Abstract
Melanin-concentrating hormone (MCH) regulates feeding and complex behaviors in mammals and pigmentation in fish. The relationship between fish and mammalian MCH systems is not well understood. Here, we identify and characterize two MCH genes in zebrafish, Pmch1 and Pmch2. Whereas Pmch1 and its corresponding MCH1 peptide resemble MCH found in other fish, the zebrafish Pmch2 gene and MCH2 peptide share genomic structure, synteny, and high peptide sequence homology with mammalian MCH. Zebrafish Pmch genes are expressed in closely associated but non-overlapping neurons within the hypothalamus, and MCH2 neurons send numerous projections to multiple MCH receptor-rich targets with presumed roles in sensory perception, learning and memory, arousal, and homeostatic regulation. Preliminary functional analysis showed that whereas changes in zebrafish Pmch1 expression correlate with pigmentation changes, the number of MCH2-expressing neurons increases in response to chronic food deprivation. These findings demonstrate that zebrafish MCH2 is the putative structural and functional ortholog of mammalian MCH and help elucidate the nature of MCH evolution among vertebrates.
Indexing terms: melanin-concentrating hormone, MCH1, MCH2, pigmentation, feeding, zebrafish
In mammals, melanin-concentrating hormone (MCH) is a key regulator of food intake and energy homeostasis (reviewed in Griffond and Baker, 2002; Pissios et al., 2006). Injection of the 19-amino acid (AA) mammalian MCH, which is normally expressed in the lateral hypothalamus (LH) and zona incerta (ZI) (Pissios et al., 2006), into the brain of rodents prompts excessive feeding (Qu et al., 1996), and overexpression of the MCH gene in the mouse increases susceptibility to obesity (Ludwig et al., 2001). Elimination of MCH in the mouse causes significant weight loss and increases energy expenditure (Kokkotou et al., 2005), and knocking out the rodent MCH receptor 1 gene, MCHR1, promotes leanness and decreases fat mass, although, curiously, these animals are hyperphagic (Chen et al., 2002; Marsh et al., 2002). MCH itself is responsive to leptin and nutritional cues: obese leptin-deficient ob/ob mice as well as fasted wild-type mice show an increase in expression of MCH mRNA (Qu et al., 1996). MCH neurons also regulate other, more complex behaviors. For example, MCHR1 knockout mice display reduced anxiety (Roy et al., 2006) and cognitive impairment (Adamantidis et al., 2005). Further, MCH neuronal activity has been correlated with REM sleep (Verret et al., 2003; Hassani et al., 2009) and sleep recovery following sleep deprivation (Verret et al., 2003; Modirrousta et al., 2005).
Interestingly, MCH was first identified in the chum salmon Oncorhynchus keta as a 17-amino acid peptide that induces skin lightening (Kawauchi et al., 1983). In various teleosts (rainbow trout, tilapia, flounder, eel, and goldfish), MCH aggregates pigment-containing melanosomes around the nucleus, thereby changing the refractive index of the fish scales and lightening skin color (Kawauchi, 2006). MCH isolated from teleosts is encoded by a one-exon gene and is expressed primarily in the lateral tuberal nucleus (NLT) in the hypothalamus, a homologous structure to the mammalian arcuate nucleus (Holmqvist and Ekström, 1991; Forlano and Cone, 2007). Additionally, in the trout (Baker et al., 1995) and goldfish (Matsuda et al., 2007), MCH expression is detected just dorsal to the lateral recesses of the third ventricle.
MCH exerts its effects on skin pigmentation, behavior, and energy homeostasis through one or multiple MCH receptor (MCHR) proteins, which are rhodopsin-like G-protein-coupled receptors (GPCR; reviewed in Griffond and Baker, 2002). Humans have two distantly related receptors for MCH, named MCHR1 (Kolakowski et al., 1996) and MCHR2 (Hill et al., 2001; Mori et al., 2001; Rodriguez et al., 2001), which are 38% identical, whereas rodents have only one receptor, MCHR1, which shares >95% sequence identity with human MCHR1 (Lakaye et al., 1998; Kokkotou et al., 2001). Studies in the rat showed that among other regions, MCHR1 is expressed in the hypothalamus, as well as in the hippocampus and the amygdala, cortical regions important for learning and memory (Saito et al., 2001). MCHR1 is also expressed in the thalamus and locus coeruleus (LC), areas important for arousal, and in areas involved in olfactory processing, such as the piriform cortex. The nucleus accumbens, known as a reward center, is yet another region with MCHR1 expression in rodents. Human MCHR2 is found in many similar brain regions, but also displays abundant expression in several peripheral sites, including the intestine (Hill et al., 2001; Rodriguez et al., 2001) and adipose tissue (Hill et al., 2001). In zebrafish Danio rerio, three MCH receptors (MCHR1A, MCHR1B, and MCHR2) have been identified (Logan et al., 2003a). Zebrafish mchr1a and mchr1b genes are both orthologous to human and mouse MCHR1 and are thought to be paralogous genes arising from a late duplication event.
How the fish MCH system relates molecularly and functionally to its mammalian counterpart is unclear. The mammalian MCH peptide is 68.4% identical to fish MCH, but is encoded by a three-exon gene. Further, it has no demonstrable role in mammalian skin pigmentation (Pissios et al., 2006). There is some evidence, however, that neuropeptide circuits similar to those of mammals may regulate food intake in teleosts (Kawauchi, 2006). Neuropeptide Y (NPY), hypocretin (HCRT), and agouti-related protein (AgRP) are orexigenic in goldfish, whereas cocaine- and amphetamine-regulated transcript (CART) inhibits feeding. The melanocortin system, including α-melanocyte-stimulating hormone (α-MSH) and AgRP-expressing neurons, has recently been characterized neuro-anatomically in the zebrafish (Forlano and Cone, 2007). For MCH in particular, it has been difficult to clearly establish a role in feeding behavior and energy regulation in fish. An increase in MCH expression is seen in brains of starved flounder, and trout, tilapia, and flounder reared on a white background have longer and heavier bodies and an increase in MCH expression compared with fish raised on a black background (Kawauchi, 2006). However, overexpression of the fish MCH in transgenic medaka lightened their color but no body weight change was observed (Kinoshita et al., 2001). In addition, unlike in mammals, direct injection of synthetic human or flounder MCH into goldfish brains caused a reduction in food intake (Matsuda et al., 2006), and injection of an antibody raised against salmon MCH increased food intake in goldfish (Matsuda et al., 2007). Thus the specific function of the teleost MCH neuropeptide system in the regulation of food intake remains unclear.
To better understand the functional and evolutionary relationship of the fish MCH neuropeptide system both among teleosts and in comparison with mammalian MCH, we investigated the MCH system in zebrafish. We found that the zebrafish and other teleost genomes encode two distinct MCH peptides, one (MCH1) similar or identical to salmonid MCH and one (MCH2) that bears a striking resemblance to mammalian MCH. The two zebrafish Pmch genes are expressed in close but distinct hypothalamic neuron populations, and MCH2 neurons send projections to widespread MCH receptor-rich CNS targets implicated in satiety, olfaction, arousal, and learning and memory. Whereas zebrafish Pmch1 expression varies primarily with changes in skin pigmentation, MCH2 peptide levels dramatically increase in fasted animals. The existence of two MCH neuropeptide systems in zebrafish and other teleosts sheds light on the evolution of the MCH system across vertebrates and identifies previously unrecognized functional links between the fish and mammalian MCH systems.
MATERIALS AND METHODS
Animals
Our research using laboratory animals is overseen and evaluated by an Institutional Animal Care and Use Committee (IACUC). At Stanford, the IACUC is known as the Administrative Panel on Laboratory Animal Care (APLAC). Stanford’s APLAC has received AAALAC accreditation (Association for Assessment and Accreditation of Laboratory Animal Care).
Larvae and young adult (6 months) wild-type zebrafish, Danio rerio (Scientific Hatcheries, Huntington Beach, CA) were used for the MCH system characterization. Zebrafish were raised and maintained in Marine Biotech (Apopka, FL) Zmod systems (28.5°C, pH 7.0, conductivity = 500 μS) in a 14-hour/10-hour light/dark cycle.
Identification and cloning of teleost MCH and MCH receptor genes
To identify putative MCH orthologs in zebrafish, we queried (tblastn) the Ensembl zebrafish whole genome assembly version 7 (Zv7) by using the mammalian MCH peptide sequence (DFDMLRCMLGRVYRPCWQV). Two strongly significant hits were identified (E-val 10−6), which were not annotated but were later confirmed by using standard molecular techniques (see below) to encode Pmch1 and Pmch2. The same strategy was used to identify MCH orthologs in fugu (Takifugu rubripes, Ensembl gene numbers ENSTRUG00000011250 and ENSTRUG00000014356 for Pmch1 and Pmch2, respectively), stickleback (Gasterosteus aculeatus, ENSGACG00000018017 and ENSGACG00000020044), and medaka (Oryzias latipes, ENSORLG00000013661 and ENSORLG00000016450). To identify MCH receptor homologs in zebrafish, we queried (tblastn) the zebrafish genome by using the human MCHR1 protein sequence (accession number NM_005297) and the mouse MCHR1 sequence (AY049011). Annotated partial gene sequences for mchr1a (ENSDARG00000037048) and mchr1b (ENSDARG00000022525) were identified. The zebrafish mchr2 sequence was as described (Logan et al., 2003b, accession number AY161859), corresponding to ENSDARG00000036117.
To clone the zebrafish Pmch1 and Pmch2 genes and to determine their genomic structure, total RNA was isolated from 3dpf larvae by using the RNeasy Mini Kit (Qiagen, Valencia, CA), and cDNA was generated by using SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA) and amplified by using polymerase chain reaction (PCR). Full transcript coverage was achieved by using 5′ and 3′ rapid amplification of cDNA ends (5′ and 3′ RACE Systems, Invitrogen), and full transcripts including partial 5′ and 3′ UTR sequences were cloned into pCRII Topo TA vector (Invitrogen) and sequenced (Pmch1 GenBank accession number 1131850 and Pmch2 accession number 1131867). These constructs were used to synthesize antisense probes for ISH. Partial cDNAs of mchr1a, mchr1b, and mchr2 sequences were similarly isolated based on available sequences, sequenced, and used to synthesize antisense probes for ISH.
