A. Introduction
Medicinal chemists today find themselves in an increasingly information-rich environment. An abundance of compound activity and affinity data is being published, and medicinal chemistry data are increasingly connected with a broader world of data from the realms of bioinformatics and systems biology. In recent years, a number of publicly accessible, chemistry-oriented databases of interest to medicinal chemists have been established to facilitate access to medicinal chemistry data and their biological links, with the aim of accelerating the discovery of new medications. In order to maximize their usefulness, it is important that researchers in pertinent fields be fully aware of these resources and exploit their full potential.
Decades of growth worldwide in the pharmaceutical industry and of academic drug discovery efforts, along with technological advances that speed compound synthesis and assays1 , and the advent and growth of the related fields of chemical biology and chemical genomics, have led to an ongoing flood of publications with valuable data regarding new compounds and their biological activities. On the order of 20,000 - 30,000 new compounds are now published per year in some of the main medicinal chemistry journals, and this rate has accelerated in recent years (as detailed below). However, publication in conventional journals traps data in a form where they are inaccessible to computer search and retrieval. For example, it is not possible to search standard scientific articles for compounds of interest or to reliably extract machine-readable representations of compounds from chemical drawings in articles. As a consequence, the conventional publishing paradigm can severely restrict the discoverability and usability of medicinal chemistry data.
The parallel growth of information technology and the emergence of the World Wide Web in the 1990’s have created important new opportunities for dissemination of data. Biologists – especially structural and molecular biologists – seized these opportunities, establishing central data resources like the Protein Data Bank2 and GenBank3 and laying the foundations for the field of bioinformatics. The first public protein-ligand database aimed at serving the drug discovery community, BindingDB, came on line in late 2000. This resource has grown substantially and has since been joined by other important databases with related scopes and goals. According to Pathguide, a web resource for online databases, there are at least 43 protein-compound interaction databases4, 5 and many other useful, yet free, chemical databases are now available6. Such resources are of increasing value not only for basic uses like finding and downloading structure-activity relationship (SAR) data for a protein target of interest, but also for emergent applications that become possible as the medicinal chemistry dataset grows to provide a comprehensive picture of small molecules in the larger biological context. For example, if a cell-based screen reveals that a new compound inhibits apoptosis, then one might seek similar compounds that bind apoptosis-related proteins, and thus hypothesize that the new compound also binds one of these targets. Similarly, if one is prioritizing several lead compounds for further development, the observation that one lead is similar to a published compound known to bind a different target might lead one to reduce its priority, to minimize off-target effects. In another scenario, marking all the proteins in a defined signaling pathway according to which ones already are targeted by FDA-approved drugs might lead to suggestions for a multidrug therapy to maximally suppress signaling.
Here, we aim first to help medicinal chemists take advantage of the growing array of freely accessible medicinal chemistry-oriented databases by discussing three central resources focused on small molecule binding and bioactivity, BindingDB, ChEMBL and PubChem, and noting as well several other small molecule databases that are also of great value. (Readers interested in additional perspectives will enjoy other recent reviews7-12). In particular, Section B seeks to help users over the initial barriers encountered when one starts to use these rather complex resources, by summarizing information their organization and methods of accessing key types of data, information that is not always easy to glean from their respective web-sites. Subsequent sections then offer broader discussions of the field, and some readers may wish to jump directly to Section C, which uses the available medicinal chemistry data to derive interesting overviews of the available medicinal chemistry data; or to Section D, which offers views towards the future of online compound databases and their applications, including the possibility of integrating related databases to minimize overlapping efforts, addressing the challenge of getting data into databases where they can be most useful, and the role of medicinal chemistry databases in systems biology and systems pharmacology.
B. Databases of small molecule binding and bioactivity
This section is intended to help medicinal chemists understand and start using BindingDB, ChEMBL and/or PubChem. It provides an overview of what each database contains and how the information is structured, since this is important for effective use; explains how to perform basic tasks; and notes special capabilities. We envision the new user accessing these web-sites with the present article as a guide. This section also includes thumbnails of a number of other medicinal chemistry-related databases that readers are likely to find useful.
