Summary
Endogenous circadian rhythms regulate many aspects of an organism’s behavior, physiology and development. These daily oscillations synchronize with the environment to generate robust rhythms, resulting in enhanced fitness and growth vigor in plants. Collective studies over the years have focused on understanding the transcription-based oscillator in Arabidopsis. Recent advances combining mechanistic data with genome-wide approaches, have contributed significantly to a more comprehensive understanding of the molecular interactions within the oscillator, and with clock controlled pathways. This review focuses on the regulatory mechanisms within the oscillator, highlighting key connections between new and existing components, and direct mechanistic links to downstream pathways that control overt rhythms in the whole plant.
Introduction
As a result of the earth’s rotation on its axis, most organisms live in environments that oscillate with a period of approximately 24 hours. The circadian clock is an intrinsic and entrainable timekeeping mechanism that has evolved in organisms, allowing them to adapt to periodic environmental fluctuations such as light and temperature [1][2]. Being a self-sustaining mechanism, the clock is able to buffer against both subtle and extreme changes, and persists in the absence environmental cues, which also contributes to setting the phase of the clock[1][3]. Anticipating these cyclic changes confers an adaptive advantage since organisms are better able to coordinate important physiological and developmental processes to occur at optimal times during the day, thus improving fitness [4][5][6].
Eukaryotic systems share similarities in the basic architecture of the oscillator in that interconnected negative feedback loops between species-specific components sustain robust rhythms [1][7][8][9]. Transcription-based interactions between these components, coupled with post-transcriptional, post-translational, and chromatin modifications are regulatory mechanisms modulating the rhythmic properties of the oscillator [10][11]. Coordinating oscillator function with this hierarchical regulatory topology is not only crucial for sustaining flexible and robust rhythms but also for targeted and temporal regulation of important biological networks.
The influence of clock control in higher plants encompasses numerous regulatory pathways. For example biological processes such as, the regulation of primary metabolism, photosynthesis, the regulation of growth, hormone levels, nutrient uptake, the developmental transition to flowering, and defense responses, are a subset of key processes regulated by the circadian clock in Arabidopsis [1][12][13][14][15]. The pervasiveness of clock control is further reflected in the circadian regulation of approximately one-third of the genes in Arabidopsis [16]. Furthermore, up to 90% of the transcriptome exhibits circadian rhythmicity under various light and temperature conditions [17]. Recent advances from the use of genome-wide approaches and functional genomics strategies are providing crucial insights into the underlying regulatory mechanisms within the oscillator, and direct mechanistic connections to clock controlled processes.
Interconnected Transcriptional Circuits in the Clock Network
Historical View of the Core Oscillator Loop
In Arabidopsis, genetics and biochemical studies were instrumental in constructing the molecular architecture of the clock. The original oscillator model was described as a transcriptional regulatory feedback loop consisting of positive and negative interactions between three components, two MYB domain containing transcription factors, CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), and a member of the PSEUDO RESPONSE REGULATOR (PRR) family TIMING OF CAB2 EXPRESSION 1 (TOC1) [18]. Together these components were considered the core oscillator as they determined the topological vulnerability of the network, such that loss of function of any of the core clock genes, results in a short period clock, and overexpression confers arrhythmicity in multiple outputs (Figure 1) [19][20][21][22]. Mechanistically, CCA1 and LHY directly repress TOC1 expression, by binding specifically to a cis-element within its promoter known as the evening element (EE), a motif that is often found in promoters of clock regulated evening expressed genes [18][23]. In turn TOC1 was proposed to induce the expression of CCA1 and LHY via an unknown mechanism [18]. This presented a simple transcription based model supported by genetic and modeling data that was critical for robust clock performance [18][24]. However, with lack of biochemical activity for TOC1, the direct transcriptional mechanism driving the core oscillator was a mystery.
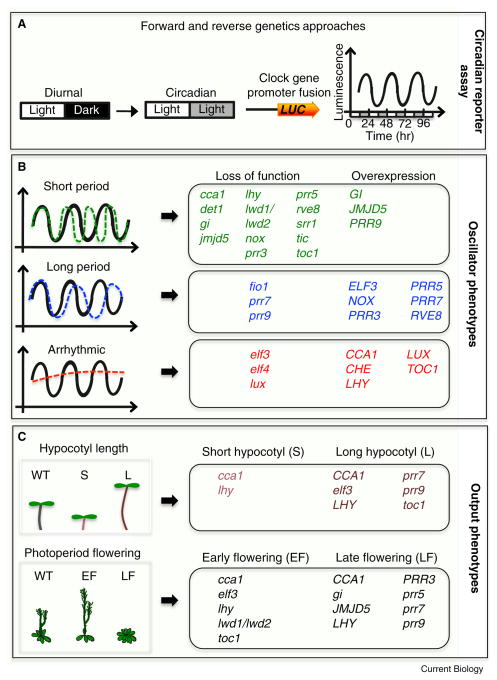
Figure 1. Monitoring clock function underlying oscillator and output phenotypes.
Endogenous circadian rhythms were first observed by daily leaf movement in plants. Genetic and biochemical approaches were then instrumental in discovering the regulatory units responsible for these rhythms. Subsequently, forward and reverse genetic approaches were used to identify additional components and monitor clock function. A) A 24 hour (24 hr) period of diurnal cycles (12 hr light: 12 hr dark) are often used to entrain the clock, and subsequently released to free running conditions (circadian) of continuous light. Clock gene promoter fusions to the firefly luciferase gene (LUC) are imaged over a period of several days to monitor altered clock phenotypes based on bioluminescence (Luminescence). B) Alteration in clock function is reflected in rhythmic changes of the 24 hr periodicity (short, long or arrhythmic). Loss of function (lower cased) or constitutive expression (upper cased - overexpression) confers these changes in period. Black sinusoidal waves represent normal circadian oscillations. Green dashed and blue dashed waves represent short and long period phenotypes respectively. Circadian oscillations can also be abolished (arrhythmic - red dashed lines). Alterations in clock gene expression are also reflected in changes in phase and amplitude. C) An altered clock confers changes in clock-controlled outputs. The circadian clock regulates hypocotyl elongation; as such loss of function or overexpression of clock genes confers short (S) or long (L) hypocotyls. In Arabidopsis, photoperiod flowering is dependent on day length. Long days (16 hr of light and 8 hr of dark) promote flowering, and short days (8 hr of light and 16 hr of dark) delay flowering. Loss of function or overexpression of clock genes confers either early flowering (EF) or late flowering (LF) relative to wild-type (WT).
Revised Model of the Core Oscillator Loop
Subsequent to these studies, numerous components were added to the oscillator, and as a result, the plant clock expanded into a complex network of interconnected feedback loops [25]. One of the most pivotal findings in the clock field comes more than a decade later with the characterization of TOC1 biochemical function. Collective contributions from targeted and rigorous molecular approaches, coupled with genome-wide expression studies, and Chromatin Immunoprecipitation followed by deep sequencing (ChIP-Seq), finally characterized the biochemical and molecular properties of TOC1 [26][27][28]. Three back to back studies conclusively showed that TOC1 is a DNA binding transcriptional repressor of CCA1 and LHY, indicating that the long held prediction that TOC1 is a positive regulator of CCA1 and LHY must be revised (Figure 2) [26][27][29]. Thus, the CCA1/LHY-TOC1 core model has been updated to one based entirely on transcriptional repression [29]. In addition to CCA1 and LHY, TOC1 also binds the promoters and inhibits the expression of existing oscillator components, PRR5, PRR7, PRR9, LUX ARRHYTHMO (LUX), GIGANTEA (GI), and EARLY FLOWERING 4 (ELF4) (Figure 2) [26][27]. Transcription factors regulate gene expression often through sequence specific DNA binding. However, the motifs enriched in the TOC1 targets share weak sequence similarity making it difficult to propose a consensus for TOC1 specificity. This suggests that TOC1 has the potential to recognize multiple cis-elements or perhaps function in combination with other transcription factors to regulate the expression of some targets [26][27]. Therefore, genome wide approaches such as protein binding microarrays coupled with structural analysis might provide the needed resolution to determine whether TOC1 is a site-specific DNA binding protein.