In situ hybridization
For only Pmch1, Pmch2, and mchr2 in situ hybridization (ISH) in 2dpf animals, larvae were raised in water containing 0.2 mM phenylthiocarbamide (PTU) to prevent pigmentation. All other experiments were conducted on animals raised in the absence of PTU. Larvae (2dpf) and brains dissected from 6-month-old adult fish were fixed in 4% paraformaldehyde (PFA) over 48 hours at 4°C. All samples were first dehydrated in 100% methanol and stored at −20°C for at least half an hour. After rehydration in phosphate-buffered saline (PBS; pH 7.4), adult brains were embedded in 2.5% agarose and cut with a Vibratome (series 1000, Sectioning System, East Granby, CT). Transverse sections were then processed and stained as free-floating slices. ISH was performed following standard protocols (Hauptmann and Gerster, 1994). Hybridization with Pmch1, Pmch2, mchr1a, mchr1b, and mchr2 digoxigenin or fluorescein-labeled antisense riboprobes were revealed with either colorimetric (BM purple) or fluorescent staining (TSA green, Fast red). Fluorescence images were obtained by using a Zeiss (Thornwood, NY) LSM 510 META laser scanning confocal microscope. The brightness and contrast of the final images were adjusted by using Adobe Photoshop CS4 (San Jose, CA).
Immunohistochemistry
Adult brains were fixed in 4% PFA overnight at 4°C and then cut as described above. Slices were blocked with 0.4% Triton X-100 and 20% normal goat serum in PBS for 1 hour at room temperature. After blocking, adult brain slices were incubated in primary antibody rabbit anti-MCH (Human, Mouse, Rat; Phoenix Pharmaceuticals, Belmont, CA, H-070-47, generated in rabbits by using MCH coupled to keyhole limpet hemocyanin), 1:400 dilution in block buffer overnight at 4°C. Slices were again washed in PBS + 0.4% Triton X-100 and blocked for an hour. Anti-MCH antibodies were then detected by using a secondary goat-anti-rabbit Alexa Fluor 488 IgG (H+L) antibody, highly cross-adsorbed (2 mg/mL, Invitrogen, Molecular Probes, A-11034). The incubation (dilution 1:400) was done for 2 hours and followed by washes in PBS. A preadsorption control using either salmon MCH (which has a sequence identical to zebrafish MCH1 peptide) or mammalian MCH peptide confirmed the specificity of this antibody to zebrafish MCH2 (see Suppl. Fig. S2). Confocal imaging was performed by using a Zeiss LSM 510 META laser scanning confocal microscope. The brightness and contrast of the final images were adjusted by using Adobe Photoshop CS4.
Neuroanatomical designations
The majority of neuroanatomical identifications in adult fish were made by using the zebrafish atlas (Wullimann et al., 1996). However, to describe the expression pattern of Pmch1-and Pmch2-expressing cells in the hypothalamus, designations from trout and goldfish (for example: Baker et al., 1995; Huesa et al., 2005; Matsuda et al., 2007) were used. These designations offered better resolution for the purposes of describing the expression pattern, and they facilitated comparison between our findings in zebrafish with other teleosts. To enable direct comparison of our study with labels found in the zebrafish atlas, however, we note that in the zebrafish atlas, what we designate in this study as NLTl is found in the zebrafish atlas as the ventral part of LH, NLTp is part of LH and Hv in the atlas, and NRLd is found in the atlas as Hd.
Background adaptation
Twenty-four wild-type adult sibling zebrafish were first raised and maintained in clear plastic tanks under a 14 hours on/10 hours off light cycle. They were then split into two groups and transferred for 24 hours to either a black or a white background tank with the same original temperature and light conditions. All the fish were simultaneously sacrificed at 12 pm; their brains were dissected and fixed immediately. Each group was subdivided into two batches of six brains processed for sectioning and Pmch1 or Pmch2 expression analysis (ISH) as described above. All the slices were processed in parallel, in the same ISH protocol conditions, and staining was stopped at the same time for comparison.
Feeding/fasting conditions and MCH2 cell count
This assay is derived from the protocol developed by Takahashi and colleagues (2004). Adult wild-type zebrafish siblings were raised and maintained together in a large tank in our facility under normal conditions on a 100% Artemia nauplia (brine shrimp) diet. For 6 months, fish were fed twice a day, every day, with an excess of brine shrimp. Then 18 fish of similar size and weight (3.8 cm and 0.6 g on average) were transferred into individual tanks on a black background. For 2 weeks, nine of them were fed twice a day with satiating amounts of brine shrimp, whereas the other nine were left completely unfed. After this period, all the fish were sacrificed at the same time, 2 pm, and their brains were dissected and fixed immediately. The two batches of nine brains were cut and processed in parallel for anti-mammalian MCH immunohistochemistry (see above). For each brain, all the sections harboring fluorescent staining were imaged with confocal microscopy, and MCH2 cell body count was performed on high-magnification pictures of each slice. One brain from the fed cohort was damaged during processing and could not be analyzed. Cell counts were repeated in a double-blind fashion by a second independent researcher.
RESULTS
Molecular characterization of Pmch1 and Pmch2 in zebrafish
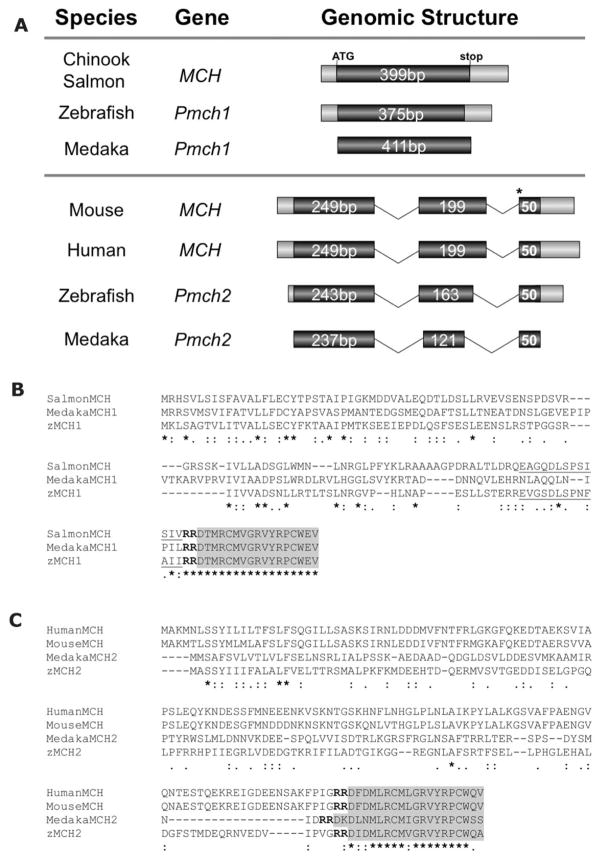
By using the known mammalian MCH peptide sequence (DFDMLRCMLGRVYRPCWQV) we queried (tblastn) the zebrafish whole genome assembly (Ensembl, Sanger Institute, Cambridge UK) for putative MCH orthologs. Two previously uncharacterized sequences were found to have high homology with mammalian MCH. We named these genes Pmch1 and Pmch2. Cloning and sequencing of the corresponding full-length Pmch1 and Pmch2 cDNAs (see Materials and Methods for details) revealed that Pmch1 and Pmch2 have distinct genomic structures (Fig. 1A). The Pmch1 cDNA (Gen-Bank accession number 1131850) contains a single exon, similar in structure to MCH found originally in salmon, whereas the Pmch2 cDNA (accession number 1131867) consists of three exons, similar to the MCH structure found in mammals (Fig. 1A). By using similar bioinformatics searches, we found that, like zebrafish, two distinct Pmch genes are predicted to exist in fugu (Takifugu rubripes, Ensembl gene numbers ENSTRUG00000011250 and ENSTRUG00000014356 for Pmch1 and Pmch2, respectively), the three-spined stickleback (Gasterosteus aculeatus, ENSGACG00000018017 and ENSGACG00000020044), and medaka (Oryzias latipes, EN-SORLG00000013661 and ENSORLG00000016450) (Fig. 1).
Figure 1.
Genomic structure of zebrafish Pmch1 and Pmch2 genes and alignment with other vertebrate MCH preproproteins. A: Schematic exon-intron organization of teleost and mammalian Pmch/MCH genes. Coding sequences are represented by dark gray boxes, untranslated regions (UTRs) are in light gray, and introns are depicted by lines. The zebrafish Pmch1 cDNA is 593 bp long and contains a single exon 468 bp in length. Zebrafish Pmch2 cDNA is 593 bp long and consists of three exons (456 bp total) that span 2.2 kb in the genome. Length of sequences contributing to the open reading frame are as indicated, although exon box and intron line sizes are approximations and not drawn to scale. As seen in salmon MCH, zebrafish and medaka Pmch1 genes are composed of a single exon (top panel). Zebrafish and medaka Pmch2 genes, however, have a three-exon gene structure similar to that seen in mouse and human MCH genes (bottom panel). The position of the 3′ splice site of intron 2 (marked by an asterisk) with respect to the MCH peptide sequence is conserved between zebrafish and medaka Pmch2 and mouse and human MCH. Sequences represented here correspond to zebrafish Pmch1 (GenBank Accession number 1131850), zebrafish Pmch2 (1131867), medaka Pmch1 (Ensembl gene sequence ENSORLG00000013661), medaka Pmch2 (ENSORLG00000016450), Chinook salmon MCH (M25754), mouse MCH (NM_029971), and human MCH (NM_002674). B: Alignment of zebrafish proMCH1 (zMCH1) and medaka proMCH1 (MedakaMCH1) with salmon proMCH (SalmonMCH). C: Alignment of zebrafish proMCH2 (zMCH2) and medaka proMCH2 (MedakaMCH2) with mouse and human proMCH. Multiple sequence alignments were performed by using ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html). The shaded box encompasses predicted mature MCH peptide sequences. Zebrafish and medaka MCH1 peptides are 100% identical to the salmon MCH peptide, whereas zebrafish MCH2 is 84% identical to the mammalian MCH. Underlined text in salmon and zMCH1 indicates a putative neuropeptide E-V, for which no function is known. This putative NEV sequence is 53.8% identical and 76.9% similar to Chinook salmon NEV and shares less significant homology (35.7% identity, 76.9% similarity) with the human NEI peptide. Stars beneath sequence indicate sequence identity across the alignment, double dots indicate conserved substitutions, and single dots semiconserved substitutions. Bold “RR” is the predicted dibasic peptide cleavage site.