1. BindingDB
History, focus and content
BindingDB (www.BindingDB.org) began in the late 1990s at the University of Maryland as apparently the first publicly accessible affinity database. Since its inception, BindingDB has collected primarily protein-small molecule binding affinity data. In particular, BindingDB focuses on quantitative data, such as Ki, Kd and IC50 measurements where there is a well-defined protein target. As of April, 2012, BindingDB contains 793,068 binding data from 5,583 protein targets and 349,917 small molecules. These holdings include about 60,000 data that have been manually extracted from journals by curators at BindingDB, including some sets that have been submitted by authors. The entries collected by BindingDB curators directly from the literature contain a particularly high level of detail regarding assay conditions such as pH, temperature, and buffer composition. A large fraction of the data in BindingDB are merged in from other open databases listed below, notably ChEMBL13, 14 and PubChem12, 15-17, as well as PDSP Ki18, 19. In each case, BindingDB carries out additional processing to ensure that all imported data meet current BindingDB criteria. For example, BindingDB imports only those measurement data from ChEMBL that include a well-defined protein target (TARGET_TYPE=’PROTEIN’). For PubChem, BindingDB imports only quantitative affinity data (i.e., Confirmatory Assays; see below). In the case of PDSP, it is sometimes necessary to supplement the existing data, such as with a manually curated protein sequence or a machine-readable representation of the ligand. It is worth noting that few if any public database projects have the internal resources to systematically check all incoming data for possible errors. BindingDB therefore sends emails to authors inviting them to check their own data as presented on the BindingDB website and report any errors for correction. Indeed, readers of this article are also invited to find their data in BindingDB at the Author page http://www.bindingdb.org/bind/ByAuthor.jsp, and to send in any corrections that may be needed.
Browsing, querying, and downloading capabilities
BindingDB offers a range of methods to find and access data; some of the most broadly useful ones are described in video tutorials available through the BindingDB homepage. One of the simplest ways to find data in BindingDB is to type any text of interest into the Full Search box at the top of the home page. This generates a powerful Google-type search for related data in compound names, protein names, article titles, assay descriptions, and author names. Wild-cards are allowed here; for example, adeny* yields hits to any word starting with “adeny”. Following the links to data in the resulting hit-list leads to a comprehensive Results Table (http://bit.ly/ws4vLt)20, where each row contains one target-ligand pair along with a rich set of links to further data on the target, the ligand, and the target-ligand combination (see below), as well as connections to further details, compound availability, and information on the origins of the data. The links on the left-hand side of the main web-page provide for more specific access to data, according to targets, compounds, citations, and protein sequence and structure. Highlights of these capabilities are now presented.
Targets
The Name link under Targets provides an alphabetical list of protein targets with direct links to data in the Results Table and to Articles. The Target list makes it easy to download an SDfile with all the compounds and affinity data for any protein target, with either 2D or computed 3D coordinates. One can, moreover, search by target name in conjunction with various conditions, such as IC50 range (http://bit.ly/AyOWyq)21, molecular weight, etc. (http://bit.ly/AAUiVz)22. Finally, BLAST sequence search23 can be used to find data for targets of interest (http://bit.ly/xuN2IY)24.
Compounds
Users may draw a compound or paste in a SMILES string with the ChemAxon plugin and then search for data in BindingDB by compound, substructure and chemical similarity. These searches may include filters by affinity range, molecular weight, target name, etc. (http://bit.ly/zL842y)25. One may query BindingDB with multiple compounds simultaneously, via the batch search page (http://bit.ly/w0A1G5)26. BindingDB also provides access to binding data based on the names of 3431 FDA-approved drugs, through cross-referencing of the Drugs@FDA database27. For example, BindingDB has about 60 measurements for nifedipine, the active ingredient of the calcium-channel blocker Adalat (http://bit.ly/wXIziD)28.
