Abstract
We briefly review the history of microRNA (miRNA) research and some of the lessons learnt. To provide some insights as to how and why miRNAs came into existence, we consider the evolution of the RNA interference machinery, miRNA genes, and their targets. We highlight the importance of systems biology approaches to integrate miRNAs as an essential subnetwork for modulating gene expression programs. Building accurate computational models that can simulate highly complex cell-specific gene expression patterns in mammals will lead to a better understanding of miRNAs and their targets in physiological and pathological situations. The impact of miRNAs on medicine, either as potential disease predisposing factors, biomarkers or therapeutics, is highly anticipated and has started to reveal itself.
Keywords: Gene expression, regulatory networks, comparative genomics, untranslated regulatory RNA, RNA interference
In eukaryotes, miRNA genes encode a class of small regulatory RNAs, 21–24 nucleotides (nt) in length, that are not translated into protein in order to function, but instead physically interact with complementary RNA sequences and Argonaute (Ago) protein complexes to mediate sequence-specific gene silencing [9]. Due to their small size and inability to code for protein, miRNAs were overlooked until recently. Nowadays, they are widely recognized as important regulators of gene expression programs and consequently a diverse array of cellular processes such as signal transduction, cell cycle, apoptosis, pluripotency, differentiation, and transformation [31, 47, 52, 59, 64, 79, 80, 107]. Thus defects in miRNA-mediated regulation could lead to human disease. To date, there are reportedly about 700 miRNAs encoded in the human genome that in turn potentially target 30–60% of the protein-coding genes [29, 32]. Accordingly, they deserve our attention as they are a major class of emerging players in broader RNA-based gene silencing processes in eukaryotes.
Studies in worms led to the discovery of the first microRNAs
Mutations in a collection of genes in the nematode worm C. elegans result in changes to the cell lineages that are genetically programmed to divide and differentiate, in a characteristic fashion, to ultimately form an adult, wildtype worm. Initially reported in 1981, one such mutant was designated lin-4 [17, 98]. Then, in 1993, Lee, Feinbaum and Ambros proposed that the lin-4 gene did not encode a protein but instead a small RNA [59]. In 2001, many more examples of such small RNAs were cloned and sequenced, and the term microRNA (miRNA) was coined to classify this distinctive class of small untranslated RNAs [3, 54, 57, 58]. We now appreciate lin-4 as the founding member of the miRNA genes.
The lin-4 gene is essential for the normal temporal control of diverse postembryonic developmental events in C. elegans. The lin-4 genetic mutation was mapped to a 693 base pair region of DNA within intron 9 of another gene, F59G1.4 [59] (Figure 1). The function of F59G1.4 is not known but is not responsible for the lin-4 mutant phenotype. Interestingly, the identified intronic genomic region is conserved not only in sequence but also in function between C. elegans, C. briggsae, C. remanei and C. vulgaris. It is unlikely that the lin-4 gene encodes a protein, since no conserved open reading frame could be identified in this genomic region. However, RNAse protection experiments revealed that two small RNAs are produced from the lin-4 gene. The approximately 61 and 22 nt long RNA species are now appreciated as the Drosha-generated precursor (pre-miRNA) in the form of a short hairpin RNA, and the Dicer-processed mature miRNA, respectively (Figure 1). The lin-4 (ma161) loss-of-function allele was particularly informative, since it revealed a C-to-A nucleotide point mutation at the evolutionarily conserved 5′ end of the lin-4 miRNA. Later studies revealed that the 5′ end of miRNAs constitute the “seed” or “nucleus” sequence that is generally required for recognition of their cognate targets [15, 53, 61, 62]. Indeed, nucleotide substitutions that disrupt seed sequence pairing diminish miRNA-mediated repression [15, 25, 51, 55].
Figure 1. A schematic of lin-4 gene, pre-miRNA, and miRNA structure.
The lin-4 miRNA was mapped to a 693 bp region (depicted by a bar) within the ninth intron of F59G1.4 (not drawn to scale) [59]. Processing of the primary lin-4 transcript (pri-miRNA) by Drosha-DGCR8 liberates the pre-miRNA. Dicer processes the pre-miRNA further into a 21 nt RNA duplex. The two miRNA strands are separated by an unknown mechanism, and typically one strand is loaded into Ago to form an active RISC, whereas the other strand (passenger strand) is excluded from RISC.
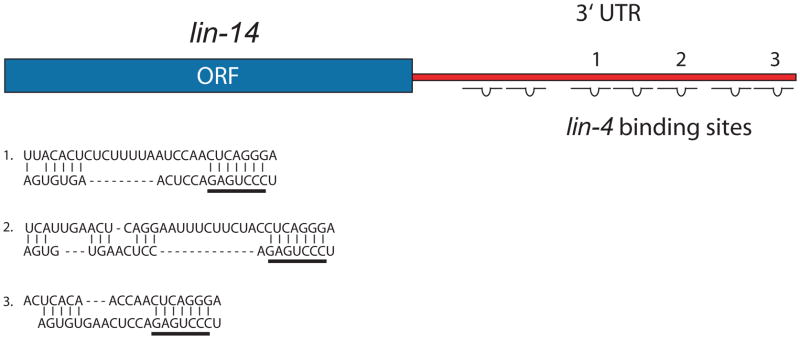
Interestingly, recessive loss-of-function lin-4 mutants displayed a similar phenotype to dominant gain-of-function lin-14 mutants [59, 109]. At the molecular level, lin-14 protein levels were increased in the aforementioned lin-4 and lin-14 mutant worms. This implied that lin-4 negatively regulated in trans the synthesis of lin-14 protein via cis element(s) linked to lin-14. The novel mechanism was proposed to be based on sense-antisense RNA hybridization, since the lin-14 3′ untranslated region (UTR) happened to contain seven sites partially complementary to lin-4 miRNA (Figure 2). Furthermore, when a region of the lin-14 3′ UTR that contained five out of the seven putative binding sites was deleted, a less severe phenotype was observed compared to a deletion that removed all seven sites [109, 110].
Figure 2. Sense-antisense binding of lin-4 miRNA to 3′ UTR of lin-14.
Schematic depicts the seven putative lin-4 binding sites in the 3′ UTR of lin-14. Three out of the seven sites are complementary to the seed sequence of lin-4, and the predicted base-pairings are depicted for illustration [56, 61, 109]. The sense (top) strand represents the 3′ UTR sequence, and the anti-sense (bottom) strand represents the mature lin-4 sequence. The seed region is underlined by a bar in each case.
Thus, there was a negative correlation between lin-4 miRNA expression and lin-14 protein levels. It appeared that lin-4 acted post-transcriptionally, since lin-14 mRNA levels were unaffected by lin-4. Therefore, it was proposed that lin-4 repressed translation of lin-14 by a novel mechanism that depended on lin-4 base pairing to complementary sites in the 3′ UTR of the lin-14 message. Furthermore, this inhibition seemed to occur at a stage following translation initiation, since lin-14 mRNA could associate with polyribosomes [82]. Subsequently, lin-4 was found to also directly repress lin-28 (an inhibitor of let-7 miRNA processing) in hypodermis; and lin-57/hbl-1 (a Zn-finger protein homologous to fly hunchback and mammalian TRIM71) in ventral nerve cord [67, 78]. It is remarkable how worm genetics can facilitate the demonstration of the functionality of miRNA-target interactions. This is currently a challenge in mammalian systems. However, in one success story, Abelson and colleagues discovered a rare single nucleotide polymorphism (SNP) in the 3′ UTR of the gene encoding Slit and Trk-like 1 (SLITRK1) that appears to interfere with direct regulation by miR-189 in Tourette’s syndrome patients [1]. Recent systematic studies suggest that SNPs in predicted miRNA-binding sites may have functional consequences [18, 93].