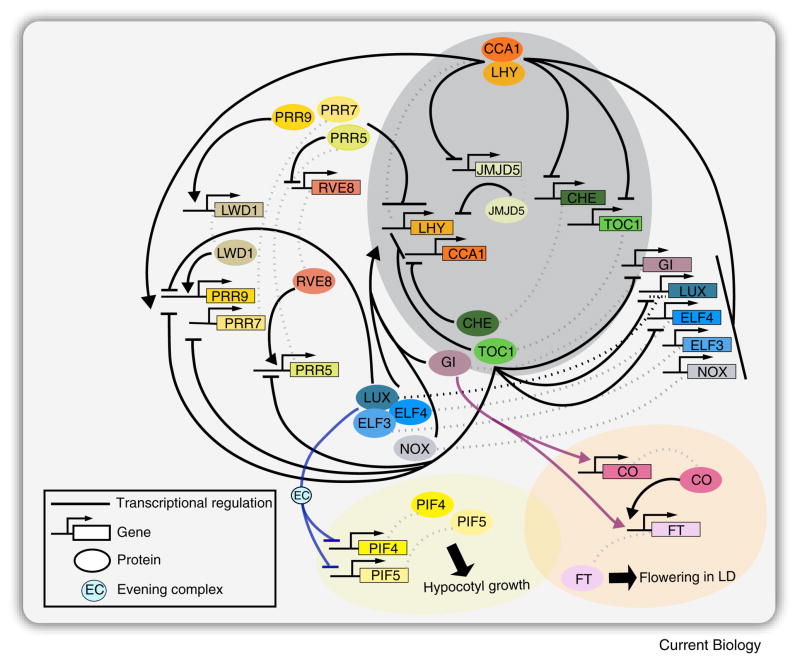
Figure 2. A model for transcription based interactions in the Arabidopsis clock network.
In vitro and invivo assays were instrumental in validating direct molecular interactions between oscillator components. The core of the oscillator consists of two Myb transcription factors CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), and TIMING OF CAB EXPRESSION 1 (TOC1). Other components expressed throughout the day interconnect with the core oscillator to form multiple feedback loops and a complex clock network. CCA1 and LHY directly repress TOC1, LUX, GI, ELF3, ELF4, CHE, JMJD5 (also known as JMJD30), and NOX (also known as BROTHER OF LUX ARRHYTHMO) by binding to their promoters. In return, TOC1, LUX, GI, ELF3, positively regulates CCA1 and LHY via an unknown mechanism. NOX directly activates CCA1 by binding to its promoter. LUX binds to its promoter and repress its own expression (indicated by black dashed lines). CHE and JMJD5 function as a direct repressor of CCA1. TOC1 inhibits the expression of CCA1, LHY, PRR9, PRR7, PRR5, LUX, ELF4, and GI. Sequential expression of PRR9, PRR7, and PRR5 directly inhibit the expression of CCA1 and LHY. In turn, PRR9 and PRR7 are positively regulated by CCA1 and LHY. PRR9 and PRR5 are also positively regulated by LWD1 and PRR5, respectively. For simplification, other components that affect clock function such as PRR3, TIC, and SRR1 are not illustrated in the figure above. Though not illustrated in the above figure, protein-protein interactions often occur between clock components and are an important mechanism regulating clock function. CCA1 and LHY physically interact. TOC1 interacts with CHE and PRR5, and interacts with JMJD5 genetically. LUX interacts with ELF3 and ELF4 to form the evening complex (EC). Direct mechanistic connections exist between clock components and modulators of physiological processes. The EC regulates hypocotyl growth by directly binding to the promoters of PIF4 and PIF5. Direct interaction between GI, CO, and FT, and GI and FT, modulates photoperiod flowering. Arrows represents transcriptional activation, and horizontal lines represent repression. Dashed lines in grey indicate the protein and gene associations.
Surprisingly, in the smallest known free-living eukaryote, the green unicellular alga Ostreococcus tauri, homologs of CCA1 and TOC1 were the only two oscillator components identified, and together they negatively regulate each other’s expression in a feedback loop [7]. It is therefore possible that this two-component system originally defined the core oscillator, and was sufficient to generate robust rhythms, and modulate clock function. Subsequently, LHY was likely added during green plant evolution as complications from multi-cellularity arose. Indeed insights into early and diverse plant circadian systems already suggests wide conservation and evidence for expansion of clock gene families resulting from genome duplication events throughout evolution [30][31][32][33][34]. Therefore contributions from comprehensive lineage specific circadian systems will be valuable to understanding how the clock model has evolved into a complex network in plants.
Interconnected Circuits
Over the years many additional clock components have been identified and positioned within the oscillator as phase specific (morning or evening expressed) reciprocal circuits [25]. In a morning specific loop, CCA1 and LHY are presumed to promote the expression of two TOC1 family members, PRR9 and PPR7 by directly binding to their promoters [35][36]. In return, PRR9, PRR7, and PRR5 function as transcriptional repressors to coregulate the expression of CCA1 and LHY (Figure 2). However the mechanism for their recruitment to target promoters is poorly understood [35][37]. A mechanistic understanding of how CCA1 and LHY function to promote the expression of PRR9 and PRR7, while directly inhibiting the expression of all other oscillator components is unclear. Perhaps exploiting inducible systems controlling CCA1 and LHY expression might help to determine the precise transcriptional effect on PRR7 and PRR9 [38]. A candidate for transcriptional activation of PRR9 is LIGHT-REGULATED WD1 (LWD1), a clock associated protein involved in the regulation of period length and photoperiodic flowering (Figure 1) [39][40]. LWD1 was shown to participate in a positive feedback loop with PRR9, and also indirectly promote the expression of CCA1, LHY, PRR5 and TOC1, suggesting that this component might function predominantly as a transcriptional activator in regulating clock function (Figure 2) [39]. In addition, based on recent microarray data LWD1 also regulates the period length of ELF4, and might form a feedback loop with TOC1, [39][26]. It was also suggested that TOC1 might assist CCA1 and LHY in positively regulating PRR9 expression [41]. However, since TOC1 directly inhibits the expression of CCA1, LHY and PRR9, this interaction needs to be further examined.
Connecting another circuit to the oscillator is LUX ARRHYTHMO (LUX), also known as PHYTOCLOCK1 (PCL1), is an evening phased component that participates in a feedback loop with CCA1 and LHY (Figure 2) [42][43]. On the negative arm of the loop, CCA1 and LHY directly bind to the EE motif within the LUX promoter and inhibit its expression [42][36]. In turn, LUX is suggested to promote the expression of CCA1 and LHY by an unknown mechanism [42]. Although LUX contains intrinsic transcription factor properties, the binding site specificity was unknown, and as such, prevented the resolution of the molecular interaction with CCA1 and LHY. However, a recent genome-wide approach, Protein Binding Microarray (PBM), coupled with targeted genetic and molecular strategies identified the LUX binding site (LBS), and demonstrated that LUX binds selectively to PRR9 and its own promoter and inhibit their expression [44][45][46]. Together, this revealed the first mechanistic link between two oscillator circuits, and the first example in plants of direct self-regulation by a clock component. It is also possible that indirect regulation of CCA1 by LUX might be mediated through its direct regulation of PRR9.
LUX belongs to a five-member gene family, for which the closest homolog, NOX (Latin word for “night”) also known as BROTHER OF LUX ARRHYTHMO (BOA), also participates in clock regulation [45][46][47]. NOX exhibits similar peak expression at night, and directly binds to the CCA1 promoter through the defined LUX binding site [47]. CCA1 expression is enhanced when NOX is constitutively expressed indicating that NOX is a transcriptional activator of CCA1, which in turn directly represses NOX. The observation that NOX also seems to promote the expression of LHY, TOC1 and GI, might suggest that this component can be classified as an activator within the oscillator [47].
Molecular Interactions and Complex Formation Underlying Oscillator Function
With the revelation that TOC1 negatively regulates the expression of CCA1 and LHY, the mechanism of CCA1 and LHY transcriptional activation remains a critical unanswered question to resolve the core oscillator dynamics. Efforts to address these questions might also be complicated by the fact that CCA1, LHY, and TOC1 also regulate their own expression, and are involved in feedback loops with other clock components [20][21][26][27]. It was proposed that TOC1 might be recruited to the CCA1 promoter through its interaction with a TCP transcription factor, CCA1 Hiking Expedition (CHE), a direct transcriptional repressor of CCA1 [48]. In addition, TOC1 also regulates the expression of two other Class1 TCPs (TCP11 and TCP23), and the expression of ~ 800 other targets (>10% of all putative transcription factors), ~ 40% of which are circadian regulated, indicating a broad role for TOC1 in the clock transcriptome, and in regulation of clock controlled targets [26]. Characterizing the functional implications of these molecular interactions will be critical to understanding the impact of TOC1 on clock function and regulation.
