, and STAT5A protein can act as a key tumor suppressor by reciprocally inhibiting expression of NPM1-ALK.14 It has been suggested that loss of STAT5A is crucial for NPM1-ALK–mediated oncogenesis as this permits uninterrupted transcription of NPM1-ALK. In human diploid fibroblasts, constitutive activation of STAT5A also induced a cell cycle arrest with the characteristics of cellular senescence including the nuclear accumulation of the p53 tumor suppressor protein and accumulated DNA damage foci and activated ATM and ATR.15 SOCS1 can link the DNA damage signals characteristic of STAT5a-expressing cells to p53, serving as a mediator for p53 phosphorylation by ATM and ATR.15,16 These results suggest that STAT5 can regulate senescence by activating the tumor suppressors SOCS1 and p53.16
STAT5, sex-specific gene expression, bile acid, drug, and steroid metabolism
STAT5 and sex-specific gene expression
The liver is a sexually dimorphic organ, with sex-dependent differential expression of more than 1,000 genes in rats and mice.17,18 In several species, including mice and rats, pulsatile secretion of GH in males is distinct from that in females, and sexual dimorphic expression patterns of genes in liver tissue have been observed.19 In males, testosterone acts on the hypothalamic-pituitary axis to drive pronounced pulsatile GH secretion. The androgenic hormones testosterone and dihydroxytestosterone convey their signals through the androgen receptor (AR).20 Recent studies have revealed that expression of the Ar gene in skeletal muscle is directly controlled by GH through STAT5A/B.21 This finding opens studies to explore links between AR, GH, and sex-specific gene expression.
Many of the sex-specific genes in liver are regulated by the GH-STAT5 pathway (Table 1).19 In liver, STAT5B is ∼20-fold more abundant than STAT5A, suggesting that the vast majority of GH signaling is mediated through STAT5B. STAT5B-deficient male mice are characterized by reduced body growth and a loss of sex-specific expression of genes encoding cytochrome P-450 (Cyp) enzymes.1 Many studies showed that male-specific genes including Cyp4a12,22-24 Gstπ,22,25 Slp,22,26 Elovl3,22,27 Mup3,22,28 Mup1/2/6/8, 22 Cyp7b1, 22,29 Slco1a1, 22,30,31 Hsd3b5, 22,32,33 and Moxdl122,34 were down-regulated in the male liver of hepatocyte Stat5a/b-deficient mice. In contrast, female-specific liver genes including Cyp2a4,22,35 Cyp39a1,22 Sult1e1,22,36 Nnmt,22,37 Cyp2b9, 22,38 and Cyp2b1322,39 were up-regulated (de-repressed). Microarray analysis showed that 90% of male-predominant genes were suppressed and 60% of female-predominant genes were induced in liver tissue of Stat5b-null male mice.17 Another study also identified several gene sets displaying sexual dimorphic expression.18 Gene expression profiling on global Stat5a- and Stat5b-null mice confirmed STAT5B as the transcription factor conveying sexual dimorphism, but a distinct role for STAT5A has been established as well. The expression of 15% of female-predominant genes was dependent on the presence of STAT5A. These studies highlight sex-specific roles of the two STAT5 isoforms, with STAT5A preferentially regulating gene expression in the female liver while STAT5B plays the predominant role in the male.
The regulation of sex-specific genes through both transcriptional activators and repressors has been investigated. STAT5b synergistically enhanced the transcriptional activity of HNF4-α toward two other male-specific liver target genes, Cyp2d9 and CYP8B1.40 These results highlight the ability of STAT5b to act in concert with HNF4-α to regulate male-specific liver Cyp genes. They also proposed that HNF-6 and HNF-3β are female-predominant, positive regulators of Cyp2c12 (female specific) and negative regulators of Cyp2a2 (male specific). The transcriptional repressor Bcl6 is a male-specific rat liver gene product.41 The DNA recognition motif of Bcl642 resembles the STAT5 consensus site TTCNNNGAA.43 This raises the possibility that Bcl6 and STAT proteins may regulate overlapping sets of genes. Bcl6 was bound to a subset of STAT5-binding sites in male liver chromatin, including a Socs2 STAT5-binding site where Bcl6 binding increased substantially between plasma GH pulses when STAT5 binding was low.44 Thus, Bcl6 and STAT5 binding was inversely coordinated by the endogenous pulses of pituitary GH release. They suggest that this male-specific transcriptional repressor modulates hepatic GH signaling to select STAT5 target genes.