In each case, we identified a Pmch gene (designated Pmch1) with a single exon similar to salmon MCH as well as a second Pmch gene (designated Pmch2) with three or more predicted exons (stickleback Pmch2 has four exons), similar to mammalian MCH. In these teleosts’ Pmch2 genes, the position of the 3′ splice site of intron 2 with respect to the MCH peptide sequence is identical to that seen in mammals (designated by an asterisk, Fig. 1A). We next looked for evidence of synteny between the teleost and mammalian MCH genes (Ensembl genome browser, www.ensembl.org). We found that the IGF1 gene lies immediately upstream of zebrafish Pmch2 on chromosome 4, as well as upstream of human and mouse MCH (data not shown). IGF1 is also immediately upstream of the predicted fugu, stickleback, and medaka Pmch2 genes. As is also seen in human and in mouse, the CCDC53, SYCP3, and MYBPC1 genes are positioned closely downstream of zebrafish Pmch2. On chromosome 2, the gene neighbors of zebrafish Pmch1 were not found within the 1-Mb flanks of the mammalian MCH genes. Thus, not only is possession of two Pmch genes with distinct genomic structures a feature found in multiple teleosts, but similarities in genomic structure and synteny suggest a close evolutionary relationship between teleost Pmch2 genes and mammalian MCH.
The predicted zebrafish MCH1 and MCH2 preproproteins contain a dibasic (RR) cleavage site near the C terminus that likely is used to generate the proMCH peptides (Fig. 1B,C). The predicted MCH1 prepropeptide is 124 amino acids long and gives rise to a 17-amino acid peptide (DTMRCMVGRVYR-PCWEV), which is identical in length to salmonid MCH. The MCH2 prepropeptide is 151 amino acids long and gives rise to a 19-amino acid peptide (DIDMLRCMVGRVYRPCWQA), which is the same length as mammalian MCH. Each peptide contains two conserved cysteines required in other species to generate the cyclic mature MCH peptide. Protein sequence alignment (Emboss Align, http://www.ebi.ac.uk/emboss/align/) revealed that the predicted zebrafish mature peptides are 68.4% identical and 78.9% similar to each other, with limited identity across the full-length preproprotein.
Multiple pairwise sequence alignment of the full-length zebrafish MCH1 and MCH2 preprohormones with full-length preprohormone sequences from human, mouse, and Chinook salmon and with predicted medaka sequences revealed low sequence identity (data not shown). However, alignment of the predicted mature zebrafish MCH peptides with these species’ MCH peptides showed high sequence conservation (Fig. 1B,C). Strikingly, the zebrafish MCH1 peptide is 100% identical to salmon MCH and to medaka MCH1 (Fig. 1B), as well as to several other fish species (tilapia, trout, and flounder). By contrast, zebrafish MCH2 peptide more closely resembles mammalian MCH (84.2% identical and 89.5% similar) than it does salmon MCH or zebrafish MCH1. Other peptides of unclear function are predicted to be generated from the MCH preprohormone in fish (neuropeptide E-V, NEV) and in mammals (neuropeptide E-I, NEI, and neuropeptide G-E, NGE) (Nahon et al., 1989; Allaeys et al., 2004; Pissios et al., 2006). None of these three sequences were readily identifiable in the zebrafish proMCH2 sequence. However, we identified a sequence in zebrafish proMCH1 (EVGSDLSPNFAII) with significant homology to Chinook salmon NEV (53.8% identical, 76.9% similar; Fig. 1B). The putative presence of the fish-specific NEV peptide in zebrafish proMCH1, the one-exon genomic structure of Pmch1, and the 100% sequence identity of zebrafish MCH1 peptide with salmon MCH tightly aligns zebrafish Pmch1 with other fish-specific MCH peptides. By contrast, although reciprocal blast searches indicate that both zebrafish MCH1 and MCH2 are orthologous to mammalian MCH, zebrafish MCH2 more closely resembles mammalian MCH in terms of genomic structure (Fig. 1A), synteny, and mature peptide sequence (Fig. 1C).
Zebrafish Pmch1 and Pmch2 are expressed in the hypothalamus
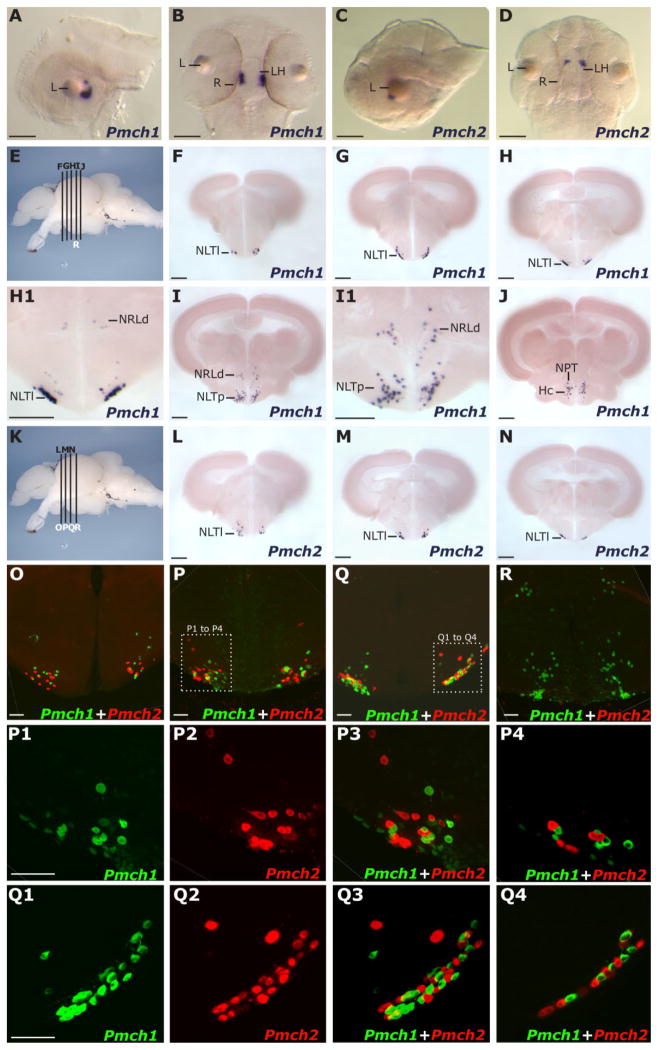
We examined the expression pattern of Pmch1 and Pmch2 during development by using whole-mount ISH (see Materials and Methods). Pmch1 and Pmch2 transcripts were first detected in 2-day post fertilization (dpf) embryos exclusively in bilateral clusters corresponding to the LH (Fig. 2B,D), as evidenced by comparison with other LH-expressed genes like HCRT (Faraco et al., 2006). A lateral view of Pmch1 expression (Fig. 2A) revealed that the Pmch1 expression domain assumes a “C” shape just caudal to the lens axis, and ventral (Fig. 2B,D) and lateral (Fig. 2A,C) views indicate that Pmch2 expression lies rostral to the Pmch1 expression domain. We next performed ISH of Pmch1 and Pmch2 in serial sections from adult zebrafish brains. Similar to what is seen in other species, the zebrafish Pmch genes displayed a highly specific and compact expression pattern within the hypothalamus (Fig. 2E–N). Pmch1-expressing cells were detected across a rostral-caudal region of ~500 μm, whereas Pmch2-expressing cells were both fewer in number and detected across a more compact field of ~300 μm. Pmch1 expression was observed bilaterally in regions corresponding to the lateral and posterior parts of the tuberal nucleus (NLTl and NLTp; Fig. 2F–I; NPTm, Fig. 2J), as well as dorsal to the lateral recesses of the third ventricle (NRL; Fig. 2H,I) (See Materials and Methods for a comparison of these specific neuroanatomical designations with those described in the zebrafish atlas [Wullimann et al., 1996].) Pmch1 expression could also be seen in the caudal zone of the periventricular hypothalamus (Hc; Fig. 2J). Pmch2-expressing cells were concentrated in the anterior portion of the Pmch1-expressing field (Fig. 2K–N). Like Pmch1, Pmch2 was expressed in the lateral tuberal nucleus. Unlike Pmch1, Pmch2 expression did not extend caudally to the posterior tuberal nucleus nor dorsally to the lateral recesses of the third ventricle. Thus, among other regions, Pmch1 and Pmch2 both displayed expression in the NLT, a region homologous to the arcuate nucleus, an important center for feeding regulation in mammals.
Figure 2.
Expression pattern of zebrafish Pmch1 and Pmch2 genes in larvae and adults. A,B: Lateral (A) and dorsal (B) views of the Pmch1 expression pattern in the brain of a 2-dpf larvae as determined by in situ hybridization (ISH). C,D: Lateral (C) and dorsal (D) views of Pmch2 expression pattern in a 2-dpf larval brain. Nose is to the left in A and in C. Note that at 2 dpf the Pmch2 staining is anterior to Pmch1 (L, lens, R, retina, LH, lateral hypothalamus). E,K: Lateral view of a dissected adult zebrafish brain, telencephalon to the left, indicating the positions of transverse sections shown in F to R. Note in K that the positions of L, M, and N sections correspond to those of O, P, and Q, respectively. F–J and associated close ups H1,I1: Serial Vibratome sections revealing, anteriorly, the compact bilateral cluster of cell bodies expressing Pmch1 mRNA in the lateral portion of the lateral tuberal nucleus (NLTl) and posteriorly, dispersed perikarya expressing Pmch1 in the posterior NLT (NLTp), dorsal to the lateral recesses of the third ventricle (NRLd), in the medial part of the posterior tuberal nucleus (NPT), and in the caudal zone of the periventricular hypothalamus (Hc). L–N: Serial Vibratome sections revealing the compact bilateral cluster of cell bodies expressing Pmch2 in the NLTl. O–R: Double fluorescent ISH as visualized by using confocal microscopy (reconstructed stacks of 0.5- or 1-μm-thick optic sections). Magenta-green copies of these panels and corresponding close-ups are available in Supplementary Figure S1. P1–P3, Q1–Q3: High-magnification pictures of Pmch1 cells (green, P1, Q1), Pmch2 cells (red, P2, Q2) and merged views (P3, Q3). P4,Q4: Representative single plane pictures demonstrating that Pmch1- and Pmch2-expressing cells are in close proximity but do not co-express both Pmch1 and Pmch2 genes. R: Double fluorescent staining reveals that Pmch1 (green), but not Pmch2 (red, cells absent), is expressed in the NLTp and NRLd. Scale bar = 50 μm in A–D; 200 μm in F–N; 50 μm in O–R.
To examine whether Pmch1 and Pmch2 were expressed by the same hypothalamic cells, we used double fluorescent ISH and confocal microscopy to analyze adult brain sections double labeled for Pmch1 and Pmch2. Although Pmch1 and Pmch2 cells closely intermingled in the lateral NLT (Fig. 2O–Q and close-ups P1–P3 and Q1–Q3), there was no significant overlap in expression pattern. First, high-resolution analysis of individual 1-μm planes showed that in areas where Pmch1-and Pmch2-expressing cells are highly intermixed and closely juxtaposed, co-localization is not observed (Fig. 2P4,Q4). Second, the confocal projections reaffirmed our above finding that Pmch1, but not Pmch2, displays prominent expression in the posterior NLT and the dorsal NRL (shown in Fig. 2R), as well as in the medial NPT and Hc (data not shown). This dense caudal field of Pmch1 expression therefore distinguishes the Pmch1 expression domain from the Pmch2 expression field, which is restricted to the lateral NLT. Although we cannot rule out the possibility that Pmch1 and Pmch2 are ever expressed in the same cell, our findings suggest that independent, yet closely associated neuronal populations within the hypothalamus express Pmch1 and Pmch2 in the zebrafish.
