Abstract
In plant-pathogen interactions, the host defends against the invading pathogen and the pathogen aims to suppress or subvert this defense. Whereas the defense suppression strategy is relatively well understood for many pathogens, the mechanisms by which pathogens can actively utilize the defense machinery of the host remain obscure. We report that Agrobacterium, a microorganism that elicits neoplastic growths on many plant species, induces expression of a plant defense-related F-box protein, VBF, which it incorporates into its own pathway for genetic transformation. Our data suggest that VBF may function to uncoat the bacterial transferred DNA from its associated virulence VirE2 and host VIP1 proteins via the SCFVBF pathway. Suppression of VBF elevates the intracellular content of VIP1, but renders the plant largely resistant to Agrobacterium, indicating that, in the infection pathway, VBF is functionally epistatic to VIP1. When expressed in Agrobacterium and exported into the plant cell, VBF facilitates tumor formation.
Host-pathogen interactions are a constant interplay between the host devising mechanisms to eliminate the invading pathogen and the pathogen evolving strategies to counteract this defense. One of the best-studied strategies employed by diverse plant pathogens is defense suppression. For example, Pseudomonas syringae has evolved avirulence proteins which suppress the plant basal defense mediated by the RIN4 protein (Ellis and Dodds, 2003), but which are counteracted by the plant R-genes (Axtell and Staskawicz, 2003; Mackey et al., 2003), forcing the pathogen to adopt alternative mechanisms to target RIN4 (Ellis and Dodds, 2003). Other pathogens, mainly plant viruses, target the plant innate immunity based on RNA silencing. Plant hosts use this response to silence the pathogen’s gene expression and destroy its genetic material whereas the pathogen encodes proteins, such as P19 of tombusviruses or HC-Pro of potyviruses, that suppress this pathogen-induced RNA silencing (Díaz-Pendón and Ding, 2008; Levy et al., 2008). A completely different and still poorly understood strategy to counteract the host defense is for the pathogen to actively subvert it for its own needs. Here, we examined such strategy using genetic transformation of plants by Agrobacterium as a model system.
Agrobacterium elicits neoplastic growths on many plant species. Moreover, although plants are the natural hosts for Agrobacterium, this microorganism can also transform a wide range of other eukaryotic species, from fungi to human cells (Lacroix et al., 2006). This genetic transformation is achieved by transporting a single-stranded copy of the bacterial transferred DNA (T-DNA) from the tumor-inducing (Ti) plasmid into the plant cell nucleus followed by integration into the host genome (Gelvin, 2000). Within the host cell, T-DNA is coated by the bacterial virulence (Vir) protein VirE2, which packages it into a nucleoprotein complex (T-complex) (Citovsky et al., 2007). In the T-complex, VirE2 associates with the host VIP1 protein that facilitates nuclear import of the T-complex and its subsequent targeting to and association with the host chromatin (Lacroix et al., 2008; Li et al., 2005; Tzfira et al., 2001).
Before integration, VirF, a bacterial F-box protein exported into the host cell (Schrammeijer et al., 2001), helps to uncoat the T-DNA from its associated VirE2 and VIP1 proteins via the SCFVirF pathway. VirF directly recognizes VIP1 and promotes proteasomal degradation of both VIP1 and its associated VirE2 (Tzfira et al., 2004). VirF itself is not essential for infection of some plant species (Hirooka et al., 1987), but its specific F-box protein function is critical for the infection process (Tzfira et al., 2004), suggesting that this function may be encoded by the host plant. Furthermore, because SCF complexes play a role in plant defense response against pathogens (Gray, 2002), F-box proteins would represent attractive candidates for a hypothetical pathogen-induced response factor of which Agrobacterium may take advantage to enhance its infection. Here, we identified an Agrobacterium-induced Arabidopsis F-box protein, VBF, that interacts with the plant ASK1 component of the SCFVBF complex. VBF also recognizes and binds VIP1 and its associated VirE2, forming ternary VBF-VIP1-VirE2 complexes. VBF then acts to destabilize both VIP1 and VirE2 via the SCFVBX pathway. Suppression of VBF expression elevated intracellular amounts of the endogenous VIP1, but rendered Arabidopsis largely resistant to Agrobacterium tumorigenicity, indicating that, in the infection pathway involving both VIP1 and VBF, VBF functions downstream of VIP1. When expressed in Agrobacterium and exported into the plant cell, VBF functionally complemented tumorigenicity of a VirF(-) Agrobacterium strain. Thus, Agrobacterium subverts a host defense SCFVBX pathway, induced in response to infection, to facilitate this infection.
RESULTS
VBF interacts with VIP1 and its expression is upregulated by Agrobacterium
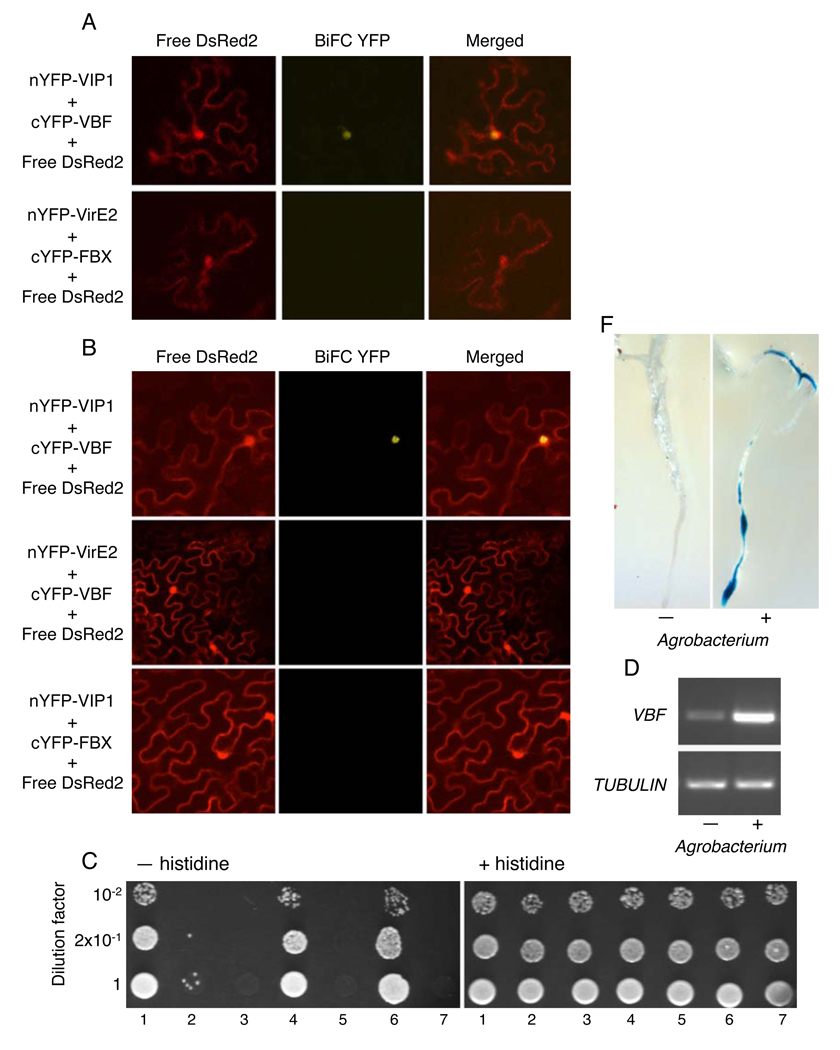
From a 694-member family of Arabidopsis F-box protein-encoding genes (Gagne et al., 2002), five genes may be upregulated by bacterial or fungal infection (Alvarez et al., 2006; Ditt et al., 2006). We tested each of them for the ability to interact with VIP1 in planta using a bimolecular fluorescence complementation (BiFC) assay (Citovsky et al., 2006). Although BiFC has inherent limitations, such as the use of fluorescent tags and relatively high levels of protein expression, it represents one of the best assays for protein-protein interactions and subcellular localization of the interacting proteins in planta. Only one tested F-box protein, encoded by At1g56250, recognized VIP1 and was designated VIP1-binding F-box (VBF) protein. Initially, the VBF-VIP1 interaction was tested both in Arabidopsis and in Nicotiana benthamiana, a choice plant for transient gene expression experiments. Figure 1A, B shows that, in both species, cYFP-tagged VBF interacted with nYFP-VIP1 in the nuclei of living plant cells, resulting in reconstruction of the YFP fluorescence, which colocalized with the nuclear DsRed2 signal. This recognition of VIP1 was specific because it was not observed with cYFP-tagged F-box proteins encoded by the Agrobacterium-induced At3g58890, At5g42350, At4g02760 (not shown), and At1g31350 genes (FBX, Figure 1A, B). Furthermore, we detected no interaction between cYFP-VBF and nYFP-VirE2 (Figure 1B and not shown), and no differences were observed between the BiFC data in Arabidopsis and N. benthamiana, allowing us to utilize the latter plant for subsequent experiments.
Figure 1. VBF interacts with VIP1 and is upregulated by Agrobacterium infection.
(A) BiFC assay for the VBF-VIP1 interaction in microbombarded Arabidopsis leaves. (B) BiFC assay for the VBF-VIP1 interaction in agroinfiltrated N. benthamiana leaves. FBX is encoded by the At1g31350 gene. Free DsRed2 labels the cell cytoplasm and the nucleus and identifies the transformed cells. All images are projections of several confocal sections. (C) Yeast two-hybrid assay for the VBF-VIP1 interaction. The indicated dilutions of cell cultures were grown either in the absence (left panel) or in the presence of histidine (right panel). Lane 1, Gal4AD-VBF+LexA-VIP1; lane 2, Gal4AD-VBF+LexA-FBX; lane 3, Gal4AD+LexA-VIP1, lane 4, Gal4AD-VirE2+LexA-VIP1; lane 5, Gal4AD-VBF+LexA-lamin C; lane 6, Gal4AD-VBF+LexA-ASK1; lane 7, Gal4AD+LexA-ASK1. (D) RT-PCR analysis of the VBF gene expression following inoculation by Agrobacterium. Constitutively-expressed TUBULIN was used as internal control. (E) Q-PCR analysis of the VBF gene expression following inoculation by Agrobacterium. The data represent average values of three independent experiments with indicated standard deviations. (F) Expression of GUS reporter from the VBF promoter in Arabidopsis roots following inoculation by Agrobacterium. (-), (+) indicate mock-inoculated or Agrobacterium-inoculated plants, respectively.