A role for combinatorial regulation via direct protein-protein interaction is suggested for CCA1 and LHY repressor activity. For example, although CCA1 and LHY are site-specific (binds the evening element) DNA binding transcriptional repressors, the actual inhibitory mechanism on TOC1 involves other interacting partners. In a recent study, it was shown that CCA1 and LHY interact with the COP10-DET1-DDB1 (CDD) complex, an evolutionarily conserved protein complex involved in repression of photomorphogenesis in Arabidopsis [49]. This molecular interaction, and specifically the corepressor function of DET1 (DE-ETIOLATED 1), is necessary for CCA1 and LHY repression of TOC1 and GI [49]. Therefore, these types of observations need to be further investigated to better understand the pervasiveness of multi-protein regulatory complexes, and how they allow the plant to effectively coordinate and maintain oscillator function.
Another aspect that confounds the ease of resolving mechanistic connections in the oscillator is the lack of known functional domains for some clock genes. In a complex network such as the circadian clock, some oscillator components likely function as coregulators, and are recruited to DNA by other DNA binding transcription factors. For example, together with LUX, two other clock genes EARLY FLOWERING 3 (ELF3) and ELF4, function in a complex known as the evening complex (EC) [50][46]. ELF3 and ELF4 lack known DNA binding properties, and therefore associate to target promoters via direct binding by LUX. As a result, ELF3 and ELF4 are also part of the regulatory machinery influencing PRR9 as both proteins associate to the PRR9 promoter [44][51][46]. In addition, ELF3 and ELF4 form a feedback loop with CCA1 and LHY, which inhibit their expression through direct binding to the EE motif within their promoters [52][53][54][55]. Similar to LUX, both ELF3 and ELF4 are also suggested to promote the expression of CCA1 and LHY, through an indirect mechanism [54][52][53]. Furthermore, the EC functions to regulate two light regulated and growth promoting transcription factors, PHYTOCHROME INTERACTING FACTOR 4 and 5 (PIF4 and PIF5) discussed in greater detail below. As part of this multi-protein complex, the EC components physically interact with each other, are coexpressed, and share multiple clock phenotypes. The identification of the EC complex provides a great mechanistic example of functional concerted regulation by a subset of clock genes in Arabidopsis.
Interestingly, CCA1 and LHY also interact in vivo, and this interaction was speculated to function as part of a complex, possibly a “morning or daytime complex” [56]. However, there is no current experimental or mechanistic evidence to support such a complex between CCA1, LHY and other morning phased components, to mediate DNA binding.
Contributions from Genome-wide and High-throughput Approaches
Of the well characterized oscillator components (~20), only CCA1, LHY, Reveille8 (RVE8), CHE, LUX and NOX have been demonstrated to bind a defined cis-element in target promoters [48][57][58][45][59][47]. It is therefore possible that direct molecular mechanisms underlying transcriptional regulation in the clock will require discovery of novel components and the use of integrative approaches. The saturation of forward genetics screens, reflected in the isolation of multiple alleles in known clock genes, and the presence of clock gene family redundancy, has limited the use of this approach. However, recent contributions from large scale functional genomics and genome wide studies have assisted in overcoming this limitation, and as a result significant progress have been made in mechanistic connections within the oscillator. For example, the identification of CHE using a large scale functional genomics approach, the identification of the LUX binding site using a genome-wide PBM approach, and the molecular and biochemical characterization of TOC1 using genome-wide deep sequencing ChIP-seq and microarray expression datasets, have all made significant mechanistic connections [48][45][26][27]. Furthermore, incorporation of creative approaches such as liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis, has also led to the discovery of novel components, and new molecular links [59]. For example, RVE8, a MYB domain transcription factor phylogenetically related to CCA1 and LHY, functions in a positive feedback loop with PRR5 [59]. RVE8 binds directly to the EE motif in PRR5 and TOC1 promoters, subsequently promoting their expression. In turn, PRR5 is suggested to inhibit the expression of RVE8 to close the loop [59][60].
The value of these studies and the approaches used are further demonstrated in the connections made in subsequent reports. For example, subsequent to the identification of CHE, other TCPs have recently been linked to the oscillator [61][62][26]. Although the clock connections with input pathways are not discussed in this review, leveraging the data from the studies described above might reveal additional direct modulators between light, temperature and the clock.
Other Layers of Regulation within the Oscillator
Multiple examples highlighting the importance of other levels of regulation such as post-transcriptional regulation, post-translational regulation, and chromatin remodeling, as critical mechanisms for normal circadian rhythmicity have emerged over the years.
RNA based Regulation
Recent interactome data from Arabidopsis revealed that TOC1 interacts with at least four genes that have been classified as RNA binding proteins (RBPs) [63]. These proteins are rhythmically expressed, and provide putative targets for formation of a TOC1 mediated ribonucleoprotein (RNP) complexes, thus connecting the clock to regulation of RNA metabolism [64]. However, the extent of post-transcriptional based regulation in the clock is best illustrated by studies on the influence of alternative splicing on oscillator function. In many organisms, alternative splicing of clock and output genes have been reported, and in several examples temperature effects on alternative splicing was shown to be important [65][66][67][68][69]. In Arabidopsis, multiple alternatively spliced isoforms of CCA1, LHY, TOC1 PRR3, PRR5, PRR7, PRR9, and GI transcripts has recently been documented (Figure 3) [68][69]. A subset of these occurrences and the effects on alternatively spliced transcript expression appears to be sensitive to temperature changes. For example, a reduction in temperature (20°C to 12°C or 4°C) results in accumulation of nonproductive transcripts of LHY, PRR7, PRR3, and TOC1, and as a consequence a reduction in expression of these genes [69]. In contrast, increase levels of CCA1, PRR9 and PRR5 transcripts when the temperature was reduced correlated with decreased accumulation of nonproductive transcripts [69][68]. As a consequence of these alternative splicing events, nonsense mediated mRNA decay (NMD) is triggered due to the accumulation of nonfunctional transcripts of LHY, PRR9, PRR7, PRR5 and TOC1. These observations suggest a functional role for RNA based regulation of oscillator components as a mechanism through which plants are able to respond and buffer temperature fluctuations.
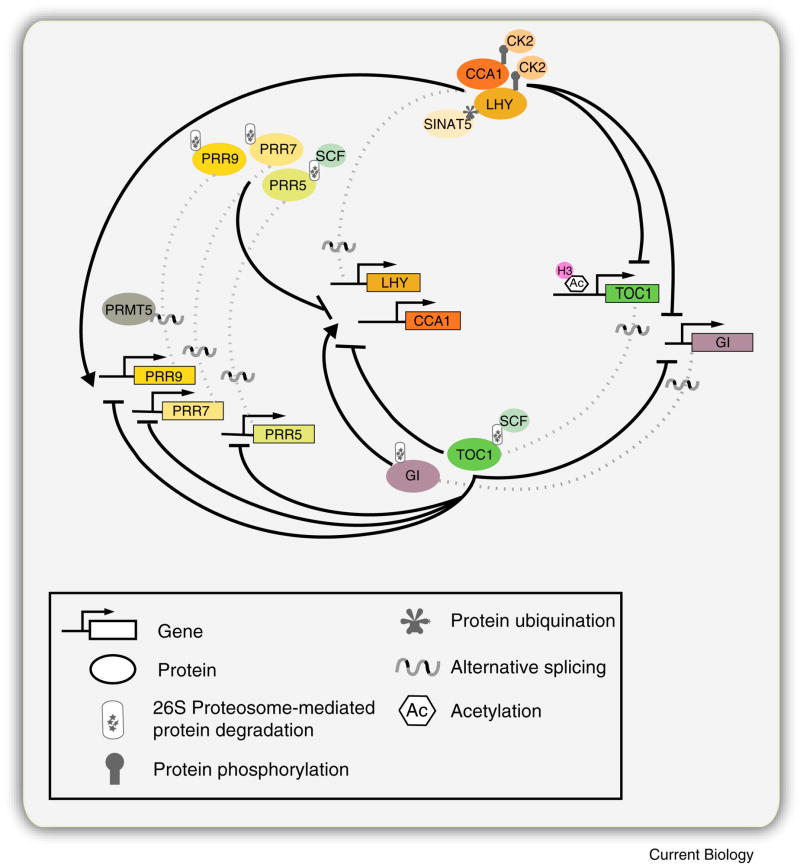
Figure 3. Multiple layers of regulation within the Oscillator.