Citations
BindingDB allows users to view all the data associated with a particular author (http://bit.ly/wHLXDl)29, article (http://bit.ly/ydpChT)30, or institution (http://bit.ly/yMYvx2)31. In addition, the pull-down menus on the Journal/Citation page provide immediate links to SDfiles with all the compounds and affinity data for each available article. Users of web-based reference managers may directly import citations for data of interest in BindingDB web-pages. As detailed on the BindingDB home page, Zotero uses a Firefox extension, Cite-U-Like uses a Bookmarklet browser plug-in, and Mendeley uses a Web Importer plug-in.
Protein structure
The Protein Data Bank (PDB)2 contains the three-dimensional structures of a number of protein-ligand complexes for which binding data are available in BindingDB, and one may search BindingDB by PDB ID or HET ID, allowing matches for either 85% or 100% BLAST 23 sequence identity (http://bit.ly/zVd6z232, http://bit.ly/x9f9Yd33, respectively).
Users may download the entire BindingDB database as an Oracle data dump, or as an SD File which includes not only the compounds but also activity data, such as targets and affinities (http://bindingdb.org/bind/chemsearch/marvin/SDFdownload.jsp). Also available on this download page are proteins in FASTA format and other specialized datasets, and opportunities are provided on various web pages within the site to download subsets of data, such as all data for a given target protein, or all data from a given article, again in the form of data-rich SDfiles. These can be imported directly into chemical viewers and spreadsheets. The data are provided under the nonrestrictive Creative Commons Attribution-ShareAlike 3.0 Unported license34.
Linking with other databases
The BindingDB Results Table provides an array of links to further information about each binding measurement and the molecules involved. In each row, links for the Target, the Ligand, or the Target and Ligand together, are presented in separate columns (e.g. http://bit.ly/zq9oW335 and see Figure 1). For example, all proteins for which structural information is available are linked to the appropriate entries in the Protein Data Bank (PDB). The biological role of each Target may be explored by following links from Targets to corresponding pathways in systems biology databases including Reactome36, KEGG37, and NCI’s Pathway Interaction Database38; the broader concept of linking compound databases with systems biology to support systems pharmacology is discussed below. Links are also provided from BindingDB data to the corresponding articles in PubMed, as well as into related databases, including many of those discussed in the present article. Finally, in 2011, BindingDB began providing links from ligands to matching compounds in the ZINC database of commercially available compounds, http://zinc.docking.org39, in order to help users obtain physical samples of compounds for further experimental study.
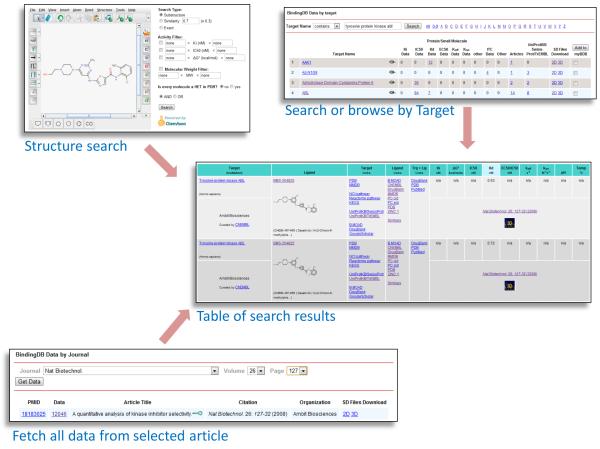
Figure 1.
Collage of selected tools for finding data in BindingDB, along with sample search results. See text for further details.
In addition, a number of databases provide links from their data to relevant data in BindingDB. For example, PDB users will find links to BindingDB from structure entries for which affinity data are available, such as PDB entry 2GQG. Similarly, one may navigate from articles in PubMed to the corresponding data in BindingDB, for viewing and download, by expanding PubMed’s LinkOut options and following the one to BindingDB, such as on the following page: http://www.ncbi.nlm.nih.gov/pubmed/17718712.