The BiFC data were then confirmed by an independent assay using the yeast two-hybrid system, in which protein interaction is indicated by histidine prototrophy (Hollenberg et al., 1995). Figure 1C shows that VBF interacted with VIP1, and that this interaction was specific because it did not occur with FBX or with lamin C, a known non-specific activator (Bartel et al., 1993). In positive control, VIP1 interacted with VirE2 (Figure 1C). Furthermore, as an F-box protein, VBF interacted with ASK1, the Arabidopsis homolog of Skp1 (Porat et al., 1998) (Figure 1C, see also below). As expected, neither VIP1 not ASK1 alone confer histidine prototrophy (Figure 1C). Under the non-selective conditions, all combinations of the tested proteins resulted in the efficient cell growth (Figure 1C).
We then demonstrated that VBF is induced by Agrobacterium. Using RT-PCR, substantially higher levels of VBF transcripts were detected in Agrobacterium-inoculated Arabidopsis tissues as compared to mock-inoculated plants (Figure 1D). Equal efficiency of the RT-PCR reactions was controlled using TUBULIN-specific transcripts. Quantitative real time PCR (Q-PCR) revealed an average of 5-fold increase in VBF expression (Figure 1E). Finally, the activation of the VBF promoter by Agrobacterium was demonstrated directly using Arabidopsis plants transgenic for the β-glucuronidase (GUS) reporter driven by the VBF regulatory sequences. Figure 1F shows that bacterial inoculation of these plants produced substantial GUS activity, detected as blue staining, in the root tissues, known as preferred substrate for Agrobacterium (Yi et al., 2002); no detectible reporter activity was observed in the same plant line in the absence of Agrobacterium. Interestingly, similar levels of VBF upregulation were observed in plants challenged with E. coli (not shown), suggesting that VBF represents a gene induced during general plant response to microbial challenge.
VBF forms ternary complexes with VIP1 and VirE2 in planta
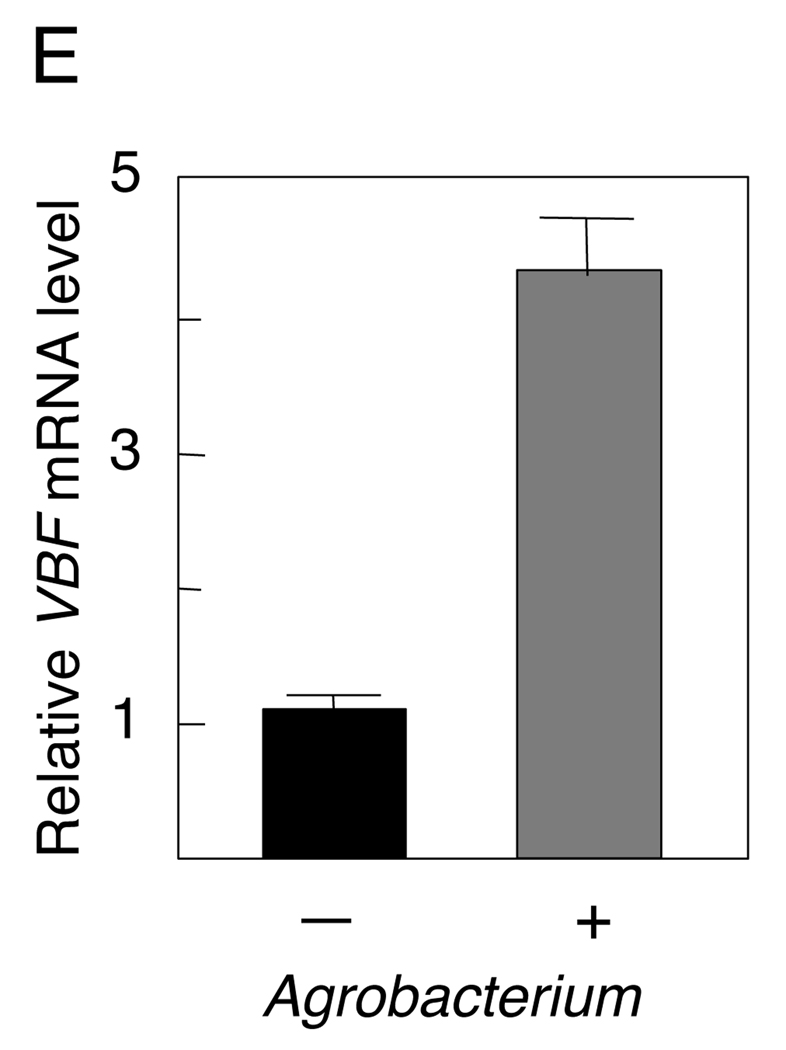
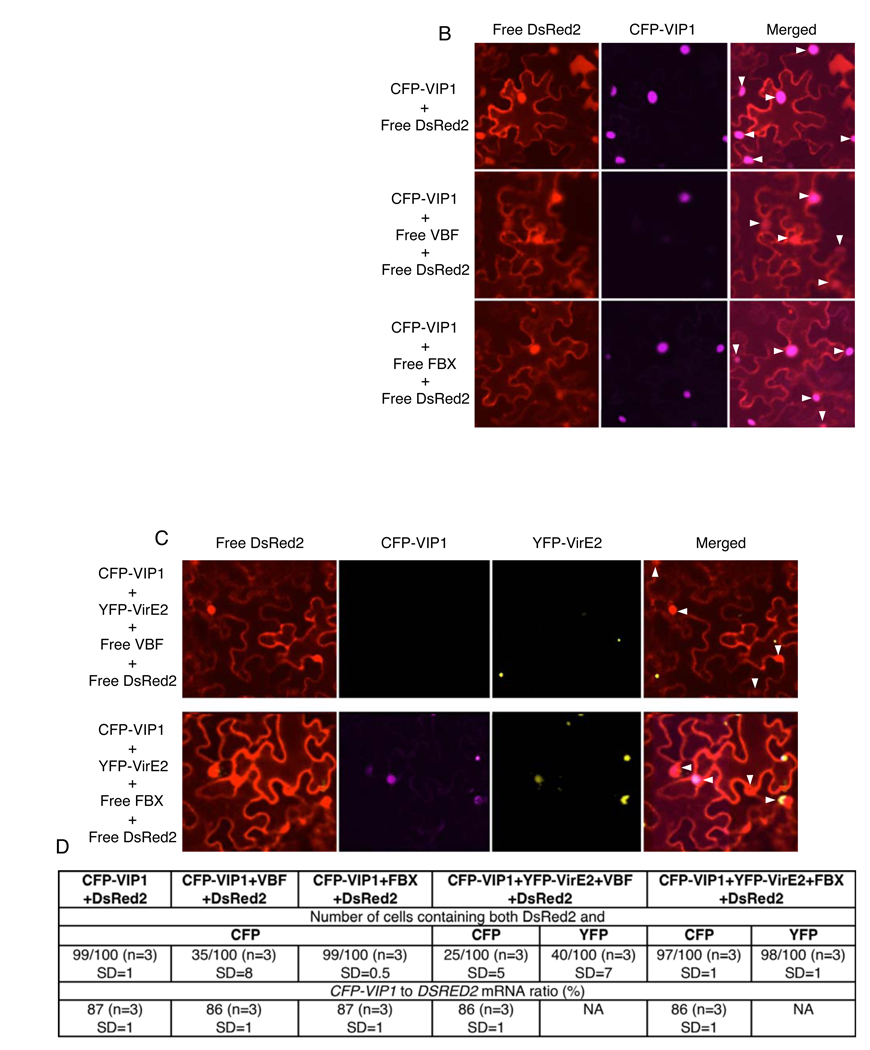
We examined whether VBF, VIP1, and VirE2 can exist in the same complex. To this end, we developed a bridge-BiFC assay in which the BiFC signal is produced when two tagged molecules that do not interact with each other, e.g., VBF and VirE2, are coexpressed with a third molecule, e.g., VIP1, that interacts with each of them and bridges between them. Initially, we tagged VBF with cCFP and VirE2 with nVenus (Lee et al., 2008). Figure 2A shows that coexpression of these proteins produced no BiFC signal. However, when they were expressed together with free VIP1, the reconstructed nVenus/cCFP fluorescence was observed, indicating formation of VBF-VIP1-VirE2 complexes which colocalized with the nuclear portion of the free DsRed2. Note that the endogenous levels of VIP1 are low (Tzfira et al., 2002), thus, its coexpression is required to enhance the detection of the BiFC signal.
Figure 2. Formation of ternary VirE2-VIP1-VBF complexes in microbombarded N. benthamiana leaves.
(A) Bridge-BiFC assay. (B) Multi-color bridge-BiFC assay. nCerulean/cCFP and nVenus/cCFP signals are indicated in blue and green, respectively; merged image represents overlay of both BiFC signals and DsRed2. Free DsRed2 labels the cytoplasm and the nucleus and identifies the transformed cells. (C) Coprecipitation. Left panel: Lane 1, GFP-VIP1+His-VBF+HA-VirE2; lane 2, GFP-VIP1+HA-VirE2. Input, the GFP-VIP1+His-VBF+HA-VirE2 sample processed without precipitation. Right panel: Lane 1, GFP-GFP+His-VBF. Input, the GFP-GFP+His-VBF sample processed without precipitation.
Next, we replaced the free VIP1 with VIP1 tagged with nCerulean and coexpressed it with nVenus-VirE2 and cCFP-VBF. In this multi-color BiFC, interactions of cCFP-tagged proteins with nVenus- or nCerulean-tagged proteins generate signals of different and specific colors (Lee et al., 2008). This approach allowed us to visualize simultaneously both the ternary VBF-VIP1-VirE2 complexes, detected as the nVenus/cCFP BiFC signal, and their constituent VIP1-VBF part, detected as the nCerulean/cCFP BiFC signal, accumulating in the nucleus of the same expressing cell (Figure 2B).
The formation of the VBF-VIP1-VirE2 complexes was confirmed by coprecipitation. We coexpressed GFP-VIP1 and HA-VirE2 and precipitated them using E. coli-produced His-VBF. Figure 2C shows that the western blot analysis of the precipitates revealed the presence of all three proteins in the precipitate whereas these precipitates were not detected in the absence of VBF, and no interaction was observed between VBF and a GFP dimer expressed in plant tissues.