Due to the high sequence conservation between zebrafish MCH2 and mammalian MCH, we used an antibody to the mammalian MCH peptide (Phoenix Pharmaceuticals; dilution 1:400) to characterize the location of MCH2 in the adult zebrafish brain. Preadsorption experiments confirmed the specificity of the mammalian MCH antibody for zebrafish MCH2 (Suppl. Fig. S2). This antibody staining revealed immunoreactive cell bodies both within the hypothalamus as well as in a dense network of fibers throughout the brain (Fig. 3). As expected, this staining was consistent with the Pmch2, but not the Pmch1 expression pattern as determined above by ISH (Fig. 2). Immunoreactive cell bodies were found only in the NLT (NLTl; Fig. 3C–E and corresponding close-ups), but not more caudally in the NLTp, NRLd (Fig. 3F,F1), Hc (Fig. 3F1), or NPTm (data not shown), regions that encompass a dense cloud of Pmch1-expressing neurons according to our ISH analysis (Fig. 2I,J,R). Given the absence of immunostaining in the remainder of the Pmch1 expression domain, the closer homology between zebrafish MCH2 and mammalian MCH peptides, and the preadsorption control (Suppl. Fig. S2), we reasoned that this mammalian MCH antibody is specifically labeling MCH2 cells.
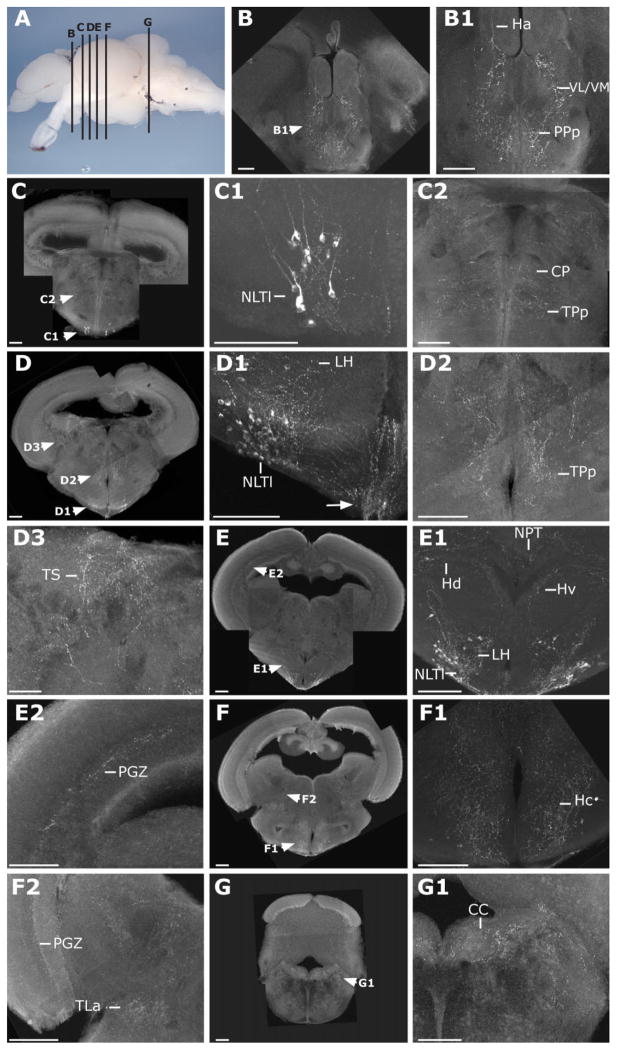
Figure 3.
Hypothalamic MCH2 cells send dense projections throughout the adult zebrafish brain. A: Lateral view of a dissected zebrafish adult brain, telencephalon to the left, indicating the positions of transverse sections shown in B–G. B–G: Confocal microscopy pictures (stacks of 0.5- or 1-μm-thick optic sections) of brain sections immunostained by a primary antibody raised against the mammalian MCH (see Materials and Methods). MCH2 cell bodies are only observed in the NLTl (C,C1,D,D1,E,E1). MCH2 fibers are present in the habenula (Ha; B1), the ventrolateral and ventromedial thalamic nucleus (VL/VM; B1), the posterior preoptic parvocellular nucleus (PPp; B1), the central posterior thalamic nuclei (CP; C2), the periventricular nucleus of the posterior tuberculum (TPp; C2,D2), the torus semicircularis (TS; D3), the dorsal (Hd; E1) and caudal (Hc; F1), zones of the periventricular hypothalamus, the lateral hypothalamus (LH; D1), the posterior tuberal nucleus (NPT; E1), the periventricular gray zone of the optic tectum (PGZ; E2,F2), the area dorsal to the lateral recesses of the third ventricle (NRLd; F1), the posterior zone of the NLT (NLTp; F1), the torus lateralis (TLa; F2), and the crista cerebellaris (CC; G1). Additional fibers were stained in the griseum centrale (GC), the locus coeruleus (LC), and the dorsal and periventricular telencephalic areas (data not shown). Scale bar = 100 μm in B–G.
Next, we used this reagent to visualize MCH2 fibers throughout the central nervous system (CNS). In the telencephalon, MCH2-immunoreactive fibers were found within the dorsal nucleus of the ventral telencephalic area (Vd; data not shown). In the diencephalon, they were found in the ventrolateral and ventromedial thalamic nuclei (VL/VM), the habenula (Ha) (Fig. 3B,B1) and the dorsal thalamus (central posterior thalamic nuclei [CP]; Fig. 3C,C2). Projections were also detected in the periventricular nucleus of the posterior tuberculum (TPp; Fig. 3D,D2), the posterior tuberal nucleus (NPT; Fig. 3E,E1) and in the torus lateralis (TLa; Fig. 3F,F2). Fibers were particularly dense in the lateral and periventricular hypothalamus (Fig. 3B–F) and in the ventralmost hypothalamic region, where fibers extended toward the pituitary gland (arrow, Fig. 3D1). In the mesencephalon, the periventricular gray zone of the optic tectum (PGZ; Fig. 3E,E2) and the torus semicircularis (TS; Fig. 3D,D3) contained immunostained fibers. Finally, in the rhombencephalon, fibers were observed in the vicinity of the griseum centrale (GC) and locus coeruleus (LC) (data not shown) as well as in the crista cerebellaris (CC; Fig. 3G,G1). Our results reveal that zebrafish MCH2 cells project widely across the brain, as seen in mammals. These projections innervate multiple regions such as the dorsal telencephalic area, the LH, and the LC known to express the MCH receptor in mammals (Saito et al., 2001 and see next section).
Zebrafish MCH receptor genes mchr1a, mchr1b, and mchr2 are expressed throughout the brain and in the skin
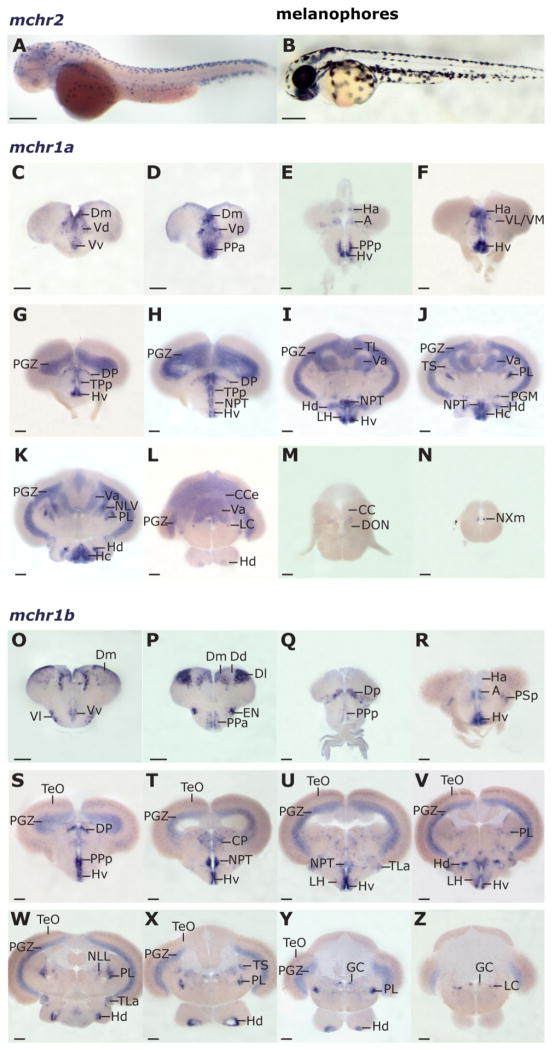
To further our understanding of the putative roles of MCH receptors in zebrafish, we investigated the expression pattern of mchr1a, mchr1b, and mchr2 genes in larvae and in the adult brain. By using available sequence data for mchr1a (Ensembl gene ENSDARG00000037048), mchr1b (ENS-DARG00000022525), and mchr2 (NCBI accession number AY161859), we generated antisense probes and performed ISH on larvae and on serial adult brain sections. Although no strong signal for mchr1a or mchr1b was detected in 2-dpf larvae, the mchr2 probe strongly labeled skin cells in the developing zebrafish larvae at 2dpf (Fig. 4A,B), suggesting a cell-autonomous role for this receptor in modulating skin pigmentation.
Figure 4.
mchr1a and mchr1b receptor gene expression in the adult zebrafish brain and mchr2 expression in the larval skin. A: mchr2 mRNA distribution (ISH) in the melanophores of a 2-dpf larva (lateral view, head to the left). Natural pigmentation was chemically inhibited by using PTU (see Materials and Methods). B: Natural pigmentation of a control larva at the same developmental stage without PTU treatment. Note how the black melanophore distribution resembles the mchr2 expression pattern. C–N: mchr1a mRNA brain distribution (ISH) in a rostrocaudal series of cross sections (telencephalon, C–E; diencephalon and mesencephalon, F–K; rhombencephalon, L–N). O–Z: mchr1b mRNA distribution (ISH) (telencephalon, O–Q; diencephalon and mesencephalon, R–X; rhombencephalon, Y,Z). No mchr1b mRNA was detected in the posterior rhombencephalon. Abbreviations of brain loci are as in the Figure 3 legend. Additional loci include the dorsal and ventral telencephalic areas (D, V), the medial, lateral, and posterior zones of D (Dm, Dl, Dp), the dorsal, medial, lateral, and ventral zones of V (Vd, Vm, Vl, Vv), the anterior thalamus (A), the anterior and posterior parvocellular preoptic nuclei (PPa, PPp), the ventrolateral and -medial thalamic nucleus (VL/VM), the valvula cerebelli (Va), the preglomerular nucleus (PGM), the perilemniscal nucleus (PL), the nucleus of the lateral lemniscus (NLL), the vagal motor neurons (NXm), the entopeduncular nucleus (EN), the optic tectum (TeO), and the descending octaval nucleus (DON). Scale bar = 100 μm in A,B; 200 μm in C–Z.