Special tools and datasets
BindingDB also provides a number of web-based tools and data subsets to help users take advantage of this large data collection. For example the Find my Compound’s Target page (http://bit.ly/zX0SfQ)40 allows one to identify possible targets of new compounds. One draws a compound or uploads a file with multiple compounds, and BindingDB reports all protein targets known to bind similar compounds. This capability can be used to predict off-target binding, and hence side-effects, of a new compound. It can also be used to generate hypotheses regarding the mechanistic targets of compounds found to be active in an empirical bioassay, such as a cell-based screen.
BindingDB also provides several online virtual screening methods allowing one to select a group of compounds in BindingDB that are known to be active against a given target protein and use them as a basis for discovering other potential actives in an uploaded compound library. The simplest and faster method, Maximum Similarity, ranks the uploaded compounds according to their maximum similarity to any of the known actives. This method uses Tanimoto similarity based upon JChem41, 42 fingerprints. A second method, Binary Kernel Discrimination (BKD), uses a training set of compounds to produce a model that can then be applied to the structures of other compounds in order to predict their likely activity43. Here, the actives are divided into reference and training sets of equal size. Each set is then supplemented with 500 other drug-like compounds presumed to be inactive, and JChem binary fingerprints are computed for all compounds. The BKD comparison is used to rank the test-set compounds based on the reference set and the enrichment of actives at the top of the ranked list is reported in order to provide the user with information on the predictivity of the BKD model. If the user wishes to proceed, based on these results, then the reference and training sets are combined into one large reference set and used to rank a large set of compounds uploaded by the user. A third method44 uses the Support Vector Machine (SVM) machine-learning approach45. This divides the first 100 actives into training and test sets, and again supplements these with 500 other compounds presumed to be inactive. Here, however, numerical descriptors, rather than binary fingerprints, are computed for each compound. The training set is used to set up an SVM model that will discriminate actives from inactives, and this model is evaluated with the test set. The results are reported, and, if the user wishes to proceed, then descriptors are computed for the user’s uploaded compounds and the SVM model is applied to rank them. It is worth noting that each of these methods has both strengths and weaknesses, and users are free to download the data from BindingDB and apply their own approaches.
In order to support the parameterization and validation of algorithms for computer-aided ligand discovery, BindingDB provides a series of validation sets manually curated from the larger data collection (http://bit.ly/yTctqN)46. Each validation set comprises a series of congeneric compounds with measured affinities for one protein target, where the crystal structure of at least one compound in the series has been solved in a complex with the target. To support more basic studies of molecular recognition, BindingDB also houses a small collection of affinity data for small, non-protein receptors and their ligands (http://bindingdb.org/bind/HostGuest.jsp).
Finally, BindingDB has also begun an initial implementation of a personalization aspect of the database, named myBDB (http://bindingdb.org/mybdb/login.jsp). This feature allows registered users to save searches for subsequent visits to the resource.
2. ChEMBL
History, focus and content
The ChEMBL database (www.ebi.ac.uk/chembl/) began as a set of commercial products known as StARlite, CandiStore, and DrugStore14 (http://chembl.blogspot.com/). With funding from The Wellcome Trust, these were essentially moved to the public domain (see below) under the aegis of the European Bioinformatics Institute, an outpost of the European Molecular Biology Laboratory near Cambridge in the UK. ChEMBL’s outsourced curation effort captures a broad range of medicinal chemistry data from the scientific literature. These include biological activities, such as cell-based assay data, and protein-ligand affinities, although ChEMBL’s curation of binding data does not include details like buffer composition and experimental conditions. About 40% of ChEMBL data are imported from PubChem, and the database also includes several large screening datasets (below). As of April, 2012, the ChEMBL database contains about 7 million measurements for 1.1 million compounds and 8,900 protein targets.
