Abstract
Mitochondrial superoxide dismutase (MnSOD) neutralizes the highly reactive superoxide radical (O2·−), the first member in a plethora of mitochondrial reactive oxygen species (ROS). Over the past decades, research has extended the prevailing view of mitochondrion well beyond the generation of cellular energy to include its importance in cell survival and cell death. In the normal state of a cell, endogenous antioxidant enzyme systems maintain the level of reactive oxygen species generated by the mitochondrial respiratory chain. Mammalian mitochondria are important to the production of reactive oxygen species, which underlie oxidative damage in many pathological conditions and contribute to retrograde redox signaling from the organelle to the cytosol and nucleus. Mitochondria are further implicated in various metabolic and aging-related diseases that are now postulated to be caused by misregulation of physiological systems rather than pure accumulation of oxidative damage. Thus, the signaling mechanisms within mitochondria, and between the organelle and its environment, have gained interest as potential drug targets. Here, we discuss redox events in mitochondria that lead to retrograde signaling, the role of redox events in disease, and their potential to serve as therapeutic targets.
Keywords: MnSOD, Retrograde signaling, Oxidative stress, Redox signaling, Apoptotic pathways, Oxidative modification, mtDNA, TOR signaling
INTRODUCTION
Reactive oxygen species (ROS), the by-products of normal aerobic metabolism, are considered to be important signaling molecules that play a role in gene expression, cell survival and growth in various cell types [1]. Various endogenous antioxidants efficiently block a cascade of reactions from generating ROS to overcome their potentially deleterious actions. However, excessive formation of ROS leads to oxidative stress, characterized by an imbalance between oxidant-producing systems and antioxidant defense mechanisms that can trigger cell damage by oxidizing macromolecules (lipids, proteins and DNA) and modifying their biological functions that ultimately cause cell death [2, 3]. Thus, depending on their concentrations, ROS can act as either beneficial or harmful biological agents.
The process by which biological systems produce superoxide from oxygen seems to be intimately associated with the processes of cellular life and death events [4]. For example, superoxide radicals, the by-product of oxidative phosphorylation, generate from mitochondria the general energy currency ATP, which is associated with electrons leaking as superoxide radicals (O2·−) during the process. It is estimated that of all the oxygen consumed for respiration 1% to 2% leaks as superoxide radicals. Thus, during normal respiration, the intramitochondrial superoxide concentration is 10−11 to 10−12 M. During hypoxia, cellular purines break down to produce xanthine and hypoxanthine. When oxygen is reintroduced, the enzyme xanthine oxidase reduces molecular oxygen and produces superoxide [4]. Superoxide itself can be toxic, especially through damage to proteins that contain iron-sulphur centers such as aconitase, succinate dehydrogenase and mitochondrial NADH-ubiquinone oxidoreductase [5]. Accordingly, enzymes have evolved with the task of detoxifying these oxygen free radicals, collectively named superoxide dismutases (SOD). This family includes copper- and zinc-containing SOD (CuZnSOD), a homodimeric enzyme found primarily in the cytoplasm and the nucleus, with small amounts within mitochondria, and a glycosylated form of CuZnSOD called extracellular SOD (ECSOD) that resides in the extracellular region of the cell. MnSOD is found exclusively in the mitochondrial matrix and exists as a homotetramer [6].
MnSOD IS ESSENTIAL FOR SURVIVAL OF AEROBIC LIFE
MnSOD is present in all oxygen-metabolizing cells but not present in most obligate anaerobes, presumably because its physiological function is to provide a defense against the potentially damaging reactivity of the O2·− generated by aerobic metabolic reactions [7]. Nevertheless, the superoxide theory of oxygen toxicity has held its ground, although there is no general agreement about why elevated levels of O2·− can be toxic, whether toxicity is caused by direct selective damage by O2·−/HO2·, or by the O2·−, dependent formation of peroxynitrite or the infamous hydroxyl radical. Is MnSOD really important? Touati [8], answered these questions using Escherichia coli (E. coli.) mutants lacking FeSOD and MnSOD where the E. coli were viable but sick. (They still had some CuZnSOD in the periplasmic space.)
Gregory and Fridovich [9] have provided substantial evidence that MnSOD is necessary for survival in all oxygen-metabolizing cells. Many studies of various model systems demonstrate the importance of MnSOD to aerobic organisms. E. coli B cells grown under 100% oxygen are much more resistant to high concentrations of oxygen compared to Bacillus subtilis (B. subtilis). Growth under 100% oxygen induces increased expression of MnSOD, which accounts for the difference in oxygen toxicity between E. coli grown under 100% oxygen and other cells tested [9].
MnSOD is vital for healthy aerobic life and the lack of this enzyme is lethal. Knock-out of MnSOD enzyme activity by creating inactive mutants or the complete elimination of MnSOD expression leads to early death in mouse [10]. Another mechanism of early death is reduced mitochondrial activity. While there are no gross changes in the mitochondrial structure of homozygous MnSOD knock-out mice (SOD2−/−), activities are significantly reduced in both succinate dehydrogenase (complex II of the electron transport chain) and aconitase (citric acid cycle enzyme) compared to wild-type mice. Van Remmen et al. [11] used heterozygous MnSOD knock-out mice to study the effects of life-long reduction in MnSOD enzyme activity. MnSOD knock-out mice have a 50% reduction in MnSOD enzyme activity in all tissues, resulting in an age-dependent increase in oxidative DNA damage (8-oxodeoxyguanidine, (8-oxodG)) in both nuclear and mitochondrial DNA compared to wild-type mice. In addition in vitro studies (mouse embryonic fibroblasts [MEFs]) from the Van Remmen lab [12] explain the possible mechanism that contributes to the early death of SOD2−/− mice. The proliferation rate of SOD2−/− MEFs was decreased significantly compared to that of wild-type MEFs, and SOD2−/− MEFs had reduced cellular ATP levels, impaired O2 consumption, reduced expression of peroxiredoxin 3 (Prdx3), and enhanced superoxide generation.
It has been suggested that MnSOD is a tumor–suppressor gene in that it inhibits the progression of cancer by detoxifying superoxide. Mutations in the promoter region and epigenetic regulation have led to decreased expression of MnSOD in cancer cells. In fact, a polymorphism in the mitochondrial import signal has been associated with an increased risk of prostate cancer [13]. Reduction or deficiency of MnSOD has been shown to promote cytotoxicity under conditions of oxidant stress [10]. A number of laboratories [14, 15] have begun to elucidate how the promoter and the intronic enhancer cause the dramatic inducible expression of MnSOD. However, the molecular and intracellular signaling pathways and the nature of the induction of MnSOD by various inflammatory mediators are still being unraveled.
REDOX STATE AND SIGNALS OF DIVERSITY
Oxygen free radicals and their byproducts that are capable of causing oxidative damage, collectively referred as ROS, may be cytotoxic when produced in excess. However, when present in moderate concentrations, intracellular ROS influence gene expression as well as posttranslational modification of proteins. To gain a better understanding of the exact mechanisms that underlie ROS-dependent disorders in biological systems, recent studies have focused on the regulation of gene expression by oxidants, antioxidants, and other determinants of the intracellular reduction-oxidation (redox) state. Over the past decade, studies have implicated ROS in the activation of transcription factors, such as nuclear factor-kappaB (NF-κB) and activator protein-1 (AP-1), that cause chromatin remodeling and gene expression of pro-inflammatory mediators [16]. These transcription factors are regulated by the activation of Iκ-B kinase, mitogen activated protein (MAP) kinase pathways and phosphoinositide 3-kinase (PI-3K) pathways. It has been shown recently that oxidative stress and redox status of the cells can also regulate nuclear chromatin remodeling (histone acetylation/deacetylation) leading to gene expression of pro-inflammatory mediators [16]. Cellular redox state may modify more than just these transcription factors, as oxidant challenge has been shown to perturb intracellular Ca2+ homeostasis [17]. Such an effect may exacerbate free radical reactions, activate endonucleases, and contribute to apoptosis [18].
Phosphorylation of IκB is a prerequisite for the activation of NF-κB [19]. The possible existence of specific IκB kinases has been hypothesized; however, definite evidence in human cells is not yet available. Numerous studies indicate that ROS may serve as common intracellular agents that contribute to the process of NF-κB activation in response to a diverse range of stimuli [20]. Baeuerle [20] and colleagues have observed that, in certain cell types (e.g., Wurzburg T cells and L6 skeletal muscle myotubes), the ROS hydrogen peroxide is a sensitive inducer of NF-κB activation and sodium hypochlorite, another oxidant, is capable of weakly inducing NF-κB activation in L6 skeletal muscle myotubes. Hypochlorite is a physiological microbiocidal oxidant produced by myeloperoxidase in stimulated neutrophils during the oxidative burst. Interleu-kin-2 (IL-2) and IL-2 receptor (IL-2Ra) genes contain a NF-κB cis-regulatory element in their enhancer regions. Los et al. [21] showed that in T lymphocytes, hydrogen peroxide induces NF-κB activation, IL-2 release, and IL-2R expression. Los et al. [22] also showed that CD-28-mediated activation of NF-κB requires the production of intracellular ROS by 5-lipoxygenase. Not only ROS but also biological derivatives such as oxidatively damaged DNA and advanced glycation endproducts have been identified as inducers of NF-κB activation [23]. Localization of the exact step (or steps) of oxidant action in NF-κB activation is unresolved. The possible presence of oxidant sensitive IκB kinase (or kinases) may not be ruled out, but concrete evidence is lacking.