Chromatin structure affected sex-dependent gene expression. The sex-related expression of the Cyp2c12 gene resulted from the inaccessibility of STAT5 to the GH-responsive element by chromatin condensation in male rat livers.45 Sex-specific differential binding affinity of STAT5 to target genes was observed in rats. Phylogenetic footprinting was used to predict functional transcription factor binding sites STAT5 in the GH-responsive genes Igf-1, SOCS2, and HNF-6.46 In female rat liver, where nuclear STAT5 activity is generally low, STAT5 binding to Igf-1 and SOCS2 was limited to high-affinity sites, while male specific gene expression was associated with male-specific STAT5 binding to multiple low-affinity STAT5 sites.
STAT5 and bile acid/drug metabolism
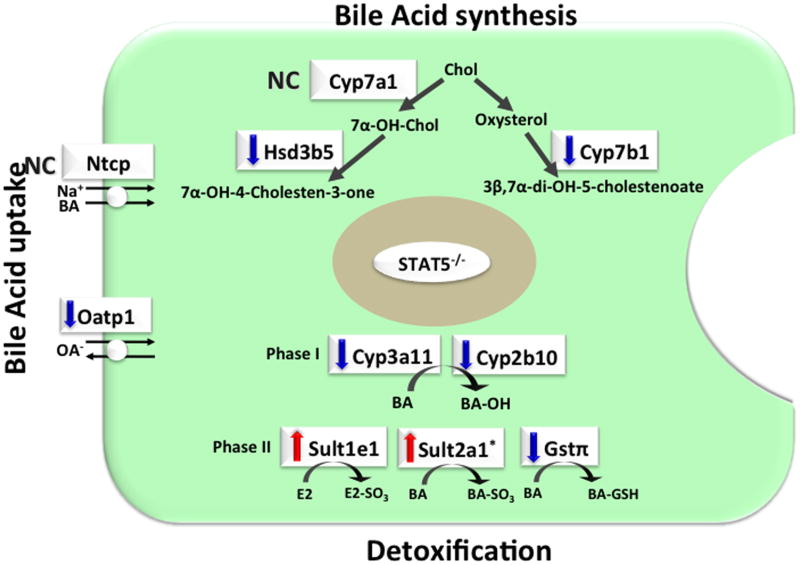
Many sex-specific genes belong to the Cyps, sulfotransferases (Sults), or glutathione transferase (Gst) family that play important roles in BA, steroid, and drug metabolism. Several studies reports that GH-STAT5 signaling regulates expression of genes associated with BA metabolism including BA synthesis, BA uptake, BA detoxification, and excretion (Fig. 2). Conversion of cholesterol to 7α-hydroxycholesterol by cyp7a1 cholesterol 7a-hydroxylase is the initial and rate-limiting step in BA synthesis.47 In addition, Cyp7b1 oxycholesterol 7α-hydroxyase catalyzes the alternative pathway of cholesterol metabolism. Expression of Cyp7b1 (oxysterol 7α-hydroxylase) was down-regulated in the liver of STAT5-deleted mice.22,29 Expression of Hsd3b5 (3βHSD; 3β-hydroxysteroid dehydrogenase type 5), which is involved in the oxidation of the 3β-OH and isomerization of the C5-C6 double bond, was also down-regulated in the liver of STAT5-deleted mice.22,32,33 Secondary BAs, lithocholic acid and deoxycholic acid, are synthesized in the gut lumen from the primary BAs by microbial enzymes and are reabsorbed into the enterohepatic circulation. Cyp3a10/6β-hydroxylase is a male-specific P450 that catalyzes 6β-hydroxylation of lithocholic acid.47,48 STAT5a/b positively mediated GH-dependent regulation of Cyp3a10 promoter activity.49
Figure 2.