VIP1 forms ternary complexes with the components of SCFVBF, VBF and ASK1, in planta
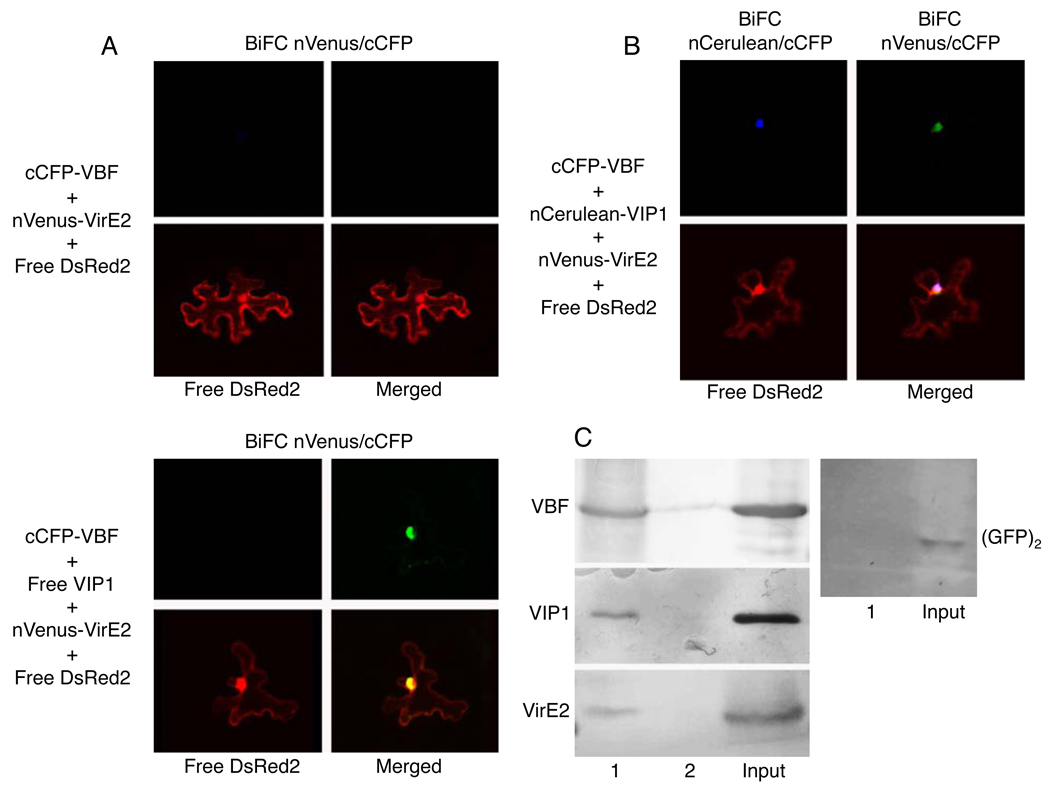
To better understand the relationship between VIP1 and the SCFVBF complex, we assayed for interactions between VIP1, VBF and ASK1. Figure 3A shows that, in the multi-color bridge-BiFC assay, VIP1 did not interact directly with ASK1. Coexpression of VBF resulted in formation of VBF-ASK1 complexes detected as the cCFP/nVenus BiFC signal, indicating that VBF indeed represents a component of the SCFVBF complex. Furthermore, ternary VIP1-VBF-ASK1 complexes were formed as indicated by the appearance of the cCFP/nCerulean signal, which accumulated predominantly in the cell nucleus (Figure 3A). These complexes were also detected using coprecipitation of His-VBF, GFP-VIP1 and HA-ASK1 (Figure 3B).
Figure 3. Formation of ternary VIP1-VBF-ASK1 complexes in microbombarded N. benthamiana leaves.
(A) Multi-color bridge-BiFC assay. nCerulean/cCFP and nVenus/cCFP signals are indicated in blue and green, respectively; merged image represents overlay of both BiFC signals and DsRed2. Free DsRed2 labels the cytoplasm and the nucleus and identifies the transformed cells. (B) Coprecipitation. Lane 1, GFP-VIP1+His-VBF+HA-ASK1; lane 2, GFP-VIP1+HA-ASK1. Input, the GFP-VIP1+His-VBF+HA-ASK1 sample processed without precipitation.
VBF destabilizes VIP1
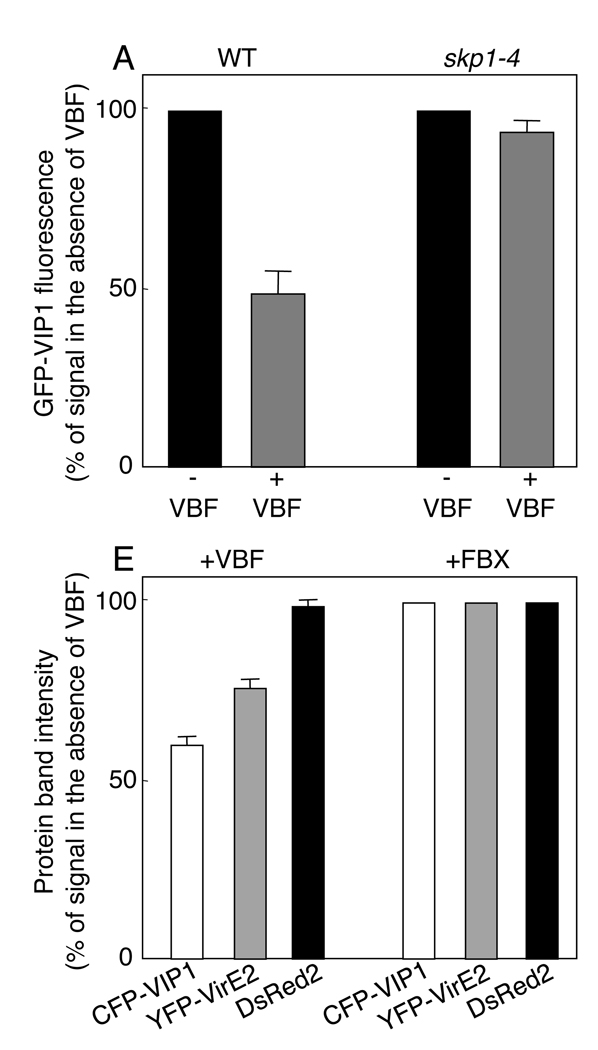
The recognition of VIP1 by SCFVBF suggests that VIP1 may be targeted for destabilization via the SCFVBF pathway. Initially, we assayed the VIP1 stability in yeast cells, known to be infected by Agrobacterium (Bundock et al., 1995). VIP1 was tagged with GFP and constitutively expressed in yeast cells together with free VBF, which was controlled by a methionine-repressible promoter (Tzfira et al., 2004). Figure 4A shows that expression of VBF depleted GFP-VIP1 by approximately one half as compared to expression without VBF induction. Furthermore, VBF expressed in a yeast temperature-sensitive mutant of Skp1, skp1-4 (Connelly and Hieter, 1996), failed to destabilize GFP-VIP1, indicating involvement of the SCF pathway.
Figure 4. VBF destabilizes VIP1 and VirE2.
(A) VBF-mediated and Skp1-dependent destabilization of GFP-VIP1 in yeast. GFP signal in the presence of VBF was calculated as percent of the signal measured in the absence of VBF expression, which was defined as 100% signal. Standard deviations are indicated. (B) VBF-mediated destabilization of CFP-VIP1 in agroinfiltrated N. benthamiana leaves. (C) VBF-mediated destabilization of CFP-VIP1 and YFP-VirE2 in agroinfiltrated N. benthamiana leaves. FBX is encoded by the At1g31350 gene. Arrows indicate cell nuclei identified by the presence of free DsRed2, which also identifies the transformed cells. (D) The effect of VBF expression on the number of transformed plant cells that accumulate CFP-VIP1 or CFP-VIP1 and YFP-VirE2 and on the amounts of the CFP-VIP1 transcript in these cells. The amounts of the CFP-VIP1 transcript were estimated by Q-PCR and expressed as percent of the amount of the coexpressed DSRED2 transcript in the same sample. The data represent three independent experiments (n=3) with indicated standard deviations (SD). NA, not applicable. (E) Quantification of VIP1 and VirE2 destabilization in N. benthamiana leaves. Amounts of each protein are represented by the intensities of the corresponding western bands and calculated as percent of those observed when VBF was replaced with FBX and defined as 100% signal. Standard deviations are indicated.
Next we examined VIP1 destabilization directly in planta. CFP-VIP1 and free DsRed2 or CFP-VIP1, free VBF and free DsRed2 were coexpressed from the same vector in leaf tissues. Figure 4B shows that CFP-VIP1 expressed only with DsRed2 efficiently accumulated in the plant cell nucleus. Indeed, coexpression of VBF significantly reduced CFP-VIP1 accumulation, showing only occasional and relatively weak signal in the cell nuclei (Figure 4B). More quantitatively, from each 100 transformed cells, i.e., those that expressed DsRed2, virtually all accumulated CFP-VIP1 in the absence of coexpressed VBF, whereas less than 40% displayed the CFP-VIP1 signal following coexpression of VBF (Figure 4D). That not all VIP1 was destabilized may be due its de novo synthesis or recalcitrance of some of the expressed protein to degradation, explaining why interactions between VBF and VIP1 could be detected in the two-hybrid system and in the BiFC assay. Note that the expression of VBF did not alter accumulation DsRed2. Furthermore, coexpression with FBX did not interfere with CFP-VIP1 accumulation (Figure 4B, D). Thus, the effect of VBF on accumulation of CFP-VIP1 was specific. Importantly, coexpression of VBF had no effect on accumulation of the CFP-VIP1 transcripts (Figure 4D), indicating that the reduction in protein amounts is not due to decreased transcription. Taken together, these data suggest that VBF destabilizes VIP1 via the SCFVBF pathway.
VBF destabilizes VirE2
Next, we examined whether VBF promotes destabilization of VirE2, the coat protein of the T-complex. Because VBF is specific for VIP1 and does not recognize VirE2 (see Figure 1A and Figure 2), it may affect VirE2 only when it is complexed with VIP1. Thus, we coexpressed VBF with CFP-VIP1, YFP-VirE2, and DsRed2. Figure 4C illustrates representative data showing that, under these conditions, no nuclear YFP-VirE2 signal was observed whereas, consistent with previous observations (Bhattacharjee et al., 2008), the only detectible YFP-VirE2 was found in a few cytoplasmic aggregates. Moreover, virtually no CFP-VIP1 signal was detected in these cells. YFP-VirE2 coexpressed with CFP-VIP1 and DsRed2 (not shown) or with CFP-VIP1, DsRed2, and FBX colocalized with CFP-VIP1 and was detected within or around the cell nuclei (Figure 4C). Comparable levels of DsRed2 accumulation in the absence and presence of VBF/FBX indicate equal transformation and expression efficiencies.
Overall, coexpression of VBF destabilized CFP-VIP1 and YFP-VirE2 in 75% and 60%, respectively, of the expressing cells (Figure 4D). This lower efficiency of VirE2 destabilization likely reflects the requirement for formation of the tripartite VBF-VIP1-VirE2 complex and is consistent with the destabilization efficiencies of VIP1 and VirE2 by VirF in yeast (Tzfira et al., 2004).
