Abstract
The A2a adenosine receptor is a member of the G-protein coupled receptor family, and its activation stimulates cyclic AMP production. To determine the residues which are involved in ligand binding, several residues in transmembrane domains 5–7 were individually replaced with alanine and other amino acids. The binding properties of the resultant mutant receptors were determined in transfected COS-7 cells. To study the expression levels in COS-7 cells, mutant receptors were tagged at their amino terminus with a hemagglutinin epitope, which allowed their immunological detection in the plasma membrane by the monoclonal antibody 12CA5. The functional properties of mutant receptors were determined by measuring stimulation of adenylate cyclase. Specific binding of [3H]CGS 21680 (15 nm) and [3H]XAC (4 nm), an A2a agonist and antagonist, respectively, was absent in the following Ala mutants: F182A, H250A, N253A, I274A, H278A, and S281A, although they were well expressed in the plasma membrane. The hydroxy group of Ser-277 is required for high affinity binding of agonists, but not antagonists. An N181S mutant lost affinity for adenosine agonists substituted at N6 or C-2, but not at C-5′. The mutant receptors I274A, S277A, and H278A showed full stimulation of adenylate cyclase at high concentrations of CGS 21680. The functional agonist potencies at mutant receptors that lacked radioligand binding were >30-fold less than those at the wild type receptor. His-250 appears to be a required component of a hydrophobic pocket, and H-bonding to this residue is not essential. On the other hand, replacement of His-278 with other aromatic residues was not tolerated in ligand binding. Thus, some of the residues targeted in this study may be involved in the direct interaction with ligands in the human A2a adenosine receptor. A molecular model based on the structure of rhodopsin, in which the 5’-NH in NECA is hydrogen bonded to Ser-277 and His-278, was developed in order to visualize the environment of the ligand binding site.
Adenosine is a ubiquitous mediator (reviewed in Jacobson et al. (1992a)) that regulates homeostasis in many organs and offers protection under conditions of stress, such as ischemia. The A1, A2a, A2b, and A3 subtypes have been cloned (Mahan et al., 1991; Maenhaut et al., 1990; Jacobson, 1995; Zhou et al., 1992). The A2a adenosine receptor activates adenylate cyclase (Hide et al., 1992) via coupling to Gs. Adenosine regulates blood pressure by centrally (Barraco et al., 1994) and peripherally mediated mechanisms (Olsson and Pearson, 1990). Activation of A2a receptors results in vasodilation, and this effect has been examined as a potential hypotensive therapy using selective A2a agonists such as CGS 216801 (Jarvis et al., 1989). A2a receptors are also present in platelets, where they inhibit aggregation, and in the liver. In the brain, A2a receptors occur primarily in the striatum, where they are involved in dopaminergic pathways (Ferré et al., 1994) and elicit locomotor depression (Nikodijević et al., 1993). Consequently therapeutic approaches, based on activating or inhibiting A2a receptors, to diseases involving the dopaminergic system, i.e. Parkinson’s disease (Kanda et al., 1994), schizophrenia (Martin et al., 1993), and Huntington’s disease (Nikodijević et al., 1993), are under investigation.
Structural studies of A2a receptors have been impeded by the lack of a selective high affinity A2a antagonist. We have shown that an 8-phenylxanthine antagonist, [3H]XAC, binds to A2a receptors with especially high affinity in the human (Ji et al., 1992) and rabbit (Jacobson et al., 1992b) striatum (XAC is non-selective in these species), and therefore this ligand was selected for use in the present study for characterizing mutant human A2a receptors.
Until the present study, site-directed mutagenesis of A2a receptors has not been reported, however, mutational studies have provided insight into the ligand binding site of A1 receptors. Olah et al. (1992) demonstrated the involvement in ligand binding of two His residues in the sixth and seventh transmembrane helical domains (TMs). In bovine A1 receptors, His-250 (TM6) has been shown to be important for antagonist binding, and His-278 (TM7) is important for both agonist and antagonist binding. Mutagenesis studies of other adenosine receptor subtypes have implicated a hexapeptide region in TM5 of rat A3 receptors (Olah et al., 1994) and Ile-274 and Ser-277 in TM7 of bovine A1 receptors (Townsend-Nicholson and Schofield, 1994; Tucker et al., 1994) in binding of adenosine derivatives.
Chemical modification studies of A2a receptors indicated that, as in A1 receptors, His residues appear to be involved in ligand binding (Jacobson et al., 1992b). Two conserved His residues are present in TM6 and TM7. Using an agonist molecular probe, the bovine A2a receptor was photoaffinity labeled (Barrington et al., 1989). The site of this labeling was later shown using peptide mapping to occur on TM5 (Piersen et al., 1994).
A molecular model of the rat A2a receptor (IJzerman et al., 1994) was deduced from the primary sequence and by computer assisted homology modeling, based on the electron density map of bacteriorhodopsin. According to the model, TM5, TM6, and TM7 are most likely involved in ligand binding. As with other receptors, the transmembrane helices tend to be amphipathic, with the more hydrophilic sides postulated to be facing the ligand binding cleft and in direct contact with the ligand. In particular, residues Phe-182, His-250, Asn-253, His-278, and Ser-281 (numbering for human sequence) were predicted to be involved in ligand binding. In the present study, these and other residues in the same transmembrane region were selected as sites for the replacement of single amino acids, to probe the influence of individual side chains on molecular recognition by the human A2a receptor.
Finally, based on the results of site-directed mutagenesis, we have utilized molecular modeling to predict the environment of the ligand binding pocket. Molecular modeling serves to test the internal consistency of the results through a unified model that is consonant with all of the pharmacological observations. The approach to receptor modeling used in the present study is based on homology with rhodopsin (Ballesteros and Weinstein, 1995), for which low resolution electron density information has been reported (Henderson et at., 1990). The homology in sequence and structure between rhodopsin and other G-protein coupled receptors has been explored (Baldwin, 1993). We have recently applied the rhodopsin-based modeling approach to ligand binding at the P2y (ATP) receptor (van Rhee et al., 1994).
EXPERIMENTAL PROCEDURES
Materials
Human A2a adenosine receptor cDNA (pSVLA2a) was provided by Dr. Marlene A. Jacobson (Merck Research Labs, West Point, PA). Taq polymerase for the polymerase chain reaction (PCR) was purchased from Perkin Elmer (Emeryville CA). All enzymes used in this study were obtained from New England Biolabs (Beverly, MA). The agonists CGS 21680, NECA, R-PIA, 2-chloroadenosine, and DPMA, and the antagonists XAC and CGS 15943 were from RBI (Natick, MA). [3H]CGS 21680 (38.3 Ci/mmol) and [3H]XAC (118 Ci/mmol) were obtained from DuPont NEN, and [3H]adenine (15 Ci/mmol) was purchased from American Research Chemicals Inc. (St. Louis, MO). IB-MECA was prepared as described (Gallo-Rodriguez et al., 1994). Fetal bovine serum and o-phenylenediamine dihydrochloride were purchased from Sigma. The Sequenase kit, ATP, and cyclic AMP were from U. S. Biochemical Corp. (Cleveland, OH). All oligonucleotides used were synthesized by Bioserve Biotechnologies (Laurel, MD). A monoclonal antibody (12CA5) against a hemagglutinin epitope (HA) was purchased from Boehringer Mannheim Biochemicals (Indianapolis, IN), and goat anti-mouse IgG (γ-chain specific) antibody conjugated with horseradish peroxidase was purchased from Sigma. DEAE-dextran was obtained from Pharmacia Biotech Inc. (Piscataway, NJ). Rolipram was a gift of Schering AG (Berlin, Germany).
Plasmid Construction and Site-directed Mutagenesis
The coding region of pSVLA2a was subcloned into the pcD cDNA expression vector (Okayama and Berg, 1983), yielding pcDA2a. All mutations were introduced into pcDA2a using standard PCR mutagenesis techniques (Higuchi, 1989). The correctness of all PCR-derived sequences was confirmed by dideoxy sequencing of the mutant plasmids (Sanger et al., 1977).
Epitope Tagging
A 9-amino acid sequence derived from the influenza virus hemagglutinin protein (TAC CCC TAC GAC GTC CCC GAC TAC GCC; peptide sequence: YPYDVPDYA) was inserted into three different locations at the extracellular NH2 terminus of the A2a adenosine receptor gene (Fig. 3A). Oligonucleotides containing the HA-tag sequence were designed and used to generate PCR fragments, which were then used to replace the homologous wild type pcDA2a sequences.
Fig. 3.
A, introduction of HA epitope tags into the amino terminus of the human A2a adenosine receptor. B, amount of absorbance detected in ELISA experiments with the wild type and the various epitope-tagged A2a receptors expressed in COS-7 cells. ELISA measurements were carried out in 96-well plates as described under “Experimental Procedures.” Data are presented as means ± S.E. of two or three independent experiments, each performed in triplicate.