Changes in hepatic gene expression of BA metabolism in STAT5-deficient mice. STAT5-liver specific knockout mice (STAT5−/ −) exhibited changes in gene expression of BA metabolism including synthesis,22,29,32,33 uptake,22,30,31 and detoxification.17,22,25 Sult2a1 gene was induced in GH-releasing hormone receptor-deficient little (lit/lit) mouse.50 Ntcp, Na+/taurocholate cotransporter; Oatp1, organic anion transporter 1; BA, bile acid; Gstπ, glutathione S-transferase pi class; Chol, cholesterol; E2, estradiol; OA-, organic anions; BA-OH, hydroxylated BA; BA-SO3, sulfated BA; GSH, glutathione. Arrows indicate changes in gene expression (up, down). NC, no change.
GH-STAT5 signaling regulates gene expression associated with BA transport. Slco1a1 (the organic anion transporter Oatp1) is an uptake transport of a variety of organic endo and exogenous compounds, such as BAs, steroid and thyroid hormones and their conjugates, and numerous drugs and toxins.30,31 Expression of BA transporter slco1a1 was down-regulated in Stat5-deleted male mice.22,30,31 To investigate the physiologic and pharmacologic roles of Oatps of the 1a and 1b subfamilies, mice were generated lacking all established and predicted mouse Oatp1a/1b transporters (referred to as Slco1a/1b−/− mice).30 Slco1a/1b−/− exhibited markedly increased plasma levels of bilirubin conjugated to glucuronide and increased plasma levels of unconjugated BAs, indicating that Oatp1a/1b transporters normally mediate extensive hepatic reuptake of glucuronidated bilirubin. Slco1a/1b–/– mice also showed marked decrease in hepatic uptake and consequently increased systemic exposure following intravenous or oral administration of the OATP substrate drugs methotrexate and fexofenadine. These data indicate that Oatp1a/1b transporters play an essential role in hepatic reuptake of conjugated bilirubin and uptake of unconjugated BAs and drugs.
The GH-STAT5 pathway also regulates gene expression associated with BA detoxification. Hydroxylation of BAs is mediated by the phase I–detoxifying Cyp enzymes Cyp3a11 and Cyp2b10.17 The expression of Cyp3a11 and Cyp2b10 genes was down-regulated in STAT5b-deficient male mice.17 Sult2a1 catalyzes the formation of BAs sulfates (BA-sulfates). Sulfation of BAs increases their solubility, decreases their intestinal absorption, and enhances their fecal and urinary excretion. Sult2a1 is female-predominant Sult in mouse liver. Expression of Sult2a1 is suppressed by androgens and male-pattern GH secretion, while it is induced by estrogens and female-pattern GH secretion.50 Hepatic expression of Sult2a1 was increased in GH-releasing hormone receptor-deficient little (lit/lit) mouse in males.50 In mice, STAT5 deletion in the liver resulted in decreased glutathione S-transferase pi class (Gstπ), which is one of phase II detoxification enzyme, catalyzing the conjugation of glutathione to BA.22,25
Recent studies pointed to the importance of GH-STAT5 signaling for maintaining BA homeostasis. Deletion of the Stat5a/b locus in hepatocyte and cholangiocytes in the multidrug resistance gene 2 knockout (Mdr2–/–), resulted in an early and severe liver fibrosis phenotype, accompanied by deregulated expression of important regulators of BA homeostasis,51 suggesting that loss of STAT5 sensitizes hepatocytes to BA-induced damage.
