Summary
While the glucose transporter-4 (GLUT4) is fundamental to insulin-regulated glucose metabolism, its dynamic spatial organization in the plasma membrane (PM) is unclear. Here, using multi-color TIRF microscopy in transfected adipose cells, we demonstrate that insulin regulates not only the exocytosis of GLUT4 storage vesicles, but also PM distribution of GLUT4 itself. In the basal state, domains (clusters) of GLUT4 molecules in PM are created by an exocytosis that retains GLUT4 at the fusion site. Surprisingly, when insulin induces a burst of GLUT4 exocytosis, it does not merely accelerate this basal exocytosis, but rather stimulates ~60-fold another mode of exocytosis that disperses GLUT4 into PM. In contradistinction, internalization of most GLUT4, regardless of insulin, occurs from pre-existing clusters via the subsequent recruitment of clathrin. The data fit a new kinetic model that features multifunctional clusters as intermediates of exocytosis and endocytosis.
Highlights.
GLUT4 exists in PM as freely-diffusing molecules and stable clusters.
Clusters/domains are generated by fusion with selective retention of GLUT4.
GLUT4 is internalized at the clusters via subsequent recruitment of clathrin.
Insulin induces a burst of GLUT4 exocytosis that disperses GLUT4 directly into PM.
Introduction
Insulin regulates glucose transport through recruitment of GLUT4 to the plasma membrane (PM) where these transporters facilitate glucose uptake. The current model of GLUT4 recycling proposes a complex regulated system of GLUT4 cycling among specialized GLUT4 storage vesicles (GSV), intracellular compartments, and PM (Cushman and Wardzala, 1980; Suzuki and Kono, 1980; Satoh et al, 1993; Holman et al, 1994; Rea and James, 1997; Xu and Kandror, 2002; Saltiel and Pessin, 2003; Martin et al., 2006). Insulin has been variably reported to regulate GLUT4 recycling in several ways including sorting and formation of the GSV (Kandror and Pilch, 1994; Shi and Kandror, 2005), intracellular untethering (Bogan et al., 2003), trafficking towards PM (Huang et al., 2007), actin cytoskeleton rearrangement (Kanzaki and Pessin, 2001; Torok et al., 2004), PM tethering (Bai et al., 2007; Lizunov et al., 2005), fusion (Jiang et al., 2008; Lizunov et al., 2005; Rea and James, 1997; Xu and Kandror, 2002), and inhibition of endocytosis (Blot and McGraw, 2008; Martin et al., 2006). However, despite this large number of processes apparently affected by insulin, recent experimental work strongly suggests that the main site of regulation of GLUT4 recycling and glucose uptake occurs at PM (Lizunov et al., 2005; Koumanov et al., 2005; Gonzalez and McGraw, 2006; Bai et al., 2007; Huang et al., 2007).
Recently, high resolution live cell microscopy techniques have been successfully applied to study GLUT4 recycling processes that take place near PM. Detection of single GLUT4 vesicles has allowed quantification of tethering and fusion of GSV in 3T3 L1 cells (Li et al., 2004; Jiang et al., 2008), and analysis of GLUT4 traffic in primary adipose cells (Lizunov et al., 2005; Lizunov et al., 2009), and muscles (Lauritzen et al., 2008; Fazakerley et al., 2009). These studies have also confirmed earlier observations that GLUT4 are distributed non-homogeneously in PM (Gustavsson et al., 1996; Parton et al., 2002). However, it remains unclear how insulin regulates GLUT4 organization in the PM and their spatial dynamics.
To probe both GLUT4 organization in PM and its relationship to insulin-regulated recycling, we investigated GLUT4 dynamics in isolated rat adipose cells. We find: 1) clusters are generated by fusion with retention of GLUT4 in nascent domains; 2) GLUT4 is internalized at these domains after subsequent recruitment of clathrin, and 3) insulin induces a burst of GLUT4 exocytosis that mostly bypasses these domains and disperses GLUT4 directly into PM.
Results
GLUT4 distribution in the vicinity of PM
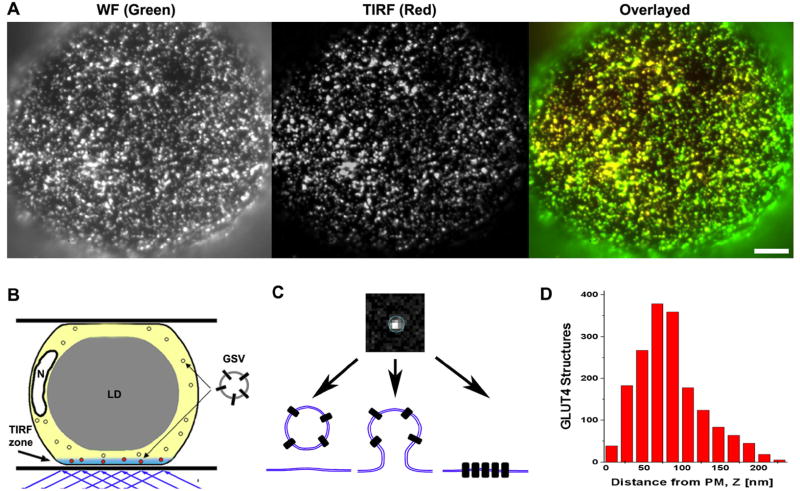
To quantify the subcellular distribution of GLUT4 near PM we employed a combination of total internal reflection fluorescence (TIRF) and wide-field fluorescence (WF) microscopy. Isolated rat adipose cells were transiently transfected with either HA-GLUT4-GFP or HA-GLUT4-mCherry, and sequential TIRF and WF images were acquired (Fig.1A). The evanescent wave illumination used in the TIRF mode decays exponentially with the distance from the water-glass interface, and thus effectively excites fluorophores only in a thin layer (TIRF-zone) near PM. Therefore TIRF microscopy selectively images GLUT4 structures that are localized in the vicinity of, or associated with, PM. WF microscopy, on the other hand, visualizes all GLUT4 structures present in the cytoplasm layer (~1 μm) between PM and the central lipid droplet (Fig. 1B). About 70% of total GLUT4 detected in the WF mode (shown in green) is visible also in the TIRF mode (shown in red) (Fig. 1A)
Figure 1. GLUT4 structures in the vicinity of PM.
(A) Isolated rat adipose cells were transiently transected with HA-GLUT4-GFP and imaged using a combination of TIRF (red) and WF (green) microscopy. The overlay image shows that the majority of HA-GLUT4-GFP is present in the TIRF-zone close to PM. Bar, 5 μm.
(B) Scheme of experiment. TIRF microscopy selectively images HA-GLUT4-GFP structures localized in the TIRF-zone (within 200 nm of PM), while the WF image comprises fluorescence from PM and the full thickness of the adjacent layer of cytoplasm (thickness of ~1 μm). N - Nucleus; LD - lipid droplet.
(C) Zoomed image of a single GLUT4 punctum detected in a TIRF image. Diffraction-limited GLUT4 puncta may represent (from left to right): GSV in the vicinity of PM (tethered or untethered), a vesicle undergoing exocytosis or endocytosis, and/or GLUT4 molecules clustered in PM.
(D) Histogram of GLUT4 structure distribution in the TIRF-zone in the absence of insulin. The distance from PM (TIRF interface) was calculated for individual GLUT4 puncta from the ratio of TIRF/WF signals. Note the significant amount of GLUT4 structures localized within 100 nm of PM.