Transient Expression of Mutant Receptors in COS-7 Cells
2 × 106 COS-7 cells were seeded into 100-mm culture dishes containing 10 ml of Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. Cells were transfected with plasmid DNA (4 µg of DNA/dish) by the DEAE-dextran method (Cullen, 1987) about 24 h later, and grown for an additional 72 h at 37 °C.
Membrane Preparation and Radioligand Binding Assay
Cells were scraped into ice-cold lysis buffer (4 ml of 50 mm Tris, pH 6.8, at room temperature, containing 10 mm MgCl2). Harvested cells were homogenized using a Polytron homogenizer and then spun at 27,000 × g for 15 min. Cell membranes (pellet) were resuspended in the same buffer.
For saturation and competition binding experiments, each tube contained 100 µl of membrane suspension, 50 µl of radioligand, and either 50 µl of buffer/competitor (50 mm Tris, pH 6.8, 10 mm MgCl2) or 50 µl of 80 µm CADO in buffer (to define nonspecific binding) and finally 2 units/ml adenosine deaminase (Boehringer Mannheim). The mixtures were incubated at 25 °C for 90 min, filtered, and washed three times with ~5 ml of ice-cold buffer each using a Brandel cell harvester. Data analysis was performed by using the GraphPad program.
Cyclic AMP Determination
cAMP levels were determined by measuring the conversion of [3H]ATP to cyclic [3H]AMP. One day after transfection, cells were transferred from 100-mm dishes into 6 well dishes (about 3 × 105 cells/well) and incubated with culture media containing 2 µCi/ml [3H]adenine. After 24 h, the cultures were washed and incubated with 1 ml/well Hank’s balanced salt solution containing 0.1 mm rolipram for 15 min at 37 °C. The cells were incubated with different concentrations of the agonist CGS 21680 (in culture media) for 30 min at 37 °C. The reaction was terminated by aspiration of the media and addition of 1 ml of ice-cold 5% trichloroacetic acid containing 1 mm ATP and 1 mm cAMP. After 30 min incubation at 4 °C, cell lysates were eluted through sequential chromatography on Dowex and alumina columns (Enjalbert and Bockaert, 1983). Cyclic AMP formation is expressed as fold-stimulation of conversion of [3H]ATP into [3H]cyclic AMP (Weiss et al., 1985).
ELISA
For indirect cellular ELISA measurements, cells were transferred to 96-well dishes (4–5 × 104 cells/well) 1 day after transfection. About 48 h after splitting, cells were fixed in 4% formaldehyde in phosphate-buffered saline for 30 min at room temperature. After washing with phosphate-buffered saline three times and blocking with Dulbecco’s modified Eagle’s medium (containing 10% fetal bovine serum), cells were incubated with HA-specific monoclonal antibody (12CA5), 20 µg/ml, for 3 h at 37 °C. Plates were washed and incubated with a 1:2000 dilution of a peroxidase-conjugated goat anti-mouse IgG antibody (Sigma) for 1 h, at 37 °C. H2O2 and o-phenylenediamine (2.5 mm in 0.1 m phosphate/citrate buffer, pH 5.0) served as substrate and chromogen, respectively. The enzymatic reaction was stopped after 30 min at room temperature with 1 m H2SO4 solution containing 0.05 m Na2SO3, and the color development was measured bichromatically in the BioKinetics reader (EL 312, Bio Tek Instruments, Inc., Winooski, VT) at 490 and 630 nm (reference wavelength).
Western Blot Analysis
Whole cell or membrane fractions (20 µg) were fractionated by denaturing SDS-polyacrylamide gel electrophoresis (12%) and were assayed by Western analysis (detailed procedure found in Bio-Rad product guide). Anti-HA antibody (20 µg/ml) was incubated for 2 h, and the secondary alkaline phosphatase-conjugated goat anti-mouse IgG was used in 1:2,000 dilution.
Molecular Modeling
A sequence alignment of 40 G-protein coupled receptors was constructed to assist in locating common amphipathic patterns in helical regions. The human A2a model was built and optimized using the InsightII/Discover modeling package (BIOSYM Technologies, San Diego, CA, versions 2.2.0 and 2.90, respectively), employing the Amber force field, running on a Silicon Graphics Indigo XZ4000 workstation (Silicon Graphics Inc., Mountain View CA), based on the methods described elsewhere (van Rhee et al., 1994; Ballesteros and Weinstein, 1995). Briefly, TMs were identified with the aid of Kyte-Doolittle hydrophobicity and Emini surface probability parameters. Transmembrane helices were built from the sequences and minimized individually. The minimized helices were then grouped together to form a helical bundle that matches the overall characteristics of the electron density map of rhodopsin (Schertler et al., 1993) and the model of rhodopsin as proposed by Baldwin (1993). The helical bundle was minimized in a stepwise process gradually releasing tethering of the backbone. Initially, 500 steps conjugate gradient were performed with the backbone of the helices tethered with a force constant of 100 kcal/Å. In consecutive runs (500 steps conjugate gradient each), the force constant was reduced to 50 kcal/Å, then 25 kcal/Å and, finally, without tethering. NECA was then docked into the helical bundle with the express purpose to explain as many experimental data as possible, regardless of the conformation of the ligand. The NECA·A2a complex was then minimized using conjugate gradient until the root mean square derivative was <0.1 kcal/mol/Å. The energy of the complex was 36.8 kcal/mol lower than the components.
RESULTS
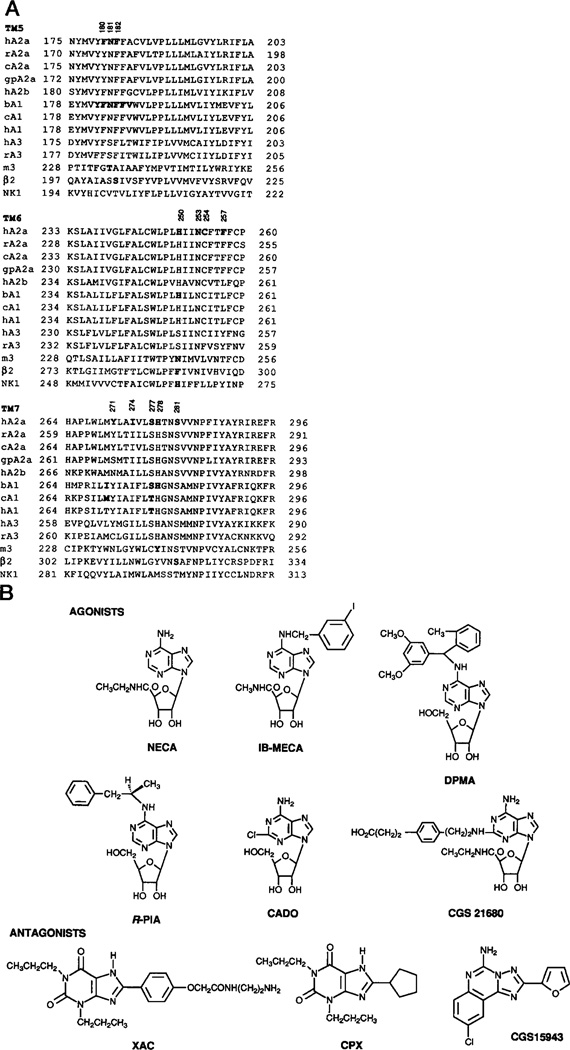
The A2a receptor has been cloned initially from a canine thyroid cDNA library (Maenhaut et al., 1990) and later in other species from the brain (Jacobson, 1995). A sequence alignment was carried out, and the fifth, sixth, and seventh transmembrane helical domains thereof (TM5-TM7) are shown in Fig. 1A. In these regions there is a high degree of sequence homology with rat, canine, and guinea pig A2a receptors and with other subtypes of adenosine receptors.
Fig. 1.
A, location of mutations carried out in this study, illustrated through an alignment of TMs 5, 6, and 7 of selected adenosine receptor subtypes. Individually mutated residues in the A2a (present study), A1 (Townsend-Nicholson and Schofield, 1994; Tucker et al., 1994), and m3 muscarinic receptors (Wess et al., 1991; Blüml et al., 1994) and a hexapeptide mutated in a chimeric bovine A1/rat A3 construct (Olah et al., 1994) are shown in bold type. Accession numbers are: hA2a (human) P29274; rA2a (rat) P30543; cA2a (canine) P11617; gpA2a (guinea-pig) U04201; hA2b (human) P29275; bAl (bovine) P28190; cAl (canine) P11616; hAl (human) P30542; hA3 (human) P33765; rA3 (rat) P28647; m3 (rat) P08483; β2 (hamster) P04274; and NK1 (human) P25103. B, structures of ligands used in this study.