Overall, loss of liver-specific STAT5 resulted in decreases in Cyp7b1 and Hsd3b5 gene expression for BA synthesis and decrease in Oapt1 gene expression for BA uptake. In addition, deletion of STAT5 gene resulted in decrease in phase I detoxifying Cyp3a11 and Cyp2b10 gene expression, whereas phase II detoxifying Sult1e1 gene expression was increased in these mice. Thus, these studies imply that the GH-STAT5 pathway has an important function in regulating BA metabolism including synthesis, transport, and detoxification. Whether these changes are either directly or indirectly regulated by GH-STAT5 signaling remains to be determined. Elucidating the direct or indirect molecular mechanisms involved in these processes should lead to a better understanding of the role of GH-STAT5 in BA metabolism, cholesterol homeostasis, and liver diseases such as cholestasis and gallstones.
STAT5 and steroid hormone metabolism
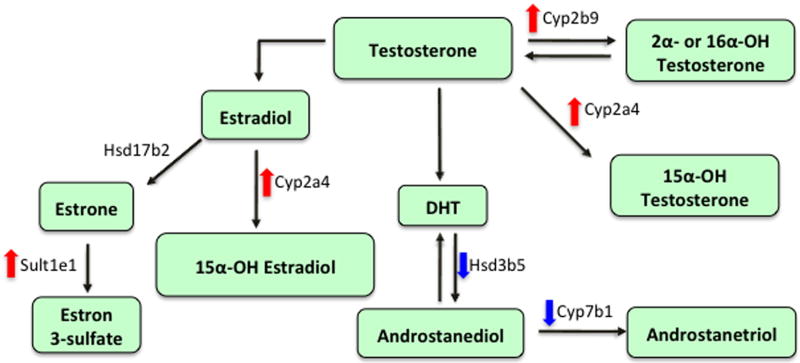
Liver is a target organ for steroid hormone metabolism. Several studies have suggested that GH-STAT5 regulates genes linked to steroid metabolism (Fig. 3). One of these genes, HSD3b5, catalyzes the formation of the relatively inactive androstanediol from the active dihydrotestosterone.33 Hsd3b5 gene expression was down-regulated in the liver of STAT5-deleted male mice.22,32,33 In contrast, another gene involved in testosterone metabolism, testosterone 16α-hydroxylase (Cyp2b9), which hydroxylates testosterone at the 16α position, was up-regulated in STAT5-deleted male mice.22 Cyp7b1 (oxysterol 7α-hydroxylase), responsible for the hydroxylation of dehydroepiandrosterone, androstenediol, androstanetriol, and 25-hydroxycholesterol, was downregulated in Stat5-deleted male mice.22,29
Figure 3.
Changes in hepatic gene expression of testosterone and estrogen metabolism in STAT5-deficient mice. STAT5-liver specific knockout mice down-regulated expression of Hsd3b522,32 and Cyp7b1 genes,22,29 whereas it up-regulated expression of Cyp2b9,22,38 Cyp2a4,22,35 and Sult1e1 genes22,36 in the liver. DHT, dihydrotestosterone; OH, hydroxylated. Arrows indicate changes in gene expression (up, down).
Genes associated with estrogen metabolism are also regulated by GH-STAT5. Sult1e1 is the major Sult isoform responsible for the inactivation of β-estradiol via sulfation at physiological concentrations.52 Expression of sult1e1 gene was up-regulated in STAT5-deficient male mice.22,36 Changes in Sult1e1 activity may alter E2 levels and E2-regulated processes in tissues and cells, including those of the liver where it is expressed at significant levels. Sult1e1 was also shown to have high affinity (nM range) for diethylstilbestrol and tamoxifen as well as for E2 and estrone.53,54 Testosterone or estradiol 15alpha-hydroxylase Cyp2a4 gene is female-specific, and this gene is up-regulated in Stat5-deleted male mice.22,35,38
Thus hepatic GH-STAT5 signaling regulates, at least in part, genes involved in hepatic testosterone/estrogen and drug metabolism. Changes in the metabolism of steroid hormones may have a role in alterations in liver function or the development of liver disease. Further studies regarding direct or indirect molecular mechanisms that regulate steroid/drug metabolism by GH-STAT5 pathway will enhance our understanding of functional significance of GH-STAT5 pathway on steroid metabolism in the liver. Overall, many studies have established that GH-STAT5 signaling is key in dictating sex differences in the expression of a large number of liver gene products, including many Cyps and other DMEs. Additional investigations are needed to elucidate mechanisms that control expression of these genes by GH-STAT5 and to understand functional roles of GH-STAT5 regulating BA, steroid, and drug metabolism and its clinical implications. Studies with hepatocyte-specific STAT5-deleted mice will allow further understanding of the functional role of sex-specific genes for controlling drug, steroid, and BA metabolism and of related liver diseases.