In 2000, a second miRNA, let-7, was reported by Gary Ruvkun and colleagues [84]. Let-7 was shown to repress lin-41 via two partially complementary sites in the 3′ UTR [89, 96]. It is expressed during L2 and L3 larval stages of C. elegans development and is required for the developmental transition between the late larval and adult stages. The let-7 (n2853) loss-of-function allele revealed a G-to-A nucleotide point mutation at the evolutionarily conserved seed region of the let-7 miRNA. Pasquinelli et al observed that humans encoded multiple copies of the let-7 gene in their genome and let-7 is widely conserved across animals [84]. This finding suggested that more miRNAs may be found in C. elegans, and that miRNAs are not an oddity of the worm, thus opening the field of miRNA biology. Indeed, in 2001, more miRNAs were identified from C. elegans, Drosophila, and humans [54, 57, 58].
Today, miRNAs are recognized as functional bona fide genetic elements present in many species from alga, sponges, plants and animals – and even some viruses [32]. In recognition of their significance in biology and medicine, the 2008 Lasker Basic Medical Research Award was given to Victor Ambros, Gary Ruvkun and David Baulcombe for their laboratories’ studies that gave birth to the field of miRNA research [2, 11, 91]. In 2006, the Nobel Prize in Physiology or Medicine was awarded to Andy Fire and Craig Mello for the discovery of RNA interference (RNAi) by double-stranded RNA (dsRNA), a pathway that, in part, mediates miRNA biogenesis and function [27, 73].
RNAi machinery mediates miRNA biogenesis and function
The common link between these award-winning discoveries is that small dsRNAs, be it short interfering RNAs (siRNAs) derived from long dsRNAs or miRNAs derived from stem-loop precursors, are generated by an endonuclease of the RNaseIII superfamily called Dicer [14, 34, 41, 49]. Dicer cleavage (“dicing”) results in the production of short dsRNA species, approximately 20–25 nt long with characteristic 2 nt 3′ overhangs. These Dicer products are then bound by Ago protein(s) and are capable of triggering sequence-specific gene silencing in different ways [26, 37].
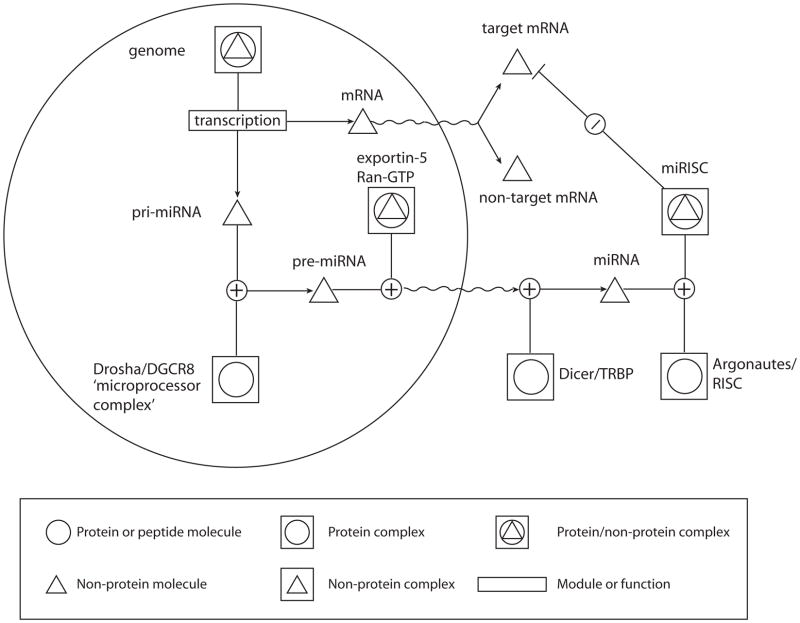
This section will focus on miRNAs, since they are currently the best understood of the mammalian Dicer products. miRNAs are encoded in our genome, sometimes as polycistronic clusters, and are evolutionarily conserved along with their putative binding sites primarily found in 3′ UTR of mRNAs [9, 50]. They are even encoded by herpesviruses such as Epstein-Barr virus and Kaposi sarcoma-associated herpesvirus [85]. In 2008, nearly 700 human miRNAs have been identified and catalogued in miRBase 12.0 [32]. Generally, miRNA genes are initially expressed from the genome as long primary miRNA (pri-miRNA) transcripts generated by RNA polymerase (pol) II. This process is regulated by transcription factors that bind the promoter and surrounding regulatory cis-elements, similar to the transcription of any other gene [50] (Figure 3). Thus, the transcription pattern of miRNA genes may be ubiquitous or highly specific. While in the nucleus, pri-miRNA transcripts are processed by the microprocessor complex (consisting of Drosha, another RNAseIII-like enzyme, and its partner DGCR8, DiGeorge Syndrome critical region gene 8) in order to excise the 60–70 nt-long short-hairpin precursor miRNA (pre-miRNA) [38]. This step may be subject to regulation. For example, Lin-28 protein was recently implicated in preventing let-7 pri-miRNA from being processed potentially by binding the loop region [108]. The Drosha-excised pre-miRNAs are exported into the cytoplasm by exportin-5, which requires the Ran-GTP gradient across the nuclear membrane [69, 114]. Once in the cytoplasm, pre-miRNAs are processed into their functional mature form by Dicer and its partner TRBP, TAR RNA-binding protein. This step may also be regulated. Again, Lin-28 protein can also inhibit the Dicer processing step by facilitating uridylation of pre-let-7 [40]. The double-stranded mature miRNA may either be unwound by a yet unidentified helicase or if the strands are highly complementary then one strand could be cleaved by the “Slicer” activity of Ago2 [72]. Ultimately, the single-stranded mature miRNA is bound by an Argonaute protein and forms the core of the multi-component RNA-induced silencing complex (RISC). In humans, there are four genes encoding Argonaute proteins (Ago1–4). The miRNA bound by each RISC guides the complex to complementary RNA sequences in the cell and mediates sequence-specific gene silencing.
Figure 3. The miRNA silencing pathway.
Transcription and pri-miRNA processing in the nucleus (large circle) are depicted, as well as miRISC assembly and silencing of target mRNAs in the cytoplasm. An interaction that results in activation is denoted by a positive symbol (+); whereas a negative symbol (−) denotes inhibition. Bottom, a legend of what the symbols represent are provided (inset box). A detailed description of the pathway can be found in the text.
Modus operandi of miRNA-mediated gene silencing
Generally, miRNAs are predicted to bind to sites in 3′ UTRs of mRNAs [61, 112]. However, there is some evidence that functional miRNA-binding sites exist in the protein-coding region of mRNAs as well [28, 61, 100]. It has been proposed that these sites tend to mediate repression to a smaller magnitude, perhaps because translating ribosomes may compete and eject RISC from their path [9, 10]. However, once translation is inhibited by miRNAs, it is conceivable that RISC can bind any accessible sites along a target mRNA without being bumped off by ribosomes.