The 12 residues of the human A2a receptor, selected as targets for site-directed mutagenesis, are shown in bold type (Fig. 1A). They include residues that are conserved among all known adenosine receptor sequences (Phe-182, Asn-253, Ile-274, His-278, and Ser-281), conserved among all A1 and A2 receptors (Phe-181 and His-250), conserved among all A1 and A3 receptors (Ser-277), present only in A2 receptors (Phe-257), or not conserved (Tyr-271). Cys-254 is conserved among nearly all adenosine receptors, and Phe-180 is conserved as Phe or Tyr. Each of these amino acid residues was individually replaced with Ala and/or other amino acids (see below). In addition, each mutant contained an epitope-tag sequence included at the NH2 terminus for immunological detection (see below), and pharmacological properties were compared with the wild type receptor similarly modified.
Ligand Binding Properties of Mutant Human A2a Adenosine Receptors
A modeling study of A2a receptors hypothesized that the residues: Phe-182 (TM5), His-250 (TM6), Asn-253 (TM6), His-278 (TM7), and Ser-281 (TM7) might be involved in direct ligand contact (IJzerman et al., 1994). Radioligand binding studies showed that mutant receptors, in which each of these residues was individually modified to Ala, were unable to bind either the agonist [3H]CGS 21680 (Table I) or the antagonist [3H]XAC (Table II). The five Ala mutant receptors displayed less than 2% (<500 cpm) of the specific binding of [3H]CGS 21680 (15 nm) observed with the wild type receptor (typically 28,000 cpm/25 µg of protein). Similarly, in these Ala mutants, the specific binding of 4 nm [3H]XAC was greatly diminished (<800 cpm/25 µg of protein for mutants versus 8800 cpm for wild type). Assuming that the Ala mutant receptors are properly expressed on the cell surface (see ELISA results below), these results indicate that the above five residues are important, either directly or indirectly, for the high affinity binding of both agonist and antagonist ligands.
Table I. Binding characteristics of wild type and mutant human A2a adenosine receptors using an agonist radioligand.
Data are presented as means ± S.E. of two or three independent experiments, each performed in duplicate. Each sample contained 7–11 µg of membrane protein/tube.
| [3H]CGS 21680 bindinga | Competitor | |||||||
|---|---|---|---|---|---|---|---|---|
| Mutantb | Kd | Bmax | Agonists | Antagonists | ||||
| CADO | DPMA | NECA | R-PIA | CGS15943 | XAC | |||
| nm | pmol/mg | nm | nm | |||||
| wt (Tag3) | 21.7 ± 2.6 | 34.5 ± 20.0 | 152 ± 10 | 244 ± 43 | 19.9 ± 8.3 | 318 ± 8.0 | 10.1 ± 2.0 | 63.2 ± 5.5 |
| F180A | 15.1 ± 1.3 | 9.6 ± 0.8 | NDc | 104 ± 19 | 46.0 ± 11.1 | ND | 13.7 ± 3.2 | 90.3 ± 7.2 |
| N181S | 24.8 ± 5.6 | 21.9 ± 0.1 | 1340 ± 200 | 981 ± 52 | 16.4 ± 2.5 | 4500 ± 1200 | 23.7 ± 2.0 | 130 ± 10.0 |
| F182Y | 57 ± 15 | 8.9 ± 1.0 | 1890 ± 170 | 1720 ± 98 | 170 ± 11 | 2800 ± 1100 | 19.2 ± 4.2 | 140 ± 20.1 |
| F182w | 66 ± 6 | 7.4 ± 3.0 | 776 ± 120 | 756 ± 102 | 144 ± 12 | 1600 ± 460 | 10.3 ± 3.3 | 66.8 ± 1.0 |
| H250F | 14.4 ± 4.0 | 3.8 ± 1.8 | 2130 ± 100 | 1650 ± 107 | 41.9 ± 12 | 7950 ± 1750 | 25.7 ± 5.0 | 60.3 ± 3.0 |
| H250Y | 14.9 ± 1.0 | 5.2 ± 0.5 | 8250 ± 1400 | 3000 ± 80 | 43.8 ± 16 | 9580 ± 280 | 8.4 ± 1.0 | 52.7 ± 11 |
| C254A | 23.7 ± 2.4 | 11.8 ± 0.6 | ND | 99.6 ± 14.0 | 18.2 ± 10.3 | ND | 19.4 ± 3.4 | 54.1 ± 8.1 |
| S277N | 25.0 ± 5.0 | 23.4 ± 3.0 | 221 ± 21 | 311 ± 32 | 43.0 ± 17.1 | 631 ± 329 | 12.8 ± 2.8 | 60.0 ± 17.4 |
| S277T | 23.6 ± 6.3 | 27.3 ± 3.2 | 201 ± 5.0 | 305 ± 20 | 39.8 ± 6.8 | 534 ± 152 | 12.2 ± 1.0 | 65.9 ± 2.2 |
| S281T | 17.1 ± 2.0 | 10.2 ± 1.0 | 13.7 ± 1.3 | 80.5 ± 22 | 6.6 ± 0.8 | 30.5 ± 8.6 | 15.6 ± 3.4 | 17.3 ± 1.6 |
Agonist and antagonist binding affinities (Ki values, structures in Jacobson et al. (1992)) were determined in [3H]CGS 21680 (15 nm) competition binding studies using membrane homogenates prepared from transiently transfected COS-7 cells, as described under “Experimental Procedures.” Ki values were calculated from IC50 values by using the GraphPad program. All constructs contain the Tag3 sequence at the NH2 terminus (Fig. 3A).
Constructs that showed that <2% of specific binding of [3H]CGS21680 (15 nm) found for wild type Tag3 receptors were: F182A, H250A, N253A, N253S, N253Q, F257A, Y271A, I274A, S277A, H278A, H278Q, H278F, H278Y, S281A.
ND, not determined.
Table II. [3H]XAC binding properties of wild type, S277A, and other mutant human A2a adenosine receptors.
Table shows mean ± S.E. of two independent experiments, each performed in duplicate. [3H]XAC saturation binding studies were carried out with membrane homogenates prepared from transiently transfected COS-7 cells as described under “Experimental Procedures.” Bmax values indicate the maximum number of binding sites/mg of membrane protein.
| Mutanta | Kd | Bmax |
|---|---|---|
| nm | pmol/mg | |
| wt (Tag3) | 9.4 ± 2.3 | 12.3 ± 2.4 |
| S277A | 6.7 ± 1.2 | 25.6 ± 3.3 |
The following constructs showed <10% specific binding of [3H]XAC (4 nm) found for wild type Tag3 receptors: F182A, H250A, N253A, N253S, N253Q, F257A, I274A, H278A, H278Q, H278F, H278Y, S281A.
To provide more detail, we mutated several additional residues (Phe-180, Cys-254, Phe-257, Tyr-271, Ile-274, and Ser-277), that were nearby in the sequence and/or predicted by molecular modeling (IJzerman et al., 1994) to be in proximity to the bound ligand. The F257A, Y271A, and I274A mutant receptors did not bind either [3H]CGS 21680 (15 nm) or [3H]XAC (4 nm). However, the F180A and C254A mutant receptors showed ligand binding properties similar to those of the wild type receptor (Table I). Competition binding studies showed that the F180A and C254A mutant receptors had the same affinities for NECA, DPMA, CGS 15943, and XAC (structures in Fig. 1B) as the wild type receptor. Therefore, the aromatic group in Phe-180 and the -SH group in Cys-254 are not required for binding of either agonists or antagonists.