Evidence suggesting the direct involvement of ROS in AP-1 activation has been obtained primarily using defined oxidative stress generating systems to challenge cultured cells. Studies from a number of laboratories have demonstrated that superoxide produced by a xanthine/xanthine oxidase system and hydrogen peroxide induce the expression of several early response genes including c-fos and c-jun, the major components of AP-1 [24, 25]. Recently it was shown that growth factor-induced AP-1 activation is also ROS-dependent. TNF-α and basic fibroblast growth factor induce ROS production, which act as a common signal to stimulate c-fos gene expression [26]. Angiotensin Il-induced AP-1 DNA binding and proliferative hypertrophic responses in skeletal muscle-derived cells are ROS mediated [27]. ROS are also involved in the activation of AP-1 by ionizing radiation [28]. Ionizing radiation and hydrogen peroxide challenge cause oxidative stress and are potent inducers of c-jun expression. This effect was observed to be protein kinase C-dependent and was evident in both normal and tumor cells [28]. It has been suggested that perturbation of cellular thiol redox status is a signal that may be implicated in the induction of c-fos and c-jun expression caused by asbestos-induced oxidative stress [29]. In support of this, high intracellular glutathione disulfide is implicated in AP-1 activation [30]. AP-1 activation under oxidative conditions may be, at least in part, mediated by phosphorylation of Jun proteins [29]. Hydrogen peroxide and TPA have an additive effect on protein kinase C stimulation, especially in Go cells [31]. Despite its remarkable effect on protein kinase C activity, hydrogen peroxide does not activate AP-1 binding to DNA. In contrast, hydrogen peroxide suppresses TPA-induced AP-1 activation. Hydrogen peroxide is able to trigger AP-1 activation in cells that were treated to down-regulate protein kinase C activity. These results suggest that hydrogen peroxide uniquely influences AP-1 activation compared to TPA-induced AP-1 activation [31]. Studies to-date on the pathway (or pathways) involved in ROS-induced AP-1 activation suggest that serum response factor (SRF) binding to serum responsive element (SRE) and ternary complex factor (TCF) phosphorylation are involved in such processes. Both SRE and AP-1 sites in the c-fos promoter have been implicated in upregulating c-fos expression in response to hydrogen peroxide challenge [32]. Consistent with the notion of the presence of ROS-sensitive kinase cascade (or cascades), oxidative stress-sensitive mitogen-activated protein (MAP) kinases and MAP kinase phosphatases exist [33].
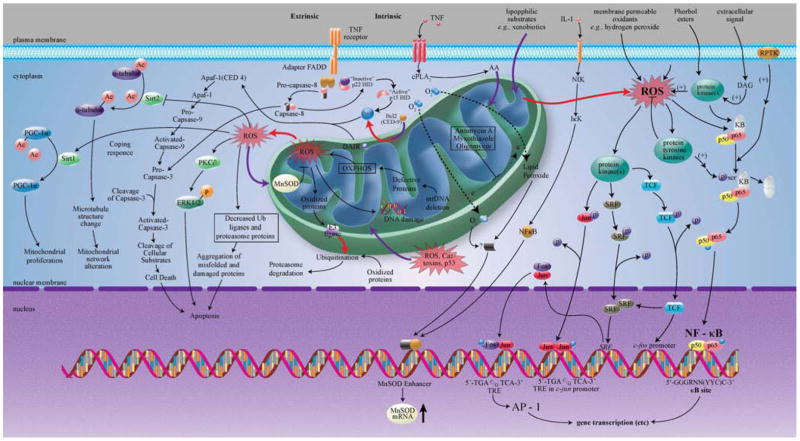
Many signaling cascades that redirect metabolism in response to stress are thought to sense changes in cellular redox status through redox-sensitive thiol-containing proteins such as thioredoxin (TR) [34], thioredoxin reductase (TRX) [35], Ref-1 [36] and AP-1 [36]. These redox-sensitive signaling proteins and downstream transcription factors, therefore, might play a central role in maintaining the steady state intracellular balance among pro-oxidant production, antioxidant capacity, and the repair of oxidative damage. Various inflammatory mediators (TNFα, IL-1 β, IL-6, and LPS) in multiple tissues elicit dramatic elevations of both the messenger RNA and protein levels of MnSOD [37]. However, the signaling pathways responsible for MnSOD expression are numerous and are still far from being fully elucidated. ROS or other mitochondrial intermediates may control both the cytotoxic and gene-regulatory effects of TNFα, thus providing a basis for a mitochondria-to-nucleus signaling pathway, which requires bidirectional communication between the nucleus and the mitochondria (discussed in detail under Retrograde Signaling section). Although the cytotoxic activity of TNFα seems to be restricted to tumor cells, nearly every cell type responds to TNFα by the activation of a wide range of different genes [38] (Fig. 1).
Fig. 1.
Schematic diagram showing the MnSOD (Signals of Diversity).
RETROGRADE SIGNALING
Research on mitochondria has evolved from bioenergetics to biogenesis, to the genetic functions of mitochondrial DNA and diseases associated with mitochondrial dysfunction. Although these areas continue to be investigated vigorously, a new era in mitochondrial research has emerged that concerns the role of this organelle in intracellular signaling—a process that is likely to have implications for human development, aging and disease. Since ROS are very short- lived molecules closely regulated by a coordinated enzyme system, they could be potential signal transducers of putative mitochondria-to-nucleus signaling pathways. Retrograde regulation is the general term for mitochondrial signaling and it is broadly defined as cellular responses to changes in the functional state of mitochondria.
The basic phenomenon of retrograde regulation was first defined in yeast. The striking fundamental similarities between yeast and mammalian mitochondria seem to justify extrapolation and allow the construction of a model of carcinogenesis. RTG genes, RTG1, RTG2, and RTG3, are responsible for this regulation in yeast and some microorganisms [39]. The products of these three genes, three regulatory proteins, have a central role in retrograde signaling. Transcription factors Rtg1p and Rtg3p interact as a heterodimer that activates transcription at target sites [40]. Rtg1p and Rtg3p are sequestered together in the cytoplasm when the retrograde system is inactive. In this state, Rtg3p is phosphorylated at multiple sites of the protein. When the retrograde pathway is activated, Rtg3p is partially dephosphorylated and translocates to the nucleus. Rtg1p probably passively follows and the proteins reassemble at the target genes to activate transcription. Rtg2p acts as a proximal sensor of mitochondrial function, relaying mitochondrial signals to the Rtg1p/Rtg3p complex that affect their intracellular localization. In the absence of Rtg2p, Rtg1p and Rtg3p remain complexed in the cytoplasm, even in cells in which the retrograde response would otherwise be activated [41].
Retrograde responses are affected by metabolic cues or by more direct routes, such as mitochondria-related changes in intracellular Ca2+ dynamics, all methods culminating in wide-ranging changes in nuclear gene expression. The outcome is usually a recasting of metabolic, regulatory, or stress-related pathways. These retrograde responses are for the most part adaptive in that they represent cellular adjustments to altered mitochondrial states. They are distinct from mitochondria-initiated apoptosis and the most extreme example of mitochondrial signaling— the release of proapoptotic factors from the organelle.
MITOCHONDRIA AS A CENTRAL REGULATOR OF INTRINSIC APOPTOTIC PATHWAYS
Intrinsic apoptotic pathways are initiated inside cells. The most important turning point in the course of the intrinsic apoptotic process occurs in the mitochondria. Although it is arguable whether the mitochondrial event triggers apoptosis, it does appear to be the key event in apoptosis. A pivotal event in the mitochondrial pathway is mitochondrial outer membrane permeabilization (MOMP). MOMP is mainly mediated and controlled by Bcl-2 family members. Once MOMP occurs, it precipitates cell death through either the release of molecules involved in apoptosis or the loss of mitochondrial functions essential for cell survival [42].
In the intrinsic pathway, death signals act directly or indirectly on the mitochondria, resulting in the release of cytochrome c and formation of the apoptosome complex. This cell death pathway is controlled by Bcl-2 family proteins (regulation of cytochrome c release), inhibitor of apoptosis proteins (IAPs) (inhibition of caspases), second mitochondrial activator of caspases (Smac), and HtrA2/Omi (negative regulator of IAPs) [43]. Expression levels of antiapoptotic proteins such as Bcl-2 and Bcl-XL have been reported to be upregulated by the transcription factor NF-κB. In addition to being a central regulator of the innate and adaptive immune response, NF-κB is commonly described as an antiapoptotic transcription factor [44, 45], although under certain circumstances NF-κB also might positively contribute to apoptosis induction [46]. NF-κB additionally transactivates a number of other antiapoptotic genes, such as the IAPs. When DNA damage is the trigger of the apoptotic response, the initially activated caspase is procaspase-2. This initiator caspase is activated within the PIDDosome complex, which includes procaspase-2, PIDD (p53-inducible protein with a death domain), and RAIDD (RIP-associated Ich-1/Ced-3-homologous protein with a death domain) [47]. Activation of caspase-2 leads to the release of cytochrome c and subsequent formation of the apoptosome complex. This is followed by the caspase cascade and the cleavage of cellular proteins leading to the biochemical and morphological alterations typical of apoptosis and, finally, cell death.
Another protein family that plays an important role in the apoptotic process is the Bcl-2 family. Proteins from this family can be divided into two groups: proapoptotic and antiapoptotic members. Importantly, a majority of these proteins fulfill their function at the level of mitochondria. On one hand, the presence of Bcl-2 or Bcl-XL prevents permeabilization of the OMM and release of proteins from the intermembrane space, thereby rescuing cells from death. On the other hand, transcriptional or posttranscriptional regulation of proapoptotic multidomain members of this family, such as Bax and/or Bak, leads to their activation by the BH3-only proteins from the same family, e.g., Bid, Puma, or Noxa. Activation of Bax/Bak is characterized by their oligomerization and insertion into the OMM, followed by permeabilization of these membranes, release of proapoptotic proteins from the mitochondria, and cell death.
Depending on which proteins are released from the intermembrane space of the mitochondria, cells might die via activation of either caspase-dependent or caspase-independent pathways. It has now been recognized that mitochondria can also release factors involved in caspase-independent cell death including apoptosis-inducing factor (AIF) [48] and Endonuclease G (EndoG) [49]. In fact, AIF is believed to play a central role in the regulation of caspase-independent cell death [48]. This mitochondrial flavoprotein was first identified and characterized in the laboratory of Kroemer in an examination of apoptotic processes in cell free systems. In these experiments, supernatants derived from mitochondria, in which permeability transition had been induced, were capable of triggering chromatin condensation in isolated HeLa nuclei [50]. The soluble factor responsible for this activity was then identified as a mitochondrial protein that translocated to the nucleus following an apoptotic stimulus to induce chromatin condensation in a caspase-independent manner [51]. Nuclear localization of AIF is linked to chromatin condensation and appearance of high-molecular-weight chromatin fragments. Importantly, an obligatory cofactor for AIF-mediated chromatin condensation is cyclophilin A; however, the precise mechanism of the chromatin condensation catalyzed by these two proteins is still unclear. The discovery of this soluble factor, AIF, revealed a new death pathway that could execute apoptosis-like cell death in the absence of caspases.
AIF cleavage and release during apoptosis is governed by multiple regulatory steps. For example, in vitro experiments using recombinant AIF protein suggest that both the respiratory and the proapoptotic functions of AIF are controlled by its interaction with NAD(P)H [52]. More recently, evidence shows that sustained intra-cellular Ca2+ increase plays a pivotal role in the activation of a mitochondrial calpain responsible for the cleavage of AIF in STS-treated cells [53]. Other studies have indicated that antioxidants can restrain cell death in experimental systems in which the AIF-mediated pathway is important [54]. The latter findings suggest that ROS formation might influence AIF processing during apoptosis.