The majority of GLUT4 structures are detected as diffraction-limited bright spots (Figs. 1A, C). These structures indeed may represent GLUT4 vesicles, intermediates of exo- and endocytosis of GLUT4, as well as domains/clusters of GLUT4 molecules in PM (Fig. 1C). GLUT4 structures detected in the TIRF-zone had a variable fluorescence intensity that was due to both variation in the amount of GLUT4 molecules and differences in the distance of GLUT4 from PM or the TIRF interface. In the basal state, the distribution of GLUT4 obtained near PM shows that ~50% of GLUT4 structures are localized within 100 nm of PM (Fig. 1D). Insulin further increased the redistribution of GLUT4 closer to PM.
By time-lapse TIRF microscopy we analyzed two populations of GLUT4 structures in the vicinity of PM: stationary structures that exhibited no apparent change of lateral position or intensity and mobile structures that exhibited lateral motion or showed significant intensity change due to vertical displacement in TIRF-zone (Video 1). In the basal state, the relative fluorescence integrated over identified stationary and mobile GLUT4 structures showed that on average 77 ± 6 % of total GLUT4 in the TIRF zone was localized in stationary structures (Figs. S1A and S1B). Most of the mobile GLUT4 were attributed to GSV as they exhibited typical microtubule-based directed motion with speeds of 1-2 μm/s. Consistent with previous studies insulin decreased the traffic of GSV and further shifted the distribution of GLUT4 toward stationary structures.
Clustered and dispersed distribution of GLUT4 in PM
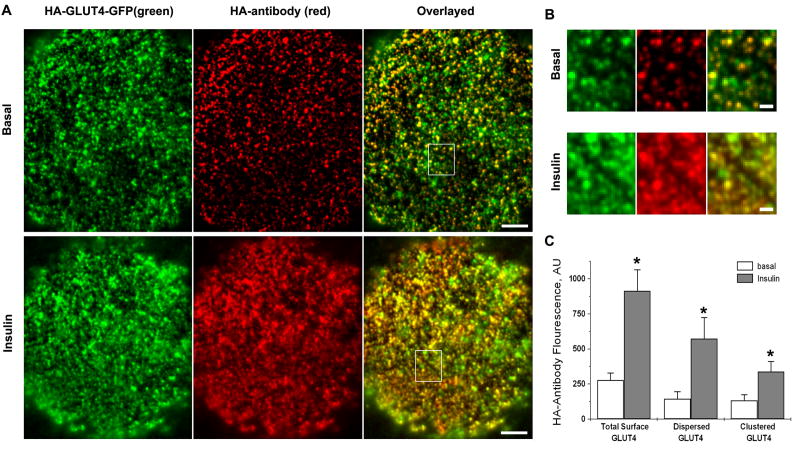
To transport glucose, the exofacial domain of GLUT4 must be exposed to the extracellular space. We tested whether the stationary GLUT4 structures were exposed to the extracellular space using an HA-antibody to label surface-exposed HA-GLUT4-GFP molecules in non-permeabilized cells. High resolution images of HA-labelling of PM showed that GLUT4 distribution in PM was highly punctate for both basal and insulin-stimulated cells (Figs. 2A and 2B). Consistent with the previous notion that GLUT4 in PM is quite inhomogeneous, these data further indicate that a significant fraction of PM GLUT4 is organized into distinct clusters. However, besides HA-labelled puncta that represent clusters, we also detected a diffuse HA-antibody labelling of the cell surface that we attributed to GLUT4 molecules dispersed in PM.
Figure 2. Insulin regulates GLUT4 exposure, identified by HA-antibody binding, and spatial distribution at the cell surface.
(A) Isolated rat adipose cells expressing HA-GLUT4-GFP were fixed and stained with HA-antibody under non-permeabilized conditions. Individual cells were imaged using multi-color TIRF microscopy. The upper panel shows a representative cell in the basal state; the lower panel shows a cell after 15 min of insulin stimulation. The green channel represents HA-GLUT-GFP fluorescence that was detected from both intracellular GLUT4 structures and GLUT4 exposed at the cell surface. HA-antibody labeling represents GLUT4 that was exposed at the cell surface and was detected with a secondary antibody conjugated to Alexa-594. Bar, 5 μm.
(B) Insulin stimulation increases HA-antibody labeling of PM and the number of GLUT4 structures labeled by HA-antibody on the surface of adipose cells. The panels show magnified regions from (A). See Video 2 in Supplementary Materials that also shows the clustered appearance of HA-antibody in puncta in live cells under the condition of blocked GLUT4 trafficking (2 mM KCN). Bars, 1 μm.
(C) Quantification of surface-exposed GLUT4 identified by HA-antibody fluorescence. The amounts of clustered and dispersed GLUT4 were obtained from HA-antibody fluorescence images that were processed by spatial filtering into two components that represented relatively uniform background and distinct puncta (clusters) (see Figs. S2B and S2C). The average intensities of the total cell-surface GLUT4-GFP, and the clustered and dispersed signals were measured for 20 basal cells (open bars) and 20 insulin-stimulated cells (filled bars) in four independent experiments. Error bars represent SEM, * Statistically different from corresponding basal values, p<0.05.
To compare the relative amounts of GLUT4 dispersed in PM and GLUT4 localized into the clusters, we analyzed the HA-antibody fluorescence intensity of uniformly-labeled regions of PM and separated it from the signal representing HA-GLUT4 clusters (Fig. 2C). Interestingly, in the basal state, the amounts of clustered and dispersed GLUT4 in PM were equal (130 ± 40 AU vs. 140 ± 50 AU, N=18). With insulin stimulation, the dispersed signal increased almost 4-fold, while the clustered signal was elevated by 2.5-fold (Fig. 2C). Importantly, the overall increase of combined HA signal (3.2-fold) was in good correspondence with the independently measured insulin-stimulated increase of glucose uptake (data not shown). This fact indicates that both GLUT4 exposed in the clusters and GLUT4 dispersed in PM are equally involved in the glucose uptake and thus represent functional glucose transporters.
Insulin stimulation significantly increased the amount of HA-GLUT4-GFP colocalized with HA-antibody, and affected both the density and intensity of individual HA-labeled GLUT4 clusters (Fig. 2B, compare upper and lower panels). On average, the fluorescence intensity of GLUT4-GFP was increased 2-fold, while the HA-antibody signal increased 3-fold (Fig. S2A), indicating that part of the GLUT4 exposed at PM came from GLUT4 structures already present in the TIRF-zone. The density of GLUT4 clusters in PM ranged from 0.21 ± 0.04 puncta/μm2 in the basal state to 0.38 ± 0.06 puncta/μm2 in the insulin-stimulated state (±SEM, N=20) and corresponded to 30-60% of the overall density of stationary GLUT4 structures detected in the TIRF-zone.
To exclude the potential artefacts of fixation and antibody aggregation, we performed HA-antibody labeling in live cells under the condition of blocked GLUT4 trafficking (2 mM KCN), and also using a different combination of primary and secondary antibodies, as well as directly conjugated HA-Alexa-594. All these experiments produced similar images with a punctate appearance of HA-GLUT4 exposed at the cell surface. We also observed individual clusters that exhibited fluorescence intensity fluctuations that could indicate exchange of GLUT4 molecules with the rest of PM (Video 2), as well as a Brownian-like motion of faint HA-labeled spots that was attributable to lateral diffusion of individual HA-GLUT4 molecules in the plane of PM.
Altogether, the data acquired with HA-antibody suggest the existence of GLUT4 in PM in at least two states: as freely-diffusing molecules and as relatively stationary clusters. Moreover, GLUT4 clustering in PM cannot be attributed to an artefact of fixation or antibody labeling, but rather represents a physiologically relevant process regulated by insulin.