Crystallographic studies of distantly related proteins from bacteria suggested that the PIWI domain of Ago proteins harbors an RNaseH-fold domain that can catalyze the “Slicer” activity [97, 116]. Indeed, it was shown that Ago2 possesses the catalytic triad (DDH motif) to carry out RNA-programmed cleavage of perfectly complementary RNA targets. Thus far, Ago1, Ago3 and Ago4 have no demonstrable “Slicer” activity, eventhough Ago3 harbors the conserved DDH motif. Thus, Ago2-containing RISC may cleave its target if it is programmed with a small RNA that is highly complementary in sequence. “Slicing” is a common mechanism for miRNA-mediated gene silencing in plants, but seems to be rarely invoked by mammalian miRNAs. Only two examples have been reported in mammals: miR-196 cleaving HOXB8 mRNA; and miR-127 and miR-136 cleaving retrotransposon-like 1 (rtl1) mRNA, a Sushi-like retrotransposon also known as paternally-expressed gene 11 (peg11) [22, 113]. In contrast, when miRNA-programmed RISCs (miRISC) anneal by imperfect base-pairing to a target, they negatively regulate translation. In parallel or in addition, miRISCs may also enhance de-adenylation and subsequent mRNA decay of their targets by recruiting the decapping and deadenylation machinery [12, 31, 111]. Furthermore, under certain conditions (e.g. stress) miRISCs become concentrated in cytoplasmic P bodies or stress granules [26]. However, it is not clear whether this is a cause or consequence of miRNA-induced mRNA decay. Thus, it was generally believed that miRNAs directly repressed gene expression by a number of mechanisms, but recent reports suggest that miRNAs may also play positive roles in gene expression. For example, it was shown that miRNAs can activate gene expression (e.g. a TNFα gene reporter construct) under certain circumstances, and miR-122 directly facilitates replication of hepatitis C virus RNA in liver cells [46, 104–106]. The molecular mechanisms and generality behind these novel phenomena need to be investigated further.
Evolutionary history of the protein apparatus involved in small untranslated RNA metabolism
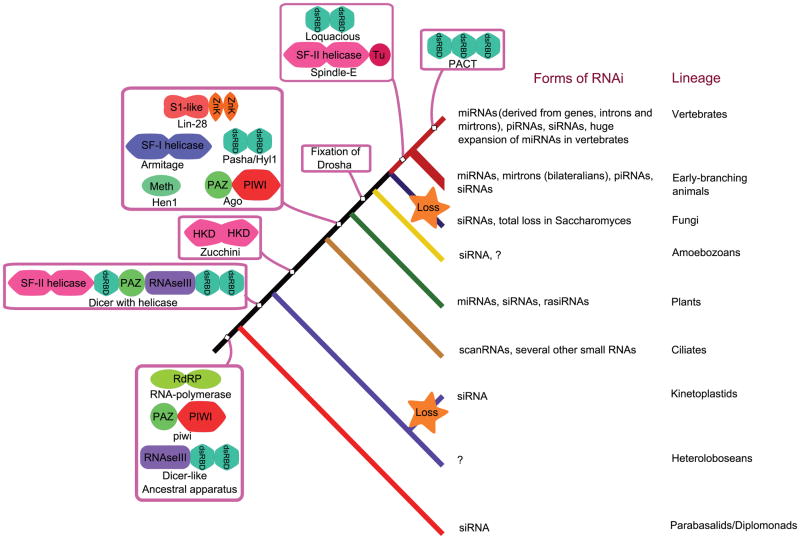
The core molecular apparatus involved in RNA-based gene silencing can be traced back to the last eukaryotic common ancestor [16, 43]. Based on the currently available data, the ancestral apparatus can be reconstructed as primarily comprising of 1) a processing RNAse (Dicer), which generates small RNAs from a longer precursor through endonucleolytic cleavage. The ancestral Dicer family of RNAses contained tandem catalytic domains of the RNAseIII superfamily linked to additional RNA-binding domains. 2) An effector nuclease which uses the small RNAs generated by the former enzyme as guides to target transcripts and other RNAs. This nuclease belongs to the Piwi-Argonaute family of the RNAseH superfamily. 3) Additionally, in several eukaryotes the siRNAs generated from transcripts might be amplified to generate further copies by means of an RNA-dependent RNA polymerase (RdRp). However, it is exclusive to the siRNA branch of the RNAi system and mammals appear to be missing RdRp orthologs. All these enzymes, comprising the core of the ancestral eukaryotic silencing system, have bacterial and archaeal antecedents [4, 42]. However, they appear to have come together as single coherent functional system only in the eukaryotes (Figure 4).
Figure 4. evolution of RNA silencing machinery.
The evolutionary scenario for proteins involved in RNA-dependent gene silencing mechanisms is superimposed on the major developments in terms of small RNAs. Details on the architectures of proteins involved in the process can be found in the text. This scenario is based on the current state of comparative genomics and may require revisions as new eukaryotic species are sequenced.
Within eukaryotes, the system has considerably diversified both in terms of the domain architecture and the recruitment of additional components [4, 16, 95]. For example, Dicers from early-branching eukaryotic lineages such as Trichomonas and Giardia contain two RNAseIII catalytic domains linked to an N-terminal PAZ domain and a divergent C-terminal dsRNA-binding domain (dsRBDs). Further duplications and domain accretions to this core appear to have happened by the time of the divergence of the heterolobosean lineage (the amoeboflagellate Naegleria), resulting in versions of Dicer with fusions to two more N-terminal modules, namely the superfamily-II helicase and an additional dsRBD [16, 43] (Figure 4). Since that point on, most eukaryotic lineages possessed two versions of the Dicer family, one with an associated helicase domain and another without. In animals and amoebozoans, a representative of the latter type, with two RNAseIII domains and a C-terminal dsRBD, seems to have been fixed as a conserved orthologous lineage of proteins prototyped by Drosha. The Piwi-Argonaute family similarly appears to have differentiated extensively in eukaryotes. The Piwi sub-group probably represents the ancestral versions of these proteins in eukaryotes, which in the crown group (animals, fungi, amoebozoans and plants) spawned a new sub-group, the Argonaute (Ago) proteins. The Ago sub-group underwent further lineage-specific expansions independently in the plants and animals [16]. In some lineages the RNAseH fold nuclease domain of Ago underwent apparent inactivation -- three of the four mammalian Ago proteins have lost catalytic activity.
Beyond these core proteins, there are additional dsRBD proteins which function as co-factors. Of these, Pasha (DiGeorge syndrome critical region gene 8; DGCR8 in mammals), a co-factor for Drosha in animals, has orthologs in plants (DRB2/Hyl1-like proteins) and Dictyostelium. Thus, such a dsRNA-binding protein is likely to have been already present prior to radiation of the eukaryotic crown group (Figure 4). In contrast, loquacious (TRBP or TARBP2 and PACT or PRKRA in mammals), the co-factor for the animal Dicer proteins with helicase domains (Dcr-1), appears to be an animal-specific innovation with no currently known orthologs in other organisms. Furthermore, the divergence of TRBP and PACT apparently occurred only in the lineage leading to vertebrates. The regulatory RNA-binding factor Lin-28, with S1-like and Zinc-knuckle RNA-binding domains, is conserved in plants and animals suggesting an origin prior to the radiation of the crown group. As mentioned previously, Lin-28 has been shown in animals to be a repressor of let-7 miRNA processing by binding its stem-loop and promoting 3′ terminal oligouridylation [40]. In plants, uridylated small RNAs appear to be actively degraded and this may also be the case in animals [63].
In addition to these proteins, studies on Piwi-interacting RNA (piRNA) maturation in Drosophila have uncovered RNA helicases (Spindle-E and Armitage), a nuclease with an HKD catalytic domain (Zucchini), and an inactive nuclease and HMG domain-containing protein (Maelstrom) [50, 83, 117]. Another protein Squash, which was implicated in this process was originally claimed to be a nuclease [83], but our analysis found no evidence for it being related to any known nuclease domain. The spindle-E type superfamily-II helicases contain a C-terminal tudor domain and are thus far restricted to animals in their distribution, although related RNA-helicases are more widely distributed across eukaryotes. The armitage family group of helicases belong to superfamily-I and appear to have emerged prior to the divergence of animals, fungi and plants [4]. The Zucchini group of nucleases is conserved in animals, ciliates and early-diverging plant lineages like chlorophyte algae (Figure 4); however, they have not been directly implicated in RNA silencing outside of Drosophila. Like Zucchini, the Maelstrom proteins have a sporadic phyletic distribution history being present in animals, Entamoeba, and trypanosomes. In the latter two taxa the nuclease domain of Maelstrom appears to be catalytically inactive unlike the version in animals [117]. MiRNAs of plants and piRNAs of animals are methylated at the 2′-OH position of the 3′ end sugar, by a RNA methylase of the Hen1 family [63, 92]. This enzyme appears to have emerged just prior to the divergence of the plants along with other components such as the Ago family, Armitage and the Pasha-like proteins [4] (Figure 4).