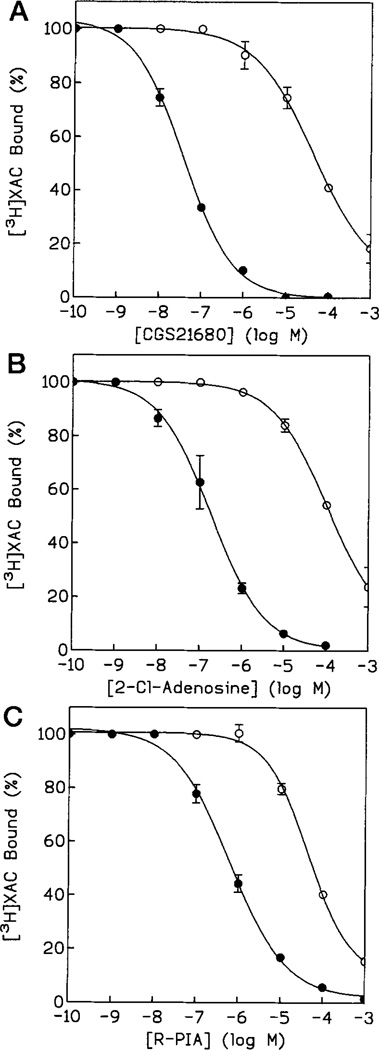
Curiously, the S277A mutant receptor was unable to bind significant amounts of [3H]CGS 21680 present at 15 nm (Table I), but showed normal affinity for [3H]XAC (Table II and Fig. 2). Thus, the hydroxy group present in the side chain of Ser-277 is required for high affinity binding of agonists, but not antagonists. [3H]XAC competition assays (Table III) showed that the affinities of the S277A mutant receptor for the agonists CGS 21680, NECA, and CADO were decreased by about 2–3 orders of magnitude. N6-Substituted agonists such as R-PIA and DPMA showed somewhat less pronounced reductions in binding affinities for the S277A mutant receptor (43- and 107-fold, respectively) than 2- and 5′-substituted adenosine derivatives. It is interesting that IB-MECA, which is both 5′- and N6-substituted, also showed a less pronounced decrease in affinity at the receptor (66-fold versus 400-fold reduction for NECA). On the other hand, the S277A substitution slightly increased the affinity for the antagonist CPX, and the affinity for the antagonist CGS 15943 remained the same. The affinity of [3H]XAC for the S277A mutant receptor was at least as high as for the wild type receptor (Table II). These results indicate that the hydroxyl group of Ser-277 is not involved in binding of antagonists, but is very important for the high affinity binding of agonists, especially N6-unsubstituted adenosine agonists.
Fig. 2. Displacement of binding of the antagonist radioligand [3H]XAC from Tag3-A2a wild type (●) and S277A (○) mutant receptors expressed in COS-7 cells.
Competitors used were CGS 21680 (A), 2-chloroadenosine (B), and N6-phenylisopropyladenosine (C). Competition binding studies were carried out using membrane homogenates prepared from transfected COS-7 cells, as described under “Experimental Procedures.” Data are presented as means ± S.E. of two or three independent experiments, each performed in duplicate.
Table III. Ligand binding properties of wild type and S277A human A2a receptors.
Table shows mean ± S.E. of two or three independent experiments, each performed in duplicate. Agonist and antagonist binding affinities (Ki values) were determined in [3H]XAC (2 nm) competition binding studies using membrane homogenates prepared from transiently transfected COS-7 cells, as described under “Experimental Procedures.” Ki values were calculated from IC50 value using the GraphPad program. About 15 µg of membrane protein/tube were used.
| Ligand | Ki | Ki (S277)/Ki (wt) | |
|---|---|---|---|
| WT | S277A | ||
| nm | -fold | ||
| Agonists | |||
| CGS 21680 | 40.7 ± 9.5 | 43600 ± 1280 | 1070 |
| CADO | 208 ± 66.4 | 81300 ± 21400 | 390 |
| DPMA | 373 ± 66.0 | 40000 ± 8240 | 107 |
| IB-MECA | 1450 ± 70.0 | 96000 ± 5700 | 66 |
| NECA | 72.5 ± 10.0 | 29100 ± 6240 | 400 |
| R-PIA | 792 ± 49.0 | 33800 ± 3710 | 43 |
| Antagonists | |||
| CGS 15943 | 20.7 ± 0.8 | 17.5 ± 4.1 | 0.8 |
| CPX | 663 ± 164 | 219 ± 31.5 | 0.3 |
All Ala mutant receptors except F180A and C254A were unable to bind specifically with high affinity either [3H]CGS 21680 (Table I) or [3H]XAC (Table II). To further probe the role of each residue in ligand-binding at the human A2a adenosine receptor, we introduced several nonalanine mutations (Table I). Substitution of Asn-181 with Ser only slightly affected the affinity of all examined antagonists and the agonist NECA (Table I), while it led to a 14- and 9-fold reduction in the affinity of the N6-substituted adenosine derivative R-PIA and 2-chloroadenosine (CADO), respectively. Substitution of Phe-182 with Tyr or Trp also resulted in 5–9-fold reductions in affinities for R-PIA or NECA, while having little effect on antagonist affinities. Substitution of Asn-253 with Ser or Gln almost completely abolished [3H]CGS 21680 or [3H]XAC binding. Substitution of Phe-257 with Arg essentially retained binding affinity for [3H]CGS 21680 (Kd = 23 nm, Bmax = 2.4 pmol/mg protein). Substitution of Ser-277 with Asn or Thr resulted in only minor changes (≤2-fold) in binding affinities of all ligands examined. Substitution of Ser-281 with Thr (present in A1 receptors) led to an about 10-fold increase in affinity for the agonists CADO and R-PIA, both of which are more potent at A1 versus A2a receptors (Jacobson et al., 1992a).
Although substitution of either His residue with Ala prevented radioligand binding, different results were obtained upon substitution with other amino acids. Substitution of His-250 with Phe or Tyr preserved radioligand binding, although the affinities of certain adenosine agonists (5′-unsubstituted) were reduced by 10–50-fold (Table I). The affinities of CGS 21680, NECA, CGS 15943, and XAC were nearly the same as those found with the wild type receptor. However, substitution of His-278 with Gln, Phe, or Tyr abolished high affinity binding of either [3H]CGS 21680 (15 nm) or [3H]XAC (4 nm).
HA-tag Construction
A strategy for the immunological detection of mutant receptors that lacked binding of either agonist or antagonist radioligands was required in order to discern whether these results were due to a reduced expression of the mutant receptors on the cell surface or to greatly decreased binding affinities. Thus, prior to mutagenesis, the human A2a receptor was tagged at the extracellular NH2 terminus with a 9-amino acid HA epitope (see “Experimental Procedures”). The HA-tag was initially inserted after the first Met residue in the amino terminus of the wild type A2a receptor (“Experimental Procedures” and Fig. 3A). The tagged receptor (Tag1-A2a) showed the same affinity for [3H]CGS 21680 as the wild type receptor (Table IV), but could not be detected by immunological techniques (ELISA, Western blotting, or immunocytochemistry) as shown in Fig. 3B.
Table IV. [3H]CGS 21680 binding properties of wild type and three epitope-tagged human A2a receptor constructs.
Table shows mean ± S.E. of two or three independent experiments, each performed in duplicate. [3H]CGS 21680 saturation binding studies were carried out with membrane homogenates prepared from transiently transfected COS-7 cells as described under “Experimental Procedures.”
| Constructa | Kd | Bmax |
|---|---|---|
| nm | pmol/mg | |
| A2a (wt) | 15.9 ± 0.3 | 26.9 ± 13.3 |
| Tag1-A2a | 24.1 ± 0.1 | 24.3 ± 12.4 |
| Tag2-A2a | 23.8 ± 2.0 | 28.0 ± 14.4 |
| Tag3-A2a | 21.7 ± 2.6 | 35.4 ± 20.0 |
The localization of the HA epitope tag in the various constructs is shown in Fig. 3A.
Interestingly, there are two Met residues in the amino-terminal segment of the human A2a adenosine receptor. The introduction of the HA-tag following the first Met might affect the initiation of protein translation, causing protein expression to start at the second Met. This would result in the deletion of the first 3 amino acids (MPI) and the HA-tag not being translated. However, the resulting mutant receptor may still be functional. We therefore introduced an HA-tag after the second Met, resulting in Tag2-A2a. This mutant receptor showed the same affinity for [3H]CGS 21680 and the same expression level (Bmax) as the non-tagged receptor (Table IV). Moreover, we could detect the Tag2-A2a receptor by immunological techniques (ELISA, but not Western blotting, Fig. 3B). To test if the first Met is important for the expression of Tag2-A2a, we deleted the first Met to construct the Tag3-A2a receptor (Fig. 3A). The Tag3-A2a receptor showed the same [3H]CGS 21680 binding properties (Table IV) and gave a similar signal in ELISA assay as the Tag2 wild type receptor (Fig. 3B). We therefore concluded that the second Met may represent the physiological translation start site in the human A2a adenosine receptor. Based on these results, the Tag3 epitope-tag was introduced into all other mutant receptors (Table I). However, it was not possible to detect Tag3-A2a receptor by Western blotting.