Unlike mchr2, mchr1a and mchr1b displayed prominent expression in the adult brain (Fig. 4). In the telencephalon, mchr1a transcript appeared in the parvocellular preoptic nucleus (PP; Fig. 4D,E) and in the dorsal and ventral telencephalic areas (D, V; Fig. 4C,D). This staining included the medial zone of the dorsal telencephalic area (Dm), which is thought to be homologous to the mammalian amygdala (Wullimann and Mueller, 2004; Northcutt, 2006), a region involved in emotional learning and memory. The habenular nucleus, a region involved with reward processing, sleep, and cognition (Lecourtier and Kelly, 2007) displayed mchr1a staining (Ha; Fig. 4E,F), as did the thalamus (A, VL/VM, DP; Fig. 4E–H), a nexus of sensory, motor, and sleep and arousal functions in mammals. mchr1a expression was also detected in several hypothalamic regions in the diencephalon, including the ventral, caudal, and dorsal zones of the periventricular hypothalamus (Hv, Hc, Hd; Fig. 4E–L), the posterior tuberal nucleus (NPT; Fig. 4H–J), and the LH (Fig. 4I). Sites of expression within the diencephalon and mesencephalon include the torus longitudinalis (TL; Fig. 4I), the periventricular gray zone of the optic tectum (PGZ; Fig. 4G–L), a location preliminarily identified as the perilemniscal nucleus (PL; Fig. 4K), and the preglomerular nucleus (PGM; Fig. 4J). Hindbrain expression was found in the corpus cerebelli (CCe; Fig. 4K,L), valvula cerebelli (Va; Fig. 4I–L), locus coeruleus (LC, Fig. 4L) and an unidentified region ventral to the crista cerebellaris (CC) and dorsal to the descending octaval nucleus (DON; Fig. 4M). Adjacent to the rhombencephalic ventricle, the vagal motor nucleus (NXm; Fig. 4N; Mueller et al., 2004) displayed mchr1a expression. The vagal motor nuclei express MCHR1 in the rat (Saito et al., 2001).
We found that mchr1b shared multiple sites of expression with mchr1a throughout the CNS, although some important distinctions were observed. In the telencephalon, mchr1b was detected in the ventral and dorsal telencephalic areas (V and D; Fig. 4O,P), including in the Dm, homologous to the mammalian amygdala. However, unlike mchr1a, mchr1b displayed very prominent expression in the lateral zone of the dorsal telencephalic area (Dl; Fig. 4P), more precisely in the dorsal region of Dl. The Dl has been proposed as homologous to the mammalian hippocampus, which is important for learning and memory (Wullimann and Mueller, 2004; Northcutt, 2006), although some studies restrict this homology to the lateroventral region of Dl (Portavella et al., 2004). Like mchr1a, in the midbrain, the thalamus (A, DP; Fig. 4R,S) and the perilemniscal nucleus (PL; Fig. 4V–Y) displayed mchr1b expression. We also observed mchr1b expression in many hypothalamic nuclei that express mchr1a, including in the preoptic area (PPa, PPp; Fig. 4P,Q,S), the LH (Fig. 4U,V), the NPT (Fig. 4T,U), and the Hv and Hd (Fig. 4R–Y). PGZ expression of mchr1b, like mchr1a, was prominent and widespread (Fig. 4S–Z). In the hindbrain, expression of mchr1b in the LC (Fig. 4Z) was noted.
In addition to these regions in common, mchr1b mRNA alone was detected within the forebrain in the entopeduncular nucleus (EN; Fig. 4P), as well as in the posterior zone of the dorsal telencephalic area (Dp; Fig. 4Q), a region thought to be homologous to the piriform cortex in mammals (Wullimann and Mueller, 2004), an important olfactory center and a site of MCHR1 expression in the rat (Saito et al., 2001). In the diencephalon, the torus lateralis (TLa), which integrates multiple sensory inputs, as well as the periventricular hypothalamus specifically displayed mchr1b expression, whereas in the mesencephalon, the torus semicircularis (TS), and the optic tectum (TeO) displayed mchr1b expression (Fig. 4S–X). Expression of mchr1b was also unique to the griseum centrale (GC; Fig. 4Y,Z) in the hindbrain. Overall, the widespread expression of mchr1a and mchr1b in brain regions implicated in food sensing and ingestion, learning and memory, arousal, and energy metabolism is consistent with the MCH system being utilized in fish for functions beyond pigmentation, perhaps in the coordination and regulation of feeding behavior.
A role for MCH1 in skin pigmentation
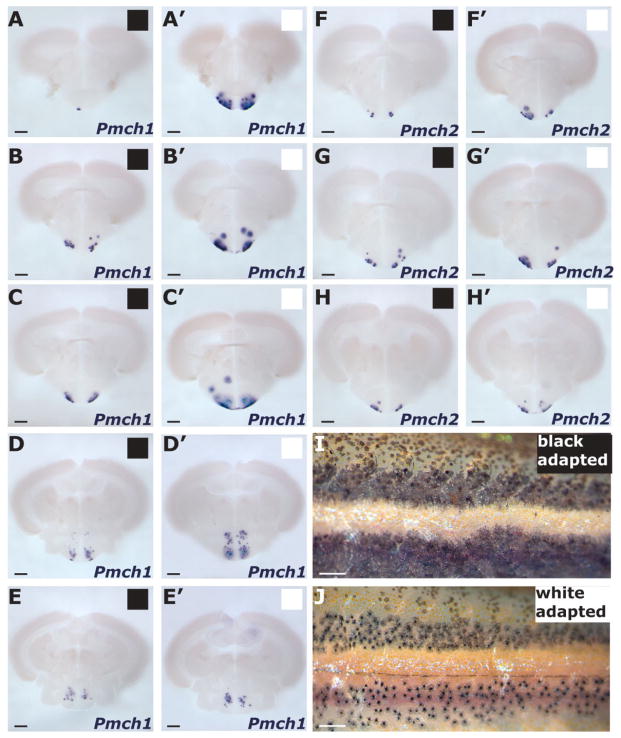
Zebrafish display melanosome aggregation within 24 hours of being exposed to a white background, and application of rat MCH directly to zebrafish scales induces melanosome aggregation (Logan et al., 2006). We hypothesized that if MCH induces melanosome aggregation in zebrafish, then conditions under which melanosome concentration is observed might correlate with an increase in Pmch expression. To examine this possibility, we compared expression of Pmch1 and Pmch2 genes in adult fish maintained in a black background (black adapted [BA]) versus in fish kept in a white background (white adapted [WA]) (see Materials and Methods). As seen in other fish species as well as in prior zebrafish studies (Kawauchi, 2006; Logan et al., 2006), we observed that adult WA zebrafish (Fig. 5J) displayed significant aggregation of skin melanosomes compared with BA fish (Fig. 5I). We next used ISH on serial brain sections to compare Pmch expression levels between six WA and six BA fish. We observed a modest difference in Pmch2 expression in WA fish compared with BA controls (Fig. 5F–H, F′–H′).
Figure 5.
Change of Pmch expression levels in response to background color. A–E, A′–E′: Comparison of Pmch1 expression (ISH) in an anteroposterior series of equivalent transverse sections in black-adapted (BA; black squares) fish (A–E) versus white-adapted (WA; white squares) fish (A′–E′). This series compares two representative brains from 12 brains examined (see Materials and Methods). Within an experiment, time of staining was identical for WA fish and BA fish. Consequently, BM purple staining would often reach saturation in the WA brains (hence the diffuse and the turquoise staining in A′–D′) before slices from BA brains would show any staining (e.g., A). F–H, F′–H′: Comparison of Pmch2 expression (ISH) in an anteroposterior series of equivalent transverse sections in BA fish (F–H) versus WA fish (F′–H′). In strong contrast with Pmch1, Pmch2 staining was either mostly similar or slightly increased in WA fish compared with BA fish. This series compares 2 representative brains from 12 brains examined. I–J: Lateral view of the trunk of BA and WA adult zebrafish. In the WA skin, melanophores display perinuclear concentration of pigment, whereas in BA skin, pigment is diffuse. Scale bar = 200 μm in A–H; 1 mm in I,J.
However, in the brains of fish with aggregated melanosomes, there was a dramatic increase in Pmch1 expression compared with that seen in fish with dispersed pigment (Fig. 5A–E). The increase in expression appeared most notable in the anterior part of the Pmch1 expression field, specifically in the lateral region of the NLT (NLTl; Fig. 5A–C, A′–C′). All parts of the Pmch1 expression field, however, including NLTp and NPTm, demonstrated some increase in Pmch1 expression. Thus, a significant increase in Pmch1 expression correlates with the aggregation of melanosomes, suggesting a role for zebrafish MCH1 in pigment regulation. In contrast, the comparatively limited response of Pmch2 expression to changes in background color may be indicative of a more modest, if any, role for zebrafish MCH2 in skin pigmentation.