Browsing, querying, and downloading capabilities
A search bar on the ChEMBL home page (https://www.ebi.ac.uk/chembl/) provides direct access to searches by name and certain database IDs for Compounds, Targets or Assays. Here, a Target may be not only a protein, but also an entire organism, such as the yeast Candida albicans in the case of an antifungal bioassay. A series of tabs along the top of the home page provide access to a range of more detailed search and browsing options, organized primarily by Compounds and Targets. Highlights of these capabilities are now summarized.
Targets The Protein Target Search tab allows for sequence-based searches with BLAST. These yield a table of Targets with their BLAST scores, with links to the UniProtKB protein database and further information in ChEMBL. The Browse Targets tab enables intuitive browsing of protein targets through a hierarchy of protein types (e.g., enzymes and ion channels), or a taxonomy of organisms, where, again, a Target may be an organism or a protein from an organism. The results of a Target search are presented in a table with UniProtKB IDs, gene names, and information on how many compounds and activity data are associated with each Target. A pull-down menu at the top right of the table allows one to access the bioactivity data, optionally filtered according to parameters such as IC50 range. Alternatively, one may click on the name of a Target of interest in order to view a richly informative Target Report Card, as described below.
Compounds
The Compound Search tab allows one to draw a compound or fragment with a choice of the JME47, Marvin48, or JDraw49 sketcher, and search ChEMBL by identity, similarity or substructure. Alternatively, one may search ChEMBL for a list of compounds by pasting multiple SMILES50 strings into a text window. Any of these searches leads to a compound table, where clicking on a compound leads to an informative Compound Report Card (below). The compound table is also equipped with a pull-down menu allowing all or selected compounds to be downloaded as an SDfile containing the molecular structures or as a table of compound IDs with various computed descriptors, such as molecular weight and computed logP estimates. The pull-down menu also provides access to the bioactivity data for the selected compounds, as described above for Targets. An appealing alternative to the compound table display is provided in the form of scatter plots of computed compound properties, with color coded data points linked back to compound data. An additional Browse Drugs tab on the ChEMBL homepage focuses on the subset of ChEMBL compounds that are marketed drugs and provides commercial and pharmaceutical information such as a compound’s approved drug name and its route of administration.
The Report Card format is a distinctive feature of the ChEMBL site (Figure 2). Thus, clicking on a Compound in a search result table leads to a Compound Report Card, which provides a range of additional information, such as names and database identifiers, links to clinical trial information, computed properties, and links to the same compound in other databases, such as DrugBank, PubChem, and the Protein Data Bank in Europe, PDBe51. Importantly, a set of pie-charts and associated links at the bottom of the Compound Report Card provide direct access to bioactivity and other data for this compound in ChEMBL. Similarly, clicking on a Target in a ChEMBL result table leads to a Target Report Card, which contains not only further Target identifiers and links, but also histograms of molecular weight, AlogP and polar surface area for the compounds tested against this Target. One may navigate to a result table for all compounds tested against this Target, or else choose the range of a compound parameter by clicking on histogram bars and then generating a table of results for only compounds within this range. Analogous Assay Report Cards and Document Report Cards provide details of assay techniques and the documents from which ChEMBL data are drawn.
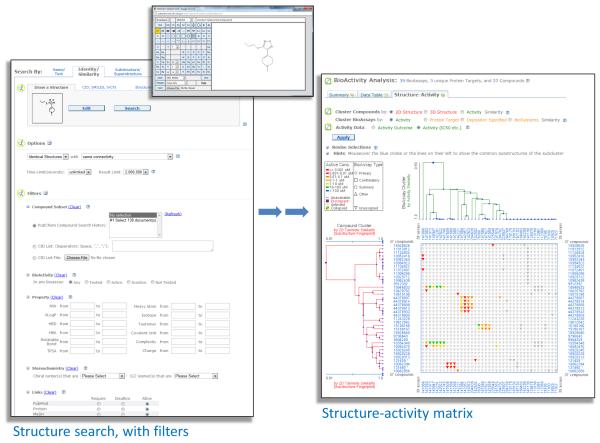
Figure 2.