The origin of miRNAs and their relation with the evolution of the protein apparatus
Early-branching eukaryotes like Giardia and Trichomonas appear to only have the system for siRNA processing, which has been shown in the former organism to function as a regulatory mechanism in antigenic variation [88]. While very little is known about the function of other small RNAs in eukaryotes outside of the crown group (Figure 4), studies on ciliates show that small RNAs known as scanRNAs are required for the targeting of internal eliminated sequences during the generation of a new macronucleus by elimination of micronuclear sequences [70, 76]. In Tetrahymena ~28 nucleotide scanRNAs are produced via the dicing action of a Dicer-like nuclease Dcl1p and associates with a Piwi-like protein Twi1p. Other distinct, shorter ~24nt RNAs have also been reported in ciliates and might be transcribed from distinct non-protein coding genes or introns [60]. Such small RNAs are conceivably comparable to the antecedents of the classical piRNAs and miRNAs of the crown group eukaryotes.
The advent of the crown group was characterized by the emergence of several distinctive RNAs, namely the miRNAs, the longer piRNAs in animals, and the functionally comparable shorter rasiRNA in plants [50]. However, among the crown group lineages this diversity is only observed in plants and animals; currently characterized fungi, which are more closely related to animals than to plants, appear to have only conventional siRNA as seen in earlier branching eukaryotes [16]. Furthermore, plant miRNAs differ from animal miRNAs in showing methylation of the 3′ end sugar and in being derived from longer stem-loop precursors processed by a single Dicer protein solely in the nucleus. In contrast, animal miRNAs are processed in two stages by two distinct nucleases: Drosha in the nucleus (pri-miRNA processing) and Dcr-1 in the cytoplasm (pre-miRNA processing). They also have distinct modes of action; the majority of plant miRNAs target mRNAs for Ago-mediated slicing, whereas only a small subset of animal miRNAs direct Ago-mediated slicing of their cognate target mRNAs [45]. This raises the question as to whether miRNAs in plants and animals are independent innovations.
However, it should be noted that despite the differences, several components of the protein apparatus involved in processing and action of miRNAs/piRNAs appear to have emerged coevally in the crown group and specifically lost in lineages like fungi that lack miRNAs (Figure 4). These include the Ago subfamily, the dsRBD cofactors, like Pasha and DRB2/Hyl1, different RNA helicases, Hen1 RNA methylase, and Lin-28, the negative regulator of let-7 biogenesis. This might imply that there was a distinct miRNA-system in the common ancestor of animals and plants, but in each lineage miRNAs have subsequently undergone a drastic diversification, repeatedly displacing ancestral versions. Consistent with this proposal, none of the miRNAs of early-branching plant lineages like the algae have counterparts in multicellular plants [77, 119]. Likewise, almost all bilateralian animal-specific miRNAs appear to be innovations with no equivalents in early-diverging animal groups like cnidarians and sponges [33]. A close sister group of the vertebrates, the urochordate Oikopleura, and the early-branching animal Trichoplax show dramatic reduction or complete loss of miRNAs [30, 33]. Thus, the miRNA system could be easily attenuated and possibly entirely lost, as was perhaps the case with Saccharomyces in the fungi.
Evolution of animal miRNAs: explosive radiation in vertebrates
Identification of about eight miRNAs in the sponge shows that the common ancestor of all multicellular animals (metazoans) possessed at least a minimal miRNA complement [33]. This miRNA repertoire vastly expanded over metazoan evolution. Cnidarians, the first metazoans with a nervous system, possess a complement of around 40 miRNAs and about 30 miRNA families have been reconstructed as being present in the last common ancestor of bilateralian animals [30, 33, 101]. However, barring the highly conserved miR-100, none of the conserved bilateralian families have representatives amongst the cnidarian miRNAs, nor are any of the bilateralian and cnidarians miRNAs discernible orthologs of their counterparts in sponges. This suggests that there have been cycles of miRNA loss and lineage-specific innovation and expansion in the course of the animal radiation. Thus, the dynamics of miRNA evolution appears to resemble that of transcription factors in the course of eukaryotic diversification [43]. Indeed a number of studies have made the case that miRNAs, like transcription factors, might be incorporated into conserved molecular regulatory networks that were central to the emergence of animal-specific structures. For example, the conserved bilateralian miR-7 and the homeodomain protein Rx (Rx3 in vertebrates) appear to cooperate in a regulatory network to specify the development of a group of extraoccular photoreceptor and chemoreceptor neurons in both invertebrates (e.g. the annelid worm Platynereis) and vertebrates (e.g. Zebrafish) [102]. These neurons appear to define the core of the hypothalamus and its homologous structures, which might have been important in regulating reproductive behavior in response to light across bilateralian animals [102]. Thus, the origin of a regulatory network including both a conventional transcription factor Rx and a miRNA might have been critical for the emergence of a key brain-structure, the hypothalamus. Interestingly, miRNAs target transcription factors in both plants and animals and this interplay might represent an ancient regulatory theme even if both of the actual players, miRNAs and transcription factors, have been rapidly turning over in the course of evolution [6, 19, 90].
Just as in the case of transcription factors, miRNAs exhibit a dramatic expansion in vertebrates – about 500–700 miRNAs have been confidently identified in mice and humans, which is almost 3–5 times the number identified in insects and nematodes. Of these, at least 41 orthologous lineages of miRNAs appear to have newly emerged in the last common ancestor of all vertebrates and probably an additional 30–40 in the ancestor of all mammals [39]. Given studies suggesting a role for miRNAs in specification of specialized structures in animals (e.g. see above), it has been postulated that the great radiation of miRNAs in vertebrates has been central to the evolution of morphological complexity in terms of cell and tissue types in vertebrates [39]. Some researchers have even correlated the complexity of the nervous system in terms of number of neurons with miRNA diversity [101]. Several circumstantial pieces of evidence support the general thrust of these arguments. Firstly, the urochordate Oikopleura, a vertebrate relative with a degenerate morphology indeed shows a marked reduction in miRNAs [30]. Secondly, several thousands of genes are predicted to be regulated by miRNAs and increasing the number of miRNA genes provides for an increased possibility of combinatorial regulation [19, 61]. Thirdly, whereas several developmental and differentiation regulators are conserved throughout animal evolution, miRNAs show much greater lineage-specific specialization, suggesting a role in morphological diversity of animals, and vertebrates in particular. Computational studies also suggest that miRNA-target relationships are rapidly diversifying. In a three-way comparison between nematodes, insects and vertebrates, only five miRNA-target relationships are preserved across all lineages, though over 250 were observed to be shared by at least a pair of lineages [20]. This observation was interpreted as an extensive ongoing rewiring of the miRNA-target interactions, with only a very few being conserved across all organisms. Thus, during evolution, even with conserved miRNAs, there is a turnover of genes that attach themselves to the miRNA as a target. Here too, evolution of miRNA-target interactions closely resembles the rewiring of interactions between transcription factors and their target genes [7].