MCH2 is upregulated in response to fasting
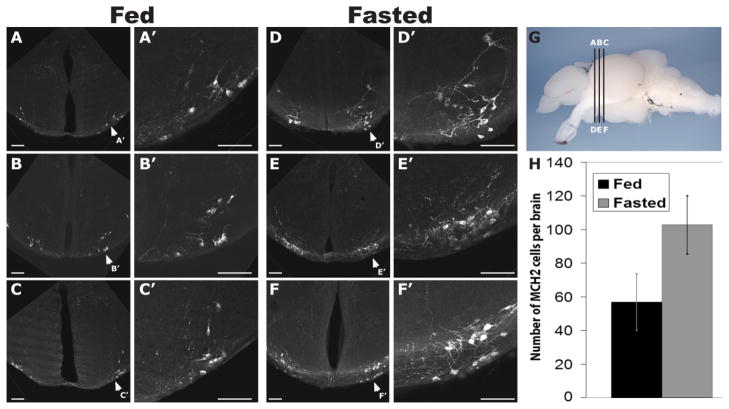
Extensive studies in mammals have characterized a role for MCH in the promotion of feeding behavior and in the regulation of energy conservation (Pissios et al., 2006). Due to the similarities between zebrafish Pmch2 gene and mammalian MCH, we asked whether zebrafish MCH2 plays a role in feeding behavior. To address this question, we compared the levels of MCH2 in the brains of nine fish fasted for 20 days with eight fed ad libitum. Using ISH, we were not able to detect any strong difference in mRNA level between the two groups (data not shown). However, by performing immunostaining using the mammalian α-MCH antibody, we found a nearly twofold increase in the number of MCH2-labeled cells within the hypothalamus of fasted fish (Fig. 6D–F and respective close-ups D′ – 6F′) compared with fed controls (Fig. 6A–C and close-ups A′ – 6C′). Whereas our antibody labeled an average of 58 MCH2 cells per brain (95% CI 40.1–75.4) in fed fish, fasted fish had an average of 102 labeled cells (95% CI 83.9 –120.1; Fig. 6H). This difference in the average number of MCH2-labeled cells per brain was statistically significant (P = 0.0038). The increase in MCH2 peptide expression was detected in the NLTl and not ectopically in other brain regions. These data support the idea that in zebrafish, as is true in mammals, MCH may function to regulate energy balance. Furthermore, its upregulation in fasted animals suggests that, like in mammals, zebrafish MCH2 may promote food intake in response to internal energy cues.
Figure 6.
Increase in the number of MCH2 peptide-expressing cells in fasted adult zebrafish. A–F and corresponding close-ups A′–F′: Confocal pictures of a series of slices immunostained with an antibody that visualizes zebrafish MCH2. Three representative sections (A–C) from the brain of a normally fed adult zebrafish and three representative sections (D–F) from the brain of an adult zebrafish fasted for 2 weeks. G: Lateral view of a dissected adult zebrafish brain, telencephalon to the left, indicating the positions of transverse sections shown in A–F. A, B, and C slices correspond to D, E, and F slices, respectively. H: Average number of MCH2 cells identified in the brains of fed and of fasted adult fish. Error bars represent the 95% CI. In fed fish (n = 8) an average of 58 MCH2 cells was labeled per brain (95% CI 40.1–75.4), whereas fasted fish (n = 9) had an average of 102 labeled cells (95% CI 83.9 –120.1). This difference in the average number of MCH2-labeled cells per brain is statistically significant (P = 0.0038, two-tailed Student’s t test). Scale bar = 50 μm in A–G.
DISCUSSION
The varied roles of MCH in mammals, from regulation of metabolic functions to modulation of complex cognitive processes like sleep and anxiety, highlights the flexibility and functional sophistication of neurologically active peptides. As MCH is only found in vertebrates, it has not been possible to apply the powerful molecular biology and microscopy tools available in simple invertebrate model systems toward understanding the fundamental properties of the MCH neuronal network. This study represents a significant step forward by characterizing the MCH system in a genetically tractable vertebrate, the zebrafish, and by identifying previously unappreciated parallels between the zebrafish and mammalian MCH systems.
Earlier work established the existence of a single MCH gene in salmonids with a simple, one-exon genomic structure (Griffond and Baker, 2002), whose peptide product induces skin lightening in fish scales by directing the aggregation of melanosomes. Multiple studies in mammals, however, failed to firmly establish a role for MCH in mammalian skin pigmentation. Furthermore, it seemed puzzling that not only did mammalian MCH have a distinct genomic structure and longer peptide sequence than salmonid MCH, but the major function of mammalian MCH in feeding behavior was entirely disparate from the role of MCH in pigmentation function in fish. Our discovery that two Pmch genes, one orthologous to MCH found originally in salmon and the other more similar to mammalian MCH, exist in zebrafish and other teleosts establishes a definitive link between the fish and mammalian MCH systems. The dynamic response of zebrafish MCH2 levels within the CNS to changes in food intake further supports this connection.
What, then, is the evolutionary relationship between the teleost and mammalian MCH systems? One simple hypothesis is that mammalian MCH and teleost Pmch2 genes arose in a common ancestor prior to the divergence of tetrapods and fishes some 450 million years ago (Volff, 2005). Unlike the teleosts described in this study, however, mammalian genomes contain a single MCH gene. Humans do have two MCH-like genes, PMCHL1 and PMCHL2, but these genes have evolved during primate evolution and are chimeric genes of unknown function (Courseaux and Nahon, 2001). Why do mammals contain one MCH gene whereas several teleosts contain two? If two MCH genes existed in the ancestor common to teleosts and tetrapods, one copy could have been lost specifically during mammalian evolution (and retained in teleosts). Another possibility, however, is that during teleost evolution, a second MCH paralog arose out of a duplication event. This latter hypothesis is appealing because multiple phylogenomic analyses in fugu, Tetraodon, and zebrafish have identified hundreds of gene pairs with single-copy orthologs in tetrapods (Volff, 2005, references within).
This pattern applies to the clusters of HOX genes, which specify cell fate along the anterior-posterior axis of a developing embryo (Amores et al., 2004), as well as for the EGF receptor (egfr) genes (Volff, 2005), supporting the widely embraced hypothesis that a whole-genome duplication event occurred during the ray-finned fish lineage subsequent to its divergence from tetrapods. Acquisition of novel functions in a duplicate gene via mutation and positive selection is one functional consequence of gene duplication. Perhaps zebrafish Pmch1 arose via a Pmch2 duplication event. Loss of introns in this paralog could have resulted in the single-exon Pmch1 gene, which was then co-opted for a novel beneficial function in pigmentation regulation. Consistent with this hypothesis, the role of MCH in lightening the skin is believed to have arisen at a later stage of ray-finned fish evolution (Griffond and Baker, 2002; Kawauchi, 2006). Further experiments will be necessary to determine whether zebrafish Pmch genes are indeed functionally distinct, with Pmch1 principally involved in pigmentation and Pmch2 responsive to changes in food availability, but our functional data support this notion. Alternatively, these genes could partition roles with respect to pigmentation and food intake regulation. One might speculate that this is the case for the zebrafish MCH receptor paralogs MCHR1A and MCHR1B, which share a significant amount of overlap in their expression patterns within the CNS (Fig. 4).
Our initial whole-system characterization of the cells (Fig. 2), projections (Fig. 3), and targets (Fig. 4) of the MCH neuropeptide system in zebrafish revealed many organizational properties reminiscent of those described in other teleost and mammalian MCH systems. For instance, rainbow trout (Naito et al., 1985; Bird and Baker, 1989; Kawauchi, 1989), seabream (Duarte et al., 2001), and goldfish (Huesa et al., 2005) also contain MCH neurons in the NLT in the basal hypothalamus. Another population of MCH neurons in rainbow trout (Baker et al., 1995) and in the goldfish (Matsuda et al., 2007) is present above the lateral recesses of the third ventricle (NRLd), a specific site where Pmch1, but not Pmch2, is expressed in the zebrafish (Fig. 2). Interestingly, a recent immunocytochemical investigation of MCH peptide expression in the goldfish revealed an expression pattern very similar to what we see in this study with zebrafish Pmch1 but not Pmch2, including expression in the NLTl, NLTp, NPTm, and NRLd (Matsuda et al., 2007). This is notable because the goldfish study used an antibody raised against the salmon MCH peptide, which is identical in sequence to zebrafish MCH1. Thus zebrafish MCH1 cells display a neuroanatomical distribution highly reminiscent of MCH expression patterns described in goldfish and other teleosts. This concordance between zebrafish Pmch1 expression pattern and MCH expression in trout and goldfish further distinguishes zebrafish MCH2 peptide from the MCH originally identified in salmonids. By using ISH and immunostaining with an antibody to mammalian MCH, we found that Pmch2 and the MCH2 peptide is expressed in the NLTl but not in the NLTp, NPTm, or NRLd.
Defining the role of the NLTl in teleosts thus is critical to understanding the role of MCH2 in this species. Recent studies in the goldfish have proposed that the NLT of teleosts is homologous to the arcuate nucleus of mammals (Cerdá-Reverter and Peter, 2003), a hypothalamic region adjacent to the LH that is dense with centrally projecting neurons involved in appetite. In goldfish, AgRP is expressed in the NLT (Cerdá-Reverter and Peter, 2003), whereas in zebrafish, MSH is expressed in the NLT (Forlano and Cone, 2007). Thus, as in the arcuate nucleus of mammals, the NLT of teleosts contains multiple cell types associated with the regulation of food intake. The presence of MCH-expressing cells within the NLT of zebrafish and the apparent increase of the number of MCH2-expressing cells in the NLT in response to food deprivation lends support to the idea that the teleost MCH system might operate within the context of a complex neuronal network dedicated to regulating food intake.
Initial analysis revealed that the relatively compact population of MCH2 cells within the NLTl sends projections to targets within all brain regions, from the telencephalon to the hind-brain. Many of the regions innervated by MCH2 fibers also contain MCH-immunoreactive fibers in the goldfish (Huesa et al., 2005). This includes, but is not limited to, the dorsal telencephalic area, preoptic nucleus, thalamus, posterior tuberal nucleus, ventral periventricular hypothalamus (Hv), locus coeruleus, and vagal motor neurons. Similarly, studies of MSH and AgRP cell projections in the zebrafish identified fibers in the tuberal nucleus, the thalamus, the preoptic area, and the torus semicircularis (Forlano and Cone, 2007). The pattern of zebrafish MCH2 projections thus resembles projection sites for other teleost neurons involved in appetite regulation. Many teleosts in which MCH has a known role in skin pigmentation display abundant MCH fibers in the posterior neurohypophysis (Kawauchi, 2006). Unfortunately, our sectioning methods do not typically retain the pituitary of the adult zebrafish. However, we did observe dense MCH2 projections in the ventralmost portion of the hypothalamus (Fig. 3D,D1), and these are reasonable candidates for projections that extend into the pituitary in intact animals.
Our examination of the CNS expression patterns of the mchr1a and mchr1b genes revealed that MCH cell targets are widespread throughout the brain, that many of these targets are consistent with a role in regulating food intake, and that many of these targets are also known sites of MCHR expression in mammals. Olfactory centers outside the olfactory bulb, including the piriform cortex, are important sites of MCHR expression in the rat (Saito et al., 2001). We detected mchr1b expression in the posterior zone of the dorsal telencephalic area (Dp), the zebrafish homolog to the piriform cortex (Fig. 4Q; Wullimann and Mueller, 2004). Cortical regions important for learning and memory, specifically the hippocampus and the basolateral amygdala, also are enriched for MCHR in rodents. It has historically been challenging to identify the telencephalic subdivisions in teleosts due to fundamental differences in early brain development between mammals and fish (telencephalic eversion versus evagination) (Wullimann and Mueller, 2004). However, anatomical and functional data compiled from various teleosts suggest that the lateral zone of the dorsal telencephalic area (Dl) is the hippocampus homolog, although some studies restrict this homology to the lateroventral region of Dl (Portavella et al., 2004), and the medial zone of the dorsal telencephalic area (Dm) is homologous to the basolateral amygdala (Wullimann and Mueller, 2004; Northcutt, 2006).