Acknowledgments
This work was funded in part through the Intramural program of the National Institutes of Health and through a grant from WCU Project (R33-10059) through the NRF and by a grant from the New BioGreen 21 Program, Rural Development Administration, Republic of Korea.
Footnotes
Conflicts of interest
References
- 1.Udy GB, Towers RP, Snell RG, et al. Requirement of STAT5b for sexual dimorphism of body growth rates and liver gene expression. Proc Natl Acad Sci U S A. 1997;94:7239–7244. doi: 10.1073/pnas.94.14.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kofoed EM, Hwa V, Little B, et al. Growth hormone insensitivity associated with a STAT5b mutation. N Engl J Med. 2003;349:1139–1147. doi: 10.1056/NEJMoa022926. [DOI] [PubMed] [Google Scholar]
- 3.Cui Y, Riedlinger G, Miyoshi K, et al. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol Cell Biol. 2004;24:8037–8047. doi: 10.1128/MCB.24.18.8037-8047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engblom D, Kornfeld JW, Schwake L, et al. Direct glucocorticoid receptor-Stat5 interaction in hepatocytes controls body size and maturation-related gene expression. Genes Dev. 2007;21:1157–1162. doi: 10.1101/gad.426007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chia DJ, Ono M, Woelfle J, et al. Characterization of distinct Stat5b binding sites that mediate growth hormone-stimulated IGF-I gene transcription. J Biol Chem. 2006;281:3190–3197. doi: 10.1074/jbc.M510204200. [DOI] [PubMed] [Google Scholar]
- 6.Eleswarapu S, Ge X, Wang Y, et al. Growth hormone-activated STAT5 may indirectly stimulate IGF-I gene transcription through HNF-3{gamma} Mol Endocrinol. 2009;23:2026–2037. doi: 10.1210/me.2009-0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chia DJ, Varco-Merth B, Rotwein P. Dispersed Chromosomal Stat5b-binding elements mediate growth hormone-activated insulin-like growth factor-I gene transcription. J Biol Chem. 2010;285:17636–17647. doi: 10.1074/jbc.M110.117697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hall LJ, Kajimoto Y, Bichell D, et al. Functional analysis of the rat insulin-like growth factor I gene and identification of an IGF-I gene promoter. DNA Cell Biol. 1992;11:301–313. doi: 10.1089/dna.1992.11.301. [DOI] [PubMed] [Google Scholar]
- 9.Chia DJ, Young JJ, Mertens AR, et al. Distinct alterations in chromatin organization of the two IGF-I promoters precede growth hormone-induced activation of IGF-I gene transcription. Mol Endocrinol. 2010;24:779–789. doi: 10.1210/me.2009-0430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan Y, Menon RK, Cohen P, et al. Liver-specific deletion of the growth hormone receptor reveals essential role of growth hormone signaling in hepatic lipid metabolism. J Biol Chem. 2009;284:19937–19944. doi: 10.1074/jbc.M109.014308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barclay JL, Nelson CN, Ishikawa M, et al. GH-dependent STAT5 signaling plays an important role in hepatic lipid metabolism. Endocrinology. 2011;152:181–192. doi: 10.1210/en.2010-0537. [DOI] [PubMed] [Google Scholar]