Sample of a ChEMBL Target Report Card (A), Compound Report Card (B) and Document Report Card (C). Only the top portion of each Card is shown, due to space limitations. See text for further details.
Each release of ChEMBL is freely available from an FTP server in a variety of formats, including Oracle 9i, 10g, 11g; MySQL; an SD file of compound structures; and a FASTA file of the target sequences. The data are provided under the nonrestrictive Creative Commons Attribution-ShareAlike 3.0 Unported license.
Linking with other databases
ChEMBL data are cross-linked with a number of other molecular databases, primarily through the various ChEMBL Report Cards (above). For example, Target Proteins are linked to three-dimensional structure data in PDBe and sequence data and annotations in Ensembl52 and UniProtKB53; Compounds are linked to ChemSpider11, 54, DrugBank3, 55, PDBe, PubChem, Wikipedia, and ChEBI56, EBI’s compound database. Articles described in Document Report Cards are linked primarily to EBI’s publicly accessible journal database CiteXplore. In turn, CiteXplore’s listing of each medicinal chemistry article includes a list of compounds in the article, each with a link to a ChEMBL Compound Report Card. Similarly, compounds in protein crystal structures are linked from PDBe to Compound Report Cards in ChEMBL.
Special tools and datasets
ChEMBL is attuned to applications in drug discovery and pharmaceutics. For example, as noted above, it provides tabular and graphical displays of a variety of computed compound properties relevant for drug design. Another unique tool is the DrugEBIlity service, which uses structural data to evaluate whether a protein can be targeted with small molecules (https://www.ebi.ac.uk/chembl/drugebility/structure). One may upload a PDB-format structure file, choose an existing PDB ID, or use BLAST to find similar proteins of known structure as a basis for this evaluation. In addition to druggability ratings, the server also provides a graphical display of the protein’s potential binding sites. ChEMBL also includes a Drug Approvals tab with information on new FDA drug approvals 2009-2011 (“Orange Book” data); and Compound Report Cards include links to clinical trials data (clinicaltrials.gov), when available. Finally, the Kinase SARfari and GPCR SARfari tools provide alternative access portals to Target, Compound and activity data for two key families of therapeutic targets that are well represented in the ChEMBL database.
ChEMBL hosts a series of special datasets related to tropical pathogens in its ChEMBL-NTD (neglected tropical diseases) pages (https://www.ebi.ac.uk/chemblntd/). The datasets, which comprise thousands of compounds, are the result of compound screening campaigns, typically against whole Plasmodium and Trypanosoma organisms, from GSK57, Novartis-GNF58, St. Jude Children’s Research Hospital59, and Drugs for Neglected Diseases Initiative (http://www.dndi.org/).
3. PubChem
History, focus and content
The PubChem database (pubchem.ncbi.nlm.nih.gov)16, 17, 60 is a United States government initiative started in 2004 by the National Institutes of Health within the National Center for Biotechnology Information (NCBI). Its broad goal is to collect and disseminate information on the biological activities of small molecules. PubChem focused initially on assay data from the high-throughput compound screening programs supported by NIH’s Molecular Libraries Roadmap Initiative. However, it also accepts chemical structures and assay data from other sources, and such depositions have substantially expanded PubChem’s data collection. For example, although the PubChem initiative does not include the extraction of activity data from journal articles, PubChem’s incorporation of the BindingDB and ChEMBL datasets allows it to provide access to a large body of literature data. PubChem currently houses about 33 million distinct chemical entries (http://pubchem.ncbi.nlm.nih.gov/help.html#faq); activity data drawn from about 4,800 NIH Molecular Libraries assays, 45,000 journal articles, and several hundred other sources, such as pharmaceutical companies and individual research groups.