GLUT4 is endocytosed from the clusters by recruitment of clathrin
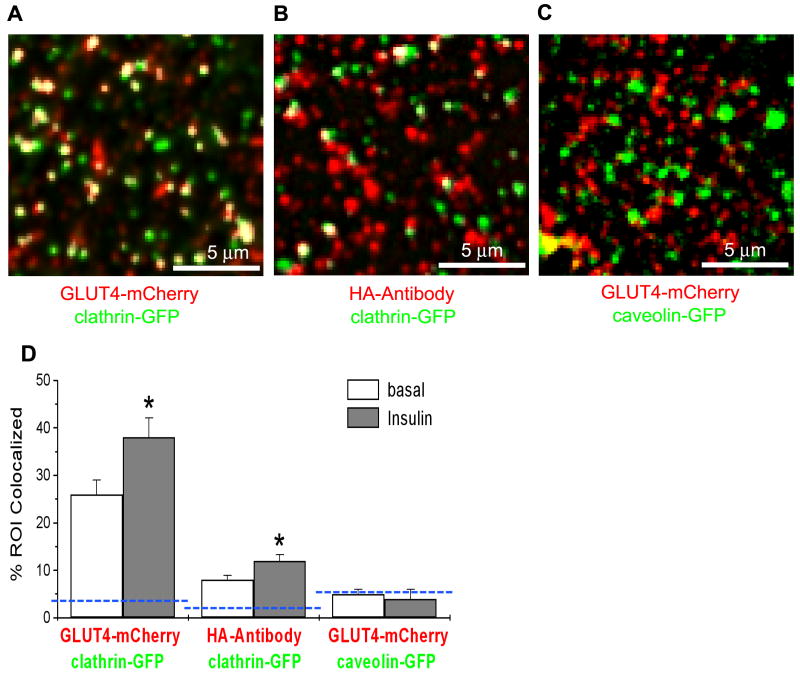
Clustering at PM could be associated with accumulation of GLUT4 in clathrin-coated pits that are implicated in internalisation of GLUT4 from PM. To test this, we transfected adipose cells with clathrin-GFP together with HA-GLUT4-mCherry, and used multi-color TIRF microscopy to resolve their dynamic colocalization and interaction. Cells expressing clathrin-GFP showed both stationary patches of clathrin at PM, and smaller, mobile clathrin-labeled compartments (Fig. 3A and Video 3). On average, we observed that ~25% of all GLUT4-mCherry was colocalized with clathrin-GFP in basal cells; in insulin-stimulated cells, this colocalization was higher and reached 30-40% (Fig. 3D). On the other hand, when we measured colocalization between clathrin-GFP and surface-exposed HA-GLUT4 labeled with HA-antibody (Fig. 3B), we found only 8 ± 2 % of HA-GLUT4 structures to be colocalized with clathrin-GFP in basal cells and 11 ± 3 % in insulin-stimulated cells (Fig. 3D). This difference between colocalization of surface-exposed GLUT4 and total GLUT4 indicates that a majority of GLUT4 structures that colocalized with clathrin is not GLUT4 accumulated in clathrin-coated pits, but rather GLUT4 associated with intracellular compartments (endosomes) localized in the vicinity of PM.
Figure 3. Colocalization of clathrin and caveolin with intracellular GLUT4 structures and PM clusters.
(A) Colocalization of clathrin and GLUT4 in the TIRF-zone. Rat adipose cells were co-transfected with clathrin-GFP (green) and HA-GLUT4-mCherry (red), and imaged using multi-color TIRF microscopy. Some of the structures were mobile and thus represented GSV (see Video 3).
(B) Colocalization of clathrin with GLUT4 clusters at the cell surface. Cells expressing clathrin-GFP (green) and HA-GLUT4 (no fluorescent tag) were fixed and stained with HA-antibody (red) under non-permeabilized conditions.
(C) Colocalization of caveolin and GLUT4 in the TIRF-zone. Rat adipose cells were co-transfected with caveolin-GFP (green) and HA-GLUT4-mCherry (red), and imaged using multi-color TIRF microscopy.
(D) Quantification of the relative number of GLUT4 structures (GLUT4-mCherry) and clusters (HA-antibody), co-localized with clathrin and caveolin (see Fig. S3). The blue dashed lines show the theoretically estimated levels of random colocalization based on the density of the structures. 10 cells were measured in each condition in five independent experiments. Error bars represent SEM, * Statistically different from corresponding basal values, p<0.05.
Since caveolae also have been implicated in the internalization of GLUT4, we co-expressed cells with GLUT4-mCherry and caveolin-GFP, the main structural protein of caveolae (Fig. 3C). The observed colocalization level (~5%) was comparable to the random overlap (4%) estimated for the given density of caveolin-GFP (~0.6 puncta/μm2) and GLUT4-mCherry (0.5-0.7 puncta/μm2). Further, insulin had no effect on the co-localization of GLUT4 and caveolin (Fig. 3D). These data suggest that overall GLUT4 clustering cannot be explained by accumulation in caveolae.
We further analyzed individual dynamic events at PM where clathrin gradually appeared and then disappeared on a time-course of tens of seconds. These events are interpreted as the assembling and disassembling of clathrin (Bellve et al., 2006), and can be attributed to formation of single endocytic vesicles when correlated with the uptake of a particular cargo (Merrifield et al., 2005). To selectively track GLUT4 endocytic events, we used HA-GLUT4 labeled at PM with HA-antibody conjugated to Alexa-594 as cargo.
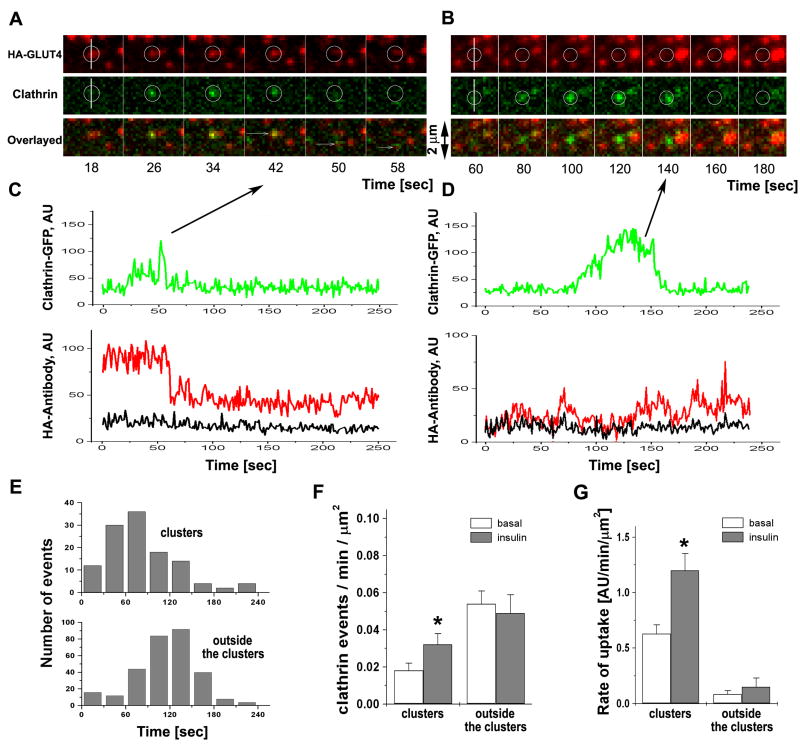
Consistent with previous studies, clathrin was found to form both relatively stationary patches at PM, as well as more dynamic puncta that appeared and disappeared, as clathrin-coated pits were formed and underwent scission from PM. These dynamic events occurred at existing GLUT4 clusters (Fig. 4A), as well in other areas of PM outside the clusters (Fig. 4B). Interestingly, the clathrin events associated with GLUT4 clusters had a shorter time span between assembly and disappearance than those occurring outside the clusters (Fig. 4E). Further, while the majority of the events (0.05 ± 0.01 events/μm2/min, 312 events, N=10 cells) took place outside the clusters, they were not associated with detectable changes of HA-GLUT4. (Figs. 4F and 4D), and their frequency was not affected by insulin (Fig. 4F). On the other hand, most of the clathrin events where HA-GLUT4 was internalized were associated with pre-existing GLUT4 clusters (Fig. 4C) and had a frequency of occurrence of 0.018 ± 0.004 events/μm2/min in the basal state and 0.032 ± 0.006 events/μm2/min (120 events, N=10 cells) in insulin-stimulated conditions (Fig. 4F). To quantify the amount of HA-GLUT4 uptake at, and outside, the clusters ,we integrated all small changes of HA-antibody associated with clathrin events in one minute of recording and normalized it to the unit area. Fig. 4G shows that the majority of HA-GLUT4 uptake was indeed associated with the clusters in both the basal and insulin-stimulated conditions. In all cases, the growth of the clathrin signal from its initial appearance was not associated with an increase of HA-GLUT4 signal, thus indicating that the assembly of clathrin did not trigger the clustering of GLUT4 at the site of pit formation. Taken together, these data suggest that clathrin mediates GLUT4 endocytosis predominantly from pre-existing clusters, but is not responsible for GLUT4 de novo cluster formation.