Despite these observations, the large number of unique miRNAs detected in mammals along with the poor conservation of miRNA-target interactions [19] does raise the issue if they are indeed all functionally important [5]. In this context it should be noted that some miRNAs are essentially intronic sequences. Interestingly, a subset of intronic miRNAs, termed “mirtrons”, bypass the conventional Drosha system of processing and are instead processed via the spliceosome as they comprise a complete small intron [13, 81]. Hence, in a sense these miRNAs are no different from regular non-coding introns other than having a peculiar structure. Furthermore, several mammalian miRNAs may have been recently derived from “selfish DNA” or transposons. For example, recent studies have shown that over 50 human miRNAs are likely to be derived from mobile elements or their fragments including the LINEs, SINEs and the miniature inverted-repeat transposable elements [86, 87]. While a regulatory role via mRNA degradation has been proposed for some of these transposon-derived miRNAs [87], it is not at all clear if all of them indeed have functional relevance. Indeed, the enormous expansion of transposable elements in various vertebrate lineages would then in part account for the proliferation of miRNAs in them. Emergence of a large number of new miRNAs from mobile elements might also explain the lineage-specific nature and lack of conservation of several miRNAs from mammals in particular. In light of this, the actual functional relevance of miRNAs in the emergence of vertebrate complexity needs to be treated with circumspection. In particular, the regulatory significance of the miRNAs needs to be carefully evaluated in comparison with conventional regulators like transcription factors that have also expanded, and chromatin proteins that have increased in architectural complexity during the provenance of vertebrates.
Towards predictive models of miRNA-regulated networks
Currently, numerous laboratories around the world are generating inventory lists of the molecular components (DNA, RNA, protein, lipids, metabolites, etc.) that make up their favorite biological systems. The current challenge for systems biologists is to determine how all these parts relate to each other, the emergent function of these inter-connected parts, and the dynamic behavior of the system in the context of the cell, tissues, organism and ecosystem. However, to date we have an incomplete, albeit extensive, parts list. For example, despite marked advances in proteomic technologies, a cell’s expressed proteome cannot be comprehensively determined at this time. Nevertheless, systems biology aims to start putting the pieces together for this immense jigsaw puzzle and models of biomolecular networks will be refined over time as better technologies and data sets become available.
As discussed, it was only recognized recently that a portion of our genome is expressed for the purpose of generating functional small RNAs that are not translated into protein, but play important roles in regulating gene expression. Moreover, deep sequencing technologies are allowing us to comprehensively discover the identities and expression levels of these small RNAs in any given cell type. Already, miRNA expression profiles appear to be useful in classifying cancers [68]. In addition, miRNAs have been found to participate in negative feedback loops [44, 64, 99], and systematic analyses of miRNA-containing circuits have begun [21, 65, 103, 115]. However, the role of miRNAs in the biology of a cell or organism remains unclear. Multiple miRNA mutants in C. elegans display no obvious phenotypes [75]. This could be due to significant redundancy of individual miRNAs or miRNA ‘families’ or the possibility that most miRNAs act to fine-tune expression of hundreds of genes. On the other hand, there are examples of miRNA mutants such as lin-4 and let-7 that display dramatic developmental phenotypes due to dysregulation of a few known key targets. In addition, the severity of knock-outs of the RNAi machinery, like Dicer, Drosha and Argonaute2 suggest that miRNAs as a system play pivotal roles in cellular differentiation. A key to understanding what miRNAs are doing will require systematic identification of the targets that they regulate. Currently, we rely on prediction algorithms that identify evolutionarily conserved miRNA seed matches to annotated 3′ UTRs [e.g. 53, 62]. However, annotation of 3′ UTRs are currently imperfect, and seed matches are not always sufficient to predict bona fide miRNA target sites [23, 24]. Furthermore, it is generally not yet possible to predict in which cell types or tissues, a miRNA-target pair might function in vivo [48, 74]. Thus, experimental strategies are needed to exhaustively map direct relationships between the small RNAs and the transcriptome [31, 36, 52, 66, 118], as well as the proteome [8, 35, 94].
Already, investigators have begun to model how miRNAs are wired into biomolecular networks [21, 65, 103, 115]. However, these modeling efforts must be interpreted with caution since they are largely unvalidated experimentally and based on predictions instead of large-scale experimental data. Recently, Martinez et al provided an example of one type of data sets required to build miRNA regulatory networks [71]. They experimentally determined for about 71 miRNA promoters (representing approximately 66% of C. elegans miRNAs) in the C. elegans genome which transcription factors can bind these putative promoter fragments, using a high throughput yeast one-hybrid screening system. As a result, they reported 347 high-confidence interactions between 116 proteins and 63 miRNA gene promoters. In the absence of experimental data, they used computationally predicted miRNA targets to model the circuitry; however, the resulting model awaits validation.
Since miRNAs regulate diverse aspects of cellular function (cell cycle, apoptosis, signal transduction, transcription), defects in such a pathway could lead to discernible phenotypes in mammals. It is safe to say that current models of regulatory gene networks are far from complete. Integration of the miRNA regulatory subnetwork will bring us one step closer to a more precise model. The resulting regulatory network maps will hopefully contribute towards a predictive model that aims to translate a cell’s genotype into its phenotype. Such models may be useful for computerized simulations that can depict the global changes that occur from healthy to disease states.
Conclusion
In summary, miRNAs can play important roles in determining cell fates by regulating the expression levels of oncoproteins, tumor suppressors, cell cycle and death regulators, signaling and transcription factors. They might also represent a mechanism by which certain viruses modulate host functions. Thus, there is a need for systematic discovery of regulatory networks involving miRNAs. However, this effort is currently limited by the availability of dependable high-throughput methods to identify direct targets of miRNAs that are functional in vivo. Furthermore, given the large number of predicted miRNA-mRNA target pairs, more efficient strategies to confirm the functionality of these interactions are needed. As more miRNA-target pairs become implicated in human diseases, therapeutic strategies can be rationally designed to restore this relationship to normal either by supplying synthetic miRNAs or antagonizing miRNA activity as appropriate.