Post-transcriptional and posttranslational-based regulation are mechanisms underlying robust clock function. Alternative splicing events have been observed for CCA1, LHY, TOC1, PRR5, PRR7, PRR9, and GI. PROTEIN ARGININE METHYL TRANSFERASE 5 (PRMT5) a protein involved in methylation of histones, RNA binding and spliceosomal proteins, is required for the alternative splicing of PRR9. Casein Kinase2 (CK2), an evolutionarily conserved serine/threonine protein kinase, phosphorylates both CCA1 and LHY; and SINAT5, a E3 ubiquitin ligase is involved in the ubiquitination of LHY. PRR5 and TOC1 are specifically targeted for proteosome dependent degradation by the E3 ubiquitin ligase Skp/Cullin/F-box (SCF) complex members ZEITLUPE (ZTL), FLAVIN BINDING KELCH F-BOX 1 (FKF1) and LOV KELCH PROTEIN 2 (LKP2). Other oscillator components PRR3, PRR7, PRR9, and GI are subjected to proteosome degradation, though the mechanism is unknown. A role for chromatin remodeling in regulating clock gene expression and function is exist for a few oscillator components. TOC1 expression correlates with histone 3 (H3) acetylation (Ac). However, TOC1, CCA1, LHY and GI expression also correlates with H3 lysine 9 acetylation (K9Ac), and H3 lysine 4 (K4) dimetylation (Me2), via an unknown mechanism. H3Ac, H3K9Ac, and H3K4Me2 are all defined as marks for gene activation. For simplicity, PRR3 is not included in the above illustration though alternative spliced transcripts have been detected for this component.
In addition, PROTEIN ARGININE METHYL TRANSFERASE 5 (PRMT5 also known as DART5/CSUL), a conserved protein among human, Drosophila and plants, that methylates histones, RNA binding and spliceosomal proteins, has also been linked to the regulation of alternative splicing of key clock genes [70][71][72]. In Arabidopsis, PRMT5 regulates the expression of CCA1, LHY, TOC1, PRR9, PRR7, and GI, and also regulates the alternative splicing of PRR9, though no evidence for epigenetic regulation was found [71][72]. Furthermore, PRMT5 is also regulated by the circadian clock implicating a putative regulatory feedback loop interconnected to the oscillator [71].
Post-translational Modifications
Eukaryotic clocks are known to integrate protein-based level of regulation to assist in sustaining robust biological rhythms [73][74]. For example, Casein Kinase 2 (CK2) is one of the few evolutionarily conserved molecular components involved in modulating the regulation of key clock genes in the mammalian, Drosophila, Neurospora and Arabidopsis circadian systems [75][76][77][78][79][80]. In Arabidopsis, CK2 phosphorylates CCA1 and LHY, and this process is considered to be important for CCA1 function, specifically the DNA binding properties and subsequent regulation of its targets within the oscillator (Figure 3) [79][80][81]. However, CK2 appears to have both agonistic and antagonistic effects on CCA1 binding to target promoters, suggesting that the precise functional role requires further mechanistic clarification. Perhaps the different effects of CCA1 transcriptional activity are dependent on the specific regulatory subunit of CK2 mediating the phosphorylation event. For example, binding of CCA1 to the PRR9, PRR7, TOC1, and LUX promoters was drastically reduced when CKB4 (a regulatory subunit of CK2) was constitutively expressed [36]. Conversely, binding of CCA1 to these targets was significantly increased when CK2 activity was decreased. Consequently, a decrease in TOC1 expression was observed when CK2 activity was decreased, suggesting that the direct repressive properties of CCA1 is mediated by CK2 activity [36]. Interestingly, both the regulatory function of CK2, and the binding of CCA1 to the TOC1 promoter were observed to be more effective at higher temperatures, suggesting a mechanism for temperature dependent protein modification in the modulating of clock function [36].
A number of key oscillator components that are subject to phosphorylation are also regulated by subsequent ubiquitination and degradation. In Arabidopsis, LHY is ubiquitinated by an evolutionarily conserved E3 ubiquitin ligase, SINAT5 (Figure 3) [82][83]. However, this activity is inhibited by DET1 which protects LHY from proteosome mediated degradation by physically interacting with SINAT5 [83]. Other components such as TOC1, PRR3, PRR5, PRR7, PRR9 and GI proteins are suggested to be regulated by 26S proteosome mediated degradation, though the precise mechanism to support some of these observations needs further investigation [84][85]. However, TOC1 and PRR5 are directly targeted for proteosomal degradation through physical interaction with members of the E3 ubiquitin ligase Skp/Cullin/F-box (SCF) complex, (ZEITLUPE) ZTL, FLAVIN BINDING KELCH F-BOX 1 (FKF1) and LOV KELCH PROTEIN 2 (LKP2) [86][87]. A phosphorylation dependent TOC1-PRR3, and TOC1-PRR5 interaction, is suggested to be critical for regulating the ZTL-mediated degradation of TOC1 [84][88]. Mechanistically, this molecular association involves the binding of PRR3 directly to the ZTL interacting domain of TOC1, thus preventing ZTL-mediated degradation [89][84]. As ZTL is localized in the cytoplasm, PRR5 directly interacts with TOC1 and promotes their localization to the nucleus, thereby escaping degradation by ZTL [84][88][90]. The dynamics of TOC1 degradation is also modulated by a light dependent interaction between GI and ZTL, as this direct interaction enhances the stabilization of ZTL during the day reinforcing the degradation of TOC1 protein [91]. Exploring genome-wide approaches such as protein modification assays will likely provide crucial mechanistic insights to protein-based regulation of the other oscillator components, and the functional consequence in the clock.
Rhythmic Chromatin Regulation
Compared to other eukaryotic systems, mechanistic insights for the extent and influence of epigenetic modifications on the clock function are poorly understood in plants. In Arabidopsis, attempts to address the role of chromatin remodeling detected a CCA1 dependent correlation between rhythmic histone acetylation (associated with actively transcribed genes), and histone deacetylation (associated with repressed genes), at the TOC1 promoter [92]. Maximum binding of CCA1 to the TOC1 promoter correlated with minimum histone 3 (H3) acetylation and vice versa, and resulting in decrease accumulation of TOC1 and H3 acetylation [92]. Furthermore, when histone deacetylation (HDAC) was inhibited, an upregulation of TOC1 was observed, together confirming that histone modification contributes to rhythmic regulation of TOC1. Another oscillator component, RVE8, also plays a role in regulation of H3 acetylation and deacetylation at the TOC1 promoter [59][60]. Other examples of associated histone modification marks at clock gene promoters are also observed for CCA1, LHY, TOC1 and GI [93]. Histone acetylation (H3K9Ac) and histone dimethylation (H3K4Me2) appear to correlate with the expression of CCA1, LHY, TOC1 and GI, though the precise nature of this association in modulating clock function is unclear [93].
In a recent study, JMJD5 (also known as JMJD30) was shown to be involved in the regulation of CCA1, LHY and TOC1 [94][95]. Interestingly, the human homolog KDM8, is also involved in clock function, and contains intrinsic histone demethylase properties [96]. Both JMJD5 and KDM8 are able to fulfill similar molecular functions in the plant and human circadian systems, but the enzymatic activity of JMJD5 in Arabidopsis is unknown [94][96][97]. Therefore, it would be interesting to determine whether JMJD5 contains histone methylase properties, and whether this activity is a conserved regulatory function in both plant and human circadian systems. Integrating targeted circadian driven epigenome data will contribute significantly to the discovery of important chromatin readers and modifiers regulating oscillator components in Arabidopsis. Coherently integrating various levels of regulatory information is absolutely crucial to understanding the underlying mechanism of clock function, and how robust oscillator performance controls an array of downstream pathways.
Interconnected Outputs from the Oscillator
Considering that plants are sessile and exposed to numerous environmental conditions and stresses, clock dependent integration of these external perturbations with downstream physiological and developmental processes is crucial for enhanced fitness and growth. The pervasive control by the interconnected clock network on virtually all known biological processes in Arabidopsis is well documented [98][16]. Significantly, major advances have been made in recent years to mechanistically connect components of the oscillator with the modulators of some of these downstream pathways.