Figure 4. Clathrin endocytic events at, but not outside of, GLUT4 clusters mediate internalization of GLUT4.
Single endocytic events were imaged in isolated rat adipose cells expressing clathrin-GFP and HA-GLUT4 (no fluorescent tag) using multi-color TIRF microscopy (Video 4).
(A) Selected frames show a characteristic transient appearance of clathrin-GFP (green) at the site of a GLUT4 cluster (red), followed by partial removal of HA-GLUT4 from PM detected by HA-antibody conjugated to Alexa-594. The position of the event is shown by a circle in both channels. Individual frames were selected from the stack four sec apart to depict the entire time course of the event.
(B) A representative clathrin event that takes place outside of a GLUT4 cluster identified by HA-antibody does not result in a change in the HA-antibody signal. Individual frames were selected from the stack 10 sec apart to depict the entire time course of the event.
(C) Graphs represent the fluorescent intensity time-courses of HA-antibody (red) and clathrin-GFP (green) for the event shown in (A). The black trace represents the average HA-antibody background intensity outside of clusters. Note that the residual fluorescence signal of HA-antibody after the appearance and disappearance of clathrin-GFP is significantly higher than the background..
(D) Graphs represent the fluorescent intensity time-courses of HA-antibody (red) and clathrin-GFP (green) for the event shown in (B). The black trace represents the average HA-antibody background intensity outside of clusters. Note that the fluorescence signal of HA-antibody is not affected by the assembly of clathrin-GFP and shows no drop at the time of clathrin-GFP removal.
(E) Histogram of the time of the onset of clathrin assembly and disappearance of clathrin-coated pits. The upper panel represents clathrin assembly events at GLUT4 clusters identified by HA-antibody and the lower panel represents clathrin assembly outside the clusters.
(F) The frequency of clathrin events at, and outside of, GLUT4 clusters in the basal and insulin-stimulated steady states. The number of events detected per unit area per min of recording was averaged for 10 cells each in the basal and insulin-stimulated states. Error bars represent SEM, * statistically different from corresponding basal values, p<0.05.
(G) Rate of HA-antibody uptake per unit area of the cell surface. The average rate was calculated from the slope of cumulative uptake measured in 12 cells in three independent experiments. Cumulative uptakes were constructed using all detected decrements of HA-antibody fluorescence corresponding to clathrin events associated with the clusters, as well as all the other clathrin assembly events outside the clusters. The decrements were calculated as the difference of the mean intensity of HA-antibody averaged for 10 frames before and after clathrin disappearance (see Fig. S4). Note that while the numbers of clathrin events at the clusters account for a fraction of the total (see Fig. 4F), they are responsible for the major part of HA-antibody internalization from the cell surface. Error bars represent SEM, * statistically different from corresponding basal values, p<0.05.
Two modes of GLUT4 exocytosis: release of GLUT4 and formation of the clusters
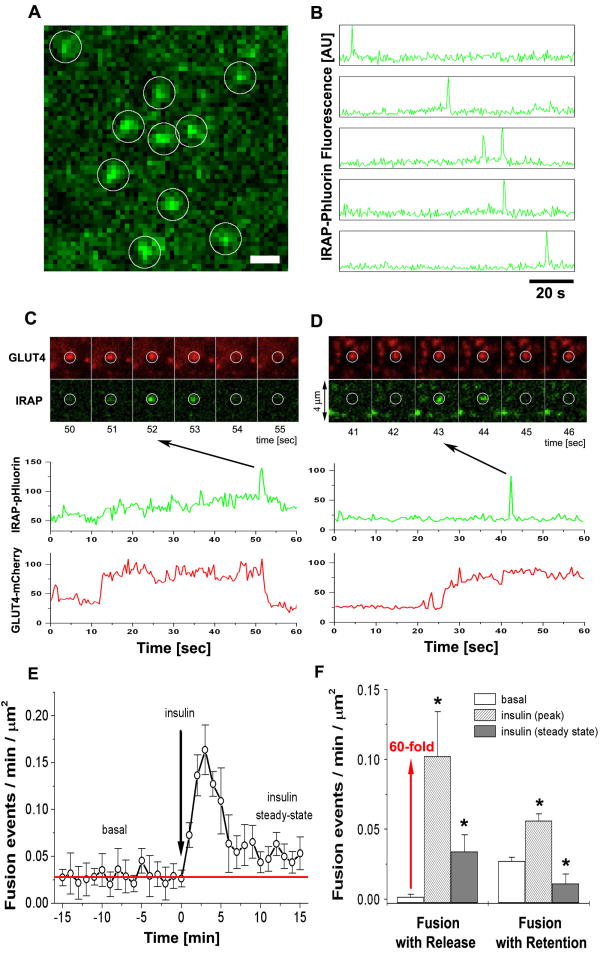
Next, in order to investigate the relationship between GLUT4 exocytosis and the GLUT4 PM clusters, we monitored the fusion of single GSV double–labeled with HA-GLUT4-mCherry and pH-sensitive IRAP-pHluorin. Fusion events were detected as characteristic flashes of IRAP-pHluorin fluorescence triggered by exposure of pHluorin to extracellular pH (Video 5). A rapid increase of the intensity of pHluorin was followed by a gradual decay associated with dispersal of IRAP molecules into PM. This was confirmed in individual cases by the analysis of intensity profile widening that corresponds to lateral diffusion (Fig. S5B).
To quantify the basal rate of GSV exocytosis, we first analyzed all fusion events marked by a characteristic flash of IRAP-pHluorin (Figs. 5A and 5B). Further, we verified that the detected events were associated with the presence of GLUT4-mCherry signal and analyzed how GLUT4 were released into PM upon fusion. Interestingly, we distinguished two different types of fusion based on the analysis of GLUT4-mCherry fluorescence at the fusion sites. After fusion, GLUT4 were either dispersed into PM, called fusion-with-release (Fig. 5C), or retained at the site of fusion, called fusion-with-retention (Fig. 5D).
Figure 5. Two modes of exocytosis, fusion-with-retention and fusion-with-release, mediate GLUT4 delivery and spatial distribution in PM.