In order to make effective use of PubChem, it is helpful for one to have a basic knowledge of its conceptual framework. First, the information in PubChem is organized into Compounds, Substances and BioAssays. A given chemical can be listed as both a Compound and a Substance, where the Compound listing is its single standard representation, while the Substance listing corresponds to the specific material used in a given BioAssay. Thus, a given Compound can correspond to multiple Substances, and there are about three times as many Substances as Compounds in PubChem. It is the Compound listings that will generally be most meaningful to PubChem users. It is also worth noting that PubChem includes many Compounds for which there are no BioAssay data. All activity and binding data, including those drawn from the literature, are represented in terms of BioAssays. There are three types of BioAssay record: Summary, Primary and Confirmatory. A Summary record contains an overview of a given experiment. A Primary record contains results of a primary screen in which each compound is listed simply as Active or Inactive at a given concentration (e.g., 10 μM). A Confirmatory record reports the effective concentrations (e.g. IC50s) of compounds found to be Active in a Primary screen, based on a multi-concentration dose-response study. For BioAssays with well-defined protein targets, target information is provided through seamless links to the NCBI protein database.
Browsing, querying, and downloading capabilities
The PubChem homepage provides immediate access to text-based searches within BioAssay, Compound and Substance listings. (As noted above, the Compound listings are in general more useful than Substances.) One may also click through to a comprehensive tool for chemical structure searches (below). An Advanced Search link on the homepage allows for more fine-tuned searching capabilities of each category (Figure 3). Clicking on the BioActivity analysis or Bioactivity summary links leads to a particularly useful BioActivity Services page (http://pubchem.ncbi.nlm.nih.gov/assay/), offering Compound-centric, Target-centric, and Assay-centric query tools. Several useful paths into this rich dataset are now described.
Figure 3.
Tools for searching PubChem by chemical structure along with a variety of filters (left), and an interactive structure-activity relationship matrix, with Compounds listed along the left and BioAssays along the top.
Target
There are at least three ways of accessing compound and activity data for a given protein target. One is to choose the Target-centric option on the BioActivity Services page, and enter the name of one’s protein of interest, such as, Rin1, into the Search by protein family text box. This search leads to a list of BioAssays for this protein target with an array of information, including the number of Active compounds by various criteria. Clicking on these numbers leads to data tables showing compounds and their activities, along with tools for downloading the data in various formats, such as comma-separated value (CSV) with a database id for each compound. The compound activity table also provides tools for plotting the data and gaining an overview of the compounds and their activities through clustering and dendrogram displays.
A second way of accessing data for a given protein target is to enter through the NCBI Protein database. For example, one may search the NCBI Protein database for Rin1. This leads to a list of hits, where one may check the box for the Homo sapiens variant and then, on the right, choose PubChem BioAssay from the Find Related Data pulldown menu. This reveals a further Option pulldown menu, where one may choose Bioassay by Target (identical proteins). This in turn leads to a list of BioAssays involving Rin1 from which one may choose the Confirmatory BioAssay and thus access a BioAssay Summary page for this quantitative dataset. Clicking on either Show Data Active or All leads again to a table of compounds and their activities, as described above.
Compound
Chemically oriented compound searches are available at the Structure Search page (http://pubchem.ncbi.nlm.nih.gov/search/), which allows queries by chemical identity, similarity, and substructure; molecular formula; and three-dimensional structure similarity. Tools are also provided to filter Compounds by many criteria, such as computed chemical properties and depositor. Results are displayed as a list of compounds with a range of navigation options. Simply clicking on a compound leads to a Compound Summary page, described below. Alternatively, one may search more directly for BioAssay data for compounds of interest via the BioActivity Services page. From here, hits go directly to a data table of compounds and activities (see above).
Bioassay
PubChem contains a wealth of information on high-throughput assay methods for various protein targets and bioactivities. For example, if one wishes to learn about assays for protein Rin1, one may type this protein name into the first text box under the Assay-centric tab on the BioActivity Services page. This leads directly to a list of high-throughput assays involving the protein of interest.