Furthermore, in certain death paradigms caspase-independent chromatin condensation can be triggered in the absence of obvious AIF or EndoG translocation [55]. Thus, there are likely additional factors that can contribute to the caspase-independent apoptotic process. One such candidate is the AIF homolog AMID (AIF-homologous mitochondrion-associated inducer of death) [56] also known as PRG3 [57]. As with AIF, AMID/PRG3 is homologous with NADH oxidoreductases/flavoproteins. Unlike AIF, however, it lacks a recognizable mitochondria localization sequence (MLS) and appears to reside primarily in the cytoplasm or to be loosely associated with the mitochondrial outer membrane. Recent studies in human leukemia Jurkat T-cells suggest increased expression and plasma membrane association of AMID after apoptosis induction.
Interestingly, AMID is induced by p53 [57] and overexpression studies have demonstrated that it is capable of inducing peripheral chromatin condensation and cell death in the absence of caspase activation [56]. Although AMID represents an intriguing candidate, its physiological importance and relationship to currently known death pathways remain to be determined. Owing to the many unanswered questions in this field, it is highly likely that additional factors will be discovered that will help to clarify these mechanisms.
In cultured mammalian cell systems, overexpression of the bcl-2 gene can protect against hydroxyl radical production and against hypoxia induced cell death [58]. In general bcl-2 is thought to be a protective gene against apoptosis. It can form heterodimers with Bax, a protein of similar structure that is pro-apoptotic. A part of the apoptotic process may involve induction of free-radical production by mitochondria [59]. Bcl-2 protein titre is increased in cultured skin fibroblasts treated with respiratory chain inhibitors [60] and in response to genetically determined respiratory chain defects, especially those in complex I. It has been shown that there seems to be a balance between induction of MnSOD and the induction of Bcl-2 by oxygen free radicals such that cell lines that have little change in redox state and MnSOD induction go on to induce Bcl-2 and indeed have high Bcl-2/Bax ratios [61].
Eukaryotic cells contain fidelity proteins that monitor the integrity of critical intracellular processes, and deletion or mutation of corresponding genes results in a cellular environment permissive for the accumulation of DNA damage [62]. Despite the central role of mitochondria in the control of apoptosis, surprisingly little is known about how mitochondrial sirtuins participate in apoptotic programs. SIRT3 plays a pro-apoptotic role in BCL2, p53 and JNK-regulated apoptosis [63]. Additionally, cells lacking SIRT3 show decreased stress-induced apoptosis, lending further support to a pro-apoptotic role for SIRT3 [64]. The mechanism for the tumor-suppressive function of SIRT3 is incompletely understood, but it involves repression of ROS and protection against DNA damage [64]. In some studies, SIRT3 has been shown to be anti-apoptotic. For example, in cellular response to DNA damage when mitochondrial NAD+ levels fall below critical levels, SIRT3 and SIRT4 display anti-apoptotic activity, protecting cells from death [65]. The mitochondria contain fidelity proteins that maintain the integrity of the mitochondria. In this regard, loss of SIRT3 results in an aging-related decrease in MnSOD, resulting in an increase in mitochondrial superoxide and, also perhaps, other ROS. This may create a cellular environment permissive for in vivo carcinogenesis including receptor-positive mammary tumors that are observed in SIRT3 knockout mice older than 13 months of age [64]. As such, SIRT3 functions as a genomically expressed, mitochondrially localized fidelity protein, in addition to meeting the criteria to be classified as a tumor suppressor gene.
ROS AND PROTEIN MODIFICATION
Oxidative Modification and Retrograde Signaling
Proteins are major targets of ROS, which can trigger multiple modifications of protein structure. ROS are highly reactive and, when generated close to cell membranes, they oxidize membrane phospholipids (lipid peroxidation) that can lead to the generation and accumulation of lipid peroxidation products, such as malondialdehyde, 4-hydroxy-2-nonenal (4-HNE), acrolein and F2-isoprostanes. 4-HNE is a highly reactive and specific diffusible end-product of lipid peroxidation, and is known to induce/regulate various cellular events such as proliferation and growth inhibition [66], T cell apoptosis [67] and activation of signaling pathways [68]. 4-HNE has a high affinity towards cysteine, histidine and lysine residues forming direct protein-adducts and thereby altering protein function. Moreover, 4-HNE activates glutathione (GSH) synthesis via induction of the g-glutamyl cysteine ligase (GCL) gene (a key enzyme for GSH synthesis) and a variety of pro-inflammatory genes, such as IL-8, MCP-1, COX-2, EGFR, MUC5AC, etc., suggesting that 4-HNE works as a signaling molecule in gene transcription [66–68].
Posttranslational modification by carbonylation might result in partial unfolding, inactivation, or proteasomal degradation [69, 70]. Questions that need to be resolved are: Do the modifications subserve a physiologic homeostatic function or disrupt cellular processes, and are the redox-based modifications simply an intrinsic function of ROS/RNS that results in protective or deleterious effects, or do they subserve cellular signaling? And to the extent that the redox regulation of protein function elicits cellular signals, are those signals: (1) physiologic and homeostatic, (2) stress-related or adaptive (e.g., upregulation of protective, proliferative responses, increased release of calcium from internal stores), or (3) maladaptive/injurious, including growth signals with malignant potential and overt cellular injury.
Excess production of ROS by defective mitochondria causes oxidative damage to cellular constituents, especially those in the mitochondria. Many studies have shown that oxidative modification of membrane lipids and proteins is increased in the mitochondria of aged tissues or in cells under oxidative stress. Weinstein et al. [71], recently showed that peroxynitrite is formed in cardiomyocytes in mice after treatment with anthracycline antibiotic adriamycin, one of the most effective antitumor agents used to treat human malignancies, and Aldieri et al. [72], have demonstrated that Dox induces nitric oxide synthesis, resulting in accumulation of nitrite in cells. Mitochondria are a primary site of oxidative/nitrative damage products and they are the organelle most extensively involved in Dox-induced cardiotoxicity. Chaiswing [73] was the first to document and quantify levels of oxidative/nitrative damage products in situ in specific subcellular locations in cardiac tissues following Dox treatment. According to Norberg et al. [74]. AIF becomes oxidatively modified (carbonylated) as a consequence of enhanced mitochondrial ROS generation during STS-induced apoptosis and this significantly promotes its cleavage by calpain. Suppression of ROS accumulation by pretreatment of the cells with antioxidants prevents carbonylation, as well as the cleavage and release of AIF [75].
p53 Activity is Affected by ROS/RNS
ROS controls the activity of a variety of signaling proteins such as protein tyrosine phosphatases [76, 77] and protein kinases [77], as well as myriad transcription factors, including NF-κB, AP-1 [78–80] and p53 [81–84], by modifying key amino acids within the transcription factor. These modifications include carbonylation of proline, arginine, threonine, and lysine [85], as well as nitration and phosphorylation of tyrosine [86, 87]. Cysteine can undergo myriad redox modifications. The thiol group of cysteine can react with hydrogen peroxide to form sulfenic acid (SOH). Once sulfenic acid is formed, it can be further oxidized to sulfinic (SO2H) and sulfonic (SO3H) acids; or it can be conjugated with low molecular weight thiols, such as glutathione; or it can undergo intrachain or interchain disulfide bond formation [88]. Several redox modifications have various effects on p53 function. Tyrosine nitration of p53 has been seen in cells treated with various compounds capable of nitrating p53. Interestingly, low concentrations of these nitrating compounds increase p53 DNA binding activity [89], while high concentrations decrease p53 DNA binding activity [90, 91]. The formation of intra chain disulfide bonds also inhibits p53 DNA binding activity [92]. The redox status of p53 affects its DNA binding activity through various mechanisms [93]. Oxidized p53 does not bind supercoiled DNA as effectively as reduced p53 does [94, 95]. Loss of DNA binding activity is not restored by reduction of oxidized p53 by dithiothreitol (DTT). However, due to the loss of Zn2+ ions in the DNA binding domain of p53, the presence of excess Zn2+ ions partially protects p53 from the effects of oxidizing agents [93, 94].
ROS/RNS-induced changes in conformation and oligomerization of p53 can also affect DNA binding activity. Reduced p53 contains 5 free thiols, while oxidized p53 has 4 free thiols, suggesting a disulfide bond may be present in oxidized p53. This disulfide bond results in noticeable conformational changes in oxidized p53 compared to reduced p53. In a study by Sun et al., the majority of oxidized p53 forms monomers, dimers, and other high molecular weight oligomers, while reduced p53 forms tetramers that are able to bind DNA [96]. Modifications of cysteine residues are important for bestowing ROS responsiveness to p53. Two groups of cysteine residues exist within the DNA binding domain of p53 [84, 97]. One group of cysteine residues (Cys 173, 235, and 239) is located within the Zn2+ binding site of the DNA binding domain. The other group of cysteine residues (Cys 121, 132, 138, and 272) is located in the loop-sheet-helix region, which confers site-specific DNA binding to p53 [84]. Sequence-specific DNA binding of p53 is also affected by redox modification of Cys277 [97]. In WS-1 human fibroblasts, UVC irradiation leads to increased p53 binding to response elements that contain thymine, while reduction of p53 with DTT leads to increased binding to cytosine-containing response elements [98]. p53 structure and DNA-binding activity are also altered by glutathione modification. Sun et al. propose that glutathionylation of p53 at Cys182 may be involved in the formation of monomers and unusual oligomers and the decreased DNA binding activity observed in oxidized p53. Several cysteine residues in the proximal DNA binding domain (Cys 124, 141, and 182) are glutathionylated, which can interfere with dimerization and DNA binding of p53 [96].
Different oxidants and antioxidants exert their effects through p53. Treatment of H1299 human lung cancer cells with selenomethionine (SeMet) results in increased p53 DNA binding activity through Ref-1-dependent reduction of p53 [99]. Other selenium-containing compounds (sodium selenite and methyl-selenic acid) induce phosphorylation of various serine and threonine residues in p53 that are important for p53-mediated apoptosis [100]. The oxidant PDTC oxidizes cysteine residues in p53, resulting in diminished UV-induced p53 nuclear translocation and decreased transactivation of p53-responsive genes [101]. Several mutations [102, 103] and polymorphisms [104, 105] identified in p53 may alter redox regulation of p53 activity and intracellular localization. p53 intracellular localization is affected by a polymorphism at codon 72 (resulting in either proline or arginine) without affecting p53-dependent gene regulation. Mitochondrial localization of p53 is more prevalent with the Arg72 variant than the Pro72 variant due to an enhanced interaction of the Arg72 variant with CRM1 and MDM2 that results in enhanced ubiquitination and nuclear export of p53. The Arg72 variant also interacts more strongly with the mitochondrial import proteins GRP75 and Hsp60 [106]. Ascertaining how codon 72 and other polymorphisms influence ROS-dependent regulation of p53 may be important for determining the effects of oxidative stress oncellular strategies in the regulation of p53 (activity, intracellular localization, and protein stabilization).