References
- 1.Abelson JF, Kwan KY, O’Roak BJ, Baek DY, Stillman AA, Morgan TM, Mathews CA, Pauls DL, Rasin MR, Gunel M, Davis NR, Ercan-Sencicek AG, Guez DH, Spertus JA, Leckman JF, Dure LSt, Kurlan R, Singer HS, Gilbert DL, Farhi A, Louvi A, Lifton RP, Sestan N, State MW. Sequence variants in SLITRK1 are associated with Tourette’s syndrome. Science. 2005 Oct 14;310(5746):317–20. doi: 10.1126/science.1116502. 310/5746/317 [pii] [DOI] [PubMed] [Google Scholar]
- 2.Ambros V. The evolution of our thinking about microRNAs. Nat Med. 2008 Oct;14(10):1036–40. doi: 10.1038/nm1008-1036. nm1008-1036 [pii] [DOI] [PubMed] [Google Scholar]
- 3.Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, Matzke M, Ruvkun G, Tuschl T. A uniform system for microRNA annotation. RNA. 2003 Mar;9(3):277–9. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anantharaman V, Koonin EV, Aravind L. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 2002 Apr 1;30(7):1427–64. doi: 10.1093/nar/30.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Axtell MJ. Evolution of microRNAs and their targets: are all microRNAs biologically relevant? Biochim Biophys Acta. 2008 Nov;1779(11):725–34. doi: 10.1016/j.bbagrm.2008.02.007. S1874-9399(08)00047-3 [pii] [DOI] [PubMed] [Google Scholar]
- 6.Axtell MJ, Bowman JL. Evolution of plant microRNAs and their targets. Trends Plant Sci. 2008 Jul;13(7):343–9. doi: 10.1016/j.tplants.2008.03.009. S1360-1385(08)00137-4 [pii] [DOI] [PubMed] [Google Scholar]
- 7.Babu MM, Iyer LM, Balaji S, Aravind L. The natural history of the WRKY-GCM1 zinc fingers and the relationship between transcription factors and transposons. Nucleic Acids Res. 2006;34(22):6505–20. doi: 10.1093/nar/gkl888. gkl888 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008 Sep 4;455(7209):64–71. doi: 10.1038/nature07242. nature07242 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004 Jan 23;116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5. S0092867404000455 [pii] [DOI] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009 Jan 23;136(2):215–33. doi: 10.1016/j.cell.2009.01.002. S0092-8674(09)00008-7 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baulcombe D. Of maize and men, or peas and people: case histories to justify plants and other model systems. Nat Med. 2008 Oct;14(10):1046–9. doi: 10.1038/nm1008-1046. nm1008-1046 [pii] [DOI] [PubMed] [Google Scholar]
- 12.Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006 Jul 15;20(14):1885–98. doi: 10.1101/gad.1424106. gad.1424106 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007 Oct 26;28(2):328–36. doi: 10.1016/j.molcel.2007.09.028. S1097-2765(07)00669-7 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001 Jan 18;409(6818):363–6. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 15.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005 Mar;3(3):e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cerutti H, Casas-Mollano JA. On the origin and functions of RNA-mediated silencing: from protists to man. Curr Genet. 2006 Aug;50(2):81–99. doi: 10.1007/s00294-006-0078-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chalfie M, Horvitz HR, Sulston JE. Mutations that lead to reiterations in the cell lineages of C. elegans. Cell. 1981 Apr;24(1):59–69. doi: 10.1016/0092-8674(81)90501-8. 0092-8674(81)90501-8 [pii] [DOI] [PubMed] [Google Scholar]
- 18.Chen K, Rajewsky N. Natural selection on human microRNA binding sites inferred from SNP data. Nat Genet. 2006 Dec;38(12):1452–6. doi: 10.1038/ng1910. ng1910 [pii] [DOI] [PubMed] [Google Scholar]
- 19.Chen K, Rajewsky N. Deep conservation of microRNA-target relationships and 3′UTR motifs in vertebrates, flies, and nematodes. Cold Spring Harb Symp Quant Biol. 2006;71:149–56. doi: 10.1101/sqb.2006.71.039. [DOI] [PubMed] [Google Scholar]
- 20.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007 Feb;8(2):93–103. doi: 10.1038/nrg1990. nrg1990 [pii] [DOI] [PubMed] [Google Scholar]
- 21.Cui Q, Yu Z, Purisima EO, Wang E. Principles of microRNA regulation of a human cellular signaling network. Mol Syst Biol. 2006;2:46. doi: 10.1038/msb4100089. msb4100089 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis E, Caiment F, Tordoir X, Cavaille J, Ferguson-Smith A, Cockett N, Georges M, Charlier C. RNAi-mediated allelic trans-interaction at the imprinted Rtl1/Peg11 locus. Curr Biol. 2005 Apr 26;15(8):743–9. doi: 10.1016/j.cub.2005.02.060. S0960-9822(05)00233-2 [pii] [DOI] [PubMed] [Google Scholar]
- 23.Didiano D, Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat Struct Mol Biol. 2006 Sep;13(9):849–51. doi: 10.1038/nsmb1138. nsmb1138 [pii] [DOI] [PubMed] [Google Scholar]
- 24.Didiano D, Hobert O. Molecular architecture of a miRNA-regulated 3′ UTR. RNA. 2008 Jul;14(7):1297–317. doi: 10.1261/rna.1082708. rna.1082708 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004 Mar 1;18(5):504–11. doi: 10.1101/gad.1184404 1184404. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008 Feb;9(2):102–14. doi: 10.1038/nrg2290. nrg2290 [pii] [DOI] [PubMed] [Google Scholar]
- 27.Fire AZ. Gene silencing by double-stranded RNA (Nobel Lecture) Angew Chem Int Ed Engl. 2007;46(37):6966–84. doi: 10.1002/anie.200701979. [DOI] [PubMed] [Google Scholar]
- 28.Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci U S A. 2008 Sep 30;105(39):14879–84. doi: 10.1073/pnas.0803230105. 0803230105 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009 Jan;19(1):92–105. doi: 10.1101/gr.082701.108. gr.082701.108 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu X, Adamski M, Thompson EM. Altered miRNA repertoire in the simplified chordate, Oikopleura dioica. Mol Biol Evol. 2008 Jun;25(6):1067–80. doi: 10.1093/molbev/msn060. msn060 [pii] [DOI] [PubMed] [Google Scholar]
- 31.Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006 Apr 7;312(5770):75–9. doi: 10.1126/science.1122689. 1122689 [pii] [DOI] [PubMed] [Google Scholar]
- 32.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008 Jan;36(Database issue):D154–8. doi: 10.1093/nar/gkm952. gkm952 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008 Oct 30;455(7217):1193–7. doi: 10.1038/nature07415. nature07415 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001 Jul 13;106(1):23–34. doi: 10.1016/s0092-8674(01)00431-7. S0092-8674(01)00431-7 [pii] [DOI] [PubMed] [Google Scholar]
- 35.Grosshans H, Filipowicz W. Proteomics joins the search for microRNA targets. Cell. 2008 Aug 22;134(4):560–2. doi: 10.1016/j.cell.2008.08.008. S0092-8674(08)01016-7 [pii] [DOI] [PubMed] [Google Scholar]
- 36.Hammell M, Long D, Zhang L, Lee A, Carmack CS, Han M, Ding Y, Ambros V. mirWIP: microRNA target prediction based on microRNA-containing ribonucleoprotein-enriched transcripts. Nat Methods. 2008 Sep;5(9):813–9. doi: 10.1038/nmeth.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001 Aug 10;293(5532):1146–50. doi: 10.1126/science.1064023 293/5532/1146. [pii] [DOI] [PubMed] [Google Scholar]
- 38.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006 Jun 2;125(5):887–901. doi: 10.1016/j.cell.2006.03.043. S0092-8674(06)00516-2 [pii] [DOI] [PubMed] [Google Scholar]
- 39.Heimberg AM, Sempere LF, Moy VN, Donoghue PC, Peterson KJ. MicroRNAs and the advent of vertebrate morphological complexity. Proc Natl Acad Sci U S A. 2008 Feb 26;105(8):2946–50. doi: 10.1073/pnas.0712259105. 0712259105 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008 Oct 24;32(2):276–84. doi: 10.1016/j.molcel.2008.09.014. S1097-2765(08)00660-6 [pii] [DOI] [PubMed] [Google Scholar]
- 41.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001 Aug 3;293(5531):834–8. doi: 10.1126/science.1062961 1062961. [pii] [DOI] [PubMed] [Google Scholar]