Expression Levels of Ala Mutant Receptors
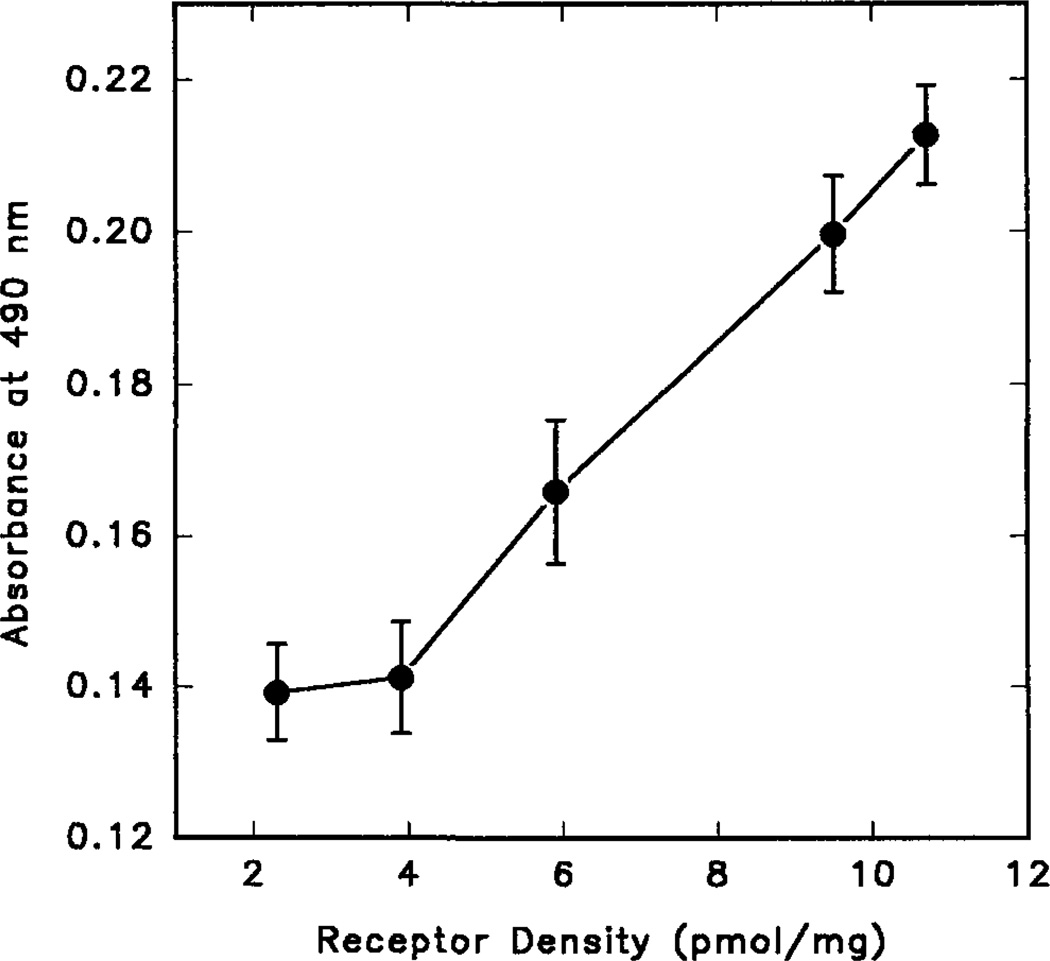
To estimate approximate levels of receptor protein present in the plasma membrane, a standard curve was constructed from different batches of transfected COS-7 cells expressing different levels of Tag3-A2a wild type receptors (Fig. 4). To decrease the density of Tag3-A2a, COS-7 cells were transfected with progressively reduced amounts of receptor DNA (supplemented with pcD vector DNA to maintain 4 µg of total DNA per dish). From the linear region of this standard curve, an equation was obtained by linear regression, and this equation (see Table V) was used to calculate the amount of receptor protein for the Ala mutants. Experiments were done with nonpermeabilized cells, therefore the ELISA assay detected only receptors in the plasma membrane. To establish this point, control experiments with nonpermeabilized COS-7 cells expressing a COOH-terminally HA-tagged form of the rat m3 muscarinic receptor were carried out. The resulting OD readings were similarly low as those found with the non-tagged versions of the m3 muscarinic and A2a adenosine receptors (data not shown). This finding, together with the observation that the COOH-terminally tagged m3 muscarinic receptor can be easily detected in permeabilized cells demonstrates that the employed ELISA procedure does not interfere with the intactness of the plasma membrane barrier.
Fig. 4. Correlation between the strength of the absorbance signal determined by the ELISA method and the density (Bmax) of receptors.
The receptor densities were determined by [3H]CGS 21680 saturation binding experiments using transfected COS-7 cells derived from the same plate as those for the ELISA experiments. About 24 h after transfections, aliquots of COS-7 cells were transferred into 96-well plates, and the remaining cells were grown for saturation binding assays. ELISA studies were carried out as described under “Experimental Procedures”.
Table V. ELISA detection of human A2a receptor mutants on the surface of COS-7 cells.
Table shows expression level as percentage of Tag3-A2a wild type (= 100%).
| Mutant | Expression levela |
|---|---|
| % | |
| F182A | 71 ± 13 |
| H250A | 58 ± 1 |
| N253A | 40 ± 6 |
| F257A | 68 ± 16 |
| Y271A | 34 ± 8 |
| I274A | 72 ± 15 |
| S277A | 25 ± 6 |
| H278A | 72 ± 24 |
| S281A | 36 ± 7 |
Expression level was calculated by an equation, y = 0.1 + 0.01x, which was obtained from Fig. 4 (standard curve).
All Ala mutant receptors including H278A were present at relatively high levels in the plasma membrane, i.e. greater than 25% compared to the wild type receptor. However, this calculation did not always match the results of saturation binding experiments. For example, we determined receptor density for the S277A mutant receptor by the ELISA method to be 25% of wild type, while by saturation binding the mutant showed increased density (25.6 pmol/mg versus 12.3 pmol/mg for wild type; Table II). This discrepancy might have resulted from different transfection efficiencies, since we did not carry out the experiments simultaneously. Since the nine Ala mutant receptors were expressed at significant levels in the plasma membrane, dramatically decreased radioligand affinity, rather than insufficient expression, is the likely source of the observed lack of radioligand binding.
Functional Assay
To determine whether Ala mutant receptors that lacked high affinity radioligand binding were still functional at high agonist concentrations, their ability to mediate increases in intracellular cyclic AMP levels was studied. Untransfected COS-7 cells showed a strong stimulation of cyclic AMP production (4-fold; Fig. 5) at >100 µm CGS 21680, using 0.1 mm rolipram as a phosphodiesterase inhibitor (Hide et al., 1992). Since at this concentration, CGS 21680 is nonselective among adenosine receptor subtypes (Hide et al., 1992), perhaps this response may occur through activation of another receptor, such as the A2b subtype, since the concentration required is above the range of A2a receptors. COS-7 cells transfected with the wild type Tag3-A2a receptor showed a pronounced stimulation of cyclic AMP production (3.2-fold) following CGS 21680 treatment, with an EC50 value of 3 nm. The fact that stimulation occurs at concentrations below the Kd value for CGS 21680 (21 nm) suggests that under these conditions only a minor fraction of the receptors needs to be activated for effective signal transduction.
Fig. 5. Stimulation of adenylyl cyclase in COS-7 cells transiently expressing Tag3-A2a wild type or mutant A2a adenosine receptors in the presence of 2 units/ml adenosine deaminase and 0.1 mm rolipram.
The following receptors were studied: wild type (●), S277A (○), I274A (■), H278A (□); untransfected COS-7 (×). Transfected COS-7 cells were incubated for 30 min at 37 °C (for details, see “Experimental Procedures”) with increasing concentrations of CGS 21680. Data are presented as fold increase in cyclic AMP above basal levels (400–600 cpm) in the absence of CGS 21680. Each curve represents the fold-stimulation average of two or three independent experiments, each carried out in duplicate. Alternately, the conversion factor (f) was calculated using the following equation, f = cAMP(cpm)/(cAMP(cpm) + ATP(cpm)). The ATP fraction gave very similar values in all samples (598,000 ± 4000 cpm). The maximum conversion factor was 0.7.
The receptors I274A, S277A, and H278A, containing mutations in TM7, also showed dose-dependent stimulation of cyclic AMP production following treatment with CGS 21680 (Fig. 5; Table VI) with EC50 values in the 10−7–10−6 m range. The agonist efficacy in these mutant receptors was at least as much as for the wild type receptor. With all other Ala mutations (Table I), we could not detect any stimulation over control up to 1 mm CGS 21680.
Table VI. CGS 21680-induced stimulation of cAMP production mediated by mutant A2a adenosine receptors.