VIP1 and VirE2 destabilization was also demonstrated by western blot analysis. Figure 4E shows that the amounts of both CFP-VIP1 and YFP-VirE2 in the expressing tissues were consistently reduced in the presence of VBF as compared to those in the presence of FBX. Collectively, our data suggest that VBF promotes VIP1-dependent destabilization of VirE2 and, by implication, the entire T-complex.
Suppression of VBF expression elevates endogenous VIP1 and the PR-1 defense protein
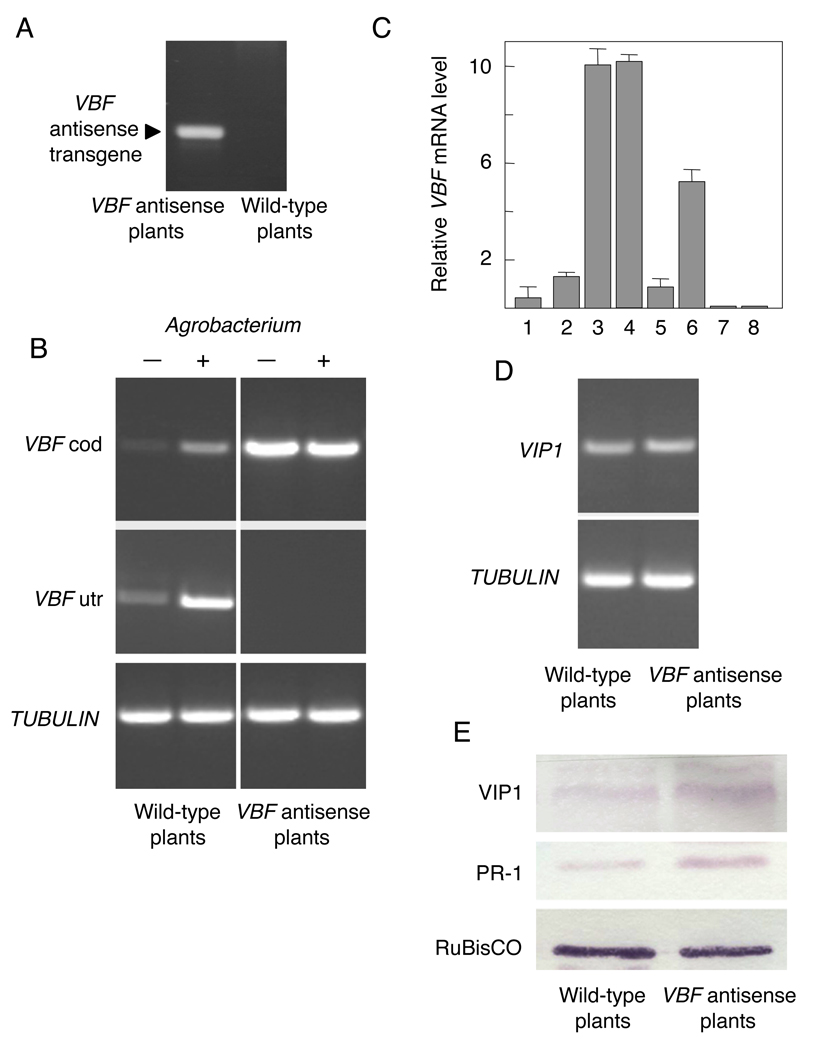
To examine further the biological role of VBF in Agrobacterium infection in planta, we generated transgenic Arabidopsis expressing the VBF coding sequence in the antisense orientation. Five independently-transformed lines were produced and analyzed as described below. Two lines had no detectible phenotypes in regard to VBF expression whereas, in the other three lines, this expression became largely suppressed (data not shown). Here, we describe a detailed analysis of one of these lines, in which we first confirmed the presence of the antisense VBF transgene (Figure 5A). Then, we examined the presence of the VBF transcript using RT-PCR (Zaltsman et al., 2005) and primers specific either for the VBF coding sequences, expected to be found both in the wild-type and in the VBF antisense plants, or for the VBF 5’ and 3’ UTRs, expected to be detected only in the wild-type, but not in the VBF antisense plants, which should not produce the endogenous VBF mRNA. Wild-type Arabidopsis exhibited relatively low levels of VBF transcripts (Figure 5B). In contrast, the VBF antisense plants accumulated high amounts of the coding sequence-specific transcripts due to antisense expression (Figure 5A), but virtually undetectable levels of the endogenous VBF mRNA containing the 5’ and 3’ UTRs (Figure 5A). Consistent with the inducibility of the VBF gene, the wild-type plants, but not the VBF antisense plants, accumulated higher amounts of VBF transcripts following inoculation with Agrobacterium (Figure 5B). Analysis of TUBULIN-specific transcripts detected similar amounts of PCR products in all samples, indicating equal efficiencies of the RT-PCR reactions (Figure 5B). These data were confirmed by Q-PCR (Figure 5C). Thus, antisense expression of VBF in Arabidopsis substantially reduced transcription of the endogenous VBF gene, most likely by an RNAi-related pathway. Phenotypically, the VBF antisense plants were indistinguishable from the wild-type plants in their morphology and seed viability (data not shown), suggesting that the expression of the VBF antisense transgene did not interfere with essential plant cellular functions.
Figure 5. Expression of the VBF and VIP1 genes and accumulation of the VIP1 and PR-1 proteins in wild-type and VBF antisense Arabidopsis plants.
(A) Detection of the VBF antisense transgene by PCR using primers specific for the 35S promoter and terminator sequences of pSAT4-35SP-MCS-35ST. (B) Detection of the VBF transcripts by PCR. VBF cod, PCR products obtained with primers specific for the VBF coding sequence; VBF utr, PCR products obtained with primers specific for the VBF 5’ and 3’ UTR sequences. (-), (+) indicate mock-inoculated or Agrobacterium-inoculated plants, respectively. (C) Detection of the VBF transcripts by Q-PCR. Bars 1–4, VBF cod; bars 5–8, VBF utr; bars 1, 2 and 5, 6, wild-type plants; bars 3, 4 and 7, 8, VBF antisense plants; bars 1, 3, 5, 7, and 2, 4, 6, 8, mock-inoculated or Agrobacterium-inoculated plants, respectively. The data represent average values of three independent experiments with indicated standard deviations. (D) Detection of the VIP1 transcripts. (E) Detection of the VIP1 and PR-1 proteins.
Next, we used the VBF antisense plants to examine the effect of suppression of the endogenous VBF expression on the cellular amounts of VIP1. Consistent with the likely posttranslational effect of VBF on VIP1, our RT-PCR analysis detected no substantial differences in the amounts of the VIP1 transcript between the wild-type and the VBF antisense plants (Figure 5D). In contrast, the western blot analysis revealed that the VBF antisense plants accumulated higher amounts of the VIP1 protein as compared to the wild-type plants (Figure 5E).
Because VIP1 is involved in the expression of the PR-1 pathogenesis-related gene (Djamei et al., 2007; Pitzschke et al., 2009), we investigated the PR-1 content in the VBF antisense plants. These experiments indicated that the PR-1 amounts essentially mirrored those of VIP1. Specifically, the wild-type uninfected Arabidopsis contained small amounts of PR-1 whereas the VBF antisense plants accumulated higher levels of this protein (Figure 5E). Detection of RuBisCO confirmed equal loading of all samples. Thus, VBF may, at least in part, regulate accumulation of PR-1 by controlling the cellular levels of VIP1.
Suppression of VBF expression inhibits Agrobacterium-induced tumor formation
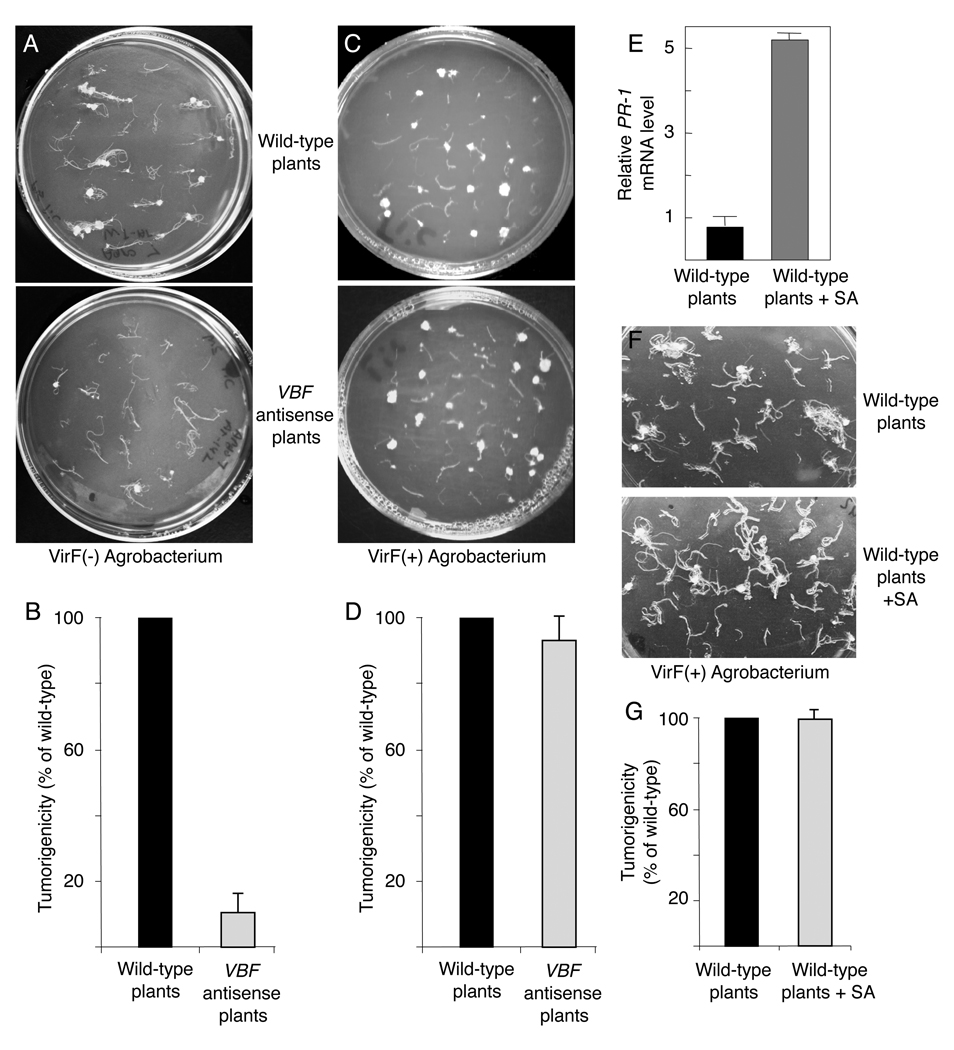
We tested the ability of the VBF antisense plants to develop tumors following inoculation with the VirF(-) oncogenic Agrobacterium LBA1517 strain (Hooykaas et al., 1984). Figure 6A shows that LBA1517 elicited numerous tumors on roots of the wild-type Arabidopsis. In contrast, Figure 6A shows that only very few tumors developed on roots from the VBF antisense plants inoculated with LBA1517.Agrobacterium infectivity was then quantified as the ratio between the total number of tumors and the root mass. Overall, the tumor-inducing activity of LBA1517 in VBF antisense plants was reduced to approximately 15% of that observed with the wild-type plants (Figure 6B). Both the wild-type plants and the VBF antisense plants were equally susceptible to the VirF(+) oncogenic Agrobacterium LBA11010 strain (Hooykaas et al., 1984) (Figure 6C, D), indicating that the recalcitrance of the VBF antisense plants to infection by the VirF(-) Agrobacterium is due to the loss of function of VBF, which can be complemented by the bacterial F-box protein VirF.