Mitochondrial ROS Regulate p53 Activity
Since mitochondria are an important source of ROS in cells and ROS regulate p53 activity, reason dictates that altered mitochondrial function that affects ROS production may also impact p53. Changes in mitochondrial membrane potential (MMP) may affect p53 activity. In MOLT-3 human leukemia cells, treatment with the ATP synthase inhibitor oligomycin decreases MMP, resulting in a decrease in etoposide-induced p53 accumulation, p53 target gene expression, and a decrease in apoptosis [107]. In JB6 mouse epithelial cells, treatment with cyclosporine A (CsA, a mitochondrial permeability transition pore inhibitor) inhibits TPA (12-O-tetradecanoylphorbol-13-acetate)-induced p53 mitochondrial translocation by impeding TPA-induced decreases in complex I activity and MMP [108]. This laboratory discovered that TPA treatment of JB6 cells results in a rapid build-up of p53 protein and localization to the nucleus, the outer membrane and the matrix of mitochondria. Interaction between p53 and MnSOD correlates with a decrease in MnSOD enzyme activity. The MnSOD mimetic, MnTE-2-PyP5+ blocks TPA induced p53 nuclear translocation and expression of the antiapoptotic protein Bax without altering TPA-stimulated mitochondrial localization of p53 [109].
Mitochondrial ROS production is also affected by depletion of mitochondrial DNA (mtDNA), and this change in generation of ROS may have important effects on p53. These effects may be cell-type specific. For example, in HeLa cells depleted of mtDNA (ρ0 cells), oxidative nuclear DNA damage increases, nuclear DNA repair decreases, and lipid peroxidation increases despite an increase in the antioxidant capacity of the cells [110]. In SK-Hep1 hepatoma cells, treatment with ethidium bromide (EtBr) to deplete the cells of mtDNA (ρ0 cells) causes an initial burst of superoxide production. Once ρ0 SK-Hep1 cells are established, there is no difference in ROS production between ρ0 and parental cells because of an increase in the antioxidant capacity of the ρ0 cells through increased expression of the mitochondrial forms of GPX (GPX1 and PHGPX) and MnSOD. The increased antioxidant capacity of the ρ0 SK-Hep1 cells makes them more resistant to paraquat, menadione, and doxorubicin (ROS-generating agents), as well as leads to increased expression of p53 [111].
Mitochondria are important oxygen sensors in the cell, and changes in oxygen concentration alter mitochondrial activity, resulting in a cellular adaptive response [112]. In embryonic chick cardiomyocytes, decreased oxygen levels cause increased ROS production. Inhibitors of oxidative phosphorylation (TTFA, rote-none) and different antioxidants (2-mercaptopropionyl glycine and 1,10-phenanthroline) block this increase in ROS production, suggesting that ROS production occurs at mitochondria [113]. In MCF-7 human breast cancer cells, hypoxia causes an increase in mitochondrial ROS production, leading to an accumulation of p53 protein [112]. In human ML-1 cells, hypoxia leads to an increase in p53 at the mitochondrial outer membrane [114]. These results imply that mitochondrial ROS may stimulate mitochondrial translocation of p53 and amplify mitochondrial oxidative stress through transcription-dependent and –independent mechanisms, enhancing apoptosis. Therefore, antioxidants may affect both mitochondrial and nuclear actions of p53 by modulating mitochondrial ROS production.
ROS-Dependent Regulation of Src Signaling
Src is part of a nine-member family of non-receptor tyrosine kinases that regulate a variety of cellular functions, including cell adhesion, growth factor signaling, cell proliferation, angiogenesis, and bone remodeling [115]. Increased expression and activity of Src family members has been observed in many cancer types, making Src an attractive target for cancer therapeutics [116]. ROS are important for regulation of Src activity [117]. However, controversy exists about whether oxidation of Src kinases leads to activation or inactivation of Src. Kemble and Sun report that oxidation of Src leads to its inactivation. Cys277, within the catalytic domain of Src, is vital for the ROS-mediated deactivation of Src through the formation of an intermolecular disulfide bond between two Src molecules [118]. Some reports suggest that oxidation of key cysteine residues activates Src. Using NIH3T3 cells, Akhand et al. [119], demonstrated that nitric oxide generators S-nitroso-N-acetyl pencillamine (SNAP) and sodium nitroprusside, as well as hydrogen peroxide, increase Src kinase activity through both the auto-phosphorylation of Src at Tyr416 and the formation of disulfide bonds, resulting in aggregation of Src molecules. Krasnowska et al. found that activation of Src by hydrogen peroxide is reversed by treatment with the reducing agent N-acetylcysteine (NAC) and, in various cancer cells tested, NAC treatment decreases Src-activating phosphorylation at Tyr419 and causes Src to localize away from the plasma membrane toward endolysosomes [120]. Src oxidation is also important for integrin-mediated cell adhesion. Inhibition of 5′-lipoxygenase using NDGA (nordihydroguaiaretic acid) results in decreased oxidation of Src, reduced autophosphorylation of Src at Tyr416, and diminished association between Src and focal adhesion kinase (FAK) during cell adhesion [121].
Mitochondria regulate Src activation through both ROS-dependent and –independent mechanisms. A-kinase anchor protein 121 (AKAP121) and AKAP84 are found in the outer membrane of mitochondria. These proteins normally attract protein kinase A (PKA) to mitochondria and focus PKA signaling on the organelle. AKAP121 and AKAP84 also bind to protein tyrosine phosphatase D1 (PTPD1), an important positive regulator of Src. When cells are transfected with PTPD1 alone, there is a Src-dependent amplification of EGF receptor signaling. When AKAP121 is transfected, Src-dependent activation of the EGF receptor is diminished because PTPD1 is recruited to mitochondria [122]. The downstream of kinase (Dok) adaptor protein family member Dok-4 also localizes to mitochondria and recruits c-Src to the organelle. Dok-4-dependent mitochondrial translocation of c-Src is important for modulation of tumor necrosis factor-α (TNF-α) mitochondrial ROS production and NF-κB activation [123]. Lluis et al. [124], found that hypoxia-induced production of mROS leads to activation of c-Src, which stimulates NF-κB activity through phosphorylation of IκB-α. Cys487 within the kinase domain of c-Src is vital for mROS-induced c-Src activation [124]. NADPH oxidase-4 (Nox4), a member of a large family of ROS producing enzymes, is localized to mitochondria [125, 126], and Nox4 is important for the regulation of angiotensin II (AngII) signaling in rat glomerular mesangial cells [127]. Small-interfering RNA (siRNA) targeted against Nox4 prevents AngII-induced activation of Src and PDK-1 (3-phosphoinositide-dependent protein kinase-1), as well as an oxidation-resistant Src mutant (Cys487Ala), suggesting that Nox4 oxidative activation of Src is an important mechanism that effects AngII in kidney cells. The pattern of protein tyrosine phosphorylation after ROS treatment has a striking similarity with that following surface immunoglobulin (slg) dependent physiological stimulation [128]. Src family protein tyrosine kinases (e.g., lck, fyn, and lyn) are activated after slg stimulation. At least two members of the src family, p56lck and p59fyn, have been found to be activated by hydrogen peroxide and also by the thiol oxidizing agent diamide [128, 129]. Spleen tyrosine kinase (Syk) protein is responsive to hydrogen peroxide, UV light, and slg stimulations, suggesting a common pathway of signal transduction. The antioxidant N-acetylcysteine (NAC) inhibits antigen-mediated syk activation in most cells.
SUMMARY AND FUTURE DIRECTIONS
Retrograde signaling is an important mechanism of communication between mitochondria and nucleus and it impacts a wide spectrum of cellular activities under both normal and pathophysiological conditions. In both yeast and mammalian cells, retrograde signaling is also linked to TOR signaling, but the precise signals and connections that interlink these pathways are unclear. In mammalian cells compromised mitochondrial function resulting from mtDNA lesions, membrane damage, insufficient O2, or nutrient supply triggers signaling cascades through altered Ca2+ dynamics, which induces activation of transcription factors. Retrograde signaling also induces invasive behavior in otherwise nontumorigenic cells implying it has a role in tumor progression. Abrogation of mitochondrial stress signaling under in vivo tumor settings through the use of specific inhibitors, such as the dominant-negative IκBβ, may offer new therapeutic intervention strategies for some cancers. Oxidative mitochondrial dysfunction is not mediated by a single mechanism, but that may be a consequence of multiple vicious circles organized within a complex functional network. Investigation of the protective roles played by MnSOD in different cell types has to be elucidated, although information from transgenic mouse models indicates that MnSOD is absolutely essential for the removal of mitochondrially produced oxygen free radicals. Similarly, the exact role of free-radical production in the process of apoptosis still needs to be clarified.
Many studies show indirect evidence that overexpressing MnSOD in several human cell lines increases the production of H2O2 [130–133]. However, the current paradigm shows that the concentrations of O2·− and H2O2 in a cell are assumed to be in a quasi-steady state, reflecting a balance between the rate of formation and the rate of removal. Thus, the steady-state level can change by either changing the rate of formation and/or the rate of removal. An increase in SOD under normal circumstances would increase the rate of removal of O2·− and thereby lower O2·−. This paradigm is based on the observation that the rate of production of H2O2 by the enzyme xanthine oxidase (XO) is not affected by SOD [134]. Furthermore, the efficient H2O2 removal system present in the cells would make it unlikely that the H2O2 generated will accumulate.
Studies conducted by Buettner et al. [134] suggest an entirely new MnSOD function as that of an enzyme that plays a role in establishing the flux of H2O2 in cells. Because H2O2 is a key to determining the redox environment of cells and tissues, MnSOD should now be viewed not only as an antioxidant enzyme but also as a key enzyme involved in the establishment of the cellular redox environment and thereby the biological status of cells and tissues. It is also possible that H2O2 generated from forced overexpression of MnSOD beyond physiological conditions may react with MnSOD and enhanced production of O2·−, not H2O2. These and other possibilities need further evaluation in vivo under physiological and/or pathological conditions.