(A) Isolated rat adipose cells were transfected with the pH-sensitive probe IRAP-pHluorin (green) and HA-GLUT4-mCherry (red), and imaged using multi-color TIRF microscopy. Single exocytosis events were detected as spikes of IRAP-pHluorin fluorescence that reflected opening of the fusion pore and equilibration of the pH inside the lumen of the fusing vesicle. The image represents a maximum projection of a time-lapse stack of 240 frames, i.e. each pixel contains the maximum value over all frames in the stack at the particular pixel location. IRAP-pHluorin events are marked with white circles. See also Fig. S5 and Video 5. Bar, 1 μm.
(B) Time-course of IRAP-pHluorin fluorescence measured in five of the circular regions shown in (A). Note characteristic transient spikes of IRAP-pHluorin marking the fusion pore openings and consequent lateral diffusion of IRAP-pHluorin into PM. At all times the detected IRAP-pHluorin events were associated with the presence of GLUT4-mCherry signal at the same site.
(C) Series of time-frames showing a characteristic fusion event with release of HA-GLUT4-mCherry (red) into PM and corresponding transient spike of IRAP-pHluorin (green) marking the moment of fusion pore opening. The graphs below represent the time-courses of HA-GLUT4-mCherry and IRAP-pHluorin fluorescence measured at the sites of fusion. Note the simultaneous decreases of the GLUT4-mCherry and IRAP-pHluorin signals.
(D) Series of time-frames showing another type of fusion event that retains HA-GLUT4-mCherry (red) at the site of fusion and leads to formation of a GLUT4 cluster. The transient spike of IRAP-pHluorin (green) marks the moment of fusion pore opening, but the HA-GLUT4-mCherry does not disperse upon fusion and remains clustered. The graphs below present the quantification of HA-GLUT4-mCherry and IRAP-pHluorin fluorescence, and show that the GLUT4-mCherry signal remains relatively unchanged after fusion.
(E) Frequency of fusion events detected with IRAP-pHluorin plotted as a function of time before and after insulin stimulation. The number of fusion events detected per unit area during one min of recording was calculated for individual cells and averaged for at least five cells for each time point. Insulin resulted in a significant increase of the fusion frequency, which typically peaked at 3 min after stimulation and then declined towards a level similar to the frequency of fusion events under the non-stimulated condition (red line). Error bars represent SEM.
(F) The frequency of fusion events with retention and release of GLUT4, measured in the basal and insulin-stimulated steady states. The number of fusion events detected per unit area per min of recording was averaged for 20 cells each in the basal and insulin-stimulated states. In insulin-treated cells, the fusion frequency was measured at 3-5 min (peak) and at 13-15 min (steady state) after insulin stimulation. 269 events were analyzed in 16 cells. Error bars represent SEM, * statistically different from corresponding basal values, p<0.05, assessed by one-way ANOVA.
In non-stimulated cells, the average fusion frequency was 0.028 ± 0.006 events/μm2/min (269 events, N=16 cells) and 95 % of the detected fusion events were not associated with GLUT4 dispersion into PM. Following insulin stimulation the overall fusion frequency increased to a maximum of 0.15 ± 0.03 events/μm2/min already after two-three minutes (Fig. 5E). This increase in the number of fusion events was due to differential stimulation of both modes of exocytosis. While insulin had only a moderate effect on the fusion-with-retention (~2-fold increase), fusion-with-release of GLUT4 was dramatically increased more than 60-fold (Fig. 5F). Of further interest, already after 4-5 min in the presence of insulin, the overall fusion frequency started to decline and stabilized at a level of 0.05 ± 0.01 events/μm2/min at around 10-15 min (Figs. 5E and 5F). This decrease in the number of fusion events after 10-15 min was not due to photodamage or other factors associated with a prolonged experiment, as the same fusion frequency was measured at a particular time point after insulin, independent of the length of the prior recording. Moreover, the transient increase in fusion events was sufficient to achieve and maintain the insulin steady-state, as judged by the elevated PM GLUT4 signal.
These data suggest that fusion-with-release represents an acute response to insulin where a significant amount of GLUT4 is delivered to, and dispersed into, PM. On the other hand, fusion-with-retention seems to mainly provide PM with GLUT4 to sustain glucose uptake in non-stimulated conditions and is also responsible for creation of GLUT4 clusters at PM.
Kinetic model of GLUT4 recycling through the clusters
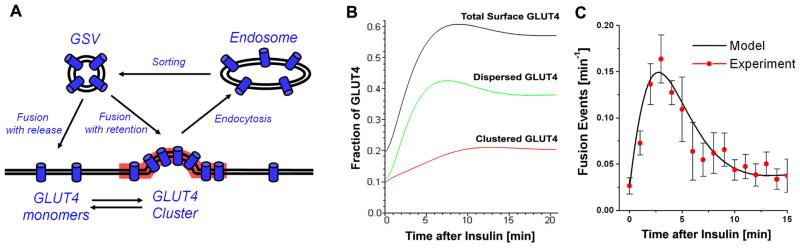
Now that the organization of GLUT4 into clusters, and the effects of insulin on the relationship between GLUT4 exocytosis and clusters have been determined, it is no longer possible to use kinetic models that assume homogeneous distributions of GLUT4 in PM or an independence of the mode of GLUT4 exocytosis from insulin. To take into account these novel details of GLUT4 recycling, we have built a new kinetic model (see a detailed description in the Supplementary Materials) that includes the existence of GLUT4 clusters, and can quantitatively explain the transition to, and maintenance of, the insulin-steady state, together with the observed transient increase of fusion events. In brief, we add to the recycling of GLUT4 among GSV, PM, and endosomes the fact that PM GLUT4 exist in two states: monomers and clusters. Thus, we added clusters as a new quasi-compartment to extend previously introduced 3-compartment models (Holman et al., 1994) GLUT4 in GSV; GLUT4 present as monomers in PM; GLUT4 in clusters; and GLUT4 in endosomes (Fig. 6A). For simplicity, we have excluded the minor amounts of GLUT4 present in other compartments and the peri-nuclear area (less than 4 %), and assume that the total amount of GLUT4 in these four compartments is constant. Based on our data and published data, we estimated the fraction of GLUT4 in GSV, monomers, endosomes, and clusters in the basal and insulin-stimulated steady-states. The other unknown parameters were found from a set of algebraic equations determining the equilibrium at the corresponding steady-states (Table S1, Supplemental Materials.).
Figure 6. Kinetic model of insulin-regulated recycling and spatial distribution of GLUT4 on the cell surface through post-fusion dispersal.
(A) Kinetic model of insulin-regulated GLUT4 recycling among intracellular GSV, endosomes, and GLUT4 dispersed and clustered in PM. GLUT4 are assumed to be cycling among four GLUT4 quasi-compartments: GLUT4-storage vesicles (GSV), monomeric GLUT4 in PM (dispersed GLUT4), clustered GLUT4, and GLUT4 in endosomes. GLUT4 endocytosis is restricted to the clusters. See also Fig. S6.
(B) Modeling of GLUT4 redistribution to PM in response to insulin. Calculated time-course of GLUT4 redistribution in response to insulin: total fraction of GLUT4 present on the cell surface (black); GLUT4 dispersed in PM as monomers (green); and GLUT4 clustered in PM domains (red).
(C) Transient increase in the amount of GLUT4 fusion events in response to insulin (black – model; red circles – experimental data; see also Fig. 5E). The frequency of the fusion events was calculated as a sum of fusion-with-release and fusion-with-retention. Error bars represent SEM, N=5.