- 12.Hosui A, Kimura A, Yamaji D, et al. Loss of STAT5 causes liver fibrosis and cancer development through increased TGF-{beta} and STAT3 activation. J Exp Med. 2009;206:819–831. doi: 10.1084/jem.20080003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu JH, Zhu BM, Wickre M, et al. The transcription factors signal transducer and activator of transcription 5A (STAT5A) and STAT5B negatively regulate cell proliferation through the activation of cyclin-dependent kinase inhibitor 2b (Cdkn2b) and Cdkn1a expression. Hepatology. 2010;52:1808–1818. doi: 10.1002/hep.23882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Q, Wang HY, Liu X, et al. STAT5A is epigenetically silenced by the tyrosine kinase NPM1-ALK and acts as a tumor suppressor by reciprocally inhibiting NPM1-ALK expression. Nat Med. 2007;13:1341–1348. doi: 10.1038/nm1659. [DOI] [PubMed] [Google Scholar]
- 15.Mallette FA, Gaumont-Leclerc MF, Ferbeyre G. The DNA damage signaling pathway is a critical mediator of oncogene-induced senescence. Genes Dev. 2007;21:43–48. doi: 10.1101/gad.1487307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calabrese V, Mallette FA, Deschênes-Simard X, et al. SOCS1 links cytokine signaling to p53 and senescence. Mol Cell. 2009;36:754–767. doi: 10.1016/j.molcel.2009.09.044. [DOI] [PubMed] [Google Scholar]
- 17.Clodfelter KH, Holloway MG, Hodor P, et al. Sex-dependent liver gene expression is extensive and largely dependent upon signal transducer and activator of transcription 5b (STAT5b): STAT5b-dependent activation of male genes and repression of female genes revealed by microarray analysis. Mol Endocrinol. 2006;20:1333–1351. doi: 10.1210/me.2005-0489. [DOI] [PubMed] [Google Scholar]
- 18.Clodfelter KH, Miles GD, Wauthier V, et al. Role of STAT5a in regulation of sex-specific gene expression in female but not male mouse liver revealed by microarray analysis. Physiol Genomics. 2007;31:63–74. doi: 10.1152/physiolgenomics.00055.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waxman DJ, O'Connor C. Growth hormone regulation of sex-dependent liver gene expression. Mol Endocrinol. 2006;20:2613–2629. doi: 10.1210/me.2006-0007. [DOI] [PubMed] [Google Scholar]
- 20.Herbst KL, Bhasin S. Testosterone action on skeletal muscle. Curr Opin Clin Nutr Metab Care. 2004;7:271–277. doi: 10.1097/00075197-200405000-00006. [DOI] [PubMed] [Google Scholar]
- 21.Klover P, Chen W, Zhu BM, et al. Skeletal muscle growth and fiber composition in mice are regulated through the transcription factors STAT5a/b: linking growth hormone to the androgen receptor. FASEB J. 2009;23:3140–3148. doi: 10.1096/fj.08-128215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holloway MG, Cui Y, Laz EV, et al. Loss of sexually dimorphic liver gene expression upon hepatocyte-specific deletion of Stat5a-Stat5b locus. Endocrinology. 2007;148:1977–1986. doi: 10.1210/en.2006-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okita RT, Okita JR. Cytochrome P450 4A fatty acid omega hydroxylases. Curr Drug Metab. 2001;2:265–281. doi: 10.2174/1389200013338423. [DOI] [PubMed] [Google Scholar]
- 24.Hardwick JP. Cytochrome P450 omega hydroxylase (CYP4) function in fatty acid metabolism and metabolic diseases. Biochem Pharmacol. 2008;75:2263–2275. doi: 10.1016/j.bcp.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 25.Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 26.Krebs CJ, Khan S, MacDonald JW, et al. Regulator of sex-limitation KRAB zinc finger proteins modulate sex-dependent and -independent liver metabolism. Physiol Genomics. 2009;38:16–28. doi: 10.1152/physiolgenomics.90391.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brolinson A, Fourcade S, Jakobsson A, et al. Steroid hormones control circadian Elovl3 expression in mouse liver. Endocrinology. 2008;149:3158–3166. doi: 10.1210/en.2007-1402. [DOI] [PubMed] [Google Scholar]