CGS 21680-stimulated cAMP production was examined in transiently transfected COS-7 cells in the presence of 0.1 mm rolipram as described under “Experimental Procedures.” In each case the maximal stimulation was 3–4-fold over control (see Fig. 5). Table shows mean ± S.E. of two or three independent experiments, each performed in duplicate.
| Receptor | EC50 |
|---|---|
| nm | |
| wt (Tag3) | 3.0 ± 1.0 |
| I274A | 100 ± 20 |
| S277A | 210 ± 35 |
| H278A | 920 ± 70 |
Construction of a Human A2a Adenosine Receptor Model: Ligand Binding Hypothesis
A human A2a receptor model was constructed, independent of the mutagenesis results, using standard computational methods (see “Experimental Procedures”) and based on rhodopsin as a template. Docking of NECA to the human A2a receptor model was carried out manually, followed by energy minimization, in a fashion that placed the ligand in the center of a region defined by the essential residues, determined by mutagenesis, shown in green in Fig. 6B.2 The orientation of the ligand was such that the adenine ring rested in a hydrophobic pocket defined by His-250, Phe-182, and Phe-257 (TM5 and TM6), and the N6-substituent would align with Asn-253 (H-bonding distance to the exocyclic NH) and Met-270. The ribose ring pointed in the direction of His-278, while the 5′-uronamide group was in proximity to Ser-277 and Ile-274 (all TM7). Although a recent study (Olah et al., 1994) found that replacement of a 6-amino acid cassette in TM5 (see Fig. 1A) affected mostly binding of 5′-substituted adenosine derivatives, we were unable to place the 5′-uronamide of NECA in direct proximity to this region in our model while maintaining other favorable interactions.
Fig. 6. Molecular model of the human A2a receptor containing NECA bound in the proposed binding site, viewed either (A) perpendicularly to the membrane from the extracellular side or (B) as a side view of the residues in TM5, TM6, and TM7 in proximity to the bound ligand.
The model was based on the structure of rhodopsin and minimized using the Discover program (BIOSYM Technologies, San Diego CA, Version 2.90) employing the Amber force field. Side chains in dark green were those residues found in mutation experiments in this study to be essential for ligand binding. Side chains in yellow were those residues found in mutation experiments in this study to be non-essential for ligand binding, thus Ala mutants at these positions were fully functional in binding of both agonist and antagonist radioligands.
Residues that are in proximity of the ribose moiety in our model are Thr-88, Gln-89, Ser-90, Ser-91, Ile-92, Ala-273, Ile-274, Ser-277, and His-278. Of these residues, Thr-88, Gln-89, Ser-90, and Ala-273 seem to be interacting with NECA primarily through their backbone atoms rather than through their side chains. Thr-88 may, in addition, show hydrogen bonding to both the 2′-O and 3′-O. The 5′-OH of adenosine, transformed into 5′-NH in NECA, is most notably hydrogen bonded to Ser-277 and His-278 in our model.
DISCUSSION
The complete understanding of ligand-receptor interactions will lead to answers to questions, such as: (i) how are receptors activated by agonist ligands and how is the binding site blocked by antagonists? (ii) Which synthetic drug analogues are suitable to control the function of the receptors? Even though mutagenesis studies lead to only a limited understanding of detailed three-dimensional structure, they are still a powerful tool for deriving an overall hypothesis.
Modeling of G-protein coupled receptors (GPCRs) has become an important tool in understanding drug-receptor interactions and in the development of new ligands for these receptors. The first widely accepted method was the homology modeling method by Hibert et al. (1991). This method involved the alignment of the receptor sequence with the sequence of bacteriorhodopsin and the subsequent mapping of the sequence onto the structure of bacteriorhodopsin that was determined by Henderson et al. (1990). This procedure was based on the assumption that even though GPCRs and bacteriorhodopsin, a proton pump in the outer membrane of Halobacterium halobium, lacked any functional or sequence homology, there would be considerable structural homology. This structural homology was inferred by the extraordinary similarity in the hydrophobicity plots, or Kyte-Doolittle plots, of the biogenic amine subfamily of GPCRs and bacteriorhodopsin. Recently, a low resolution electron density map of rhodopsin, a true member of the GPCR superfamily, was published (Schertler et al., 1993). The low sequence homology with bacteriorhodopsin, the structural differences that must arise from the different placement of proline residues (causing bends in the helices) in bacteriorhodopsin and GPCR sequences, and the availability of an electron density map of a true member of the GPCR superfamily prompted us (van Rhee et al. 1994) to adapt a new method to build models of GPCRs (Ballesteros and Weinstein, 1995) that is based on a computational approach rather than strict compliance with the atomic coordinates of a distantly related protein, albeit with higher resolution.
Although molecular models for the canine A1 (IJzerman et al., 1992), rat A3 (van Galen et al., 1994), and rat A2a adenosine receptor (IJzerman et al., 1994) have been published, we thought it worthwhile to construct a model of the human A2a adenosine receptor based on the electron density map of rhodopsin rather than on the atomic coordinates of bacteriorhodopsin. Using rhodopsin as a template allowed us to better interpret the mutation studies described in this paper. We have derived a human A2a receptor model mainly through a computational approach, and docked in the binding site the high affinity agonist ligand, NECA, in a fashion that is consistent with all of the available pharmacological data. The docking of antagonist ligands and other modified agonists will be the subject of future studies. The region of ligand binding (TM5-TM7) is similar to that predicted in the bacteriorhodopsin-based modeling study of IJzerman et al. (1994), although novel specific interactions are proposed.
Histidine Side Chains Involved in Ligand Binding
The His residues of TM6 and TM7, which have been previously implicated through mutagenesis in ligand binding to A1 receptors (Olah et al., 1992), oriented according to both present and previous adenosine receptor models in the direction of the binding cleft. However, even for A1 receptors we have had little information to understand precisely the noncovalent bonding interactions. It was proposed in the modeling study by IJzerman et al. (1994) that His-278 coordinates both the 2′- and 3′-hydroxyl groups of the ribose moiety, but in our model the 5′-hydroxyl group of adenosine (or amide NH of NECA) is in hydrogen bonding proximity. It is probably this residue that is referred to as “the agonist histidine” in the literature (Jacobson et al., 1992b). The homologous His residue has also been postulated to play a role in ribose coordination in the A3 receptor (van Galen et al., 1994). More recently, Askalan and Richardson (1994) provided support for the assumption that the “agonist histidine” directly interacts with the 2′-hydroxyl group of the ribose moiety. We found it impossible to dock the ligand into the receptor in such a fashion that His-278 interacts directly with the 2′-hydroxyl group while maintaining all the other interactions. Analysis of various mutants with several (radiolabeled) ribose-modified ligands could, perhaps, further elucidate this matter.
According to the present mutagenesis results, substitution of either His-250 or His-278 with Ala in human A2a receptors dramatically reduced affinity of both agonist and antagonist radioligands. These mutant receptors were expressed in the plasma membrane, at levels not qualitatively different from that of the wild type receptors (Table V). In a functional assay CGS 21680 was able to stimulate cAMP production in the H278A mutant receptor but with a ~300-fold decreased potency. These results are in contrast to a previous study of bovine A1 receptors (Olah et al., 1992), in which mutation of the corresponding His in TM7 to Leu similarly abolished binding, but mutation of the His of TM6 only somewhat reduced affinity for XAC and did not affect agonist binding. Even though His-278 is adjacent to Ser-277 in TM7 (see below), the H278A mutant receptor did not bind either [3H]CGS 21680 or [3H]XAC, while S277A failed to bind only [3H]CGS 21680, an agonist (Table I). The fact that His-278 is located in the putative ribose binding region and also affects antagonist binding suggests that there is a possibility that His-278 may be involved in other interactions as well as ligand binding. This possibility is supported by the effects of the following other mutations: substitution of His-278 with Gln, Phe, and Tyr also prevented binding of either [3H]CGS 21680 or [3H]XAC, suggesting that His-278 may be essential for maintaining the conformation of A2a adenosine receptors.
The H250A mutant receptor showed significant plasma membrane expression (58%; Table V), but unlike H278A did not show any detectable stimulation by 1 mm CGS 21680 of cyclic AMP-production over control levels, using rolipram to inhibit phosphodiesterases. These results suggest that the H250A mutant receptor either has even lower affinity for CGS 21680 than H278A, or is defective in G-protein coupling. Substitution of His-250 with Phe, which does not readily form hydrogen bonds, diminished affinities of certain ligands but did not preclude binding, suggesting that the interaction between His-250 and ligands does not primarily involve hydrogen bonding. Substitution of the imidazole group with other aromatic side chains was particularly detrimental to the affinity of R-PIA and DPMA, which have hydrophobic, aromatic N6-substituents, and of CADO (Table I). The affinities of 5′-substituted adenosine analogues and various antagonists (Table I) were nearly unaffected. It is possible that the 5′-uronamide group may form a better anchor than 5′-hydroxyl derivatives by favorable interactions with TM7 (see below), thus reducing dependence on the imidazole interactions. In summary, these data suggest that mainly aromatic (hydrophobic) interactions in the region of His-250 are important for the binding of both agonists and antagonists.