Insulin stimulation was modeled by increases of the two rates of GLUT4 exocytosis: a) fusion-with-release and b) fusion-with-retention. We assumed that all other parameters remained relatively unchanged in response to insulin and verified if the model can explain the experimentally obtained GLUT4 distributions in the insulin steady-state, as well as the kinetics of the transient increases of fusion events. Figure 6B shows that the increase of the rate of fusion-with-release (~60-fold) and fusion-with-retention (~2-fold) can account for the experimentally observed increases of a) total surface-exposed GLUT4 (Fig. 6B, black; compare with Fig. 2C), b) dispersed GLUT4 (Fig. 6B, green), and c) clustered GLUT4 (Fig. 6B, red). Moreover, our model shows a similar kinetics of GLUT4 redistribution to that measured by independent methods (Holman et al., 1994; Lizunov et al., 2005) with cells reaching steady-state by 10-15min. Further, we calculated the frequencies of the fusion events with release/and retention as functions of time, and compared them with the experimental data (Fig. 5E). As shown in Fig. 6C, the model reproduces the transient increase of the frequency of the fusion events delivering GLUT4 to PM in response to insulin.
Discussion
The relationship between the spatial and temporal organization of plasma membrane (PM) glucose transporters is key to the regulation of cell metabolism. In addition to the complex recycling of GLUT4 among endosomal compartments, GLUT4-storage vesicles (GSV), and PM, we now know that their spatial organization in PM, where they facilitate glucose transport, depends upon insulin in a time-dependent fashion. The predominance in the basal state of PM GLUT4 cluster formation upon exocytosis of GSV gives way with insulin stimulation to a shift in GSV fusion to GLUT4 dispersal in PM. Further, GLUT4 clusters are found to nucleate clathrin assembly where most of the GLUT4 internalization from the cell surface then takes place. Thus, GLUT4 clusters represent a molecular organization mediating the transition between GLUT4 delivery and withdrawal from PM, depending upon insulin.
GLUT4 molecules in PM are thought to function as monomeric transporters, and unlike GLUT1 that forms oligomers (Zottola 1995), GLUT4 do not appear to oligomerize. However, GLUT4 at PM have repeatedly been observed to be distributed inhomogenously (Gustavsson et al., 1996; Parton et al., 2002), i.e. concentrated, clustered at certain regions of PM. Previous microscopic studies of GLUT4 distribution in muscle (Lauritzen et al., 2008), isolated adipose cells (Malide et al., 1997; Lizunov et al., 2005), and 3T3 L1 adipocytes (Li et al., 2004; Blot and McGraw, 2008) report a punctate staining of GLUT4 at PM. In this study, we further verify the presence of both GLUT4 monomers and GLUT4 clusters in PM; quantify the relative amount of GLUT4 sequestered in each state; and report differential effects of insulin on GLUT4 spatial distribution.
While no molecular mechanism for GLUT4 clustering has been established, it was often attributed to accumulation of GLUT4 either in clathrin-coated pits or caveolae. Caveolar structures have been proposed to play some intermediate role in GLUT4 internalization (Ros-Baro et al., 2001) and GLUT4 exocytosis (Gustavsson et al., 1996; Saltiel and Pessin, 2003). In our experiments we did not detect any significant co-localization of GLUT4 and caveolin in either basal or insulin-stimulated cells, which is consistent with another recent study arguing against a direct role for caveolae in GLUT4 recycling (Antonescu et al., 2008).
While the role of clathrin in the recycling of GLUT4 is well supported, no evidence exists suggesting direct involvement of clathrin in GLUT4 clustering. Huang et al 2007 observed an overall increase of GLUT4 co-localization with clathrin in response to insulin. Yet, this study did not distinguish whether this co-localization was due to specific accumulation of GLUT4 in the clathrin-coated pits, co-localization in endosomes, or a non-specific increase in the background of PM GLUT4. In our study, we were able to separately measure the overall co-localization of GLUT4 and clathrin, as well as co-localization of surface-exposed GLUT4 with clathrin. Surprisingly, we found that surface-exposed GLUT4 have much less co-localization with clathrin than total GLUT4. Thus, a specific accumulation of GLUT4 in clathrin-coated pits is unlikely; it is more likely that these two molecules co-localize in intracellular compartments. Together with the fact that the majority of GLUT4 clusters exists at PM without showing any co-localization with clathrin, our data indicate that clathrin itself cannot account for the existence and formation of GLUT4 clusters at PM.
The delivery of GLUT4 to PM through insulin-regulated exocytosis has been well documented by both biochemistry and live-cell imaging. Using TIRF microscopy, a number of groups detected single GSV fusion events (Burchfield et al., 2010; Bai et al., 2007; Huang et al 2007) using as a criterion for fusion the post-fusion dispersal of fluorescently labeled GLUT4-GFP. However, the number of fusion events measured microscopically was fewer than the number expected from biochemical and physiological approaches. Recently, the use of IRAP-pHluorin targeted into GSV has allowed more robust fusion detection, significantly increasing the number of fusion events detected (Jiang et al., 2008). However, due to spectral overlap with IRAP-pHluorin, GLUT4-GFP could not be used in the same experiment and thus the post fusion-dispersal of GLUT4 could not be monitored simultaneously. In our study, by combining the IRAP-pHluorin detection system with red fluorescent GLUT4-mCherry, we were able to overcome these limitations. Surprisingly, we found that many fusion events were not associated with dispersal of GLUT4 from the site of fusion, explaining why the number of fusion events detected only by GLUT4 dispersal was underestimated. Fusion-with-retention was predominant in the basal state where we observed that almost all fusion events detected by IRAP-pHluorin flash were not accompanied by dispersal of GLUT4 into PM. The result of this fusion-with-retention is the creation of de novo GLUT4 clusters at PM.
IRAP-pHluorin fluorescence decay after flash was attributed in most cases to IRAP dispersal via lateral diffusion, which was confirmed in control experiments where widening of the intensity profile was measured. However, a transient pH change or rapid reacidification of the lumen of partially opened fusing vesicles could also result in fluorescence decay. This sequence of events could represent a “frustrated fusion”, where a vesicle rapidly opens and closes. But if a vesicle closes shortly after a fusion attempt, it would not expose GLUT4 long enough to be detected efficiently by HA-antibody binding. In the case of fusion-with-retention, the average time between the increase and decrease of pHluorin fluorescence is about two-three seconds (~0.05 of min) and the fusion rate is ~ 0.03 events/min/μm2. Therefore, in the extreme case, if all fusion-with-retention events represented frustrated fusion, the estimated density of opened vesicles at any moment would be given by the product of 0.03 events/min/μm2 * 0.05 min = 0.0015 per μm2. This estimate is at least two orders of magnitude less than the experimentally detected density of GLUT4 clusters (~0.2 per μm2). Therefore, frustrated fusion could account for only a minor fraction of fusion events detected by IRAP-pHluorin.
Consistent with previous reports (Jiang et al., 2008), insulin increased the overall number of fusion events. Interestingly, insulin not only affected the number of fusion events, but also dramatically shifted the mode of fusion towards fusion with dispersal of GLUT4 into PM. This finding is also consistent with the pronounced increase of diffuse HA-antibody staining of PM and corresponding shift in the relative amount of GLUT4 from clusters to monomers. This observation further implies that while clustered and monomeric GLUT4 co-exist in a steady state, the relative amounts of GLUT4 in these pools can be differentially regulated by insulin.
Interestingly, the insulin-stimulated increase of GLUT4 fusion was transient, and after a pronounced peak at 2-3 min, the fusion frequency declined to a level only slightly above the basal. While old models of GLUT4 recycling predict an over-all increase of GLUT4 recycling in response to insulin, our results fit the prediction of a quantum release model (Coster et al., 2004). However, while the Coster et al. (2004) model considers insulin to regulate the size of the active pool available for GLUT4 recycling, our model includes all GLUT4 in the recycling process and addresses the existence of GLUT4 clusters as a distinct pool. One important feature of our model, based on the experimental observations, is the restriction of GLUT4 internalization to the clusters. Our data suggest that the major part of GLUT4 internalization occurs from clusters via recruitment of clathrin, while other clathrin-coated pits outside the clusters do not efficiently endocytose GLUT4. This organization of GLUT4 recycling provides flexibility to upregulate PM GLUT4 (in monomeric states) separately from GLUT4 available for endocytosis (clustered).