- 28.Chamero P, Marton TF, Logan DW, et al. Identification of protein pheromones that promote aggressive behaviour. Nature. 2007;450:899–902. doi: 10.1038/nature05997. [DOI] [PubMed] [Google Scholar]
- 29.Schwarz M, Lund EG, Rusell DW. Two 7 alpha-hydroxylase enzymes in bile acid biosynthesis. Curr Opin Lipidol. 1998;9:113–118. doi: 10.1097/00041433-199804000-00006. [DOI] [PubMed] [Google Scholar]
- 30.van de Steeg E, Wagenaar E, van der Kruijssen CM, et al. Organic anion transporting polypeptide 1a/1b-knockout mice provide insights into hepatic handling of bilirubin, bile acids, and drugs. J Clin Invest. 2010;120:2942–2952. doi: 10.1172/JCI42168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalliokoski A, Niemi M. Impact of OATP transporters on pharmacokinetics. Br J Pharmacol. 2009;158:693–705. doi: 10.1111/j.1476-5381.2009.00430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mason JI, Keeney DS, Bird IM, et al. The regulation of 3 beta-hydroxysteroid dehydrogenase expression. Steroids. 1997;62:164–168. doi: 10.1016/s0039-128x(96)00176-6. [DOI] [PubMed] [Google Scholar]
- 33.Abbaszade IG, Clarke TR, Park CH, et al. The mouse 3 beta-hydroxysteroid dehydrogenase multigene family includes two functionally distinct groups of proteins. Mol Endocrinol. 1995;9:1214–1222. doi: 10.1210/mend.9.9.7491113. [DOI] [PubMed] [Google Scholar]
- 34.Xin X, Mains RE, Eipper BA. Monooxygenase X, a member of the copper-dependent monooxygenase family localized to the endoplasmic reticulum. J Biol Chem. 2004;279:48159–48167. doi: 10.1074/jbc.M407486200. [DOI] [PubMed] [Google Scholar]
- 35.Lavery DJ, Lopez-Molina L, Margueron R, et al. Circadian expression of the steroid 15 alpha-hydroxylase (Cyp2a4) and coumarin 7-hydroxylase (Cyp2a5) genes in mouse liver is regulated by the PAR leucine zipper transcription factor DBP. Mol Cell Biol. 1999;19:6488–6499. doi: 10.1128/mcb.19.10.6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li L, He D, Wilborn TW, et al. Increased SULT1E1 activity in HepG2 hepatocytes decreases growth hormone stimulation of STAT5b phosphorylation. Steroids. 2009;74:20–29. doi: 10.1016/j.steroids.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weinshilboum R. Methyltransferase pharmacogenetics. Pharmacol Ther. 1989;43:77–90. doi: 10.1016/0163-7258(89)90048-x. [DOI] [PubMed] [Google Scholar]
- 38.Sakuma T, Kitajima K, Nishiyama M, et al. Suppression of female-specific murine Cyp2b9 gene expression by growth or glucocorticoid hormones. Biochem Biophy Res Commun. 323:776–781. doi: 10.1016/j.bbrc.2004.08.158. [DOI] [PubMed] [Google Scholar]
- 39.Lakso M, Masaki R, Noshiro M, et al. Structures and characterization of sex-specific mouse cytochrome P-450 genes as members within a large family. Duplication boundary and evolution. Eur J Biochem. 1991;195:477–486. doi: 10.1111/j.1432-1033.1991.tb15728.x. [DOI] [PubMed] [Google Scholar]
- 40.Wiwi CA, Waxman DJ. Role of hepatocyte nuclear factors in transcriptional regulation of male-specific CYP2A2. J Biol Chem. 2005;280:3259–3268. doi: 10.1074/jbc.M409294200. [DOI] [PubMed] [Google Scholar]