Other Aromatic Residues Involved in Ligand Binding
The molecular model predicts that there is a hydrophobic/aromatic binding pocket, putatively the site of coordination of the adenine moiety, defined by His-250 and Phe-257 (TM6) and Phe-182 (TM5). Mutation of residue Phe-182 or Phe-257 to Ala resulted in loss of specific binding of either [3H]CGS 21680 or [3H]XAC, indicating the importance of these side chains in ligand recognition. Mutation of Phe-182 to Trp reduced agonist binding affinities 3—10-fold, whereas antagonist affinity was not affected. Replacement of the same residue with Tyr, however, decreased agonist affinity 8–10-fold, while antagonist affinity was decreased only 2-fold. The propensity for hydrogen bond formation, increasing as residue 182 is changed from Phe to Trp to Tyr, inversely correlates with binding affinity. This notion supports the aromatic interaction hypothesis.
Phe-182 may also be involved in helix-helix packing, according to our model in which TM5 is rotated in a fashion consistent with rhodopsin in the model proposed by Baldwin (1993). Phe-182 is located close to the adenosine C2 binding region in the model (viewed as in Fig. 6A). If TM5 were rotated clockwise by several degrees, then Phe-182 would be in even closer contact with the adenine moiety.
Aliphatic and Hydrophilic Side Chains Involved in Ligand Binding
Met-270 (common to both human A2a and canine A1 adenosine receptors; Fig. 1A) was shown to be important for species differences, i.e. the lower affinity of N6-substituted adenosine derivatives at canine versus bovine A1 receptors (Tucker et al., 1994). The sulfur atom of this residue was positioned at 6.0 Å from the N6 atom in NECA in our model, thus consistent with direct contact with the N6-substituent as an explanation for the observed effect. Met-270 is conserved among species in the A2a (Fig. 4A) and A2b adenosine receptors, and may be responsible for the generally low affinity of N6-substituted derivatives at A2 versus A1 adenosine receptors.
A hydrogen bond between the exocyclic NH and His-250 was proposed by IJzerman et al. (1994). In our model the distance is too large for such a bond, however, there is a possibility of H-bonding between the exocyclic NH and Asn-253. It has been suggested that N6- and C-2-substituents occupy overlapping positions in the receptor-bound conformations of adenosine agonists (IJzerman et al. (1994) and references therein). The present results support this concept, since N6- (R-PIA and DPMA, having aromatic bulk) and C-2-modified (CADO, hydrophobic substituent) adenosine analogues (all 5′-CH2OH) behaved similarly. The side chain carbonyl of Asn-253 is favorably located at 4.3 Å from the N6 atom in NECA according to the model. Mutation of this residue, even to Gln, led to a total loss of specific [3H]CGS 21680 and [3H]XAC binding, thus the structural requirements for adenosine and xanthine derivatives at this site are highly restrictive. Interaction of Asn-253 with the N1 atom of adenine, as suggested by IJzerman et al. (1994) for the rat A2a adenosine receptor, is less likely in our model.
Substituting Asn-181 with Ser (Table I) indicated that this amino acid selectively affects the affinity of N6-substituted agonists and CADO. N181S had slightly decreased (only ≤2-fold decrease) affinity for CGS 21680, NECA, CGS 15943, and XAC, but 20-fold decreased affinity for R-PIA. Residue Asn-181, although not hydrophobic, according to our model is facing the lipid bilayer. If TM5 were rotated in the model (see above), Asn-181 would then be located in the inter-helical contact region, adjacent to Pro-139 of TM4. Thus, it is possible that the side chain of this Asn residue is involved in hydrogen bonding to the backbone. Disturbance of this hydrogen bond by mutation may affect overall receptor structure, and indirectly ligand binding. Such an assumption is consistent with the effects observed here.
In the case of the human A1 adenosine receptor, previous studies have shown that Thr-277 in TM7 is critical for the interaction with the ribose ring, especially for binding of a subset of agonist ligands, i.e. those containing the 5′-uronamide modification, such as NECA (Townsend-Nicholson and Schofield, 1994). This residue has also been indicated to be involved in species-dependent ligand binding at Ax receptors (Tucker et al., 1994). In the A2a receptor the corresponding residue, Ser-277, conserved across species (see Fig. 1A), was found in this study to contain an hydroxyl group that is essential for agonist binding. The S277A mutation only slightly changed the affinity for the antagonists, indicating that the Ser hydroxyl group does not contribute energetically to antagonist binding (Table II). The affinity of N6-substituted agonists was less affected by the S277A substitution, perhaps as a result of additional stabilization of the N6-substituent by neighboring hydrophobic residues. Hence, the interaction of the ribose ring with Ser-277 may be relatively weak compared to other interactions at the N6-region. Furthermore, mutation of Ser-277 to either hydrogen bonding residues Thr or Asn affected agonist affinity only moderately (Table I). These results corroborate a model where the ribose moiety, essential for agonist activity, occupies a predominantly hydrophilic region of the receptor that is not occupied by antagonists.
Ser-281 (Ser-276 in the rat A2a adenosine receptor) was implicated by IJzerman et al. (1994) to be involved in binding the 5′-hydroxy region of adenosine. The S281A mutant receptor indeed does not display high affinity for [3H]CGS 21680. In our model the side chain oxygen of Ser-281 is separated from the 5′ nitrogen of NECA by 8.2 Å, a distance too large to accommodate hydrogen bonds. However, our model also suggests the possibility of a conjugated hydrogen bond from Ser-281 through His-278 to the 5′ region. Such a conjugated hydrogen bond system would be disrupted by the mutation and explain the observed loss of ligand affinity. Of further interest is residue Ile-274. In the ribose-binding region, according to the model, the side chain of Ile-274 is in close proximity of the terminal methyl group of the 5′-uronamide of NECA (3.2 Å), a ligand region that has strict steric requirements (Jacobson et al., 1992a). The dramatically decreased affinity of the I274A mutant receptor for [3H]CGS 21680 and [3H]XAC might be related to the loss of hydrophobicity resulting from this mutation. However, this receptor was well expressed (72% of wild type), and showed good stimulation of cAMP-production with a 33-fold decreased affinity for CGS 21680.
Substitution of Asn-253 with Ala, Gln, and Ser prevented binding of [3H]CGS 21680 (Table I, footnote b), suggesting that Asn-253 is also essential for ligand recognition. According to the present model Asn-253 is within hydrogen bonding distance from the N6H of NECA. Substitution of Ser-281 with Thr showed increased affinity for some agonists, but S281A was unable to bind either [3H]CGS 21680 or [3H]XAC in spite of significant plasma membrane expression (36%; Table V). Taken together, we conclude that Ser-277 and Ser-281 might be involved in the interaction with the ribose ring of agonists.
Side Chains That Are Not Involved in Ligand Binding
The molecular model predicts that Phe-180 and Cys-254 (shown in yellow in Fig. 6) are pointing away from the ligand binding cleft into the lipid bilayer. If indeed located in the lipid bilayer, mutation of these residues should have little effect on ligand affinity, unless the receptor structure itself is corrupted. Indeed, mutagenesis studies are consistent in that Ala substitutions at these sites have no effect on ligand affinity. Other residues found to be essential in ligand recognition were generally predicted by the model to be facing in the direction of the putative ligand binding site (shown in green in Fig. 6) or in the inter-helical contact regions.
The model predicts that Tyr-271 is at the interface between TM7 and TM1, and when mutated to Ala, ligand affinity was greatly decreased. Mutation of Tyr-271 to other aromatic residues (Phe or His) or to Arg preserves binding of both agonists and antagonists (data not shown).
Comparison to Essential Side Chains in Other Receptors
Residues Asn-181, His-250, and His-278 of A2a receptors align with Thr-234, Asn-507, and Tyr-533, respectively, in the rat m3 muscarinic receptor (Fig. 1A), which have been shown by mutagenesis to be involved in ligand binding (Wess et al., 1991; Blüml et al., 1994). Residues Phe-182 and Ser-281 correspond to serines 204 and 319 in the hamster β2 adrenergic receptor, which are also involved in ligand recognition (Strader et al., 1989). His-250 is equivalent to His-265 in the NK1 receptor. Mutation of this residue to alanine influenced the selectivity of antagonists, but did not affect the affinity of agonists (Fong et al., 1994).
Acknowledgments
We thank Dr. Ad IJzerman (Leiden University) and Prof. Gary Stiles, Dr. Mark Olah, and Dr. Timothy Palmer (Duke University) for helpful discussions, and Dr. Marlene Jacobson (Merck) for providing the human A2a plasmid. We thank Dr. Xiao-duo Ji, Dr. Neli Melman, and Patricia Evans for assisting with binding experiments and Carola Gallo-Rodriguez for preparing IB-MECA.
Footnotes
Supported by a grant from the Cystic Fibrosis Foundation.