Direct Molecular Interactions Regulating Hypocotyl Growth
In Arabidopsis, the regulation of hypocotyl growth is influenced by the circadian clock and numerous other external cues [99][13]. Two clock regulated transcription factors PIF4 and PIF5 were identified as key modulators of hypocotyl growth in Arabidopsis [13][100]. PIF4 and PIF5 negatively regulate light signaling and promote growth in a mechanism that requires the clock [101][102][103]. However, since mis-regulation of these factors did not affect oscillator function, the precise mechanism of this interaction was poorly understood. Recently, a rigorous genetic and molecular study resolved a direct mechanistic connection between the oscillator and hypocotyl growth. The EC (LUX, ELF3 and ELF4) transcriptionally represses PIF4 and PIF5 via direct binding of LUX to their promoters (Figure 2) [103]. Consequently hypocotyl growth is inhibited in the early evening but later in the night as this repression is relieved hypocotyl elongation occurs. This study created a new direct link between oscillator function and hypocotyl growth [50].
Since the biological pathways underlying physiological and developmental processes are known to intersect, genome-wide approaches are invaluable in revealing the extent and key players modulating these connections. For example, in addition to mediating rhythmic hypocotyl growth, recent genome-wide approaches revealed a direct role for PIF4 and PIF5 in modulating the clock control of hormone signaling, specifically auxin related pathways [104][105].
Direct Molecular Interactions Regulating Photoperiod Flowering
Equally insightful were advances made in the clock control of photoperiodic flowering. In Arabidopsis the onset of flowering occurs under long-day (LD) conditions, in a complex mechanism involving the clock and other environmental stimulus [106]. While mis-regulation of several oscillator components results in altered flowering phenotypes, the precise molecular interaction between the clock and photoperiodic flowering is still poorly understood [21][107][108][20]. Mechanistically, the simplified model connecting the oscillator to photoperiod control of flowering is through GIGANTEA (GI), and the flowering regulators CONSTANS (CO) and FLOWERING LOCUS T (FT) [109][110]. Under LD conditions, GI directly activates CO, and CO in turn activates FT to trigger flowering. A light dependent complex formation between GI and FKF1 is required for stabilization of CO and proper timing of CO expression [111]. This GI-FKF1 complex degrades CYCLING DOF FACTOR 1 (CDF1), a key CO repressor. As a result, CO positively regulates FT and this mechanism induces flowering. However, recent analysis of the ectopic expression of GI showed that GI could also directly activate FT expression to promote flowering [112]. This direct GI-FT interaction has been observed in both vasculature bundles and mesophyll tissue, whereas endogenous CO induction is known to occur only in vascular bundles. Interestingly, though the regulatory role of spatial expression on oscillator components has not been well studied, evidence suggests that organ, tissue and cell specific rhythmic variations might occur frequently within the oscillator [113][114][115][116]. For example, in Arabidopsis, of the known clock components, only CCA1, LHY, PRR7, and PRR9 transcripts oscillate in both root and shoots, while the transcripts of all other components tested oscillate only in the shoot [115]. Recent analysis indicates that rhythms in stomatal guard cells are different from rhythms observed in surrounding epidermal and mesophyll leaf cells [115]. For example, the rhythmic expression of GI exhibits a longer period, and peaks later in guard cells compared with whole leaves [115].
These studies concerning flowering and tissue specificity, suggest a critical role for GI as the master mediator between oscillator function and photoperiod flowering. Leveraging modeling techniques a recent study linked GI as a modulator of the clock response to sucrose, making yet another important connection to the clock control of metabolism [117]. Since GI lacks a DNA binding domain, interactome data coupled with spatial-temporal co-expression data, will be key to gaining deeper understanding of GI function in the clock and downstream pathways. Therefore, by combining mechanistic knowledge gained from the oscillator studies, with genome-wide approaches, rapid progress can be made to comprehensively map the interconnected multi-loop oscillator network in Arabidopsis.
Conclusions and Perspectives
The wealth of mechanistic information that has emerged over the years has provided insight into the underlying regulatory mechanism of the plant clock. Future advances in plant circadian research will be significantly influenced by multi-scale integrative approaches. For example, spatial-temporal guided genome-wide datasets, coupled with functional genomics approaches, will assist in overcoming the bottleneck created by transcription factor family redundancy, promoter complexity, and saturation of forward genetics screens. Furthermore, leveraging information from transcriptome, proteomic, and epigenomic datasets, will enable direct molecular connections between clock components and hierarchal levels of regulation to be mapped. Understanding at the molecular level, how the clock mechanistically controls key biological pathways such as immunity, hormone signaling, metabolism, photosynthesis, development and growth will also require combinatorial approaches. Ultimately, integrating mechanistic data with systems biology approaches will allow direct molecular connections to be established between clock function, the clock response to environmental stresses, and the clock control of regulatory pathways. In conclusion, while the collective knowledge gained from recent and future circadian studies will help to understand the role of the clock in enhanced growth and fitness in Arabidopsis, this mechanistic information can potentially be translated to other eukaryotic systems.
Acknowledgments
We thank Colleen Doherty, Jose Pruneda-Paz and Mariko Sawa for critical reading of the manuscript. This work was supported by National Institutes of Health (NIH) Grants GM067837 and GM056006, and Department of Energy grant DE-SC0006621 to S.A.K, and National Institutes of Health under Ruth L. Kirschstein National Research Service Award F32GM090375 to DHN. Due to space constraints we apologize for not citing all relevant publications by our colleagues.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, Zoran MJ. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yerushalmi S, Green RM. Evidence for the adaptive significance of circadian rhythms. Ecol Lett. 2009;12:970–981. doi: 10.1111/j.1461-0248.2009.01343.x. [DOI] [PubMed] [Google Scholar]
- 3.Hotta CT, Gardner MJ, Hubbard KE, Baek SJ, Dalchau N, Suhita D, Dodd AN, Webb AAR. Modulation of environmental responses of plants by circadian clocks. Plant Cell Environ. 2007;30:333–349. doi: 10.1111/j.1365-3040.2006.01627.x. [DOI] [PubMed] [Google Scholar]
- 4.Dodd AN, Salathia N, Hall A, Kévei E, Tóth R, Nagy F, Hibberd JM, Millar AJ, Webb AAR. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- 5.Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH. Resonating circadian clocks enhance fitness in cyanobacteria. Proc Natl Acad Sci USA. 1998;95:8660–8664. doi: 10.1073/pnas.95.15.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green RM, Tingay S, Wang ZY, Tobin EM. Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol. 2002;129:576–584. doi: 10.1104/pp.004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corellou F, Schwartz C, Motta JP, Djouani-Tahri EB, Sanchez F, Bouget FY. Clocks in the green lineage: comparative functional analysis of the circadian architecture of the picoeukaryote ostreococcus. Plant Cell. 2009;21:3436–3449. doi: 10.1105/tpc.109.068825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harmer SL. The circadian system in higher plants. Annu Rev Plant Biol. 2009;60:357–377. doi: 10.1146/annurev.arplant.043008.092054. [DOI] [PubMed] [Google Scholar]