- 42.Iyer LM, Koonin EV, Aravind L. Evolutionary connection between the catalytic subunits of DNA-dependent RNA polymerases and eukaryotic RNA-dependent RNA polymerases and the origin of RNA polymerases. BMC Struct Biol. 2003 Jan 28;3:1. doi: 10.1186/1472-6807-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iyer LM, Anantharaman V, Wolf MY, Aravind L. Comparative genomics of transcription factors and chromatin proteins in parasitic protists and other eukaryotes. Int J Parasitol. 2008 Jan;38(1):1–31. doi: 10.1016/j.ijpara.2007.07.018. S0020-7519(07)00300-1 [pii] [DOI] [PubMed] [Google Scholar]
- 44.Johnston RJ, Jr, Chang S, Etchberger JF, Ortiz CO, Hobert O. MicroRNAs acting in a double-negative feedback loop to control a neuronal cell fate decision. Proc Natl Acad Sci U S A. 2005 Aug 30;102(35):12449–54. doi: 10.1073/pnas.0505530102. 0505530102 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 46.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005 Sep 2;309(5740):1577–81. doi: 10.1126/science.1113329. 309/5740/1577 [pii] [DOI] [PubMed] [Google Scholar]
- 47.Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005 Feb 15;19(4):489–501. doi: 10.1101/gad.1248505. 19/4/489 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C, Nagel R, Voorhoeve PM, van Duijse J, Orom UA, Lund AH, Perrakis A, Raz E, Agami R. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007 Dec 28;131(7):1273–86. doi: 10.1016/j.cell.2007.11.034. S0092-8674(07)01537-1 [pii] [DOI] [PubMed] [Google Scholar]
- 49.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001 Oct 15;15(20):2654–9. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009 Feb;10(2):126–39. doi: 10.1038/nrm2632. nrm2632 [pii] [DOI] [PubMed] [Google Scholar]
- 51.Kloosterman WP, Wienholds E, Ketting RF, Plasterk RH. Substrate requirements for let-7 function in the developing zebrafish embryo. Nucleic Acids Res. 2004;32(21):6284–91. doi: 10.1093/nar/gkh968. 32/21/6284 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, Jensen K, Cobb BS, Merkenschlager M, Rajewsky N, Rajewsky K. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008 Mar 7;132(5):860–74. doi: 10.1016/j.cell.2008.02.020. S0092-8674(08)00268-7 [pii] [DOI] [PubMed] [Google Scholar]
- 53.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005 May;37(5):495–500. doi: 10.1038/ng1536. ng1536 [pii] [DOI] [PubMed] [Google Scholar]
- 54.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001 Oct 26;294(5543):853–8. doi: 10.1126/science.1064921 294/5543/853. [pii] [DOI] [PubMed] [Google Scholar]
- 55.Lai EC, Tam B, Rubin GM. Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev. 2005 May 1;19(9):1067–80. doi: 10.1101/gad.1291905. gad.1291905 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lall S, Grun D, Krek A, Chen K, Wang YL, Dewey CN, Sood P, Colombo T, Bray N, Macmenamin P, Kao HL, Gunsalus KC, Pachter L, Piano F, Rajewsky N. A genome-wide map of conserved microRNA targets in C. elegans. Curr Biol. 2006 Mar 7;16(5):460–71. doi: 10.1016/j.cub.2006.01.050. S0960-9822(06)01059-1 [pii] [DOI] [PubMed] [Google Scholar]
- 57.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001 Oct 26;294(5543):858–62. doi: 10.1126/science.1065062 294/5543/858. [pii] [DOI] [PubMed] [Google Scholar]
- 58.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001 Oct 26;294(5543):862–4. doi: 10.1126/science.1065329 294/5543/862. [pii] [DOI] [PubMed] [Google Scholar]
- 59.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993 Dec 3;75(5):843–54. doi: 10.1016/0092-8674(93)90529-y. 0092-8674(93)90529-Y [pii] [DOI] [PubMed] [Google Scholar]
- 60.Lee SR, Collins K. Two classes of endogenous small RNAs in Tetrahymena thermophila. Genes Dev. 2006 Jan 1;20(1):28–33. doi: 10.1101/gad.1377006. gad.1377006 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005 Jan 14;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. S0092867404012607 [pii] [DOI] [PubMed] [Google Scholar]
- 62.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003 Dec 26;115(7):787–98. doi: 10.1016/s0092-8674(03)01018-3. S0092867403010183 [pii] [DOI] [PubMed] [Google Scholar]
- 63.Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol. 2005 Aug 23;15(16):1501–7. doi: 10.1016/j.cub.2005.07.029. S0960-9822(05)00773-6 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li X, Carthew RW. A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye. Cell. 2005 Dec 29;123(7):1267–77. doi: 10.1016/j.cell.2005.10.040. S0092-8674(05)01327-9 [pii] [DOI] [PubMed] [Google Scholar]
- 65.Liang H, Li WH. MicroRNA regulation of human protein protein interaction network. RNA. 2007 Sep;13(9):1402–8. doi: 10.1261/rna.634607. rna.634607 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005 Feb 17;433(7027):769–73. doi: 10.1038/nature03315. nature03315 [pii] [DOI] [PubMed] [Google Scholar]
- 67.Lin SY, Johnson SM, Abraham M, Vella MC, Pasquinelli A, Gamberi C, Gottlieb E, Slack FJ. The C elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev Cell. 2003 May;4(5):639–50. doi: 10.1016/s1534-5807(03)00124-2. S1534580703001242 [pii] [DOI] [PubMed] [Google Scholar]
- 68.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005 Jun 9;435(7043):834–8. doi: 10.1038/nature03702. nature03702 [pii] [DOI] [PubMed] [Google Scholar]
- 69.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004 Jan 2;303(5654):95–8. doi: 10.1126/science.1090599 1090599. [pii] [DOI] [PubMed] [Google Scholar]
- 70.Malone CD, Anderson AM, Motl JA, Rexer CH, Chalker DL. Germ line transcripts are processed by a Dicer-like protein that is essential for developmentally programmed genome rearrangements of Tetrahymena thermophila. Mol Cell Biol. 2005 Oct;25(20):9151–64. doi: 10.1128/MCB.25.20.9151-9164.2005. 25/20/9151 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martinez NJ, Ow MC, Barrasa MI, Hammell M, Sequerra R, Doucette-Stamm L, Roth FP, Ambros VR, Walhout AJ. A C. elegans genome-scale microRNA network contains composite feedback motifs with high flux capacity. Genes Dev. 2008 Sep 15;22(18):2535–49. doi: 10.1101/gad.1678608. 22/18/2535 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005 Nov 18;123(4):607–20. doi: 10.1016/j.cell.2005.08.044. S0092-8674(05)00922-0 [pii] [DOI] [PubMed] [Google Scholar]
- 73.Mello CC. Return to the RNAi world: rethinking gene expression and evolution (Nobel Lecture) Angew Chem Int Ed Engl. 2007;46(37):6985–94. doi: 10.1002/anie.200701713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mishima Y, Giraldez AJ, Takeda Y, Fujiwara T, Sakamoto H, Schier AF, Inoue K. Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr Biol. 2006 Nov 7;16(21):2135–42. doi: 10.1016/j.cub.2006.08.086. S0960-9822(06)02154-3 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Miska EA, Alvarez-Saavedra E, Abbott AL, Lau NC, Hellman AB, McGonagle SM, Bartel DP, Ambros VR, Horvitz HR. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 2007 Dec;3(12):e215. doi: 10.1371/journal.pgen.0030215. 07-PLGE-RA-0722 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell. 2002 Sep 20;110(6):689–99. doi: 10.1016/s0092-8674(02)00909-1. S0092867402009091 [pii] [DOI] [PubMed] [Google Scholar]
- 77.Molnar A, Schwach F, Studholme DJ, Thuenemann EC, Baulcombe DC. miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature. 2007 Jun 28;447(7148):1126–9. doi: 10.1038/nature05903. nature05903 [pii] [DOI] [PubMed] [Google Scholar]
- 78.Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997 Mar 7;88(5):637–46. doi: 10.1016/s0092-8674(00)81906-6. S0092-8674(00)81906-6 [pii] [DOI] [PubMed] [Google Scholar]
- 79.Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005 Jul 18;202(2):261–9. doi: 10.1084/jem.20050678. jem.20050678 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005 Jun 9;435(7043):839–43. doi: 10.1038/nature03677. nature03677 [pii] [DOI] [PubMed] [Google Scholar]