Figure 6. Reduced tumor formation in Agrobacterium-infected VBF antisense Arabidopsis plants.
(A, C) Tumor formation in roots from the wild-type and VBF antisense plants infected with VirF(-) or VirF(+) Agrobacterium strains LBA1517 or LBA1010, respectively. (B, D) Quantification of LBA1517 and LBA1010 tumorigenicity, respectively, in the wild-type (black bars) and VBF antisense plants (gray bars). Standard deviations are indicated. (E) RT-PCR analysis of the PR-1 gene expression following SA treatment of the wild-type plants. (F) Tumor formation in roots from untreated or SA-treated wild-type plants infected with VirF(+) Agrobacterium strain LBA1010. (G) Quantification of LBA1010 tumorigenicity in untreated (black bars) and SA-treated wild-type plants (gray bars). Standard deviations are indicated.
Expression of PR genes, notably PR-1, is often associated with resistance to microbial pathogens (Durrant and Dong, 2004). Potentially, the elevated levels of PR-1 in the VBF antisense plants may contribute to the tumor-resistant phenotype. This possibility was ruled out in experiments where the PR-1 content was elevated independently of Agrobacterium by pretreating the wild-type Arabidopsis roots with salicylic acid (SA), known to induce the PR-1 gene expression (Durrant and Dong, 2004) (Figure 6E). Subsequent inoculation of these roots with Agrobacterium resulted in tumorigenicity comparable to that observed in untreated roots (Figure 6F, G). Previous experiments (Lee et al., 2009; Yuan et al., 2007) suggested that SA inhibits Agrobacterium tumorigenicity. In Arabidopsis, however, this effect is due to the direct inhibition by SA of the bacterial VirA protein that functions as a sensor of the vir gene-inducing signals, rather than to accumulation of the PR proteins (Lee et al., 2009; Yuan et al., 2007).
VBF extends the host range of Agrobacterium
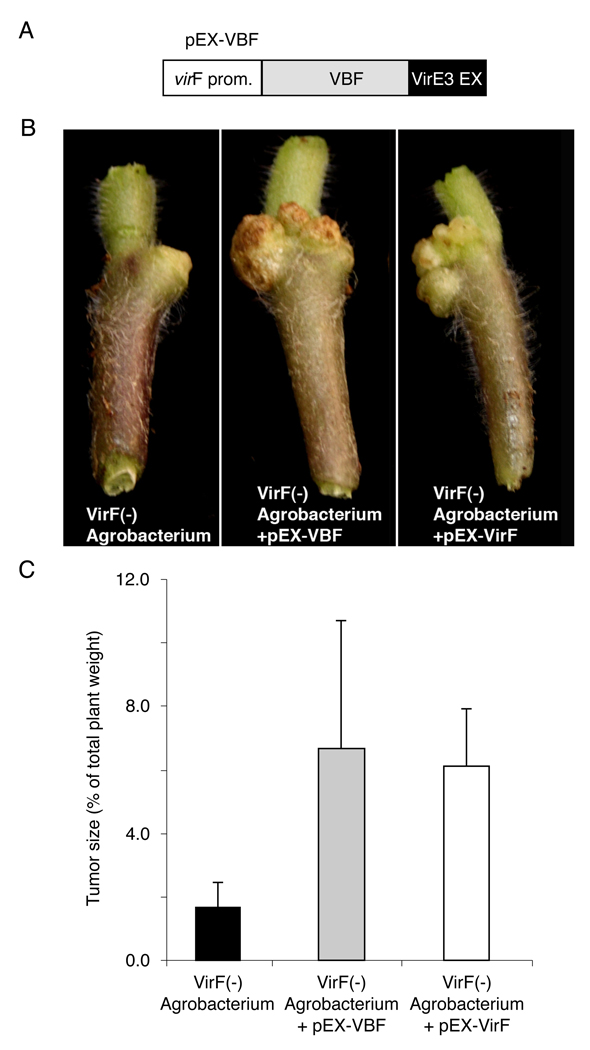
While some plant species, such as Arabidopsis and tobacco, do not require VirF for genetic transformation by Agrobacterium, other plants, notably the agronomically important crop tomato, are poorly transformed by Agrobacterium strains that lack VirF (Regensburg-Tuink and Hooykaas, 1993). We examined whether VBF can extend the host range of such an Agrobacterium strain to include tomato. W e constructed a derivative of VBF that is exported to plant cells when expressed within Agrobacterium by fusing a C-terminal export sequence of VirE3 (EX) (Schrammeijer et al., 2003) to the C-terminus of VBF. The resulting plasmid, pEX-VBF (Figure 7A), was introduced into the VirF(-) LBA1517 strain of Agrobacterium, and both the parental LBA1517 strain and the VBF-expressing/exporting LBA1517/pEX-VBF strain were inoculated onto stems of tomato plants. Figure 7B shows that LBA1517 elicited very small tumors, indicating low efficiency of genetic transformation. In contrast, inoculation with LBA1517/pEX-VBF exhibited efficient tumorigenicity (Figure 7B). Tumorigenicity was then evaluated by the relative biomass of tumors, which although not a bona fide quantitative measure of virulence, represents a well-established criterion of plant susceptibility to Agrobacterium-mediated genetic transformation (Tzfira et al., 2002; Zhu et al., 2003). Figure 7C shows that LBA1517 elicited small tumors only of about 1.7% of the total plant weight whereas LBA1517/pEX-VBF induced tumors of much larger biomass with an average of 6.7% of the total plant weight. These latter tumors were comparable to tumors elicited in positive control experiments using LBA1517 that expresses VirF with its own export signal, instead of VBF-EX, from the same vector (LBA1517/pEX-VirF) (Figure 7B, C). Thus, VBF functionally complemented the VirF function in determination of the Agrobacterium host range.
Figure 7. VBF enhances tumorigenesis in tomato infected by a VirF(-) strain of Agrobacterium.
(A) Schematic structure of the VBF expression cassette in pEX-VBF. VBF expression in Agrobacterium is directed by the virF promoter, and VBF export from the bacterial cell is promoted by the VirE3 export signal (EX) fused to the C-terminus of VBF. (B) Representative images of tumors developed on tomato stems following infection by VirF(-) Agrobacterium strains LBA1517, LBA1517 that expresses VBF from pEX-VBF, or LBA1517 that expresses VirF from pEX-VirF. (C) Quantitation of tumorigenicity of LBA1517 (black bar), VBF-expressing LBA1517 (gray bar), or VirF-expressing LBA1517 (positive control, white bar) Agrobacterium strains in tomato plants. Standard deviations are indicated.
DISCUSSION
Recently, Agrobacterium has been shown to utilize the host MAPK defense signaling pathway to target its T-complex into the plant cell nucleus (Djamei et al., 2007); however, it has remained unknown whether the T-complex can recognize the host defense proteins induced by the infection and incorporate them into its own pathways for genetic transformation. VBF represents the first example of such host factors. VBF is a nuclear F-box protein, the expression of which is induced by diverse pathogens, from bacteria to fungi (Alvarez et al., 2006; Ditt et al., 2006). VBF directly interacts with ASK1, another component of SCFVBF. One of the specific substrates of SCFVBF is VIP1. VBF recognizes VIP1 and promotes its destabilization in yeast and plant cells. Thus, VBF most likely functions to regulate the amount of VIP1 via proteasomal degradation. VIP1, in turn, regulates expression of the PR-1 defense response gene (Djamei et al., 2007), and induction of VBF expression during the same response may represent a mechanism to control and modulate this VIP1 activity. Indeed, suppression of VBF expression in transgenic plants, resulted in modestly, but consistently elevated levels of both VIP1 and PR-1. Potentially, among the almost 700 F-box protein genes in Arabidopsis (Gagne et al., 2002), some may partly overlap the VBF specificity toward VIP1.
Agrobacterium has deeply insinuated into this VIP1/VBF-based defense response of its plant hosts utilizing it for some of the most critical stages of the genetic transformation process. We propose that VIP1 is used to facilitate nuclear import of the T-complex (Tzfira et al., 2001), and VBF is subsequently used to uncoat the T-complex of its protein components. In these reactions, VIP1 most likely serves as a molecular link between VirE2, the “coat protein” of the T-complex, and either importin α of the nuclear import machinery (Citovsky et al., 2004) or SCFVBF of the proteasomal degradation machinery. Indeed, VIP1 has been shown to promote nuclear import of VirE2 via the importin α-mediated pathway (Tzfira et al., 2001, 2002) whereas this study demonstrates that VIP1 is required for destabilization of VirE2 via the SCFVBF pathway. Suppressing expression of either VIP1 (Tzfira et al., 2001) or VBF substantially reduced the plant susceptibility to the Agrobacterium-mediated genetic transformation. Interestingly, the absence of VBF also resulted in elevated levels of VIP1, indicating that excess of VIP1 cannot alleviate the lack of VBF. Thus, VBF is functionally epistatic to VIP1 in the infection pathway. This notion makes biological sense as the T-complex uncoating, which requires both VIP1 and VBF, must occur after its nuclear import, which involves VIP1 (Tzfira et al., 2001, 2002), but not VBF. That this uncoating occurs mainly in the host nucleus is further supported by nuclear localization of the VirE2-VIP1-VBF and VIP1-VBF-ASK1 complexes. It is important to emphasize that uncoating of the T-complex by proteasomal degradation is a notion based on VBF-dependent degradation of VIP1 and VirE2, and its ultimate proof awaits experiments using the entire T-complex as the SCFVBF substrate.