- 9.Wijnen H, Naef F, Boothroyd C, Claridge-Chang A, Young MW. Control of daily transcript oscillations in Drosophila by light and the circadian clock. PLoS Genet. 2006;2:e39. doi: 10.1371/journal.pgen.0020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stratmann T, Más P. Chromatin, photoperiod and the Arabidopsis circadian clock: a question of time. Semin Cell Dev Biol. 2008;19:554–559. doi: 10.1016/j.semcdb.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 11.Nakahata Y, Grimaldi B, Sahar S, Hirayama J, Sassone-Corsi P. Signaling to the circadian clock: plasticity by chromatin remodeling. Curr Opin Cell Biol. 2007;19:230–237. doi: 10.1016/j.ceb.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 12.Fukushima A, Kusano M, Nakamichi N, Kobayashi M, Hayashi N, Sakakibara H, Mizuno T, Saito K. Impact of clock-associated Arabidopsis pseudo-response regulators in metabolic coordination. Proc Natl Acad Sci USA. 2009;106:7251–7256. doi: 10.1073/pnas.0900952106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, Maloof JN. Rhythmic growth explained by coincidence between internal and external cues. Nature. 2007;448:358–361. doi: 10.1038/nature05946. [DOI] [PubMed] [Google Scholar]
- 14.Mizuno T, Yamashino T. Comparative transcriptome of diurnally oscillating genes and hormone-responsive genes in Arabidopsis thaliana: insight into circadian clock-controlled daily responses to common ambient stresses in plants. Plant Cell Physiol. 2008;49:481–487. doi: 10.1093/pcp/pcn008. [DOI] [PubMed] [Google Scholar]
- 15.Wang W, Barnaby JY, Tada Y, Li H, Tör M, Caldelari D, Lee D, Fu XD, Dong X. Timing of plant immune responses by a central circadian regulator. Nature. 2011;470:110–114. doi: 10.1038/nature09766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 2008;9:R130. doi: 10.1186/gb-2008-9-8-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michael TP, Mockler TC, Breton G, McEntee C, Byer A, Trout JD, Hazen SP, Shen R, Priest HD, Sullivan CM, et al. Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet. 2008;4:e14. doi: 10.1371/journal.pgen.0040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293:880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- 19.Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Más P, Panda S, Kreps JA, Kay SA. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science. 2000;289:768–771. doi: 10.1126/science.289.5480.768. [DOI] [PubMed] [Google Scholar]
- 20.Wang ZY, Tobin EM. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell. 1998;93:1207–1217. doi: 10.1016/s0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
- 21.Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carré IA, Coupland G. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell. 1998;93:1219–1229. doi: 10.1016/s0092-8674(00)81465-8. [DOI] [PubMed] [Google Scholar]
- 22.Millar AJ, Carré IA, Strayer CA, Chua NH, Kay SA. Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science. 1995;267:1161–1163. doi: 10.1126/science.7855595. [DOI] [PubMed] [Google Scholar]
- 23.Harmer SL, Kay SA. Positive and negative factors confer phase-specific circadian regulation of transcription in Arabidopsis. Plant Cell. 2005;17:1926–1940. doi: 10.1105/tpc.105.033035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Locke JCW, Millar AJ, Turner MS. Modelling genetic networks with noisy and varied experimental data: the circadian clock in Arabidopsis thaliana. J Theor Biol. 2005;234:383–393. doi: 10.1016/j.jtbi.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 25.Pruneda-Paz JL, Kay SA. An expanding universe of circadian networks in higher plants. Trends Plant Sci. 2010;15:259–265. doi: 10.1016/j.tplants.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gendron JM, Pruneda-Paz JL, Doherty CJ, Gross AM, Kang SE, Kay SA. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc Natl Acad Sci USA. 2012;109:3167–3172. doi: 10.1073/pnas.1200355109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang W, Pérez-García P, Pokhilko A, Millar AJ, Antoshechkin I, Riechmann JL, Mas P. Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science. 2012;336:75–79. doi: 10.1126/science.1219075. [DOI] [PubMed] [Google Scholar]
- 28.Pokhilko A, Hodge SK, Stratford K, Knox K, Edwards KD, Thomson AW, Mizuno T, Millar AJ. Data assimilation constrains new connections and components in a complex, eukaryotic circadian clock model. Mol Syst Biol. 2010;6:416. doi: 10.1038/msb.2010.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pokhilko A, Fernández AP, Edwards KD, Southern MM, Halliday KJ, Millar AJ. The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol Syst Biol. 2012;8:574. doi: 10.1038/msb.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takata N, Saito S, Saito CT, Nanjo T, Shinohara K, Uemura M. Molecular phylogeny and expression of poplar circadian clock genes, LHY1 and LHY2. New Phytol. 2009;181:808–819. doi: 10.1111/j.1469-8137.2008.02714.x. [DOI] [PubMed] [Google Scholar]
- 31.Takata N, Saito S, Saito CT, Uemura M. Phylogenetic footprint of the plant clock system in angiosperms: evolutionary processes of pseudo-response regulators. BMC Evol Biol. 2010;10:126. doi: 10.1186/1471-2148-10-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holm K, Källman T, Gyllenstrand N, Hedman H, Lagercrantz U. Does the core circadian clock in the moss Physcomitrella patens (Bryophyta) comprise a single loop? BMC Plant Biol. 2010;10:109. doi: 10.1186/1471-2229-10-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murakami M, Ashikari M, Miura K, Yamashino T, Mizuno T. The evolutionarily conserved OsPRR quintet: rice pseudo-response regulators implicated in circadian rhythm. Plant Cell Physiol. 2003;44:1229–1236. doi: 10.1093/pcp/pcg135. [DOI] [PubMed] [Google Scholar]
- 34.Kim JA, Yang TJ, Kim JS, Park JY, Kwon SJ, Lim MH, Jin M, Lee SC, Lee SI, Choi BS, et al. Isolation of circadian-associated genes in Brassica rapa by comparative genomics with Arabidopsis thaliana. Mol Cells. 2007;23:145–153. [PubMed] [Google Scholar]
- 35.Farré EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA. Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol. 2005;15:47–54. doi: 10.1016/j.cub.2004.12.067. [DOI] [PubMed] [Google Scholar]
- 36.Portolés S, Más P. The functional interplay between protein kinase CK2 and CCA1 transcriptional activity is essential for clock temperature compensation in Arabidopsis. PLoS Genet. 2010;6:e1001201. doi: 10.1371/journal.pgen.1001201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua NH, Sakakibara H. PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell. 2010;22:594–605. doi: 10.1105/tpc.109.072892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knowles SM, Lu SX, Tobin EM. Testing time: can ethanol-induced pulses of proposed oscillator components phase shift rhythms in Arabidopsis? J Biol Rhythms. 2008;23:463–471. doi: 10.1177/0748730408326749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, Wu JF, Nakamichi N, Sakakibara H, Nam HG, Wu SH. LIGHT-REGULATED WD1 and PSEUDO-RESPONSE REGULATOR9 form a positive feedback regulatory loop in the Arabidopsis circadian clock. Plant Cell. 2011;23:486–498. doi: 10.1105/tpc.110.081661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu JF, Wang Y, Wu SH. Two new clock proteins, LWD1 and LWD2, regulate Arabidopsis photoperiodic flowering. Plant Physiol. 2008;148:948–959. doi: 10.1104/pp.108.124917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ding Z, Doyle MR, Amasino RM, Davis SJ. A complex genetic interaction between Arabidopsis thaliana TOC1 and CCA1/LHY in driving the circadian clock and in output regulation. Genetics. 2007;176:1501–1510. doi: 10.1534/genetics.107.072769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hazen SP, Schultz TF, Pruneda-Paz JL, Borevitz JO, Ecker JR, Kay SA. LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc Natl Acad Sci USA. 2005;102:10387–10392. doi: 10.1073/pnas.0503029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Onai K, Ishiura M. PHYTOCLOCK 1 encoding a novel GARP protein essential for the Arabidopsis circadian clock. Genes Cells. 2005;10:963–972. doi: 10.1111/j.1365-2443.2005.00892.x. [DOI] [PubMed] [Google Scholar]