Acknowledgments
We are very thankful to The Teaching and Academic Support Center (TASC) for helping us in sketching the Fig. (1). The authors are supported by NIH grants CA 143428, CA049797, CA073599, CA139843, CA 115801, AG005119, and T32 ES007266.
ABBREVIATIONS
- MnSOD
Manganese Superoxide Dismutase
- ROS
Reactive oxygen species
- Sirt
Sirtuin
- PGC-1α
Peroxisome proliferator-activated receptor-γ co-activator 1α
- PKC
Protein Kinase C
- CED
Cholesterol-enriched diet
- FADD
Fas-Associated protein with Death Domain
- cPLA2
cytosolic phospholipase A2
- TNF
Tumor necrosis Factor
- IL
Interleukin
- DAG
diacylglycerol
- RPTK
Transmembrane receptor tyrosine protein kinases
- TCF
Ternary complex factor
- SRF
Serum response factor
- SRE
Serum responsive element
- ERK
Extracellular signal-regulated kinase-1
- NIK
nuclear transcription factor κB (NF-κB)-inducing kinase
- NF-κB
Nuclear transcription factor κB
- DNA
Deoxyribonucleic acid
- mtDNA
Mitochondrial DNA
References
- 1.Brown DI, Griendling KK. Nox proteins in signal transduction. Free Radic Biol Med. 2009;47(9):1239–1253. doi: 10.1016/j.freeradbiomed.2009.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gillespie MN, Pastukh V, Ruchko MV. Oxidative DNA modifications in hypoxic signaling. Ann N Y Acad Sci. 2009;1177:140–150. doi: 10.1111/j.1749-6632.2009.05036.x. [DOI] [PubMed] [Google Scholar]
- 3.Andersen JK. Oxidative stress in neurodegeneration: cause or consequence? Nat Med. 2004;10(Suppl):S18–25. doi: 10.1038/nrn1434. [DOI] [PubMed] [Google Scholar]
- 4.Fridovich I. Superoxide radical and superoxide dismutases. Annu Rev Biochem. 1995;64:97–112. doi: 10.1146/annurev.bi.64.070195.000525. [DOI] [PubMed] [Google Scholar]
- 5.Flint DH, Tuminello JF, Emptage MH. The inactivation of Fe-S cluster containing hydro-lyases by superoxide. J Biol Chem. 1993;268(30):22369–22376. [PubMed] [Google Scholar]
- 6.Zelko IN, Mariani TJ, Folz RJ. Superoxide dismutase multigene family: a comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic Biol Med. 2002;33(3):337–349. doi: 10.1016/s0891-5849(02)00905-x. [DOI] [PubMed] [Google Scholar]
- 7.McCord JM, Keele BB, Jr, Fridovich I. An enzyme-based theory of obligate anaerobiosis: the physiological function of superoxide dismutase. Proc Natl Acad Sci U S A. 1971;68(5):1024–1027. doi: 10.1073/pnas.68.5.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Touati D. The molecular genetics of superoxide dismutase in E. coli. An approach to understanding the biological role and regulation of SODS in relation to other elements of the defence system against oxygen toxicity. Free Radic Res Commun. 1989;8(1):1–9. doi: 10.3109/10715768909087967. [DOI] [PubMed] [Google Scholar]
- 9.Gregory EM, Fridovich I. Oxygen toxicity and the superoxide dismutase. J Bacteriol. 1973;114(3):1193–1197. doi: 10.1128/jb.114.3.1193-1197.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Huang TT, Carlson EJ, Melov S, Ursell PC, Olson JL, Noble LJ, Yoshimura MP, Berger C, Chan PH, Wallace DC, Epstein CJ. Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nat Genet. 1995;11(4):376–381. doi: 10.1038/ng1295-376. [DOI] [PubMed] [Google Scholar]
- 11.Van Remmen H, Ikeno Y, Hamilton M, Pahlavani M, Wolf N, Thorpe SR, Alderson NL, Baynes JW, Epstein CJ, Huang TT, Nelson J, Strong R, Richardson A. Life-long reduction in MnSOD activity results in increased DNA damage and higher incidence of cancer but does not accelerate aging. Physiol Genomics. 2003;16(1):29–37. doi: 10.1152/physiolgenomics.00122.2003. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Zhang HM, Shi Y, Lustgarten M, Li Y, Qi W, Zhang BX, Van Remmen H. Loss of manganese superoxide dismutase leads to abnormal growth and signal transduction in mouse embryonic fibroblasts. Free Radic Biol Med. 2010;49(8):1255–1262. doi: 10.1016/j.freeradbiomed.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Y, Krishnan A, Wan XS, Majima H, Yeh CC, Ludewig G, Kasarskis EJ, St Clair DK. Mutations in the promoter reveal a cause for the reduced expression of the human manganese superoxide dismutase gene in cancer cells. Oncogene. 1999;18(1):93–102. doi: 10.1038/sj.onc.1202265. [DOI] [PubMed] [Google Scholar]
- 14.Melov S, Schneider JA, Day BJ, Hinerfeld D, Coskun P, Mirra SS, Crapo JD, Wallace DC. A novel neurological phenotype in mice lacking mitochondrial manganese superoxide dismutase. Nat Genet. 1998;18(2):159–163. doi: 10.1038/ng0298-159. [DOI] [PubMed] [Google Scholar]
- 15.Jones PL, Ping D, Boss JM. Tumor necrosis factor alpha and interleukin-1beta regulate the murine manganese superoxide dismutase gene through a complex intronic enhancer involving C/EBP-beta and NF-kappaB. Mol Cell Biol. 1997;17(12):6970–6981. doi: 10.1128/mcb.17.12.6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richter C, Gogvadze V, Laffranchi R, Schlapbach R, Schweizer M, Suter M, Walter P, Yaffee M. Oxidants in mitochondria: from physiology to diseases. Biochim Biophys Acta. 1995;1271(1):67–74. doi: 10.1016/0925-4439(95)00012-s. [DOI] [PubMed] [Google Scholar]
- 17.Trump BF, Berezesky IK. The role of cytosolic Ca2+ in cell injury, necrosis and apoptosis. Curr Opin Cell Biol. 1992;4(2):227–232. doi: 10.1016/0955-0674(92)90037-d. [DOI] [PubMed] [Google Scholar]
- 18.Aagaard-Tillery KM, Jelinek DF. Differential activation of a calcium-dependent endonuclease in human B lymphocytes. Role in ionomycin-induced apoptosis. J Immunol. 1995;155(7):3297–3307. [PubMed] [Google Scholar]
- 19.Thanos D, Maniatis T. NF-kappa B: a lesson in family values. Cell. 1995;80(4):529–32. doi: 10.1016/0092-8674(95)90506-5. [DOI] [PubMed] [Google Scholar]
- 20.Baeuerle PA, Henkel T. Function and activation of NF-kappa B in the immune system. Annu Rev Immunol. 1994;12:141–179. doi: 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- 21.Los M, Droge W, Stricker K, Baeuerle PA, Schulze-Osthoff K. Hydrogen peroxide as a potent activator of T lymphocyte functions. Eur J Immunol. 1995;25(1):159–165. doi: 10.1002/eji.1830250127. [DOI] [PubMed] [Google Scholar]
- 22.Los M, Schenk H, Hexel K, Baeuerle PA, Droge W, Schulze-Osthoff K. IL-2 gene expression and NF-kappa B activation through CD28 requires reactive oxygen production by 5-lipoxygenase. EMBO J. 1995;14(15):3731–3740. doi: 10.1002/j.1460-2075.1995.tb00043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Legrand-Poels S, Bours V, Piret B, Pflaum M, Epe B, Rentier B, Piette J. Transcription factor NF-kappa B is activated by photosensitization generating oxidative DNA damages. J Biol Chem. 1995;270(12):6925–6934. doi: 10.1074/jbc.270.12.6925. [DOI] [PubMed] [Google Scholar]
- 24.Devary Y, Gottlieb RA, Lau LF, Karin M. Rapid and preferential activation of the c-jun gene during the mammalian UV response. Mol Cell Biol. 1991;11(5):2804–28011. doi: 10.1128/mcb.11.5.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nose K, Shibanuma M, Kikuchi K, Kageyama H, Sakiyama S, Kuroki T. Transcriptional activation of early-response genes by hydrogen peroxide in a mouse osteoblastic cell line. Eur J Biochem. 1991;201(1):99–106. doi: 10.1111/j.1432-1033.1991.tb16261.x. [DOI] [PubMed] [Google Scholar]
- 26.Lo YY, Cruz TF. Involvement of reactive oxygen species in cytokine and growth factor induction of c-fos expression in chondrocytes. J Biol Chem. 1995;270(20):11727–11730. doi: 10.1074/jbc.270.20.11727. [DOI] [PubMed] [Google Scholar]
- 27.Puri PL, Avantaggiati ML, Burgio VL, Chirillo P, Collepardo D, Natoli G, Balsano C, Levrero M. Reactive oxygen intermediates (ROIs) are involved in the intracellular transduction of angiotensin II signal in C2C12 cells. Ann N Y Acad Sci. 1995;752:394–405. doi: 10.1111/j.1749-6632.1995.tb17447.x. [DOI] [PubMed] [Google Scholar]
- 28.Datta R, Hallahan DE, Kharbanda SM, Rubin E, Sherman ML, Huberman E, Weichselbaum RR, Kufe DW. Involvement of reactive oxygen intermediates in the induction of c-jun gene transcription by ionizing radiation. Biochemistry (Mosc) 1992;31(35):8300–8306. doi: 10.1021/bi00150a025. [DOI] [PubMed] [Google Scholar]
- 29.Janssen YM, Heintz NH, Mossman BT. Induction of c-fos and c-jun proto-oncogene expression by asbestos is ameliorated by N-acetyl-L-cysteine in mesothelial cells. Cancer Res. 1995;55(10):2085–2089. [PubMed] [Google Scholar]
- 30.Galter D, Mihm S, Droge W. Distinct effects of glutathione disulphide on the nuclear transcription factor kappa B and the activator protein-1. Eur J Biochem. 1994;221(2):639–648. doi: 10.1111/j.1432-1033.1994.tb18776.x. [DOI] [PubMed] [Google Scholar]
- 31.Stauble B, Boscoboinik D, Tasinato A, Azzi A. Modulation of activator protein-1 (AP-1) transcription factor and protein kinase C by hydrogen peroxide and D-alpha-tocopherol in vascular smooth muscle cells. Eur J Biochem. 1994;226(2):393–402. doi: 10.1111/j.1432-1033.1994.tb20064.x. [DOI] [PubMed] [Google Scholar]