The critical nature of the T-complex nuclear import and its presumed uncoating for successful infection suggests that Agrobacterium may not rely exclusively on its hosts to provide the protein machinery for these stages of infection. Agrobacterium, therefore, has evolved a “backup” system composed of Vir proteins that are exported into the host cells, but are not absolutely essential for tumor formation (Hirooka and Kado, 1986; Schrammeijer et al., 2003; Stachel and Nester, 1986; Winans et al., 1987). For example, VirE3, at least partially, mimics the VIP1 function and facilitates nuclear import of VirE2 and, by implication, the T-complex (Lacroix et al., 2005). T-complex uncoating is facilitated by VirF, the bacterial functional homolog of VBF (Lacroix et al., 2008; Tzfira et al., 2004). VBF can substitute for VirF when expressed in Agrobacterium and exported into the host cell. This strategy of Agrobacterium likely reflects a general ability of pathogenic microorganisms to encode and export protein functions normally provided by the host eukaryotic cell (Nagai and Roy, 2003). Our present observations indicate that the host factors that the pathogen uses for infection also include those that the host initially produces to defend itself from the very same infection.
MATERIALS AND METHODS
Plants
Wild-type Arabidopsis thaliana (Columbia ecotype), N. benthamiana, and Lycopersicon esculentum (Micro-Tom tomato) were grown in soil in an environment-controlled chamber at 22–24°C. All plants were maintained under long day conditions of 16 h white light (70–80 µmol photons m-2 s-1) and 8 h dark. At least 10 plants were used per each experimental condition, and all the experiments were repeated three times. For assaying their PR-1 protein content, plants were grown on MS (Murashige and Skoog, 1962) aseptic medium to avoid exposure to other pathogens.
Bacterial challenge and RT-PCR and Q-PCR analyses
Three to four weeks-old Arabidopsis plants were inoculated (Kapila et al., 1997; Wroblewski et al., 2005) with Agrobacterium LBA1010 strain (Hooykaas et al., 1984), or E. coli, or mock-inoculated with the bacterial growth medium. Nine hours after inoculation, total RNA was extracted from tissue samples using Tri-Reagent (Sigma-Aldrich) and treated with RNase-free DNase I (Fermentas). The absence of contaminating genomic DNA was confirmed by PCR using TUBULIN-specific primers that flank an intron sequence, to distinguish between PCR products derived from DNA and mRNA templates (Zaltsman et al., 2005). The RT reaction was then performed using 500 ng of the purified RNA and the Superscript II reverse transcriptase (Stratagene). The resulting cDNA was PCR-amplified for 32 cycles using primers specific for the tested gene or for TUBULIN as an internal control of a constitutively expressed gene. The following primer pairs were used: VBF coding sequence 5’ATGTTACCAGAAGCATGCATAGCC3’/5’TTATGTTTTAGGCCTCACTTCAATAC3’, VBF 5’ and 3’ UTRs 5’CTCGGCAAAAGAAGAAGAAGATG3’/5’ACACATTCACACAACCCCTGAGT 3’, respectively, VIP1 5’GGAAGGTTCAGACACTTCAGAATGA3’/5’CATCAAATATTGCAGCCCGAAA3’, PR-1 5’ATGAATTTTACTGGCTATTCTCGATTTT3’/5’TTAGTATGGCTTCTCGTTCACATAATTC3’, and TUBULIN 5’CTCACTCACTCGCCTGAACATCTC3’/5’AGATTCTTCACATCCAGGGTGGTC3’.
Q-PCR was performed using the same cDNA preparations in Light Cycler 480 Real-Time PCR System (Roche) with SYBR-green I Master mix (Roche) and primers specific for VBF (5’TGGGAAAAGTTTCTACCATCGG3’/5’TCGATGAGAAGAGAGTCACATAAACA3’) or PR-1(5’GATAATCTTGTGGGCTATCTTGAGC3’/5’ATGAATTTTACTGGCTATTCTCGATTTT3’) and ACTIN8 (5’TGTATGTTGCCATTCAAGCTGTTC3’/5’GAAACCCTCGTAGATAGGCACAGTG3’) or CFP-VIP1 (5’AAGCTGACCCTGAAGTTCATCTGC3’/5’GAAGAAGTCGTGCTGCTTCATGTG3’) and DSRED2 (5’GCCACTACCTGGTGGAGTTCAAGT3’/5’GTAGTCCTCGTTGTGGGAGGTGA T3’). Relative abundance of the VBF or PR-1 mRNA-specific products was normalized to the amount of the product specific for ACTIN8, which represented an internal control of a constitutively expressed gene whereas relative abundance of the CFP-VIP1 mRNA-specific product was normalized to the amount of the product specific for DSRED2, which represented an internal control of a coexpressed transgene.
Agroinfiltration and microbombardment
For agroinfiltration, binary plasmids were introduced into the Agrobacterium GV3101 strain (Tzfira et al., 1997), grown overnight at 25°C, and infiltrated into intact N. benthamiana leaves also as described (Kapila et al., 1997; Wroblewski et al., 2005). For biolistic delivery, DNA preparations of the indicated constructs were mixed at a 1:1 w/w ratio, and 100 µg DNA was adsorbed onto 10 mg of 1-µm gold particles (Bio-Rad). The particles were microbombarded into the leaf epidermis of Arabidopsis or N. benthamiana at a pressure of 90–150 psi, using a portable Helios gene gun system (Model PDS-1000/He, Bio-Rad). Agroinfiltrated or microbombarded tissues were analyzed 36 to 48 h after transformation.
BiFC, bridge-BiFC, and multi-color BiFC
For BIFC, coding sequences of VIP1 or VirE2 were cloned into the SalI-BamHI or XhoI-XbaI sites, respectively, of pSAT1-nEYFP-C1 (Citovsky et al., 2006), and the VBF coding sequence was cloned into the PstI-SalI sites of pSAT6-cEYFP-C1 (Citovsky et al., 2006). The resulting expression cassettes were excised with AscI or PI-PspI from pSAT1- or pSAT6-based vectors, respectively, and inserted into pRS2-DsRed2, the pPZP-RCS2 binary vector (Goderis et al., 2002; Tzfira et al., 2005) with a pSAT4-based DsRed2 expression cassette (Tzfira et al., 2005) in its I-SceI site. For bridge-BiFC and multi-color BIFC, coding sequences of VIP1 or ASK1 (NM_100969) were cloned into the SalI-BamHI or EcoRI-BamHI sites, respectively of pSAT6-nCerulean-C (Lee et al., 2008), and VirE2 and VBF coding sequences were cloned into the BglII-XbaI and XhoI-XbaI sites of pSAT1-nVenus-C and pSAT4-cCFP-C (Lee et al., 2008), respectively. For expression of free VIP1, its coding sequence was cloned into the SalI-BamHI sites of pSAT6-MCS (Tzfira et al., 2005). Free DsRed2 was expressed from pSAT6-DsRed2-C1 (Tzfira et al., 2005). The multigene expression binary constructs or the individual bridge-BiFC and multi-color BiFC constructs were transiently expressed in N. benthamiana leaves following agroinfiltration or microbombardment, respectively. In Arabidopsis leaves, the BiFC constructs were expressed following microbombardment. BiFC was detected using a Zeiss LSM 5 Pascal confocal microscope (Citovsky et al., 2006). All experiments were repeated at least three times. All images are projections of several confocal sections.
Yeast two-hybrid
VBF or VirE2 were fused to GAL4 activation domain by subcloning their coding sequences into the PstI-EcoRI or BamHI-PstI sites, respectively,of pGAD424 (LEU2+, Clontech). For fusion to LexA, the VIP1 coding sequence was cloned into the BamHI-SalI sites of pSTT91 (TRP1+, Sutton et al., 2001), and coding sequences of At1g31350 (FBX) or ASK1 were cloned into the EcoRI-PstI sites of pSTT91. Lamin C fused to LexA has been described (Tzfira et al., 2004). Protein interaction was assayed in the Saccharomyces cerevisiae strain TAT7 [L40 (Hollenberg et al., 1995)-ura3] (SenGupta et al., 1996) by growing cells for 2 days at 30°C on a leucine-, tryptophan-, and histidine-deficient medium.
Protein destabilization in yeast
Coding sequences of GFP-VIP1 (Tzfira et al., 2004) and VBF were cloned under galactose-inducible (GAL1) and methionine-repressible (MET25) promoters, respectively (Tzfira et al., 2004). Both constructs were expressed in the wild-type and skp1-4 (Connelly and Hieter, 1996) yeast cells in the presence of 5% galactose with or without 100 mM methionine (Tzfira et al., 2004). GFP fluorescence was expressed as percent of the total signal measured in the presence of methionine. All experiments were repeated at least three times.
Coprecipitation and western blot analysis
For coprecipitation, VBF was fused to the 6xHis tag by inserting its coding sequence into the EcoRI-XhoI sites of pET28a(+) (Novagen) and expressed in E. coli using standard protocols (Citovsky et al., 2004). N. benthamiana leaves were agroinfiltrated with constructs expressing GFP-VIP1 (Tzfira et al., 2001) and HA-VirE2 or HA-ASK1, made by subcloning their coding sequences into the EcoRV-BglII sites of pSAT6-HA-C1 (Dafny-Yelin and Tzfira, 2007). After 48 h, the tissues (5 g) were ground in 15 ml extraction buffer (30mM NaCl, 0.1% NP-40, 1 mM PMSF, 50 mM sodium phosphate buffer, pH 8.0), and clarified by centrifugation at 14,000 rpm for 30 min. The resulting extracts (5 ml) were incubated for 1 h at 4°C with 5 ml of E. coli extracts containing His-VBF. Ni-NTA agarose beads (1 ml, Qiagen) were added to the mixture, which was then gently rocked for 6 h at 4°C. For negative controls, His-VBF was either omitted or incubated with cell extracts from plants expressing a GFP dimer, instead of GFP-VIP1. After three washes in the extraction buffer, the captured protein complexes were released by mixing with 50 µl SDS-PAGE loading buffer and boiling for 5 min and analyzed by western blotting, using anti-GFP (dilution 1:1,000, Santa Cruz Biotechnology), anti-His (dilution 1:1,000, Bethyl Laboratories), and anti-HA antibodies (dilution 1:1,000, ICL, Inc.), followed by alkaline phosphatase-conjugated secondary antibody (1:1,000 dilution, Sigma-Aldrich) and detection by chromogenic staining with BCIP and NBT.