Taken together, the data presented in this study suggest that GLUT4 clusters may function as intermediate hubs from the time of GLUT4 exocytosis until their internalization. In the basal state, these domains appear to play the major role in regulating the recycling of GLUT4 between PM and the intracellular pool of GSV. In the insulin-stimulated state, a rapid increase of PM GLUT4 is achieved by an increase in GSV fusion, particularly events with full and immediate release of GLUT4 molecules diffusely into PM. However, the rate of internalization and recycling of GLUT4 into GSV must now include a new parameter, the time it takes for GLUT4 to reach an uptake site; the rate-limiting step in this trafficking process remains to be determined. This kinetics, through GLUT4 hubs, must ultimately determine the new equilibrium GLUT4 activity level set by insulin stimulation. Thus, they are of particular interest in pathological states in which the relationship between insulin blood levels and GLUT4 activity is disrupted.
Experimental Procedures
Reagents
Mouse anti-HA antibody (HA.11) was from Berkeley Antibody Co. (Richmond, CA). Clathrin-GFP, Caveolin-GFP and Tubulin-GFP were kindly provided by Dr. J. Lippincott-Schwartz. Construction of the HA-GLUT4-mCherry has been described previously (Lizunov et al., 2009). Bovine serum albumin was from Intergen (fraction V). DMEM, Insulin, Alexa-conjugated antibodies were all from Invitrogen.
Cell Culture and Transfection
Preparation of isolated rat adipose cells from male rats (CD strain, Charles River Laboratories, MD), electroporation of rat adipose cells, and the cell-surface antibody-binding assay were performed as described previously (Al-Hasani et al., 1998; Lizunov et al., 2005). All plasmids were used at a final concentration of 4 μg/ml. Transfected cells were kept in culture overnight and optimal protein expression level was achieved at 20-24 h after electroporation. Insulin stimulation was performed by addition of 70 nM insulin for 30 min at 37 °C.
Live Cell Imaging and Immunoflourescence Microscopy
For live cell imaging, isolated adipose cells were kept in KRBH buffer with 1% BSA, pH 7.4, maintained at 37°C using a temperature-controlled stage and Delta-T environmental chamber (Bioptechs). For immunoflourescence microscopy, the isolated cells in suspension were either fixed with 4% formaldehyde in phosphate-buffered saline (PBS) for 10 min, or incubated with 2 mM KCN to deplete ATP and inhibit GLUT4 recycling (Satoh et al., 1993). The cells were then washed with PBS and transferred to KRBH with 1% BSA for incubation with antibodies in the presence of KCN. Cells were imaged using the TIRFM setup built around an Axiovert 200 microscope (Zeiss) equipped with a 100×1.45 NA objective. A TIRF slider (Till Photonics) was used to pass laser beams from an AOTF-controlled combiner system (LSM Technologies) equipped with 405/488/561 nm lasers (Coherent). Penetration depth of the evanescent field was measured to be 110 ± 20 nm by a calibration procedure with 40-nm fluorescent beads attached to the piezo-driven micropipette. Fluorescence was separated from the excitation light using a multi-band dichroic and emission filter set (GFP/DsRed-2X-A, Semrock), and passed to an electron-multiplying CCD camera (Ixon, Andor). For WF microscopy, a fiber-coupled light source (X-cite 120, EXFO) was used together with a filter-wheel (Lambda 10-B, Sutter) to sequentially excite GFP/pHluorin and mCherry/Alexa-594 using appropriate filters. Laser-switching AOTF, shutters, filter-wheels, microscope, and EMCCD camera were synchronized and controlled using μManager v1.2.38 (http://www.micro-manager.org).
Image analysis
A set of automated image processing macro/subroutines was developed based on existing algorithms of ImageJ (Rolling-Ball Background Subtraction, Gaussian and Granulometric filters, Z-project, Image5D, Find Maxima, and Particle Tracker). Individual diffraction-limited fluorescent structures were segmented within representative regions of interest (ROI). The following criteria were used to detect individual structures: i) fluorescence had a local maximum; ii) integrated pixel intensity was at least 2-fold above the standard deviation of pixel intensity; and iii) 70% of the peak intensity was contained within a circular ROI of five pixels. Density was measured as the number of structures per square micron; frequency of dynamic events of fission and fusion was calculated as the number of events per square micron per minute.
Particle tracking was carried out using a custom-modified Particle Tracker algorithm (Sbalzarini and Koumoutsakos, 2005) that utilized spatial moment analysis to detect redistribution of GLUT4 molecules during fusion. To quantify populations of stationary and mobile structures we utilized a time-projection method (Lizunov et al., 2009). Data acquired using multi-color TIRF were processed to measure colocalization of individual structures. Simulated data were used to estimate a percent of random overlap for given densities of structures in each channel. All data are represented as means ± SEM. Statistical significance was analyzed using Student’s t-test or ANOVA. Further details of the data analysis and modeling are described in Supplemental Materials.
Supplementary Material
Acknowledgments
The authors thank Dr. Kamran Melikov for useful discussions and Dena R. Yver for expert technical assistance. This work was supported in part by a Postdoctoral Fellowship to KGS from the Swedish Research Council, and by the intramural research programs of NIDDK and NICHD, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al-Hasani H, Hinck CS, Cushman SW. Endocytosis of the glucose transporter GLUT4 is mediated by the GTPase dynamin. J Biol Chem. 1998;273:17504–10. doi: 10.1074/jbc.273.28.17504. [DOI] [PubMed] [Google Scholar]
- Antonescu CN, Diaz M, Femia G, Planas JV, Klip A. Clathrin-dependent and independent endocytosis of glucose transporter 4 (GLUT4) in myoblasts: regulation by mitochondrial uncoupling. Traffic. 2008;9:1173–90. doi: 10.1111/j.1600-0854.2008.00755.x. [DOI] [PubMed] [Google Scholar]
- Bai L, Wang Y, Fan J, Chen Y, Ji W, Qu A, Xu P, James DE, Xu T. Dissecting multiple steps of GLUT4 trafficking and identifying the sites of insulin action. Cell Metab. 2007;5:47–57. doi: 10.1016/j.cmet.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Bellve KD, Leonard D, Standley C, Lifshitz LM, Tuft RA, Hayakawa A, Corvera S, Fogarty KE. Plasma membrane domains specialized for clathrin-mediated endocytosis in primary cells. J Biol Chem. 2006;281:16139–46. doi: 10.1074/jbc.M511370200. [DOI] [PubMed] [Google Scholar]
- Blot V, McGraw TE. Molecular mechanisms controlling GLUT4 intracellular retention. Mol Biol Cell. 2008;19:3477–87. doi: 10.1091/mbc.E08-03-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogan JS, Hendon N, McKee AE, Tsao TS, Lodish HF. Functional cloning of TUG as a regulator of GLUT4 glucose transporter trafficking. Nature. 2003;425:727–33. doi: 10.1038/nature01989. [DOI] [PubMed] [Google Scholar]