- 44.Dixon LE, Knox K, Kozma-Bognar L, Southern MM, Pokhilko A, Millar AJ. Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis. Curr Biol. 2011;21:120–125. doi: 10.1016/j.cub.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Helfer A, Nusinow DA, Chow BY, Gehrke AR, Bulyk ML, Kay SA. LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr Biol. 2011;21:126–133. doi: 10.1016/j.cub.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chow BY, Helfer A, Nusinow DA, Kay SA. ELF3 recruitment to the PRR9 promoter requires other Evening Complex members in the Arabidopsis circadian clock. [Accessed April 27, 2012];Plant Signaling & Behavior. 2012 7 doi: 10.4161/psb.18766. Available at: http://www.ncbi.nlm.nih.gov/pubmed/22307044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dai S, Wei X, Pei L, Thompson RL, Liu Y, Heard JE, Ruff TG, Beachy RN. BROTHER OF LUX ARRHYTHMO is a component of the Arabidopsis circadian clock. Plant Cell. 2011;23:961–972. doi: 10.1105/tpc.111.084293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pruneda-Paz JL, Breton G, Para A, Kay SA. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science. 2009;323:1481–1485. doi: 10.1126/science.1167206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lau OS, Huang X, Charron JB, Lee JH, Li G, Deng XW. Interaction of Arabidopsis DET1 with CCA1 and LHY in mediating transcriptional repression in the plant circadian clock. Mol Cell. 2011;43:703–712. doi: 10.1016/j.molcel.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farré EM, Kay SA. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature. 2011;475:398–402. doi: 10.1038/nature10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Herrero E, Kolmos E, Bujdoso N, Yuan Y, Wang M, Berns MC, Uhlworm H, Coupland G, Saini R, Jaskolski M, et al. EARLY FLOWERING4 Recruitment of EARLY FLOWERING3 in the Nucleus Sustains the Arabidopsis Circadian Clock. Plant Cell. 2012;24:428–443. doi: 10.1105/tpc.111.093807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hicks KA, Albertson TM, Wagner DR. EARLY FLOWERING3 encodes a novel protein that regulates circadian clock function and flowering in Arabidopsis. Plant Cell. 2001;13:1281–1292. doi: 10.1105/tpc.13.6.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kikis EA, Khanna R, Quail PH. ELF4 is a phytochrome-regulated component of a negative-feedback loop involving the central oscillator components CCA1 and LHY. Plant J. 2005;44:300–313. doi: 10.1111/j.1365-313X.2005.02531.x. [DOI] [PubMed] [Google Scholar]
- 54.Doyle MR, Davis SJ, Bastow RM, McWatters HG, Kozma-Bognár L, Nagy F, Millar AJ, Amasino RM. The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature. 2002;419:74–77. doi: 10.1038/nature00954. [DOI] [PubMed] [Google Scholar]
- 55.Li G, Siddiqui H, Teng Y, Lin R, Wan X, Li J, Lau OS, Ouyang X, Dai M, Wan J, et al. Coordinated transcriptional regulation underlying the circadian clock in Arabidopsis. Nat Cell Biol. 2011;13:616–622. doi: 10.1038/ncb2219. [DOI] [PubMed] [Google Scholar]
- 56.Lu SX, Knowles SM, Andronis C, Ong MS, Tobin EM. CIRCADIAN CLOCK ASSOCIATED1 and LATE ELONGATED HYPOCOTYL function synergistically in the circadian clock of Arabidopsis. Plant Physiol. 2009;150:834–843. doi: 10.1104/pp.108.133272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Michael TP, McClung CR. Phase-specific circadian clock regulatory elements in Arabidopsis. Plant Physiol. 2002;130:627–638. doi: 10.1104/pp.004929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang ZY, Kenigsbuch D, Sun L, Harel E, Ong MS, Tobin EM. A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell. 1997;9:491–507. doi: 10.1105/tpc.9.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rawat R, Takahashi N, Hsu PY, Jones MA, Schwartz J, Salemi MR, Phinney BS, Harmer SL. REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genet. 2011;7:e1001350. doi: 10.1371/journal.pgen.1001350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Farinas B, Mas P. Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. Plant J. 2011;66:318–329. doi: 10.1111/j.1365-313X.2011.04484.x. [DOI] [PubMed] [Google Scholar]
- 61.Hervé C, Dabos P, Bardet C, Jauneau A, Auriac MC, Ramboer A, Lacout F, Tremousaygue D. In vivo interference with AtTCP20 function induces severe plant growth alterations and deregulates the expression of many genes important for development. Plant Physiol. 2009;149:1462–1477. doi: 10.1104/pp.108.126136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Giraud E, Ng S, Carrie C, Duncan O, Low J, Lee CP, Van Aken O, Millar AH, Murcha M, Whelan J. TCP transcription factors link the regulation of genes encoding mitochondrial proteins with the circadian clock in Arabidopsis thaliana. Plant Cell. 2010;22:3921–3934. doi: 10.1105/tpc.110.074518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Braun P, Carvunis AR, Charloteaux B, Dreze M, Ecker JR, Hill DE, Roth FP, Vidal M, Galli M, Balumuri P, et al. Evidence for network evolution in an Arabidopsis interactome map. Science. 2011;333:601–607. doi: 10.1126/science.1203877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mockler TC, Michael TP, Priest HD, Shen R, Sullivan CM, Givan SA, McEntee C, Kay SA, Chory J. The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb Symp Quant Biol. 2007;72:353–363. doi: 10.1101/sqb.2007.72.006. [DOI] [PubMed] [Google Scholar]
- 65.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Diernfellner A, Colot HV, Dintsis O, Loros JJ, Dunlap JC, Brunner M. Long and short isoforms of Neurospora clock protein FRQ support temperature-compensated circadian rhythms. FEBS Lett. 2007;581:5759–5764. doi: 10.1016/j.febslet.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Colot HV, Loros JJ, Dunlap JC. Temperature-modulated alternative splicing and promoter use in the Circadian clock gene frequency. Mol Biol Cell. 2005;16:5563–5571. doi: 10.1091/mbc.E05-08-0756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, Wong WK, Mockler TC. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010;20:45–58. doi: 10.1101/gr.093302.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.James AB, Syed NH, Bordage S, Marshall J, Nimmo GA, Jenkins GI, Herzyk P, Brown JWS, Nimmo HG. Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. Plant Cell. 2012;24:961–981. doi: 10.1105/tpc.111.093948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Deng X, Gu L, Liu C, Lu T, Lu F, Lu Z, Cui P, Pei Y, Wang B, Hu S, et al. Arginine methylation mediated by the Arabidopsis homolog of PRMT5 is essential for proper pre-mRNA splicing. Proc Natl Acad Sci USA. 2010;107:19114–19119. doi: 10.1073/pnas.1009669107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hong S, Song HR, Lutz K, Kerstetter RA, Michael TP, McClung CR. Type II protein arginine methyltransferase 5 (PRMT5) is required for circadian period determination in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2010;107:21211–21216. doi: 10.1073/pnas.1011987107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sanchez SE, Petrillo E, Beckwith EJ, Zhang X, Rugnone ML, Hernando CE, Cuevas JC, Godoy Herz MA, Depetris-Chauvin A, Simpson CG, et al. A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature. 2010;468:112–116. doi: 10.1038/nature09470. [DOI] [PubMed] [Google Scholar]
- 73.Más P. Circadian clock function in Arabidopsis thaliana: time beyond transcription. Trends Cell Biol. 2008;18:273–281. doi: 10.1016/j.tcb.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 74.Mehra A, Baker CL, Loros JJ, Dunlap JC. Post-translational modifications in circadian rhythms. Trends Biochem Sci. 2009;34:483–490. doi: 10.1016/j.tibs.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mizoguchi T, Putterill J, Ohkoshi Y. Kinase and phosphatase: the cog and spring of the circadian clock. Int Rev Cytol. 2006;250:47–72. doi: 10.1016/S0074-7696(06)50002-6. [DOI] [PubMed] [Google Scholar]
- 76.Tsuchiya Y, Akashi M, Matsuda M, Goto K, Miyata Y, Node K, Nishida E. Involvement of the protein kinase CK2 in the regulation of mammalian circadian rhythms. Sci Signal. 2009;2:ra26. doi: 10.1126/scisignal.2000305. [DOI] [PubMed] [Google Scholar]
- 77.Lin JM, Kilman VL, Keegan K, Paddock B, Emery-Le M, Rosbash M, Allada R. A role for casein kinase 2alpha in the Drosophila circadian clock. Nature. 2002;420:816–820. doi: 10.1038/nature01235. [DOI] [PubMed] [Google Scholar]
- 78.Mehra A, Shi M, Baker CL, Colot HV, Loros JJ, Dunlap JC. A role for casein kinase 2 in the mechanism underlying circadian temperature compensation. Cell. 2009;137:749–760. doi: 10.1016/j.cell.2009.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sugano S, Andronis C, Ong MS, Green RM, Tobin EM. The protein kinase CK2 is involved in regulation of circadian rhythms in Arabidopsis. Proc Natl Acad Sci USA. 1999;96:12362–12366. doi: 10.1073/pnas.96.22.12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Daniel X, Sugano S, Tobin EM. CK2 phosphorylation of CCA1 is necessary for its circadian oscillator function in Arabidopsis. Proc Natl Acad Sci USA. 2004;101:3292–3297. doi: 10.1073/pnas.0400163101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lu SX, Liu H, Knowles SM, Li J, Ma L, Tobin EM, Lin C. A role for protein kinase casein kinase2 α-subunits in the Arabidopsis circadian clock. Plant Physiol. 2011;157:1537–1545. doi: 10.1104/pp.111.179846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Song HR, Carré IA. DET1 regulates the proteasomal degradation of LHY, a component of the Arabidopsis circadian clock. Plant Mol Biol. 2005;57:761–771. doi: 10.1007/s11103-005-3096-z. [DOI] [PubMed] [Google Scholar]