- 32.Amstad PA, Krupitza G, Cerutti PA. Mechanism of c-fos induction by active oxygen. Cancer Res. 1992;52(14):3952–3960. [PubMed] [Google Scholar]
- 33.Pahl HL, Baeuerle PA. Oxygen and the control of gene expression. Bioessays. 1994;16(7):497–502. doi: 10.1002/bies.950160709. [DOI] [PubMed] [Google Scholar]
- 34.Kirkpatrick DL, Ehrmantraut G, Stettner S, Kunkel M, Powis G. Redox active disulfides: the thioredoxin system as a drug target. Oncol Res. 1997;9(6–7):351–356. [PubMed] [Google Scholar]
- 35.Hirota K, Matsui M, Murata M, Takashima Y, Cheng FS, Itoh T, Fukuda K, Yodoi J. Nucleoredoxin, glutaredoxin, and thioredoxin differentially regulate NF-kappaB, AP-1, and CREB activation in HEK293 cells. Biochem Biophys Res Commun. 2000;274(1):177–182. doi: 10.1006/bbrc.2000.3106. [DOI] [PubMed] [Google Scholar]
- 36.Wei SJ, Botero A, Hirota K, Bradbury CM, Markovina S, Laszlo A, Spitz DR, Goswami PC, Yodoi J, Gius D. Thioredoxin nuclear translocation and interaction with redox factor-1 activates the activator protein-1 transcription factor in response to ionizing radiation. Cancer Res. 2000;60(23):6688–6695. [PubMed] [Google Scholar]
- 37.Visner GA, Dougall WC, Wilson JM, Burr IA, Nick HS. Regulation of manganese superoxide dismutase by lipopolysaccharide, interleukin-1, and tumor necrosis factor. Role in the acute inflammatory response. J Biol Chem. 1990;265(5):2856–2864. [PubMed] [Google Scholar]
- 38.Lee TH, Lee GW, Ziff EB, Vilcek J. Isolation and characterization of eight tumor necrosis factor-induced gene sequences from human fibroblasts. Mol Cell Biol. 1990;10(5):1982–1988. doi: 10.1128/mcb.10.5.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jia Y, Rothermel B, Thornton J, Butow RA. A basic helix-loop-helix-leucine zipper transcription complex in yeast functions in a signaling pathway from mitochondria to the nucleus. Mol Cell Biol. 1997;17(3):1110–1117. doi: 10.1128/mcb.17.3.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liao X, Butow RA. RTG1 and RTG2: two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell. 1993;72(1):61–71. doi: 10.1016/0092-8674(93)90050-z. [DOI] [PubMed] [Google Scholar]
- 41.Sekito T, Thornton J, Butow RA. Mitochondria-to-nuclear signaling is regulated by the subcellular localization of the transcription factors Rtg1p and Rtg3p. Mol Biol Cell. 2000;11(6):2103–2115. doi: 10.1091/mbc.11.6.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Green DR, Kroemer G. The pathophysiology of mitochondrial cell death. Science. 2004;305(5684):626–629. doi: 10.1126/science.1099320. [DOI] [PubMed] [Google Scholar]
- 43.Cory S, Huang DC, Adams JM. The Bcl-2 family: roles in cell survival and oncogenesis. Oncogene. 2003;22(53):8590–8607. doi: 10.1038/sj.onc.1207102. [DOI] [PubMed] [Google Scholar]
- 44.Heckman CA, Mehew JW, Boxer LM. NF-kappaB activates Bcl-2 expression in t(14;18) lymphoma cells. Oncogene. 2002;21(24):3898–3908. doi: 10.1038/sj.onc.1205483. [DOI] [PubMed] [Google Scholar]
- 45.Karin M, Lin A. NF-kappaB at the crossroads of life and death. Nat Immunol. 2002;3(3):221–227. doi: 10.1038/ni0302-221. [DOI] [PubMed] [Google Scholar]
- 46.Grimm S, Bauer MK, Baeuerle PA, Schulze-Osthoff K. Bcl-2 down-regulates the activity of transcription factor NF-kappaB induced upon apoptosis. J Cell Biol. 1996;134(1):13–23. doi: 10.1083/jcb.134.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chou JJ, Matsuo H, Duan H, Wagner G. Solution structure of the RAIDD CARD and model for CARD/CARD interaction in caspase-2 and caspase-9 recruitment. Cell. 1998;94(2):171–180. doi: 10.1016/s0092-8674(00)81417-8. [DOI] [PubMed] [Google Scholar]
- 48.Susin SA, Daugas E, Ravagnan L, Samejima K, Zamzami N, Loeffler M, Costantini P, Ferri KF, Irinopoulou T, Prevost MC, Brothers G, Mak TW, Penninger J, Earnshaw WC, Kroemer G. Two distinct pathways leading to nuclear apoptosis. J Exp Med. 2000;192(4):571–580. doi: 10.1084/jem.192.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li LY, Luo X, Wang X. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature. 2001;412(6842):95–99. doi: 10.1038/35083620. [DOI] [PubMed] [Google Scholar]
- 50.Susin SA, Zamzami N, Castedo M, Hirsch T, Marchetti P, Macho A, Daugas E, Geuskens M, Kroemer G. Bcl-2 inhibits the mitochondrial release of an apoptogenic protease. J Exp Med. 1996;184(4):1331–1341. doi: 10.1084/jem.184.4.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Susin SA, Lorenzo HK, Zamzami N, Marzo I, Brenner C, Larochette N, Prevost MC, Alzari PM, Kroemer G. Mitochondrial release of caspase-2 and -9 during the apoptotic process. J Exp Med. 1999;189(2):381–394. doi: 10.1084/jem.189.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Churbanova IY, Sevrioukova IF. Redox-dependent changes in molecular properties of mitochondrial apoptosis-inducing factor. J Biol Chem. 2008;283(9):5622–5631. doi: 10.1074/jbc.M709147200. [DOI] [PubMed] [Google Scholar]
- 53.Norberg E, Gogvadze V, Ott M, Horn M, Uhlen P, Orrenius S, Zhivotovsky B. An increase in intracellular Ca2+ is required for the activation of mitochondrial calpain to release AIF during cell death. Cell Death Differ. 2008;15(12):1857–1864. doi: 10.1038/cdd.2008.123. [DOI] [PubMed] [Google Scholar]
- 54.Zhou M, Baudry M. EUK-207, a superoxide dismutase/catalase mimetic, is neuroprotective against oxygen/glucose deprivation-induced neuronal death in cultured hippocampal slices. Brain Res. 2009;1247:28–37. doi: 10.1016/j.brainres.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arnoult D, Gaume B, Karbowski M, Sharpe JC, Cecconi F, Youle RJ. Mitochondrial release of AIF and EndoG requires caspase activation downstream of Bax/Bak-mediated permeabilization. EMBO J. 2003;22(17):4385–4399. doi: 10.1093/emboj/cdg423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu M, Xu LG, Li X, Zhai Z, Shu HB. AMID, an apoptosis-inducing factor-homologous mitochondrion-associated protein, induces caspase-independent apoptosis. J Biol Chem. 2002;277(28):25617–25623. doi: 10.1074/jbc.M202285200. [DOI] [PubMed] [Google Scholar]
- 57.Ohiro Y, Garkavtsev I, Kobayashi S, Sreekumar KR, Nantz R, Higashikubo BT, Duffy SL, Higashikubo R, Usheva A, Gius D, Kley N, Horikoshi N. A novel p53-inducible apoptogenic gene, PRG3, encodes a homologue of the apoptosis-inducing factor (AIF) FEBS Lett. 2002;524(1–3):163–171. doi: 10.1016/s0014-5793(02)03049-1. [DOI] [PubMed] [Google Scholar]
- 58.Hockenbery DM, Oltvai ZN, Yin XM, Milliman CL, Korsmeyer SJ. Bcl-2 functions in an antioxidant pathway to prevent apoptosis. Cell. 1993;75(2):241–51. doi: 10.1016/0092-8674(93)80066-n. [DOI] [PubMed] [Google Scholar]
- 59.Zhu W, Cowie A, Wasfy GW, Penn LZ, Leber B, Andrews DW. Bcl-2 mutants with restricted subcellular location reveal spatially distinct pathways for apoptosis in different cell types. EMBO J. 1996;15(16):4130–4141. [PMC free article] [PubMed] [Google Scholar]
- 60.Wolvetang EJ, Johnson KL, Krauer K, Ralph SJ, Linnane AW. Mitochondrial respiratory chain inhibitors induce apoptosis. FEBS Lett. 1994;339(1–2):40–44. doi: 10.1016/0014-5793(94)80380-3. [DOI] [PubMed] [Google Scholar]
- 61.Robinson BH, Luo XP, Pitkanen S, Bratinova S, Bourgeois J, Lehotay DC, Raha S. Diagnosis of mitochondrial energy metabolism defects in tissue culture. Induction of MnSOD and bcl-2 in mitochondria from patients with complex I (NADH-CoQ reductase) deficiency. Biofactors. 1998;7(3):229–230. doi: 10.1002/biof.5520070314. [DOI] [PubMed] [Google Scholar]
- 62.Hunter T. Oncoprotein networks. Cell. 1997;88(3):333–346. doi: 10.1016/s0092-8674(00)81872-3. [DOI] [PubMed] [Google Scholar]
- 63.Allison SJ, Milner J. SIRT3 is pro-apoptotic and participates in distinct basal apoptotic pathways. Cell Cycle. 2007;6(21):2669–2677. doi: 10.4161/cc.6.21.4866. [DOI] [PubMed] [Google Scholar]
- 64.Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, van der Meer R, Nguyen P, Savage J, Owens KM, Vassilopoulos A, Ozden O, Park SH, Singh KK, Abdulkadir SA, Spitz DR, Deng CX, Gius D. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell. 2010;17(1):41–52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A, de Cabo R, Sauve AA, Sinclair DA. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130(6):1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu W, Akhand AA, Kato M, Yokoyama I, Miyata T, Kurokawa K, Uchida K, Nakashima I. 4-hydroxynonenal triggers an epidermal growth factor receptor-linked signal pathway for growth inhibition. J Cell Sci. 1999;112(Pt 14):2409–2417. doi: 10.1242/jcs.112.14.2409. [DOI] [PubMed] [Google Scholar]