- Burchfield JG, Lopez JA, Mele K, Vallotton P, Hughes WE. Exocytotic Vesicle Behaviour Assessed by Total Internal Reflection Fluorescence Microscopy. Traffic. 2010;11(4):429–439. doi: 10.1111/j.1600-0854.2010.01039.x. [DOI] [PubMed] [Google Scholar]
- Coster AC, Govers R, James DE. Insulin stimulates the entry of GLUT4 into the endosomal recycling pathway by a quantal mechanism. Traffic. 2004;5(10):763–71. doi: 10.1111/j.1600-0854.2004.00218.x. [DOI] [PubMed] [Google Scholar]
- Fazakerley DJ, Lawrence SP, Lizunov VA, Cushman SW, Holman GD. A common trafficking route for GLUT4 in cardiomyocytes in response to insulin, contraction and energy-status signalling. J Cell Sci. 2009;122:727–34. doi: 10.1242/jcs.041178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez E, McGraw TE. Insulin signaling diverges into Akt-dependent and -independent signals to regulate the recruitment/docking and the fusion of GLUT4 vesicles to the plasma membrane. Mol Biol Cell. 2006;17:4484–93. doi: 10.1091/mbc.E06-07-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustavsson J, Parpal S, Stralfors P. Insulin-stimulated glucose uptake involves the transition of glucose transporters to a caveolae-rich fraction within the plasma membrane: implications for type II diabetes. Mol Med. 1996;2:367–72. [PMC free article] [PubMed] [Google Scholar]
- Holman GD, Lo Leggio L, Cushman SW. Insulin-stimulated GLUT4 glucose transporter recycling. J Biol Chem. 1994;269(26):17516–24. [PubMed] [Google Scholar]
- Huang S, Lifshitz LM, Jones C, Bellve KD, Standley C, Fonseca S, Corvera S, Fogarty KE, Czech MP. Insulin stimulates membrane fusion and GLUT4 accumulation in clathrin coats on adipocyte plasma membranes. Mol Cell Biol. 2007;27:3456–69. doi: 10.1128/MCB.01719-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Fan J, Bai L, Wang Y, Chen Y, Yang L, Chen L, Xu T. Direct quantification of fusion rate reveals a distal role for AS160 in insulin-stimulated fusion of GLUT4 storage vesicles. J Biol Chem. 2008;283:8508–16. doi: 10.1074/jbc.M708688200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandror K, Pilch PF. Identification and isolation of glycoproteins that translocate to the cell surface from GLUT4-enriched vesicles in an insulin-dependent fashion. J Biol Chem. 1994;269:138–42. [PubMed] [Google Scholar]
- Kanzaki M, Pessin JE. Insulin-stimulated GLUT4 translocation in adipocytes is dependent upon cortical actin remodeling. J Biol Chem. 2001;276:42436–44. doi: 10.1074/jbc.M108297200. [DOI] [PubMed] [Google Scholar]
- Koumanov F, Jin B, Yang J, Holman GD. Insulin signaling meets vesicle traffic of GLUT4 at a plasma-membrane-activated fusion step. Cell Metab. 2005;2:179–89. doi: 10.1016/j.cmet.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Lauritzen HP, Galbo H, Brandauer J, Goodyear LJ, Ploug T. Large GLUT4 vesicles are stationary while locally and reversibly depleted during transient insulin stimulation of skeletal muscle of living mice: imaging analysis of GLUT4-enhanced green fluorescent protein vesicle dynamics. Diabetes. 2008;57:315–24. doi: 10.2337/db06-1578. [DOI] [PubMed] [Google Scholar]
- Li CH, Bai L, Li DD, Xia S, Xu T. Dynamic tracking and mobility analysis of single GLUT4 storage vesicle in live 3T3-L1 cells. Cell Res. 2004;14:480–6. doi: 10.1038/sj.cr.7290251. [DOI] [PubMed] [Google Scholar]
- Lizunov VA, Lisinski I, Stenkula K, Zimmerberg J, Cushman SW. Insulin regulates fusion of GLUT4 vesicles independent of Exo70-mediated tethering. J Biol Chem. 2009;284:7914–9. doi: 10.1074/jbc.M806460200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizunov VA, Matsumoto H, Zimmerberg J, Cushman SW, Frolov VA. Insulin stimulates the halting, tethering, and fusion of mobile GLUT4 vesicles in rat adipose cells. J Cell Biol. 2005;169:481–9. doi: 10.1083/jcb.200412069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malide D, Dwyer NK, Blanchette-Mackie EJ, Cushman SW. Immunocytochemical evidence that GLUT4 resides in a specialized translocation post-endosomal VAMP2-positive compartment in rat adipose cells in the absence of insulin. J Histochem Cytochem. 1997;45:1083–96. doi: 10.1177/002215549704500806. [DOI] [PubMed] [Google Scholar]
- Martin OJ, Lee A, McGraw TE. GLUT4 distribution between the plasma membrane and the intracellular compartments is maintained by an insulin-modulated bipartite dynamic mechanism. J Biol Chem. 2006;281:484–90. doi: 10.1074/jbc.M505944200. [DOI] [PubMed] [Google Scholar]
- Merrifield CJ, Perrais D, Zenisek D. Coupling between clathrin-coated-pit invagination, cortactin recruitment, and membrane scission observed in live cells. Cell. 2005;121:593–606. doi: 10.1016/j.cell.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Parton RG, Molero JC, Floetenmeyer M, Green KM, James DE. Characterization of a distinct plasma membrane macrodomain in differentiated adipocytes. J Biol Chem. 2002;277:46769–78. doi: 10.1074/jbc.M205683200. [DOI] [PubMed] [Google Scholar]
- Rea S, James DE. Moving GLUT4: the biogenesis and trafficking of GLUT4 storage vesicles. Diabetes. 1997;46:1667–77. doi: 10.2337/diab.46.11.1667. [DOI] [PubMed] [Google Scholar]
- Ros-Baro A, Lopez-Iglesias C, Peiro S, Bellido D, Palacin M, Zorzano A, Camps M. Lipid rafts are required for GLUT4 internalization in adipose cells. Proc Natl Acad Sci U S A. 2001;98:12050–5. doi: 10.1073/pnas.211341698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltiel AR, Pessin JE. Insulin signaling in microdomains of the plasma membrane. Traffic. 2003;4:711–6. doi: 10.1034/j.1600-0854.2003.00119.x. [DOI] [PubMed] [Google Scholar]
- Satoh S, Nishimura H, Clark AE, Kozka IJ, Vannucci SJ, Simpson IA, Quon MJ, Cushman SW, Holman GD. Use of bismannose photolabel to elucidate insulin-regulated GLUT4 subcellular trafficking kinetics in rat adipose cells. J Biol Chem. 1993;268:17820–9. [PubMed] [Google Scholar]
- Sbalzarini IF, Koumoutsakos P. Feature point tracking and trajectory analysis for video imaging in cell biology. J Struct Biol. 2005;151:182–95. doi: 10.1016/j.jsb.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Shi J, Kandror KV. Sortilin is essential and sufficient for the formation of Glut4 storage vesicles in 3T3-L1 adipocytes. Dev Cell. 2005;9:99–108. doi: 10.1016/j.devcel.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Torok D, Patel N, Jebailey L, Thong FS, Randhawa VK, Klip A, Rudich A. Insulin but not PDGF relies on actin remodeling and on VAMP2 for GLUT4 translocation in myoblasts. J Cell Sci. 2004;117:5447–55. doi: 10.1242/jcs.01421. [DOI] [PubMed] [Google Scholar]
- Xu Z, Kandror KV. Translocation of small preformed vesicles is responsible for the insulin activation of glucose transport in adipose cells. J Biol Chem. 2002;277:47972–5. doi: 10.1074/jbc.C200486200. [DOI] [PubMed] [Google Scholar]
- Zottola RJ, Cloherty EK, Coderre PE, Hansen A, Hebert DN, Carruthers A. Glucose transporter function is controlled by transporter oligomeric structure. Biochemistry. 1995;34(30):9734–47. doi: 10.1021/bi00030a011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.