- 41.Wauthier V, Waxman DJ. Sex-specific early growth hormone response genes in rat liver. Mol Endocrinol. 2008;22:1962–1974. doi: 10.1210/me.2007-0549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seyfert VL, Allman D, He Y, et al. Transcriptional repression by the proto-oncogene BCL-6. Oncogene. 1996;12:2331–2342. [PubMed] [Google Scholar]
- 43.Ehret GB, Reichenbach P, Schindler U, et al. DNA binding specificity of different STAT proteins. Comparison of in vitro specificity with natural target sites. J Biol Chem. 2001;276:6675–6688. doi: 10.1074/jbc.M001748200. [DOI] [PubMed] [Google Scholar]
- 44.Meyer RD, Laz EV, Su T, et al. Male-specific hepatic Bcl6: growth hormone-induced block of transcription elongation in females and binding to target genes inversely coordinated with STAT5. Mol Endocrinol. 2009;23:1914–1926. doi: 10.1210/me.2009-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Endo M, Takahashi Y, Sasaki Y, et al. Novel gender-related regulation of CYP2C12 gene expression in rats. Mol Endocrinol. 2005;19:1181–1190. doi: 10.1210/me.2004-0063. [DOI] [PubMed] [Google Scholar]
- 46.Laz EV, Sugathan A, Waxman DJ. Dynamic in vivo binding of STAT5 to growth hormone-regulated genes in intact rat liver. Sex-specific binding at low- but not high-affinity STAT5 sites. Mol Endocrinol. 2009;23:1242–1254. doi: 10.1210/me.2008-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teixeira J, Gil G. Cloning, expression, and regulation of lithocholic acid 6b-hydroxylase. J Biol Chem. 1991;266:21030–21036. [PubMed] [Google Scholar]
- 48.Chang TKH, Teixeira J, et al. The lithocholic acid 6b-hydroxylase cytochrome P-450, CYP 3A10, is an active catalyst of steroid-hormone 6b-hydroxylation. Biochem J. 1993;291:429–434. doi: 10.1042/bj2910429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Subramanian A, Wang J, Gil G. STAT 5 and NF-Y are involved in expression and growth hormone-mediated sexually dimorphic regulation of cytochrome P450 3A10/lithocholic acid 6beta-hydroxylase. Nucleic Acids Res. 1998;26:2173–2178. doi: 10.1093/nar/26.9.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alnouti Y, Klaassen CD. Mechanisms of gender-specific regulation of mouse sulfotransferases (Sults) Xenobiotica. 2011;41:187–197. doi: 10.3109/00498254.2010.535923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blaas L, Kornfeld JW, Schramek D, et al. Disruption of the growth hormone--signal transducer and activator of transcription 5-insulinlike growth factor 1 axis severely aggravates liver fibrosis in a mouse model of cholestasis. Hepatology. 2010;51:1319–1326. doi: 10.1002/hep.23469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Falany JL, Greer H, Kovacs T, et al. Elevation of hepatic sulphotransferase activities in mice with resistance to cystic fibrosis. Biochem J. 2002;364:115–120. doi: 10.1042/bj3640115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Falany CN. Enzymology of human cytosolic SULTs. FASEB J. 1997;11:206–216. doi: 10.1096/fasebj.11.4.9068609. [DOI] [PubMed] [Google Scholar]
- 54.Falany CN, Krasnykh V, Falany JL. Bacterial expression and characterization of a cDNA for human liver estrogen sulfotransferase. J Steroid Biochem Molec Biol. 1995;52:529–539. doi: 10.1016/0960-0760(95)00015-r. [DOI] [PubMed] [Google Scholar]