Our ISH analysis revealed that both mchr1a and mchr1b are expressed in the Dm/amygdala, although mchr1a expression was more pronounced (Fig. 4D). mchr1b alone displayed a remarkably prominent and distinctive expression domain in the lateral zone of the dorsal telencephalic area (Dl; Fig. 4P). Outside these proto-cortical regions of the zebrafish forebrain, receptor expression was detected in multiple regions of the thalamus, which integrates sensory information and coordinates arousal behavior, the locus coeruleus, and various hypothalamic areas, including the Hv, the LH, and the tuberal nucleus, and brainstem motor neurons. These expression data are consistent with the regulation of a function such as nutritional homeostasis, which would require coordination of basic arousal and motor functions related to eating with higher order functions encompassing sensory integration, reward, and learning. Importantly, these regions are known to express MCHR in rats, in which MCH’s major functional role is to promote food ingestion and energy conservation. Interestingly, we found that the zebrafish mchr2 probe distinctively labeled melanophores in the developing embryo, but displayed no detectable expression in the adult CNS (Fig. 4). Perhaps MCHR2 alone is required for the regulation of skin pallor in response to environmental cues. Alternatively, MCH receptors expressed in the CNS might somehow contribute to the coordination of the dynamic pigmentation response.
Our data indicate that food deprivation leads to a nearly twofold increase in the number of cells expressing MCH2 in the hypothalamus (Fig. 6). This finding indicates that increased MCH2 peptide levels correlate with starvation, and implies that MCH2 could play a role in regulating food intake in zebrafish, although the exact mechanism has not been determined. In other teleosts studied, the role of MCH peptides in appetite and energy homeostasis has been unclear (Pissios et al., 2006). For example, overexpression of the salmon MCH gene in medaka caused skin lightening but produced no change in body mass (Kinoshita et al., 2001). These data, however, should be reconsidered in light of our finding that the medaka genome, like in zebrafish, contains two Pmch genes, only one of which is predicted to produce an MCH peptide (MCH1) identical to that found in salmon. If in medaka, hypothetically, receptors for MCH2, but not receptors for MCH1, play a role in food intake, MCH1 overexpression indeed might have no effect on body mass. In the goldfish Carassius auratus (Matsuda et al., 2006, 2007), an anorexigenic role for MCH has been proposed. Studies of multiple appetite-influencing neuropeptides in the goldfish have shown that, like in mammals, NPY, galanin, HCRT, AgRP, and ghrelin are orexigenic, whereas CART, bombesin, CCK, melanocortin, and CRF are anorexigenic (Kawauchi, 2006). Unlike these other neuropeptides, goldfish MCH could assume a role in appetite opposite to that seen in mammals. However, if, as in zebrafish and other teleosts, multiple MCH genes exist in the goldfish genome, these putative MCH genes could perhaps have distinct or even shared roles in pigment and food intake regulation, with potentially multiple receptors present to potentiate these signals. ICV injection of MCH peptide or other overexpression techniques could conceivably create spurious cross-reactions or dominant negative effects. Efforts in goldfish, zebrafish, and other teleosts must be made to create appropriate MCH loss-of-function strains to query the natural role for these peptides in food intake and pigmentation.
Finally, our characterization of the fundamental properties of the MCH system in zebrafish has important implications for future studies of the cognitive and obesity syndromes already attributed to these neuropeptides. First, in zebrafish, studies of the early stages of MCH cell specification and differentiation are possible, as well as a detailed characterization of the development of MCH cell projections and wiring of the overall system. It is indeed possible that failure or modification of stereotypical MCH neuronal network development programs could underlie some of the disease processes linked to the MCH system. Second, the molecular toolkit available in zebrafish to interrogate neuronal structure and function, including fluorescent transgenic markers and channelrhodopsin technology (Faraco et al., 2006; Douglass et al., 2008), will provide powerful strategies for dissecting the functional contributions of MCH1 and MCH2 neurons and their respective targets. Third, zebrafish are increasingly regarded as an optimal system for drug studies. The isolation or functional analysis of small molecule antagonists of MCHR1, for example, could be pursued in zebrafish. Further studies of the MCH system in zebrafish will elucidate the molecular basis of this system in fish and in mammals and give valuable context to the evolution of sophisticated behavioral modulation by neuropeptides in vertebrates.
Supplementary Material
Acknowledgments
Grant sponsor: Howard Hughes Medical Institute; Grant sponsor: National Institutes of Health; Grant number: R01 NS062798.
The authors thank Antoine Adamantidis for fruitful discussion and critical reading of the manuscript, and Laura Alexandre for excellent care of the animals.
LITERATURE CITED
- Adamantidis A, Thomas E, Foidart A, Tyhon A, Coumans B, Minet A, Tirelli E, Seutin V, Grisar T, Lakaye B. Disrupting the melanin-concentrating hormone receptor 1 in mice leads to cognitive deficits and alterations of NMDA receptor function. Eur J Neurosci. 2005;21:2837–2844. doi: 10.1111/j.1460-9568.2005.04100.x. [DOI] [PubMed] [Google Scholar]
- Allaeys I, Bouyer K, Loudes C, Faivre-Bauman A, Petit F, Ortola C, Cardinaud B, Epelbaum J, Nahon JL. Characterization of MCH-gene-overprinted-polypeptide-immunoreactive material in hypothalamus reveals an inhibitory role of pro-somatostatin1–64 on somatostatin secretion. Eur J Neurosci. 2004;19:925–936. doi: 10.1111/j.0953-816x.2004.03187.x. [DOI] [PubMed] [Google Scholar]
- Amores A, Suzuki T, Yan YL, Pomeroy J, Singer A, Amemiya C, Postlethwait JH. Developmental roles of pufferfish Hox clusters and genome evolution in ray-fin fish. Genome Res. 2004;14:1–10. doi: 10.1101/gr.1717804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker B, Levy A, Hall L, Lightman S. Cloning and expression of melanin-concentrating hormone genes in the rainbow trout brain. Neuroendocrinology. 1995;61:67–76. doi: 10.1159/000126814. [DOI] [PubMed] [Google Scholar]
- Bird DJ, Baker BI. An immunological study of the secretory activity of neurons producing melanin-concentrating hormone in a teleost. Neuroscience. 1989;28:245–251. doi: 10.1016/0306-4522(89)90248-0. [DOI] [PubMed] [Google Scholar]
- Cerdá-Reverter JM, Peter RE. Endogenous melanocortin antagonist in fish: structure, brain mapping, and regulation by fasting of the goldfish agouti-related protein gene. Endocrinology. 2003;144:4552–4561. doi: 10.1210/en.2003-0453. [DOI] [PubMed] [Google Scholar]
- Chen Y, Hu C, Hsu CK, Zhang Q, Bi C, Asnicar M, Hsiung HM, Fox N, Slieker LJ, Yang DD, Heiman ML, Shi Y. Targeted disruption of the melanin-concentrating hormone receptor-1 results in hyperphagia and resistance to diet-induced obesity. Endocrinology. 2002;143:2469–2477. doi: 10.1210/endo.143.7.8903. [DOI] [PubMed] [Google Scholar]
- Courseaux A, Nahon JL. Birth of two chimeric genes in the Hominidae lineage. Science. 2001;291:1293–1297. doi: 10.1126/science.1057284. [DOI] [PubMed] [Google Scholar]
- Douglass AD, Kraves S, Deisseroth K, Schier AF, Engert F. Escape behavior elicited by single, channelrhodopsin-2-evoked spikes in zebrafish somatosensory neurons. Curr Biol. 2008;18:1133–1137. doi: 10.1016/j.cub.2008.06.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte G, Segura-Noguera MM, Martin del Rio MP, Mancera JM. The hypothalamo-hypophyseal system of the white seabream Diplodus sargus: immunocytochemical identification of arginine-vasotocin, isotocin, melanin-concentrating hormone and corticotropin-releasing factor. Histochem J. 2001;33:569–578. doi: 10.1023/a:1014912110318. [DOI] [PubMed] [Google Scholar]
- Faraco JH, Appelbaum L, Marin W, Gaus SE, Mourrain P, Mignot E. Regulation of hypocretin (orexin) expression in embryonic zebrafish. J Biol Chem. 2006;281:29753–29761. doi: 10.1074/jbc.M605811200. [DOI] [PubMed] [Google Scholar]
- Forlano PM, Cone RD. Conserved neurochemical pathways involved in hypothalamic control of energy homeostasis. J Comp Neurol. 2007;505:235–248. doi: 10.1002/cne.21447. [DOI] [PubMed] [Google Scholar]
- Griffond B, Baker BI. Cell and molecular cell biology of melanin-concentrating hormone. Int Rev Cytol. 2002;213:233–277. doi: 10.1016/s0074-7696(02)13016-6. [DOI] [PubMed] [Google Scholar]
- Hassani OK, Lee MG, Jones BE. Melanin-concentrating hormone neurons discharge in a reciprocal manner to orexin neurons across the sleep-wake cycle. Proc Natl Acad Sci U S A. 2009;106:2418–2422. doi: 10.1073/pnas.0811400106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauptmann G, Gerster T. Two-color whole-mount in situ hybridization to vertebrate and Drosophila embryos. Trends Genet. 1994;10:266. doi: 10.1016/0168-9525(90)90008-t. [DOI] [PubMed] [Google Scholar]