Protein destabilization in planta
VIP1 or VirE2 coding sequences were cloned into the NotI-SalI sites of pSAT1-ECFP-C1 or NotI-XhoI sites of pSAT6-EYFP-C1, respectively (Tzfira et al., 2005). The resulting expression cassettes were excised with AscI or PI-PspI, respectively, and inserted separately or together into pRS2-DsRed2, producing pRCS2-DsRed2-ECFP-VIP1, pRCS2-DsRed2-EYFP-VirE2, and pRCS2-DsRed2-ECFP-VIP1-EYFP-VirE2. Coding sequences of VBF or FBX were cloned into the NotI-XhoI sites of pSAT6-MCS (Tzfira et al., 2005), the expression cassettes were excised with PI-PspI and inserted into pRCS2-DsRed2-ECFP-VIP1, pRCS2-DsRed2-EYFP-VirE2, and pRCS2-DsRed2-ECFP-VIP1-EYFP-VirE2. Different combinations of these constructs were transiently expressed in N. benthamiana, following agroinfiltration, and analyzed by confocal microscopy. The transformed cells were identified by the presence of DsRed2 whereas VIP1 and VirE2 degradation in these cells was determined by reduction or disappearance of the CFP and YFP signals. For quantification, the number of transformed plant cells that accumulated CFP and/or YFP was determined per 100 transformed cells. All experiments were repeated at least three times, with the entire infiltrated leaf area examined in each experiment.
For quantification of protein destabilization, 72 h after agroinfiltration with a construct coexpressing either ECFP-VIP1, EYFP-VirE2, DsRed2 and VBF or ECFP-VIP1, EYFP-VirE2, DsRed2 and FBX, N. benthamiana leaves were extracted and subjected to western blot analysis using anti-GFP antibodies and anti-DsRed2 antibodies (dilution 1:2,000, MBL International) followed by detection with secondary antibody conjugated to HRP (dilution 1:10,000, Pierce) and an ECL kit (Pierce). The anti-GFP antibody detects both ECFP-VIP1 and EYFP-VirE2, and the resulting protein bands were distinguished from each other based on their specific electrophoretic mobilities. Protein amounts were estimated by scanning densitometry of the corresponding western bands using the ImageJ 1.43 software (NIH) and normalized to the amount of DsRed2 accumulated in the presence of FBX.
Generation of transgenic Arabidopsis plants
For production of plants that express GUS from the VBF promoter, we utilized VBF sequences containing 1.25 kb upstream of the ATG codon, based on the size of the predicted VBF intergenic region. This region was amplified from the wild-type Arabidopsis genomic DNA with the forward 5’TCTCGAGACCGGTCAAAGGACTCGATGTTTGGTGTC3’ and reverse 5’TTCCCGGGGCTAACCGGAAAACCGAAGAGACCT3’ primers and cloned into the AgeI-XmaI sites of pSAT4-35SP-MCS-35ST (Chung et al., 2005), replacing the 35S promoter. Then, the GUS reporter gene was subcloned into the XbaI(filled-in)-BamHI sites downstream of the VBF regulatory sequences.
For production of VBF antisense plants, the VBF coding sequence from the VBF cDNA (ABRC stock U61789, GenBank accession number BT011915) was inserted into the EcoRI-NdeI sites of pBluescript II (Stratagene) and then transferred into the SalI-XmaI sites of pSAT4-35SP-MCS-35ST. Expression cassettes from each of the pSAT4-based vectors were transferred into the I-SceI site of the pPZP-RCS2 binary vector, containing the bar gene for BASTA selection in its XhoI-BamHI sites.
The resulting binary constructs were introduced into the Agrobacterium EHA105 strain, used to transform the wild-type Arabidopsis plants by flower dipping (Kim et al., 2003), and transformants were obtained using BASTA selection. BASTA-resistant T3 plants were used for further analyses.
GUS activity and tumorigenesis in Arabidopsis
Transgenic 10–14 day-old Arabidopsis seedlings aseptically grown in baby food jars in MS agar with 5 mg/l BASTA (Nam et al., 1999) were inoculated with 50 µl Agrobacterium culture (OD600=0.1) or mock-inoculated with the bacterial growth medium. After 12 h, the plantlets were assayed histochemically for GUS activity (Nam et al., 1999), and recorded under a Leica MZ FLIII stereoscope.
For induction of tumors, root segments from aseptically grown 10–14 day-old Arabidopsis seedlings were submerged in liquid culture (OD600=0.1) of Agrobacterium strains LBA1010 [VirF(+)] or LBA1517 [VirF(-)] (Hooykaas et al., 1984), incubated for 10 min at 25°C, cultivated for 48 h at 25°C in hormone-free MS (HFMS) medium, washed, and cultured for 4 weeks in HFMS with 100 µg/ml timentin and scored for tumors. For treatment with SA, root segments were incubated with 250 µM SA for 24 h before inoculation with Agrobacterium. Each experiment was repeated three times.
Tumorigenesis in tomato
pPCB302 (Xiang et al., 1999) lacking the T-DNA and T-DNA border sequences was amplified using PCR and the primer pair 5’CTCACCGGGCTGGTTGCCCT3’/5’ACTGACCCCACAAGGCCCTAG3’, and blunt-end ligated to the virF promoter and to the sequence coding for the VirE3 C-terminal export signal (EX) (Schrammeijer et al., 2003) and a stop codon, which were PCR-amplified from the genomic DNA of the wild-type Agrobacterium C58 strain using the primer pair 5’AGAGCTCGGTTCGGATCGCCATCT3’/5’AACCGCGGTGCATGCTCCTTCTTTCT3’/5’AAGGTACCTTAGAAACCTCTGGAGGTGGAACG3’/5’AACTCGAGTTGCTGAATCAATTGCTTAGTGTGC3’, resulting in pEX302 (GeneBank accession number FJ386489). Then, the VBF coding sequence without the stop codon was inserted into the SacII-XhoI sites of pEX302 as a translational fusion to EX. Alternatively the VirF coding sequence with its own export signal was inserted into the SacII-BamHI sites of pEX302, introducing a stop codon between VirF and EX and producing pEX-VirF. Finally, each of these plasmids was introduced into the Agrobacterium LBA1517 strain.
For standardized inoculation of three-week-old Micro-Tom plants with Agrobacterium, stems were punctured to 1-mm depth with a 27G needle to create identical wounds onto which 2 µl of an overnight-grown Agrobacterium culture diluted (OD600=0.2) was applied. Plants were scored for tumors after one month of growth. Each experiment was repeated three times.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Stanton Gelvin for his kind gifts of LBA1010, LBA1517 and multi-color BiFC vectors, and Dr. Robert Dietrich for his generous gift of the anti-Arabidopsis PR-1 antibody. A.Z. was supported in part by a postdoctoral fellowship from the U.S.-Israel Binational Research and Development Fund (BARD). The work in our laboratory is supported by grants from USDA National Institute of Food and Agriculture, NIH, NSF, BARD, and BSF to VC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alvarez JP, Pekker I, Goldshmidt A, Blum E, Amsellem Z, Eshed Y. Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell. 2006;18:1134–1151. doi: 10.1105/tpc.105.040725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Staskawicz BJ. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell. 2003;112:369–377. doi: 10.1016/s0092-8674(03)00036-9. [DOI] [PubMed] [Google Scholar]
- Bartel P, Chien C, Sternglanz R, Fields S. Elimination of false positives that arise in using the two-hybrid system. Biotechniques. 1993;14:920–924. [PubMed] [Google Scholar]
- Bhattacharjee S, Lee LY, Oltmanns H, Cao H, Veena, Cuperus J, Gelvin SB. IMPa-4, an Arabidopsis importin alpha isoform, is preferentially involved in Agrobacterium-mediated plant transformation. Plant Cell. 2008;20:2661–2680. doi: 10.1105/tpc.108.060467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundock P, den Dulk-Ras A, Beijersbergen A, Hooykaas PJJ. Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J. 1995;14:3206–3214. doi: 10.1002/j.1460-2075.1995.tb07323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung SM, Frankman EL, Tzfira T. A versatile vector system for multiple gene expression in plants. Trends Plant Sci. 2005;10:357–361. doi: 10.1016/j.tplants.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Citovsky V, Kapelnikov A, Oliel S, Zakai N, Rojas MR, Gilbertson RL, Tzfira T, Loyter A. Protein interactions involved in nuclear import of the Agrobacterium VirE2 protein in vivo and in vitro. J Biol Chem. 2004;279:29528–29533. doi: 10.1074/jbc.M403159200. [DOI] [PubMed] [Google Scholar]
- Citovsky V, Kozlovsky SV, Lacroix B, Zaltsman A, Dafny-Yelin M, Vyas S, Tovkach A, Tzfira T. Biological systems of the host cell involved in Agrobacterium infection. Cell Microbiol. 2007;9:9–20. doi: 10.1111/j.1462-5822.2006.00830.x. [DOI] [PubMed] [Google Scholar]
- Citovsky V, Lee LY, Vyas S, Glick E, Chen MH, Vainstein A, Gafni Y, Gelvin SB, Tzfira T. Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J Mol Biol. 2006;362:1120–1131. doi: 10.1016/j.jmb.2006.08.017. [DOI] [PubMed] [Google Scholar]
- Connelly C, Hieter P. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell. 1996;86:275–285. doi: 10.1016/S0092-8674(00)80099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dafny-Yelin M, Tzfira T. Delivery of multiple transgenes to plant cells. Plant Physiol. 2007;145:1118–1128. doi: 10.1104/pp.107.106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Pendón JA, Ding SW. Direct and indirect roles of viral suppressors of RNA silencing in pathogenesis. Annu Rev Phytopathol. 2008;46:303–326. doi: 10.1146/annurev.phyto.46.081407.104746. [DOI] [PubMed] [Google Scholar]
- Ditt RF, Kerr KF, de Figueiredo P, Delrow J, Comai L, Nester EW. The Arabidopsis thaliana transcriptome in response to Agrobacterium tumefaciens. Mol Plant-Microbe Interact. 2006;19:665–681. doi: 10.1094/MPMI-19-0665. [DOI] [PubMed] [Google Scholar]
- Djamei A, Pitzschke A, Nakagami H, Rajh I, Hirt H. Trojan horse strategy in Agrobacterium transformation: abusing MAPK defense signaling. Science. 2007;318:453–456. doi: 10.1126/science.1148110. [DOI] [PubMed] [Google Scholar]
- Durrant WE, Dong X. Systemic acquired resistance. Annu Rev Phytopathol. 2004;42:185–209. doi: 10.1146/annurev.phyto.42.040803.140421. [DOI] [PubMed] [Google Scholar]
- Ellis J, Dodds P. Plant pathology: monitoring a pathogen-targeted host protein. Curr Biol. 2003;13:R400–R402. doi: 10.1016/s0960-9822(03)00321-x. [DOI] [PubMed] [Google Scholar]