The abbreviations and trivial names used are: CGS 21680, 2-[4-[(2-carboxyethyl)phenyl]ethylamino]-5′-N-ethylcarboxamidoadenosine; CADO, 2-chloroadenosine; CGS 15943, 9-chloro-2-(furyl)[l,2,4]triazolo [l,5-c]quinazolin-5-amine; CPX, l,3-dipropyl-8-cyclopentylxanthine; DPMA, N6-[2-(3,5-dimethoxyphenyl)-2-(2-methylphenyl)ethyl]adenosine; ELISA, enzyme-linked immunosorbent assay; GPCR, G-protein coupled receptor; HA, hemagglutinin; IB-MECA, N6-(3-iodobenzyl)adenosine-5′-N-methyluronamide; NECA, 5′-N-ethylcarboxamidoadenosine; PCR, polymerase chain reaction; R-PIA, R-N6-phenylisopropylad-enosine; TM, transmembrane helical domain; XAC, 8-[4- [[[[(2-amino-ethyl)amino]carbonyl]methyl]oxy]phenyl]-l,3-dipropylxanthine.
A list of distances between atoms of NECA and key amino acids is available from the authors.
REFERENCES
- Askalan R, Richardson RJ. J. Neurochem. 1994;63:1477–1484. doi: 10.1046/j.1471-4159.1994.63041477.x. [DOI] [PubMed] [Google Scholar]
- Baldwin JM. EMBO J. 1993;12:1693–1703. doi: 10.1002/j.1460-2075.1993.tb05814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros JA, Weinstein H. Methods Neurosci. 1995;25:366–428. [Google Scholar]
- Barraco RA, Martens K, Parizon M, Normile HJ. Prog. Neuropsychopharmacol. Biol. Psychiatry. 1994;18:545–553. doi: 10.1016/0278-5846(94)90011-6. [DOI] [PubMed] [Google Scholar]
- Barrington WW, Jacobson KA, Hutchison AJ, Williams M, Stiles GL. Proc. Natl. Acad. Sci. U. S. A. 1989;86:6572–6576. doi: 10.1073/pnas.86.17.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blüml K, Mutschler E, Wess J. J. Biol. Chem. 1994;269:18870–18876. [PubMed] [Google Scholar]
- Cullen BR. Methods Enzymol. 1987;152:684–704. doi: 10.1016/0076-6879(87)52074-2. [DOI] [PubMed] [Google Scholar]
- Enjalbert A, Bockaert J. Mol. Pharmacol. 1983;23:576–584. [PubMed] [Google Scholar]
- Ferré S, O’Connor WT, Snaprud P, Ungerstedt U, Fuxe K. Neuroscience. 1994;63:765–773. doi: 10.1016/0306-4522(94)90521-5. [DOI] [PubMed] [Google Scholar]
- Fong TM, Yu H, Cascieri MA, Underwood U, Swain CJ, Strader CD. J. Biol. Chem. 1994;269:2728–2732. [PubMed] [Google Scholar]
- Gallo-Rodriguez C, Ji XD, Melman N, Siegman BD, Sanders LH, Orlina J, Fischer B, Pu QL, Olah ME, van Galen PJM, Stiles GL, Jacobson KA. J. Med. Chem. 1994;37:636–646. doi: 10.1021/jm00031a014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R, Baldwin JM, Ceska TA, Zemlin F, Beckmann E, Downing KH. J. Mol. Biol. 1990;213:899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- Hibert MF, Trumpp-Kallmeyer S, Bruinvels A, Hoflack J. Mol. Pharmacol. 1991;40:8–15. [PubMed] [Google Scholar]
- Hide I, Padgett WL, Jacobson KA, Daly JW. Mol. Pharmacol. 1992;41:352–359. [PMC free article] [PubMed] [Google Scholar]
- Higuchi R. In: PCR Technology. Ehrlich HA, editor. New York: Stockton Press; 1989. pp. 61–70. [Google Scholar]
- IJzerman AP, van Galen PJM, Jacobson KA. Drug Design Discov. 1992;9:49–67. [PMC free article] [PubMed] [Google Scholar]
- IJzerman AP, Van Galen PJM, Jacobson KA. Eur. J. Pharmacol. Mol. Pharmacol. Sect. 1994;268:95–104. doi: 10.1016/0922-4106(94)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson M. In: Adenosine and Adenine Nucleotides: From Molecular Biology to Integrative Physiology. Belardinelli L, Pelleg A, editors. Norwell, MA: Kluver; 1995. [Google Scholar]
- Jacobson KA, van Galen P, Williams M. J. Med. Chem. 1992a;35:407–422. doi: 10.1021/jm00081a001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson KA, Stiles GL, Ji XD. Mol. Pharmacol. 1992b;42:123–133. [PMC free article] [PubMed] [Google Scholar]
- Jarvis MF, Schulz R, Hutchison AJ, Do UH, Sills MA, Williams M. J. Pharmacol. Exp. Ther. 1989;251:888–893. [PubMed] [Google Scholar]
- Ji X-D, Stiles GL, van Galen PJM, Jacobson KA. J. Recept. Res. 1992;12:149–169. doi: 10.3109/10799899209074789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda T, Shiozaki S, Shimada J, Suzuki F, Nakamura J. Eur. J. Pharmacol. 1994;256:263–268. doi: 10.1016/0014-2999(94)90551-7. [DOI] [PubMed] [Google Scholar]
- Maenhaut C, Sande JV, Libert F, Abramowicz M, Parmentier M, Vanderhaegen JJ, Dumont JE, Vassart G, Schiffmann S. Biochem. Biophys. Res. Commun. 1990;173:1169–1178. doi: 10.1016/s0006-291x(05)80909-x. [DOI] [PubMed] [Google Scholar]
- Mahan LC, McVittie LD, Smyk-Randall EM, Nakata H, Monsma FJ, Jr, Gerfen CR, Sibley DR. Mol. Pharmacol. 1991;40:1–7. [PubMed] [Google Scholar]
- Martin GE, Rossi DJ, Jarvis MF. Pharmacol. Biochem. Behav. 1993;45:951–958. doi: 10.1016/0091-3057(93)90146-k. [DOI] [PubMed] [Google Scholar]
- Nikodijević O, Jacobson KA, Daly JW. Drug Dev. Res. 1993;30:121–128. doi: 10.1002/ddr.430300209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okayama H, Berg P. Mol. Cell. Biol. 1983;3:280–289. doi: 10.1128/mcb.3.2.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olah ME, Ren HZ, Ostrowski J, Jacobson KA, Stiles GL. J. Biol. Chem. 1992;267:10764–10770. [PMC free article] [PubMed] [Google Scholar]
- Olah ME, Jacobson KA, Stiles GL. J. Biol. Chem. 1994;269:18016–18020. [PMC free article] [PubMed] [Google Scholar]
- Olsson RA, Pearson JD. Pharmacol. Rev. 1990;3:761–845. doi: 10.1152/physrev.1990.70.3.761. [DOI] [PubMed] [Google Scholar]
- Piersen CE, True CD, Wells JN. Mol. Pharmacol. 1994;45:871–877. [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. Proc. Natl. Acad. Sci. U. S. A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schertler GF, Villa C, Henderson R. Nature. 1993;362:770–772. doi: 10.1038/362770a0. [DOI] [PubMed] [Google Scholar]
- Strader CD, Candelore MR, Hill WS, Sigal IS, Dixon RAF. J. Biol. Chem. 1989;264:13572–13578. [PubMed] [Google Scholar]
- Townsend-Nicholson A, Schofield PR. J. Biol. Chem. 1994;269:2373–2376. [PubMed] [Google Scholar]
- Tucker A, Robeva AS, Taylor HE, Holeton D, Bockner M, Lynch KR, Linden J. J. Biol. Chem. 1994;269:27900–27906. [PubMed] [Google Scholar]
- van Galen PJM, van Bergen AH, Gallo-Rodriguez C, Melman N, Olah ME, IJzerman AP, Stiles GL, Jacobson KA. Mol. Pharmacol. 1994;45:1101–1111. [PMC free article] [PubMed] [Google Scholar]
- van Rhee AM, Fischer B, Jacobson KA. ACS Abstracts 208th National Meeting, August 21–25 1994, Washington, D. C. Washington, D. C.: American Chemical Society; 1994. MEDI 0205. [Google Scholar]
- Weiss S, Sebben M, Garcia-Sainz JA, Bockaert J. Mol. Pharmacol. 1985;27:595–599. [PubMed] [Google Scholar]
- Wess J, Gdula D, Brann MR. EMBO J. 1991;10:3729–3734. doi: 10.1002/j.1460-2075.1991.tb04941.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou QY, Li CY, Olah ME, Johnson RA, Stiles GL, Civelli O. Proc. Natl. Acad. Sci. U. S. A. 1992;89:7432–7436. doi: 10.1073/pnas.89.16.7432. [DOI] [PMC free article] [PubMed] [Google Scholar]