- 81.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007 Jul 13;130(1):89–100. doi: 10.1016/j.cell.2007.06.028. S0092-8674(07)00795-7 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999 Dec 15;216(2):671–80. doi: 10.1006/dbio.1999.9523. S0012-1606(99)99523-4 [pii] [DOI] [PubMed] [Google Scholar]
- 83.Pane A, Wehr K, Schupbach T. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell. 2007 Jun;12(6):851–62. doi: 10.1016/j.devcel.2007.03.022. S1534-5807(07)00147-5 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000 Nov 2;408(6808):86–9. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 85.Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, Russo JJ, Ju J, Randall G, Lindenbach BD, Rice CM, Simon V, Ho DD, Zavolan M, Tuschl T. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005 Apr;2(4):269–76. doi: 10.1038/nmeth746. nmeth746 [pii] [DOI] [PubMed] [Google Scholar]
- 86.Piriyapongsa J, Jordan IK. A family of human microRNA genes from miniature inverted-repeat transposable elements. PLoS ONE. 2007;2(2):e203. doi: 10.1371/journal.pone.0000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Piriyapongsa J, Marino-Ramirez L, Jordan IK. Origin and evolution of human microRNAs from transposable elements. Genetics. 2007 Jun;176(2):1323–37. doi: 10.1534/genetics.107.072553. genetics.107.072553 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Prucca CG, Slavin I, Quiroga R, Elias EV, Rivero FD, Saura A, Carranza PG, Lujan HD. Antigenic variation in Giardia lamblia is regulated by RNA interference. Nature. 2008 Dec 11;456(7223):750–4. doi: 10.1038/nature07585. nature07585 [pii] [DOI] [PubMed] [Google Scholar]
- 89.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000 Feb 24;403(6772):901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 90.Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007 Dec;17(12):1850–64. doi: 10.1101/gr.6597907. gr.6597907 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ruvkun G. The perfect storm of tiny RNAs. Nat Med. 2008 Oct;14(10):1041–5. doi: 10.1038/nm1008-1041. nm1008-1041 [pii] [DOI] [PubMed] [Google Scholar]
- 92.Saito K, Sakaguchi Y, Suzuki T, Siomi H, Siomi MC. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi- interacting RNAs at their 3′ ends. Genes Dev. 2007 Jul 1;21(13):1603–8. doi: 10.1101/gad.1563607. 21/13/1603 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Saunders MA, Liang H, Li WH. Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci U S A. 2007 Feb 27;104(9):3300–5. doi: 10.1073/pnas.0611347104. 0611347104 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008 Sep 4;455(7209):58–63. doi: 10.1038/nature07228. nature07228 [pii] [DOI] [PubMed] [Google Scholar]
- 95.Shi H, Tschudi C, Ullu E. An unusual Dicer-like1 protein fuels the RNA interference pathway in Trypanosoma brucei. RNA. 2006 Dec;12(12):2063–72. doi: 10.1261/rna.246906. rna.246906 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 2000 Apr;5(4):659–69. doi: 10.1016/s1097-2765(00)80245-2. S1097-2765(00)80245-2 [pii] [DOI] [PubMed] [Google Scholar]
- 97.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004 Sep 3;305(5689):1434–7. doi: 10.1126/science.1102514 1102514. [pii] [DOI] [PubMed] [Google Scholar]
- 98.Sulston JE, Horvitz HR. Abnormal cell lineages in mutants of the nematode Caenorhabditis elegans. Dev Biol. 1981 Feb;82(1):41–55. doi: 10.1016/0012-1606(81)90427-9. 0012-1606(81)90427-9 [pii] [DOI] [PubMed] [Google Scholar]
- 99.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006 Aug 15;103(33):12481–6. doi: 10.1073/pnas.0605298103. 0605298103 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008 Oct 23;455(7216):1124–8. doi: 10.1038/nature07299. nature07299 [pii] [DOI] [PubMed] [Google Scholar]
- 101.Technau U. Evolutionary biology: Small regulatory RNAs pitch in. Nature. 2008 Oct 30;455(7217):1184–5. doi: 10.1038/4551184a. 4551184a [pii] [DOI] [PubMed] [Google Scholar]
- 102.Tessmar-Raible K, Raible F, Christodoulou F, Guy K, Rembold M, Hausen H, Arendt D. Conserved sensory-neurosecretory cell types in annelid and fish forebrain: insights into hypothalamus evolution. Cell. 2007 Jun 29;129(7):1389–400. doi: 10.1016/j.cell.2007.04.041. S0092-8674(07)00605-8 [pii] [DOI] [PubMed] [Google Scholar]
- 103.Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell. 2007 Jun 8;26(5):753–67. doi: 10.1016/j.molcel.2007.05.018. S1097-2765(07)00319-X [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007 Mar 23;128(6):1105–18. doi: 10.1016/j.cell.2007.01.038. S0092-8674(07)00201-2 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007 Dec 21;318(5858):1931–4. doi: 10.1126/science.1149460. 1149460 [pii] [DOI] [PubMed] [Google Scholar]
- 106.Vasudevan S, Tong Y, Steitz JA. Cell-cycle control of microRNA-mediated translation regulation. Cell Cycle. 2008 Jun 1;7(11):1545–9. doi: 10.4161/cc.7.11.6018. 6018 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, Jaenisch R, Sharp PA, Jacks T. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008 Mar 7;132(5):875–86. doi: 10.1016/j.cell.2008.02.019. S0092-8674(08)00267-5 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008 Apr 4;320(5872):97–100. doi: 10.1126/science.1154040. 1154040 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993 Dec 3;75(5):855–62. doi: 10.1016/0092-8674(93)90530-4. 0092-8674(93)90530-4 [pii] [DOI] [PubMed] [Google Scholar]
- 110.Wightman B, Burglin TR, Gatto J, Arasu P, Ruvkun G. Negative regulatory sequences in the lin-14 3′-untranslated region are necessary to generate a temporal switch during Caenorhabditis elegans development. Genes Dev. 1991 Oct;5(10):1813–24. doi: 10.1101/gad.5.10.1813. [DOI] [PubMed] [Google Scholar]
- 111.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci U S A. 2006 Mar 14;103(11):4034–9. doi: 10.1073/pnas.0510928103. 0510928103 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005 Mar 17;434(7031):338–45. doi: 10.1038/nature03441. nature03441 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004 Apr 23;304(5670):594–6. doi: 10.1126/science.1097434 304/5670/594. [pii] [DOI] [PubMed] [Google Scholar]
- 114.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003 Dec 15;17(24):3011–6. doi: 10.1101/gad.1158803 1158803. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yu X, Lin J, Zack DJ, Mendell JT, Qian J. Analysis of regulatory network topology reveals functionally distinct classes of microRNAs. Nucleic Acids Res. 2008 Nov;36(20):6494–503. doi: 10.1093/nar/gkn712. gkn712 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yuan YR, Pei Y, Ma JB, Kuryavyi V, Zhadina M, Meister G, Chen HY, Dauter Z, Tuschl T, Patel DJ. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol Cell. 2005 Aug 5;19(3):405–19. doi: 10.1016/j.molcel.2005.07.011. S1097-2765(05)01475-9 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang D, Xiong H, Shan J, Xia X, Trudeau VL. Functional insight into Maelstrom in the germline piRNA pathway: a unique domain homologous to the DnaQ-H 3′-5′ exonuclease, its lineage-specific expansion/loss and evolutionarily active site switch. Biol Direct. 2008;3:48. doi: 10.1186/1745-6150-3-48. 1745-6150-3-48 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhang L, Ding L, Cheung TH, Dong MQ, Chen J, Sewell AK, Liu X, Yates JR, 3rd, Han M. Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 proteins AIN-1 and AIN-2. Mol Cell. 2007 Nov 30;28(4):598–613. doi: 10.1016/j.molcel.2007.09.014. S1097-2765(07)00626-0 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhao T, Li G, Mi S, Li S, Hannon GJ, Wang XJ, Qi Y. A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes Dev. 2007 May 15;21(10):1190–203. doi: 10.1101/gad.1543507. gad.1543507 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]