- Gagne JM, Downes BP, Shiu SH, Durski AM, Vierstra RD. The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci USA. 2002;99:11519–11524. doi: 10.1073/pnas.162339999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelvin SB. Agrobacterium and plant genes involved in T-DNA transfer and integration. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:223–256. doi: 10.1146/annurev.arplant.51.1.223. [DOI] [PubMed] [Google Scholar]
- Goderis IJ, De Bolle MF, Francois IE, Wouters PF, Broekaert WF, Cammue BP. A set of modular plant transformation vectors allowing flexible insertion of up to six expression units. Plant Mol Biol. 2002;50:17–27. doi: 10.1023/a:1016052416053. [DOI] [PubMed] [Google Scholar]
- Gray WM. Plant defense: a new weapon in the arsenal. Curr Biol. 2002;12:R352–R354. doi: 10.1016/s0960-9822(02)00857-6. [DOI] [PubMed] [Google Scholar]
- Hirooka T, Kado CI. Location of the right boundary of the virulence region on Agrobacterium tumefaciens plasmid pTiC58 and a host specifying gene next to the boundary. J Bacteriol. 1986;168:237–243. doi: 10.1128/jb.168.1.237-243.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirooka T, Rogowsky PM, Kado CI. Characterization of the virE locus of Agrobacterium tumefaciens plasmid pTiC58. J Bacteriol. 1987;169:1529–1536. doi: 10.1128/jb.169.4.1529-1536.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenberg SM, Sternglanz R, Cheng PF, Weintraub H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooykaas PJJ, Hofker M, den Dulk-Ras H, Schilperoort RA. A comparison of virulence determinants in an octopine Ti plasmid, a nopaline Ti plasmid, and an Ri plasmid by complementation analysis of Agrobacterium tumefaciens mutants. Plasmid. 1984;11:195–205. doi: 10.1016/0147-619x(84)90026-x. [DOI] [PubMed] [Google Scholar]
- Kapila J, De Rycke R, Van Montagu M, Angenon G. An Agrobacterium-mediated transient gene expression system for intact leaves. Plant Sci. 1997;122:101–108. [Google Scholar]
- Kim JY, Yuan Z, Jackson D. Developmental regulation and significance of KNOX protein trafficking in Arabidopsis. Development. 2003;130:4351–4362. doi: 10.1242/dev.00618. [DOI] [PubMed] [Google Scholar]
- Lacroix B, Loyter A, Citovsky V. Association of the Agrobacterium T-DNA-protein complex with plant nucleosomes. Proc Natl Acad Sci USA. 2008;105:15429–15434. doi: 10.1073/pnas.0805641105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix B, Tzfira T, Vainstein A, Citovsky V. A case of promiscuity: Agrobacterium’s endless hunt for new partners. Trends Genet. 2006;22:29–37. doi: 10.1016/j.tig.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Lacroix B, Vaidya M, Tzfira T, Citovsky V. The VirE3 protein of Agrobacterium mimics a host cell function required for plant genetic transformation. EMBO J. 2005;24:428–437. doi: 10.1038/sj.emboj.7600524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CW, Efetova M, Engelmann JC, Kramell R, Wasternack C, Ludwig-Muller J, Hedrich R, Deeken R. Agrobacterium tumefaciens promotes tumor induction by modulating pathogen defense in Arabidopsis thaliana. Plant Cell. 2009;21:2948–2962. doi: 10.1105/tpc.108.064576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LY, Fang MJ, Kuang LY, Gelvin SB. Vectors for multi-color bimolecular fluorescence complementation to investigate protein-protein interactions in living plant cells. Plant Methods. 2008;4:24. doi: 10.1186/1746-4811-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy A, Dafny-Yelin M, Tzfira T. Attacking the defenders: plant viruses fight back. Trends Microbiol. 2008;5:194–197. doi: 10.1016/j.tim.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Li J, Krichevsky A, Vaidya M, Tzfira T, Citovsky V. Uncoupling of the functions of the Arabidopsis VIP1 protein in transient and stable plant genetic transformation by Agrobacterium. Proc Natl Acad Sci USA. 2005;102:5733–5738. doi: 10.1073/pnas.0404118102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey D, Belkhadir Y, Alonsos JM, Ecker JR, Dangl JL. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell. 2003;112:743–754. doi: 10.1016/s0092-8674(03)00040-0. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiolog Plant. 1962;15:473–497. [Google Scholar]
- Nagai H, Roy CR. Show me the substrates: modulation of host cell function by type IV secretion systems. Cell Microbiol. 2003;5:373–383. doi: 10.1046/j.1462-5822.2003.00285.x. [DOI] [PubMed] [Google Scholar]
- Nam J, Mysore KS, Zheng C, Knue MK, Matthysse AG, Gelvin SB. Identification of T-DNA tagged Arabidopsis mutants that are resistant to transformation by Agrobacterium. Mol Gen Genet. 1999;261:429–438. doi: 10.1007/s004380050985. [DOI] [PubMed] [Google Scholar]
- Pitzschke A, Djamei A, Teige M, Hirt H. VIP1 response elements mediate mitogen-activated protein kinase 3-induced stress gene expression. Proc Nat Acad Sci USA. 2009;106:18414–18419. doi: 10.1073/pnas.0905599106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porat R, Lu P, O'Neill SD. Arabidopsis SKP1, a homologue of a cell cycle regulator gene, is predominantly expressed in meristematic cells. Planta. 1998;204:345–351. doi: 10.1007/s004250050265. [DOI] [PubMed] [Google Scholar]
- Regensburg-Tuink AJ, Hooykaas PJJ. Transgenic N. glauca plants expressing bacterial virulence gene virF are converted into hosts for nopaline strains of A. tumefaciens. Nature. 1993;363:69–71. doi: 10.1038/363069a0. [DOI] [PubMed] [Google Scholar]
- Schrammeijer B, den Dulk-Ras A, Vergunst AC, Jurado Jácome E, Hooykaas PJJ. Analysis of Vir protein translocation from Agrobacterium tumefaciens using Saccharomyces cerevisiae as a model: evidence for transport of a novel effector protein VirE3. Nucleic Acids Res. 2003;31:860–868. doi: 10.1093/nar/gkg179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrammeijer B, Risseeuw E, Pansegrau W, Regensburg-Tuïnk TJG, Crosby WL, Hooykaas PJJ. Interaction of the virulence protein VirF of Agrobacterium tumefaciens with plant homologs of the yeast Skp1 protein. Curr Biol. 2001;11:258–262. doi: 10.1016/s0960-9822(01)00069-0. [DOI] [PubMed] [Google Scholar]
- SenGupta DJ, Zhang B, Kraemer B, Pochart P, Fields S, Wickens M. A three-hybrid system to detect RNA-protein interactions in vivo. Proc Natl Acad Sci USA. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachel SE, Nester EW. The genetic and transcriptional organization of the vir region of the A6 Ti plasmid of Agrobacterium. EMBO J. 1986;5:1445–1454. doi: 10.1002/j.1460-2075.1986.tb04381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton A, Heller RC, Landry J, Choy JS, Sirko A, Sternglanz R. A novel form of transcriptional silencing by Sum1-1 requires Hst1 and the origin recognition complex. Mol Cell Biol. 2001;21:3514–3522. doi: 10.1128/MCB.21.10.3514-3522.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T, Jensen CS, Wangxia W, Zuker A, Altman A, Vainstein A. Transgenic Populus: a step-by-step protocol for its Agrobacterium-mediated transformation. Plant Mol Biol Rep. 1997;15:219–235. [Google Scholar]
- Tzfira T, Tian GW, Lacroix B, Vyas S, Li J, Leitner-Dagan Y, Krichevsky A, Taylor T, Vainstein A, Citovsky V. pSAT vectors: a modular series of plasmids for fluorescent protein tagging and expression of multiple genes in plants. Plant Mol Biol. 2005;57:503–516. doi: 10.1007/s11103-005-0340-5. [DOI] [PubMed] [Google Scholar]
- Tzfira T, Vaidya M, Citovsky V. VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO J. 2001;20:3596–3607. doi: 10.1093/emboj/20.13.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T, Vaidya M, Citovsky V. Increasing plant susceptibility to Agrobacterium infection by overexpression of the Arabidopsis VIP1 gene. Proc Natl Acad Sci USA. 2002;99:10435–10440. doi: 10.1073/pnas.162304099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T, Vaidya M, Citovsky V. Involvement of targeted proteolysis in plant genetic transformation by Agrobacterium. Nature. 2004;431:87–92. doi: 10.1038/nature02857. [DOI] [PubMed] [Google Scholar]
- Winans SC, Allenza P, Stachel SE, McBride KE, Nester EW. Characterization of the virE operon of the Agrobacterium Ti plasmid pTiA6. Nucleic Acids Res. 1987;15:825–837. doi: 10.1093/nar/15.2.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wroblewski T, Tomczak A, Michelmore R. Optimization of Agrobacterium-mediated transient assays of gene expression in lettuce, tomato and Arabidopsis. Plant Biotech J. 2005;3:259–273. doi: 10.1111/j.1467-7652.2005.00123.x. [DOI] [PubMed] [Google Scholar]
- Xiang C, Han P, Lutziger I, Wang K, Oliver DJ. A mini binary vector series for plant transformation. Plant Mol Biol. 1999;40:711–717. doi: 10.1023/a:1006201910593. [DOI] [PubMed] [Google Scholar]
- Yi H, Mysore KS, Gelvin SB. Expression of the Arabidopsis histone H2A-1 gene correlates with susceptibility to Agrobacterium transformation. Plant J. 2002;32:285–298. doi: 10.1046/j.1365-313x.2002.01425.x. [DOI] [PubMed] [Google Scholar]
- Yuan ZC, Edlind MP, Liu P, Saenkham P, Banta LM, Wise AA, Ronzone E, Binns AN, Kerr K, Nester EW. The plant signal salicylic acid shuts down expression of the vir regulon and activates quormone-quenching genes in Agrobacterium. Proc Natl Acad Sci USA. 2007;104:11790–11795. doi: 10.1073/pnas.0704866104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaltsman A, Feder A, Adam Z. Developmental and light effects on the accumulation of FtsH protease in Arabidopsis chloroplasts - implications for thylakoid formation and photosystem II maintenance. Plant J. 2005;42:609–617. doi: 10.1111/j.1365-313X.2005.02401.x. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Nam J, Humara JM, Mysore KS, Lee LY, Cao H, Valentine L, Li J, Kaiser A, Kopecky A, et al. Identification of Arabidopsis rat mutants. Plant Physiol. 2003;132:494–505. doi: 10.1104/pp.103.020420. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.