- Hill J, Duckworth M, Murdock P, Rennie G, Sabido-David C, Ames RS, Szekeres P, Wilson S, Bergsma DJ, Gloger IS, Levy DS, Chambers JK, Muir AI. Molecular cloning and functional characterization of MCH2, a novel human MCH receptor. J Biol Chem. 2001;276:20125–20129. doi: 10.1074/jbc.M102068200. [DOI] [PubMed] [Google Scholar]
- Holmqvist BI, Ekström P. Galanin-like immunoreactivity in the brain of teleosts: distribution and relation to substance P, vasotocin, and isotocin in the Atlantic salmon (Salmo salar) J Comp Neurol. 1991;306:361–381. doi: 10.1002/cne.903060302. [DOI] [PubMed] [Google Scholar]
- Huesa G, van den Pol AN, Finger TE. Differential distribution of hypocretin (orexin) and melanin-concentrating hormone in the goldfish brain. J Comp Neurol. 2005;488:476–491. doi: 10.1002/cne.20610. [DOI] [PubMed] [Google Scholar]
- Kawauchi H. Melanin concentrating hormone. II. Structure and biosynthesis of melanin-concentrating hormone. Life Sci. 1989;45:1133–1140. doi: 10.1016/0024-3205(89)90500-6. [DOI] [PubMed] [Google Scholar]
- Kawauchi H. Functions of melanin-concentrating hormone in fish. J Exp Zool A Comp Exp Biol. 2006;305:751–760. doi: 10.1002/jez.a.310. [DOI] [PubMed] [Google Scholar]
- Kawauchi H, Kawazoe I, Tsubokawa M, Kishida M, Baker BI. Characterization of melanin-concentrating hormone in chum salmon pituitaries. Nature. 1983;305:321–323. doi: 10.1038/305321a0. [DOI] [PubMed] [Google Scholar]
- Kinoshita M, Morita T, Toyohara H, Hirata T, Sakaguchi M, Ono M, Inoue K, Wakamatsu Y, Ozato K. Transgenic medaka overexpressing a melanin-concentrating hormone exhibit lightened body color but no remarkable abnormality. Mar Biotechnol (NY) 2001;3:536–543. doi: 10.1007/s10126-001-0061-y. [DOI] [PubMed] [Google Scholar]
- Kokkotou E, Jeon JY, Wang X, Marino FE, Carlson M, Trombly DJ, Maratos-Flier E. Mice with MCH ablation resist diet induced obesity through strain specific mechanisms. Am J Physiol Regul Integr Comp Physiol. 2005;289:R117–124. doi: 10.1152/ajpregu.00861.2004. [DOI] [PubMed] [Google Scholar]
- Kokkotou EG, Tritos NA, Mastaitis JW, Slieker L, Maratos-Flier E. Melanin-concentrating hormone receptor is a target of leptin action in the mouse brain. Endocrinology. 2001;142:680–686. doi: 10.1210/endo.142.2.7981. [DOI] [PubMed] [Google Scholar]
- Kolakowski LF, Jr, Jung BP, Nguyen T, Johnson MP, Lynch KR, Cheng R, Heng HH, George SR, O’Dowd BF. Characterization of a human gene related to genes encoding somatostatin receptors. FEBS Lett. 1996;398:253–258. doi: 10.1016/s0014-5793(96)01160-x. [DOI] [PubMed] [Google Scholar]
- Lakaye B, Minet A, Zorzi W, Grisar T. Cloning of the rat brain cDNA encoding for the SLC-1 G protein-coupled receptor reveals the presence of an intron in the gene. Biochim Biophys Acta. 1998;1401:216–220. doi: 10.1016/s0167-4889(97)00135-3. [DOI] [PubMed] [Google Scholar]
- Lecourtier L, Kelly PH. A conductor hidden in the orchestra? Role of the habenular complex in monoamine transmission and cognition. Neurosci Biobehav Rev. 2007;31:658–672. doi: 10.1016/j.neubiorev.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Logan DW, Bryson-Richardson RJ, Pagan KE, Taylor MS, Currie PD, Jackson IJ. The structure and evolution of the melanocortin and MCH receptors in fish and mammals. Genomics. 2003a;81:184–191. doi: 10.1016/s0888-7543(02)00037-x. [DOI] [PubMed] [Google Scholar]
- Logan DW, Bryson-Richardson RJ, Taylor MS, Currie P, Jackson IJ. Sequence characterization of teleost fish melanocortin receptors. Ann N Y Acad Sci. 2003b;994:319–330. doi: 10.1111/j.1749-6632.2003.tb03196.x. [DOI] [PubMed] [Google Scholar]
- Logan DW, Burn SF, Jackson IJ. Regulation of pigmentation in zebrafish melanophores. Pigment Cell Res. 2006;19:206–213. doi: 10.1111/j.1600-0749.2006.00307.x. [DOI] [PubMed] [Google Scholar]
- Ludwig DS, Tritos NA, Mastaitis JW, Kulkarni R, Kokkotou E, Elmquist J, Lowell B, Flier JS, Maratos-Flier E. Melanin-concentrating hormone overexpression in transgenic mice leads to obesity and insulin resistance. J Clin Invest. 2001;107:379–386. doi: 10.1172/JCI10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh DJ, Weingarth DT, Novi DE, Chen HY, Trumbauer ME, Chen AS, Guan XM, Jiang MM, Feng Y, Camacho RE, Shen Z, Frazier EG, Yu H, Metzger JM, Kuca SJ, Shearman LP, Gopal-Truter S, MacNeil DJ, Strack AM, MacIntyre DE, Van der Ploeg LH, Qian S. Melanin-concentrating hormone 1 receptor-deficient mice are lean, hyperactive, and hyperphagic and have altered metabolism. Proc Natl Acad Sci U S A. 2002;99:3240–3245. doi: 10.1073/pnas.052706899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda K, Shimakura S, Maruyama K, Miura T, Uchiyama M, Kawauchi H, Shioda S, Takahashi A. Central administration of melanin-concentrating hormone (MCH) suppresses food intake, but not locomotor activity, in the goldfish, Carassius auratus. Neurosci Lett. 2006;399:259–263. doi: 10.1016/j.neulet.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Matsuda K, Shimakura S, Miura T, Maruyama K, Uchiyama M, Kawauchi H, Shioda S, Takahashi A. Feeding-induced changes of melanin-concentrating hormone (MCH)-like immunoreactivity in goldfish brain. Cell Tissue Res. 2007;328:375–382. doi: 10.1007/s00441-006-0347-5. [DOI] [PubMed] [Google Scholar]
- Modirrousta M, Mainville L, Jones BE. Orexin and MCH neurons express c-Fos differently after sleep deprivation vs. recovery and bear different adrenergic receptors. Eur J Neurosci. 2005;21:2807–2816. doi: 10.1111/j.1460-9568.2005.04104.x. [DOI] [PubMed] [Google Scholar]
- Mori M, Harada M, Terao Y, Sugo T, Watanabe T, Shimomura Y, Abe M, Shintani Y, Onda H, Nishimura O, Fujino M. Cloning of a novel G protein-coupled receptor, SLT, a subtype of the melanin-concentrating hormone receptor. Biochem Biophys Res Commun. 2001;283:1013–1018. doi: 10.1006/bbrc.2001.4893. [DOI] [PubMed] [Google Scholar]
- Mueller T, Vernier P, Wullimann MF. The adult central nervous cholinergic system of a neurogenetic model animal, the zebrafish Danio rerio. Brain Res. 2004;1011:156–169. doi: 10.1016/j.brainres.2004.02.073. [DOI] [PubMed] [Google Scholar]
- Nahon JL, Presse F, Bittencourt JC, Sawchenko PE, Vale W. The rat melanin-concentrating hormone messenger ribonucleic acid encodes multiple putative neuropeptides coexpressed in the dorsolateral hypothalamus. Endocrinology. 1989;125:2056–2065. doi: 10.1210/endo-125-4-2056. [DOI] [PubMed] [Google Scholar]
- Naito N, Nakai Y, Kawauchi H, Hayashi Y. Immunocytochemical identification of melanin-concentrating hormone in the brain and pituitary gland of the teleost fishes Oncorhynchus keta and Salmo gairdneri. Cell Tissue Res. 1985;242:41–48. [Google Scholar]
- Northcutt RG. Connections of the lateral and medial divisions of the goldfish telencephalic pallium. J Comp Neurol. 2006;494:903–943. doi: 10.1002/cne.20853. [DOI] [PubMed] [Google Scholar]
- Pissios P, Bradley RL, Maratos-Flier E. Expanding the scales: the multiple roles of MCH in regulating energy balance and other biological functions. Endocr Rev. 2006;27:606–620. doi: 10.1210/er.2006-0021. [DOI] [PubMed] [Google Scholar]
- Portavella M, Torres B, Salas C, Papini MR. Lesions of the medial pallium, but not of the lateral pallium, disrupt spaced-trial avoidance learning in goldfish (Carassius auratus) Neurosci Lett. 2004;362:75–78. doi: 10.1016/j.neulet.2004.01.083. [DOI] [PubMed] [Google Scholar]
- Qu D, Ludwig DS, Gammeltoft S, Piper M, Pelleymounter MA, Cullen MJ, Mathes WF, Przypek R, Kanarek R, Maratos-Flier E. A role for melanin-concentrating hormone in the central regulation of feeding behaviour. Nature. 1996;380:243–247. doi: 10.1038/380243a0. [DOI] [PubMed] [Google Scholar]
- Rodriguez M, Beauverger P, Naime I, Rique H, Ouvry C, Souchaud S, Dromaint S, Nagel N, Suply T, Audinot V, Boutin JA, Galizzi JP. Cloning and molecular characterization of the novel human melanin-concentrating hormone receptor MCH2. Mol Pharmacol. 2001;60:632–639. [PubMed] [Google Scholar]
- Roy M, David NK, Danao JV, Baribault H, Tian H, Giorgetti M. Genetic inactivation of melanin-concentrating hormone receptor subtype 1 (MCHR1) in mice exerts anxiolytic-like behavioral effects. Neuropsychopharmacology. 2006;31:112–120. doi: 10.1038/sj.npp.1300805. [DOI] [PubMed] [Google Scholar]
- Saito Y, Cheng M, Leslie FM, Civelli O. Expression of the melanin-concentrating hormone (MCH) receptor mRNA in the rat brain. J Comp Neurol. 2001;435:26–40. doi: 10.1002/cne.1191. [DOI] [PubMed] [Google Scholar]
- Takahashi A, Tsuchiya K, Yamanome T, Amano M, Yasuda A, Yamamori K, Kawauchi H. Possible involvement of melanin-concentrating hormone in food intake in a teleost fish, barfin flounder. Peptides. 2004;25:1613–1622. doi: 10.1016/j.peptides.2004.02.022. [DOI] [PubMed] [Google Scholar]
- Verret L, Goutagny R, Fort P, Cagnon L, Salvert D, Leger L, Boissard R, Salin P, Peyron C, Luppi PH. A role of melanin-concentrating hormone producing neurons in the central regulation of paradoxical sleep. BMC Neurosci. 2003;4:19. doi: 10.1186/1471-2202-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volff JN. Genome evolution and biodiversity in teleost fish. Heredity. 2005;94:280–294. doi: 10.1038/sj.hdy.6800635. [DOI] [PubMed] [Google Scholar]
- Wullimann MF, Mueller T. Teleostean and mammalian forebrains contrasted: evidence from genes to behavior. J Comp Neurol. 2004;475:143–162. doi: 10.1002/cne.20183. [DOI] [PubMed] [Google Scholar]
- Wullimann MF, Rupp B, Reichert H. A topological atlas. Basel: Birkäuser Verlag; 1996. Neuroanatomy of the zebrafish brain. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.