- 67.Liu W, Kato M, Akhand AA, Hayakawa A, Suzuki H, Miyata T, Kurokawa K, Hotta Y, Ishikawa N, Nakashima I. 4-hydroxynonenal induces a cellular redox status-related activation of the caspase cascade for apoptotic cell death. J Cell Sci. 2000;113(Pt 4):635–641. doi: 10.1242/jcs.113.4.635. [DOI] [PubMed] [Google Scholar]
- 68.Uchida K, Shiraishi M, Naito Y, Torii Y, Nakamura Y, Osawa T. Activation of stress signaling pathways by the end product of lipid peroxidation. 4-hydroxy-2-nonenal is a potential inducer of intracellular peroxide production. J Biol Chem. 1999;274(4):2234–2242. doi: 10.1074/jbc.274.4.2234. [DOI] [PubMed] [Google Scholar]
- 69.Orrenius S, Gogvadze V, Zhivotovsky B. Mitochondrial oxidative stress: implications for cell death. Annu Rev Pharmacol Toxicol. 2007;47:143–183. doi: 10.1146/annurev.pharmtox.47.120505.105122. [DOI] [PubMed] [Google Scholar]
- 70.Wong CM, Cheema AK, Zhang L, Suzuki YJ. Protein carbonylation as a novel mechanism in redox signaling. Circ Res. 2008;102(3):310–318. doi: 10.1161/CIRCRESAHA.107.159814. [DOI] [PubMed] [Google Scholar]
- 71.Weinstein DM, Mihm MJ, Bauer JA. Cardiac peroxynitrite formation and left ventricular dysfunction following doxorubicin treatment in mice. J Pharmacol Exp Ther. 2000;294(1):396–401. [PubMed] [Google Scholar]
- 72.Aldieri E, Bergandi L, Riganti C, Costamagna C, Bosia A, Ghigo D. Doxorubicin induces an increase of nitric oxide synthesis in rat cardiac cells that is inhibited by iron supplementation. Toxicol Appl Pharmacol. 2002;185(2):85–90. doi: 10.1006/taap.2002.9527. [DOI] [PubMed] [Google Scholar]
- 73.Chaiswing L, Cole MP, St Clair DK, Ittarat W, Szweda LI, Oberley TD. Oxidative damage precedes nitrative damage in adriamycin-induced cardiac mitochondrial injury. Toxicol Pathol. 2004;32(5):536–547. doi: 10.1080/01926230490502601. [DOI] [PubMed] [Google Scholar]
- 74.Norberg E, Gogvadze V, Vakifahmetoglu H, Orrenius S, Zhivotovsky B. Oxidative modification sensitizes mitochondrial apoptosis-inducing factor to calpain-mediated processing. Free Radic Biol Med. 2010;48(6):791–797. doi: 10.1016/j.freeradbiomed.2009.12.020. [DOI] [PubMed] [Google Scholar]
- 75.Norberg E, Orrenius S, Zhivotovsky B. Mitochondrial regulation of cell death: processing of apoptosis-inducing factor (AIF) Biochem Biophys Res Commun. 2010;396(1):95–100. doi: 10.1016/j.bbrc.2010.02.163. [DOI] [PubMed] [Google Scholar]
- 76.Meng TC, Fukada T, Tonks NK. Reversible oxidation and inactivation of protein tyrosine phosphatases in vivo. Mol Cell. 2002;9(2):387–399. doi: 10.1016/s1097-2765(02)00445-8. [DOI] [PubMed] [Google Scholar]
- 77.Burgoyne JR, Madhani M, Cuello F, Charles RL, Brennan JP, Schroder E, Browning DD, Eaton P. Cysteine redox sensor in PKGIa enables oxidant-induced activation. Science. 2007;317(5843):1393–1397. doi: 10.1126/science.1144318. [DOI] [PubMed] [Google Scholar]
- 78.Oktyabrsky ON, Smirnova GV. Redox regulation of cellular functions. Biochemistry (Mosc) 2007;72(2):132–145. doi: 10.1134/s0006297907020022. [DOI] [PubMed] [Google Scholar]
- 79.Klatt P, Molina EP, De Lacoba MG, Padilla CA, Martinez-Galesteo E, Barcena JA, Lamas S. Redox regulation of c-Jun DNA binding by reversible S-glutathiolation. FASEB J. 1999;13(12):1481–1490. doi: 10.1096/fasebj.13.12.1481. [DOI] [PubMed] [Google Scholar]
- 80.Pineda-Molina E, Klatt P, Vazquez J, Marina A, Garcia de Lacoba M, Perez-Sala D, Lamas S. Glutathionylation of the p50 subunit of NF-kappaB: a mechanism for redox-induced inhibition of DNA binding. Biochemistry (Mosc) 2001;40(47):14134–14142. doi: 10.1021/bi011459o. [DOI] [PubMed] [Google Scholar]
- 81.Liu B, Chen Y, St Clair DK. ROS and p53: a versatile partnership. Free Radic Biol Med. 2008;44(8):1529–1535. doi: 10.1016/j.freeradbiomed.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Marshall HE, Merchant K, Stamler JS. Nitrosation and oxidation in the regulation of gene expression. FASEB J. 2000;14(13):1889–1900. doi: 10.1096/fj.00.011rev. [DOI] [PubMed] [Google Scholar]
- 83.Sun Y, Oberley LW. Redox regulation of transcriptional activators. Free Radic Biol Med. 1996;21(3):335–348. doi: 10.1016/0891-5849(96)00109-8. [DOI] [PubMed] [Google Scholar]
- 84.Rainwater R, Parks D, Anderson ME, Tegtmeyer P, Mann K. Role of cysteine residues in regulation of p53 function. Mol Cell Biol. 1995;15(7):3892–3903. doi: 10.1128/mcb.15.7.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ghezzi P. Oxidoreduction of protein thiols in redox regulation. Biochem Soc Trans. 2005;33(Pt 6):1378–1381. doi: 10.1042/BST0331378. [DOI] [PubMed] [Google Scholar]
- 86.Yeo WS, Lee SJ, Lee JR, Kim KP. Nitrosative protein tyrosine modifications: biochemistry and functional significance. BMB Rep. 2008;41(3):194–203. doi: 10.5483/bmbrep.2008.41.3.194. [DOI] [PubMed] [Google Scholar]
- 87.Monteiro HP, Arai RJ, Travassos LR. Protein tyrosine phosphorylation and protein tyrosine nitration in redox signaling. Antioxid Redox Signal. 2008;10(5):843–889. doi: 10.1089/ars.2007.1853. [DOI] [PubMed] [Google Scholar]
- 88.Paulsen CE, Carroll KS. Orchestrating redox signaling networks through regulatory cysteine switches. ACS Chem Biol. 2010;5(1):47–62. doi: 10.1021/cb900258z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chazotte-Aubert L, Hainaut P, Ohshima H. Nitric oxide nitrates tyrosine residues of tumor-suppressor p53 protein in MCF-7 cells. Biochem Biophys Res Commun. 2000;267(2):609–613. doi: 10.1006/bbrc.1999.2003. [DOI] [PubMed] [Google Scholar]
- 90.Cobbs CS, Samanta M, Harkins LE, Gillespie GY, Merrick BA, MacMillan-Crow LA. Evidence for peroxynitrite-mediated modifications to p53 in human gliomas: possible functional consequences. Arch Biochem Biophys. 2001;394(2):167–172. doi: 10.1006/abbi.2001.2540. [DOI] [PubMed] [Google Scholar]
- 91.Cobbs CS, Whisenhunt TR, Wesemann DR, Harkins LE, Van Meir EG, Samanta M. Inactivation of wild-type p53 protein function by reactive oxygen and nitrogen species in malignant glioma cells. Cancer Res. 2003;63(24):8670–8673. [PubMed] [Google Scholar]
- 92.Delphin C, Cahen P, Lawrence JJ, Baudier J. Characterization of baculovirus recombinant wild-type p53. Dimerization of p53 is required for high-affinity DNA binding and cysteine oxidation inhibits p53 DNA binding. Eur J Biochem. 1994;223(2):683–692. doi: 10.1111/j.1432-1033.1994.tb19041.x. [DOI] [PubMed] [Google Scholar]
- 93.Hainaut P, Milner J. Redox modulation of p53 conformation and sequence-specific DNA binding in vitro. Cancer Res. 1993;53(19):4469–4473. [PubMed] [Google Scholar]
- 94.Fojta M, Kubicarova T, Vojtesek B, Palecek E. Effect of p53 protein redox states on binding to supercoiled and linear DNA. J Biol Chem. 1999;274(36):25749–25755. doi: 10.1074/jbc.274.36.25749. [DOI] [PubMed] [Google Scholar]
- 95.Fojta M, Pivonkova H, Brazdova M, Nemcova K, Palecek J, Vojtesek B. Investigations of the supercoil-selective DNA binding of wild type p53 suggest a novel mechanism for controlling p53 function. Eur J Biochem. 2004;271(19):3865–3876. doi: 10.1111/j.1432-1033.2004.04323.x. [DOI] [PubMed] [Google Scholar]
- 96.Sun XZ, Vinci C, Makmura L, Han S, Tran D, Nguyen J, Hamann M, Grazziani S, Sheppard S, Gutova M, Zhou F, Thomas J, Momand J. Formation of disulfide bond in p53 correlates with inhibition of DNA binding and tetramerization. Antioxid Redox Signal. 2003;5(5):655–665. doi: 10.1089/152308603770310338. [DOI] [PubMed] [Google Scholar]
- 97.Chene P. Mutations at position 277 modify the DNA-binding specificity of human p53 in vitro. Biochem Biophys Res Commun. 1999;263(1):1–5. doi: 10.1006/bbrc.1999.1294. [DOI] [PubMed] [Google Scholar]
- 98.Buzek J, Latonen L, Kurki S, Peltonen K, Laiho M. Redox state of tumor suppressor p53 regulates its sequence-specific DNA binding in DNA-damaged cells by cysteine 277. Nucleic Acids Res. 2002;30(11):2340–2348. doi: 10.1093/nar/30.11.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Seo YR, Kelley MR, Smith ML. Selenomethionine regulation of p53 by a ref1-dependent redox mechanism. Proc Natl Acad Sci U S A. 2002;99(22):14548–14553. doi: 10.1073/pnas.212319799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Smith ML, Lancia JK, Mercer TI, Ip C. Selenium compounds regulate p53 by common and distinctive mechanisms. Anticancer Res. 2004;24(3a):1401–1408. [PubMed] [Google Scholar]
- 101.Wu HH, Momand J. Pyrrolidine dithiocarbamate prevents p53 activation and promotes p53 cysteine residue oxidation. J Biol Chem. 1998;273(30):18898–18905. doi: 10.1074/jbc.273.30.18898. [DOI] [PubMed] [Google Scholar]
- 102.Petitjean A, Achatz MI, Borresen-Dale AL, Hainaut P, Olivier M. TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene. 2007;26(15):2157–2165. doi: 10.1038/sj.onc.1210302. [DOI] [PubMed] [Google Scholar]