- 83.Park BS, Eo HJ, Jang IC, Kang HG, Song JT, Seo HS. Ubiquitination of LHY by SINAT5 regulates flowering time and is inhibited by DET1. Biochem Biophys Res Commun. 2010;398:242–246. doi: 10.1016/j.bbrc.2010.06.067. [DOI] [PubMed] [Google Scholar]
- 84.Fujiwara S, Wang L, Han L, Suh SS, Salomé PA, McClung CR, Somers DE. Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J Biol Chem. 2008;283:23073–23083. doi: 10.1074/jbc.M803471200. [DOI] [PubMed] [Google Scholar]
- 85.David KM, Armbruster U, Tama N, Putterill J. Arabidopsis GIGANTEA protein is post-transcriptionally regulated by light and dark. FEBS Lett. 2006;580:1193–1197. doi: 10.1016/j.febslet.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 86.Más P, Kim WY, Somers DE, Kay SA. Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature. 2003;426:567–570. doi: 10.1038/nature02163. [DOI] [PubMed] [Google Scholar]
- 87.Baudry A, Ito S, Song YH, Strait AA, Kiba T, Lu S, Henriques R, Pruneda-Paz JL, Chua NH, Tobin EM, et al. F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell. 2010;22:606–622. doi: 10.1105/tpc.109.072843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang L, Fujiwara S, Somers DE. PRR5 regulates phosphorylation, nuclear import and subnuclear localization of TOC1 in the Arabidopsis circadian clock. EMBO J. 2010;29:1903–1915. doi: 10.1038/emboj.2010.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Para A, Farré EM, Imaizumi T, Pruneda-Paz JL, Harmon FG, Kay SA. PRR3 Is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock. Plant Cell. 2007;19:3462–3473. doi: 10.1105/tpc.107.054775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kiba T, Henriques R, Sakakibara H, Chua NH. Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by an SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell. 2007;19:2516–2530. doi: 10.1105/tpc.107.053033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kim WY, Fujiwara S, Suh SS, Kim J, Kim Y, Han L, David K, Putterill J, Nam HG, Somers DE. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature. 2007;449:356–360. doi: 10.1038/nature06132. [DOI] [PubMed] [Google Scholar]
- 92.Perales M, Más P. A functional link between rhythmic changes in chromatin structure and the Arabidopsis biological clock. Plant Cell. 2007;19:2111–2123. doi: 10.1105/tpc.107.050807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ni Z, Kim ED, Ha M, Lackey E, Liu J, Zhang Y, Sun Q, Chen ZJ. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature. 2009;457:327–331. doi: 10.1038/nature07523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jones MA, Covington MF, DiTacchio L, Vollmers C, Panda S, Harmer SL. Jumonji domain protein JMJD5 functions in both the plant and human circadian systems. Proc Natl Acad Sci USA. 2010;107:21623–21628. doi: 10.1073/pnas.1014204108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lu SX, Knowles SM, Webb CJ, Celaya RB, Cha C, Siu JP, Tobin EM. The Jumonji C domain-containing protein JMJ30 regulates period length in the Arabidopsis circadian clock. Plant Physiol. 2011;155:906–915. doi: 10.1104/pp.110.167015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hsia DA, Tepper CG, Pochampalli MR, Hsia EYC, Izumiya C, Huerta SB, Wright ME, Chen HW, Kung HJ, Izumiya Y. KDM8, a H3K36me2 histone demethylase that acts in the cyclin A1 coding region to regulate cancer cell proliferation. Proc Natl Acad Sci USA. 2010;107:9671–9676. doi: 10.1073/pnas.1000401107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jones MA, Harmer S. JMJD5 Functions in concert with TOC1 in the arabidopsis circadian system. Plant Signal Behav. 2011;6:445–448. doi: 10.4161/psb.6.3.14654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yakir E, Hilman D, Harir Y, Green RM. Regulation of output from the plant circadian clock. FEBS J. 2007;274:335–345. doi: 10.1111/j.1742-4658.2006.05616.x. [DOI] [PubMed] [Google Scholar]
- 99.Dowson-Day MJ, Millar AJ. Circadian dysfunction causes aberrant hypocotyl elongation patterns in Arabidopsis. Plant J. 1999;17:63–71. doi: 10.1046/j.1365-313x.1999.00353.x. [DOI] [PubMed] [Google Scholar]
- 100.Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 2008;53:312–323. doi: 10.1111/j.1365-313X.2007.03341.x. [DOI] [PubMed] [Google Scholar]
- 101.Huq E, Quail PH. PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 2002;21:2441–2450. doi: 10.1093/emboj/21.10.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Khanna R, Huq E, Kikis EA, Al-Sady B, Lanzatella C, Quail PH. A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell. 2004;16:3033–3044. doi: 10.1105/tpc.104.025643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fujimori T, Yamashino T, Kato T, Mizuno T. Circadian-controlled basic/helix-loop-helix factor, PIL6, implicated in light-signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 2004;45:1078–1086. doi: 10.1093/pcp/pch124. [DOI] [PubMed] [Google Scholar]
- 104.Nozue K, Harmer SL, Maloof JN. Genomic analysis of circadian clock-, light-, and growth-correlated genes reveals PHYTOCHROME-INTERACTING FACTOR5 as a modulator of auxin signaling in Arabidopsis. Plant Physiol. 2011;156:357–372. doi: 10.1104/pp.111.172684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kunihiro A, Yamashino T, Nakamichi N, Niwa Y, Nakanishi H, Mizuno T. Phytochrome-interacting factor 4 and 5 (PIF4 and PIF5) activate the homeobox ATHB2 and auxin-inducible IAA29 genes in the coincidence mechanism underlying photoperiodic control of plant growth of Arabidopsis thaliana. Plant Cell Physiol. 2011;52:1315–1329. doi: 10.1093/pcp/pcr076. [DOI] [PubMed] [Google Scholar]
- 106.Imaizumi T, Kay SA. Photoperiodic control of flowering: not only by coincidence. Trends Plant Sci. 2006;11:550–558. doi: 10.1016/j.tplants.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 107.Koornneef M, Alonso-Blanco C, Blankestijn-de Vries H, Hanhart CJ, Peeters AJ. Genetic interactions among late-flowering mutants of Arabidopsis. Genetics. 1998;148:885–892. doi: 10.1093/genetics/148.2.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Somers DE, Webb AA, Pearson M, Kay SA. The short-period mutant, toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development. 1998;125:485–494. doi: 10.1242/dev.125.3.485. [DOI] [PubMed] [Google Scholar]
- 109.Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, Coupland G, Putterill J. GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 1999;18:4679–4688. doi: 10.1093/emboj/18.17.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature. 2001;410:1116–1120. doi: 10.1038/35074138. [DOI] [PubMed] [Google Scholar]
- 111.Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;303:1003–1006. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- 112.Sawa M, Kay SA. GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2011;108:11698–11703. doi: 10.1073/pnas.1106771108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Thain SC, Murtas G, Lynn JR, McGrath RB, Millar AJ. The circadian clock that controls gene expression in Arabidopsis is tissue specific. Plant Physiol. 2002;130:102–110. doi: 10.1104/pp.005405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.James AB, Monreal JA, Nimmo GA, Kelly CL, Herzyk P, Jenkins GI, Nimmo HG. The circadian clock in Arabidopsis roots is a simplified slave version of the clock in shoots. Science. 2008;322:1832–1835. doi: 10.1126/science.1161403. [DOI] [PubMed] [Google Scholar]
- 115.Yakir E, Hassidim M, Melamed-Book N, Hilman D, Kron I, Green RM. Cell autonomous and cell-type specific circadian rhythms in Arabidopsis. Plant J. 2011;68:520–531. doi: 10.1111/j.1365-313X.2011.04707.x. [DOI] [PubMed] [Google Scholar]
- 116.Wenden B, Toner DLK, Hodge SK, Grima R, Millar AJ. Spontaneous spatiotemporal waves of gene expression from biological clocks in the leaf. Proc Natl Acad Sci USA. 2012;109:6757–6762. doi: 10.1073/pnas.1118814109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dalchau N, Baek SJ, Briggs HM, Robertson FC, Dodd AN, Gardner MJ, Stancombe MA, Haydon MJ, Stan GB, Gonçalves JM, et al. The circadian oscillator gene GIGANTEA mediates a long-term response of the Arabidopsis thaliana circadian clock to sucrose. Proc Natl Acad Sci USA. 2011;108:5104–5109. doi: 10.1073/pnas.1015452108. [DOI] [PMC free article] [PubMed] [Google Scholar]