- 103.Bolshakov S, Walker CM, Strom SS, Selvan MS, Clayman GL, El-Naggar A, Lippman SM, Kripke ML, Ananthaswamy HN. p53 mutations in human aggressive and nonaggressive basal and squamous cell carcinomas. Clin Cancer Res. 2003;9(1):228–234. [PubMed] [Google Scholar]
- 104.Murphy ME. Polymorphic variants in the p53 pathway. Cell Death Differ. 2006;13(6):916–920. doi: 10.1038/sj.cdd.4401907. [DOI] [PubMed] [Google Scholar]
- 105.Pietsch EC, Humbey O, Murphy ME. Polymorphisms in the p53 pathway. Oncogene. 2006;25(11):1602–1611. doi: 10.1038/sj.onc.1209367. [DOI] [PubMed] [Google Scholar]
- 106.Dumont P, Leu JI, Della Pietra AC, 3rd, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet. 2003;33(3):357–365. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- 107.Karawajew L, Rhein P, Czerwony G, Ludwig WD. Stress-induced activation of the p53 tumor suppressor in leukemia cells and normal lymphocytes requires mitochondrial activity and reactive oxygen species. Blood. 2005;105(12):4767–4775. doi: 10.1182/blood-2004-09-3428. [DOI] [PubMed] [Google Scholar]
- 108.Liu J, St Clair DK, Gu X, Zhao Y. Blocking mitochondrial permeability transition prevents p53 mitochondrial translocation during skin tumor promotion. FEBS Lett. 2008;582(9):1319–1324. doi: 10.1016/j.febslet.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhao Y, Chaiswing L, Velez JM, Batinic-Haberle I, Colburn NH, Oberley TD, St Clair DK. p53 translocation to mitochondria precedes its nuclear translocation and targets mitochondrial oxidative defense protein-manganese superoxide dismutase. Cancer Res. 2005;65(9):3745–3750. doi: 10.1158/0008-5472.CAN-04-3835. [DOI] [PubMed] [Google Scholar]
- 110.Delsite RL, Rasmussen LJ, Rasmussen AK, Kalen A, Goswami PC, Singh KK. Mitochondrial impairment is accompanied by impaired oxidative DNA repair in the nucleus. Mutagenesis. 2003;18(6):497–503. doi: 10.1093/mutage/geg027. [DOI] [PubMed] [Google Scholar]
- 111.Park SY, Chang I, Kim JY, Kang SW, Park SH, Singh K, Lee MS. Resistance of mitochondrial DNA-depleted cells against cell death: role of mitochondrial superoxide dismutase. J Biol Chem. 2004;279(9):7512–7520. doi: 10.1074/jbc.M307677200. [DOI] [PubMed] [Google Scholar]
- 112.Chandel NS, Vander Heiden MG, Thompson CB, Schumacker PT. Redox regulation of p53 during hypoxia. Oncogene. 2000;19(34):3840–3848. doi: 10.1038/sj.onc.1203727. [DOI] [PubMed] [Google Scholar]
- 113.Duranteau J, Chandel NS, Kulisz A, Shao Z, Schumacker PT. Intracellular signaling by reactive oxygen species during hypoxia in cardiomyocytes. J Biol Chem. 1998;273(19):11619–11624. doi: 10.1074/jbc.273.19.11619. [DOI] [PubMed] [Google Scholar]
- 114.Sansome C, Zaika A, Marchenko ND, Moll UM. Hypoxia death stimulus induces translocation of p53 protein to mitochondria. Detection by immunofluorescence on whole cells. FEBS Lett. 2001;488(3):110–115. doi: 10.1016/s0014-5793(00)02368-1. [DOI] [PubMed] [Google Scholar]
- 115.Aleshin A, Finn RS. SRC: a century of science brought to the clinic. Neoplasia. 2010;12(8):599–607. doi: 10.1593/neo.10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Johnson FM, Gallick GE. SRC family nonreceptor tyrosine kinases as molecular targets for cancer therapy. Anticancer Agents Med Chem. 2007;7(6):651–659. doi: 10.2174/187152007784111278. [DOI] [PubMed] [Google Scholar]
- 117.Giannoni E, Taddei ML, Chiarugi P. Src redox regulation: again in the front line. Free Radic Biol Med. 2010;49(4):516–527. doi: 10.1016/j.freeradbiomed.2010.04.025. [DOI] [PubMed] [Google Scholar]
- 118.Kemble DJ, Sun G. Direct and specific inactivation of protein tyrosine kinases in the Src and FGFR families by reversible cysteine oxidation. Proc Natl Acad Sci U S A. 2009;106(13):5070–5075. doi: 10.1073/pnas.0806117106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Akhand AA, Pu M, Senga T, Kato M, Suzuki H, Miyata T, Hamaguchi M, Nakashima I. Nitric oxide controls src kinase activity through a sulfhydryl group modification-mediated Tyr-527-independent and Tyr-416-linked mechanism. J Biol Chem. 1999;274(36):25821–2586. doi: 10.1074/jbc.274.36.25821. [DOI] [PubMed] [Google Scholar]
- 120.Krasnowska EK, Pittaluga E, Brunati AM, Brunelli R, Costa G, De Spirito M, Serafino A, Ursini F, Parasassi T. N-acetyl-l-cysteine fosters inactivation and transfer to endolysosomes of c-Src. Free Radic Biol Med. 2008;45(11):1566–1572. doi: 10.1016/j.freeradbiomed.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 121.Giannoni E, Buricchi F, Raugei G, Ramponi G, Chiarugi P. Intracellular reactive oxygen species activate Src tyrosine kinase during cell adhesion and anchorage-dependent cell growth. Mol Cell Biol. 2005;25(15):6391–6403. doi: 10.1128/MCB.25.15.6391-6403.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cardone L, Carlucci A, Affaitati A, Livigni A, DeCristofaro T, Garbi C, Varrone S, Ullrich A, Gottesman ME, Avvedimento EV, Feliciello A. Mitochondrial AKAP121 binds and targets protein tyrosine phosphatase D1, a novel positive regulator of src signaling. Mol Cell Biol. 2004;24(11):4613–4626. doi: 10.1128/MCB.24.11.4613-4626.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Itoh S, Lemay S, Osawa M, Che W, Duan Y, Tompkins A, Brookes PS, Sheu SS, Abe J. Mitochondrial Dok-4 recruits Src kinase and regulates NF-kappaB activation in endothelial cells. J Biol Chem. 2005;280(28):26383–26396. doi: 10.1074/jbc.M410262200. [DOI] [PubMed] [Google Scholar]
- 124.Lluis JM, Buricchi F, Chiarugi P, Morales A, Fernandez-Checa JC. Dual role of mitochondrial reactive oxygen species in hypoxia signaling: activation of nuclear factor-{kappa}B via c-SRC and oxidant-dependent cell death. Cancer Res. 2007;67(15):7368–7377. doi: 10.1158/0008-5472.CAN-07-0515. [DOI] [PubMed] [Google Scholar]
- 125.Graham KA, Kulawiec M, Owens KM, Li X, Desouki MM, Chandra D, Singh KK. NADPH oxidase 4 is an oncoprotein localized to mitochondria. Cancer Biol Ther. 2010;10(3):223–231. doi: 10.4161/cbt.10.3.12207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Block K, Gorin Y, Abboud HE. Subcellular localization of Nox4 and regulation in diabetes. Proc Natl Acad Sci U S A. 2009;106(34):14385–14390. doi: 10.1073/pnas.0906805106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Block K, Eid A, Griendling KK, Lee DY, Wittrant Y, Gorin Y. Nox4 NAD(P)H oxidase mediates Src-dependent tyrosine phosphorylation of PDK-1 in response to angiotensin II: role in mesangial cell hypertrophy and fibronectin expression. J Biol Chem. 2008;283(35):24061–24076. doi: 10.1074/jbc.M803964200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hayashi T, Ueno Y, Okamoto T. Oxidoreductive regulation of nuclear factor kappa B. Involvement of a cellular reducing catalyst thioredoxin. J Biol Chem. 1993;268(15):11380–11388. [PubMed] [Google Scholar]
- 129.Valle A, Kinet JP. N-acetyl-L-cysteine inhibits antigen-mediated Syk, but not Lyn tyrosine kinase activation in mast cells. FEBS Lett. 1995;357(1):41–44. doi: 10.1016/0014-5793(94)01329-y. [DOI] [PubMed] [Google Scholar]
- 130.Li S, Yan T, Yang JQ, Oberley TD, Oberley LW. The role of cellular glutathione peroxidase redox regulation in the suppression of tumor cell growth by manganese superoxide dismutase. Cancer Res. 2000;60(14):3927–3939. [PubMed] [Google Scholar]
- 131.Zhang HJ, Zhao W, Venkataraman S, Robbins ME, Buettner GR, Kregel KC, Oberley LW. Activation of matrix metalloproteinase-2 by overexpression of manganese superoxide dismutase in human breast cancer MCF-7 cells involves reactive oxygen species. J Biol Chem. 2002;277(23):20919–20926. doi: 10.1074/jbc.M109801200. [DOI] [PubMed] [Google Scholar]
- 132.Kim KH, Rodriguez AM, Carrico PM, Melendez JA. Potential mechanisms for the inhibition of tumor cell growth by manganese superoxide dismutase. Antioxid Redox Signal. 2001;3(3):361–373. doi: 10.1089/15230860152409013. [DOI] [PubMed] [Google Scholar]
- 133.Rodriguez AM, Carrico PM, Mazurkiewicz JE, Melendez JA. Mitochondrial or cytosolic catalase reverses the MnSOD-dependent inhibition of proliferation by enhancing respiratory chain activity, net ATP production, and decreasing the steady state levels of H(2)O(2) Free Radic Biol Med. 2000;29(9):801–813. doi: 10.1016/s0891-5849(00)00362-2. [DOI] [PubMed] [Google Scholar]
- 134.Buettner GR, Ng CF, Wang M, Rodgers VG, Schafer FQ. A new paradigm: manganese superoxide dismutase influences the production of H2O2 in cells and thereby their biological state. Free Radic Biol Med. 2006;41(8):1338–1350. doi: 10.1016/j.freeradbiomed.2006.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]