PubChem provides an informative Summary page for each Compound and BioAssay, similar in spirit to the ChEMBL Report Cards (above). A Compound Summary page (e.g., http://1.usa.gov/x3eREX)61 provides 2D and sometimes 3D representations of the Compound, along with a wealth of additional information and links. These include alternative identifiers, such as synonyms, InChI Identifier and SMILES; computed characteristics such as molecular weight, XlogP, number of H-bonding groups; links to BioAssays results for this Compound, toxicity information from the National Library of Medicine ChemIDplus resource, and representations of the Compound in vendor catalogs and other databases; and links to similar Compounds within PubChem. The precise content of a BioAssay Summary page depends upon the assay type. In general, a BioAssay Summary (e.g., http://1.usa.gov/x2nfRP)62 provides a direct link to the assay data from a Show Data link near the top of the page, followed potentially by information on the protein Target and information on Compounds tested and found active, and information on the assay itself, often including a detailed protocol. An array of links lead to further information, such as related BioAssays and Targets.
Many PubChem pages offer downloads of data subsets, while an FTP server (ftp://ftp.ncbi.nlm.nih.gov/pubchem) allows users to download complete listings of Compounds, Substances, BioAssays, and associated information. Users are referred to the originally submitters of the various dataset for any possible license terms (ftp://ftp.ncbi.nlm.nih.gov/pubchem/README).
Linking with other databases
As a component of the NCBI, PubChem is tightly integrated with the other bioinformatics databases available at the NCBI website, such as those for gene sequence, protein sequence and structure, gene expression, and the scientific literature, via bidirectional links which allow seamless navigation across NCBI resources. The relatively new BioSystems component of NCBI63 (http://www.ncbi.nlm.nih.gov/biosystems) places protein Targets into the context of biomolecular pathways and other functional groupings, such as structural complexes, and includes links to external resources like KEGG64, 65 and Reactome36, 66. Protein targets are also linked to the curated NCBI Conserved Domain Database (CDD)67 (http://www.ncbi.nlm.nih.gov/cdd) and to three-dimensional structures of closely related proteins contained in the NCBI Molecular Modeling Database (MMDB)68 (http://www.ncbi.nlm.nih.gov/structure). Such links help to identify and characterize conserved binding sites in proteins. Many other external links are also provided. For example, Compound Summary pages provide links to external information in categories like Use and Manufacturing, Safety and Handling, Chemical Vendors, etc., when available. For data imported to PubChem from other databases, such as ChEMBL and BindingDB, PubChem includes links to the corresponding information in those resources.
Special tools and datasets
PubChem offers a unique set of tools for analyzing groups of Compounds. For example, a Compound search (e.g., by similarity to a drawn structure and optionally with filters according to activities and computed properties; http://pubchem.ncbi.nlm.nih.gov/search/) leads to a page with a list of Compounds meeting the search criteria. One may then use check-boxes to select any or all of these compounds, and then, on the right-hand side of the page, choose BioActivity Analysis, Structure Clustering, or a link to biomolecular pathways involving the selected Compounds. Choosing BioActivity Analysis, and then the Structure-Activity tab, leads to an interactive heat-plot showing the activities of the Compounds against multiple BioAssays, along with a hierarchical clustering of Compounds by chemical similarity and of BioAssays by Compound activity profiles17 (Figure 3).
As noted above, PubChem focuses in particular on high-throughput screening data from the Molecular Libraries Screening Centers Network (MLSCN), ten centers with a diverse set of screening platform technologies. The MLSCN is a component of the NIH Molecular Libraries Roadmap, and along with the Molecular Libraries Probe Production Centers Network (MLPCN), with nine centers, offers biomedical researchers access to their large-scale screening capabilities, along with medicinal chemistry and informatics aimed at discovering chemical probes to explore the functions of genes and signaling pathways in health and disease69. The molecular libraries centers are NIH’s New Pathways to Discovery initiative, which aims to advance the understanding of biological systems. The unique high-throughput assay data in PubChem obtained directly from these screening centers are not typically present in the published literature.
4. Other Small Molecule Databases of Interest
There are dozens more chemically oriented databases of potential interest to medicinal chemists. Several noteworthy ones are summarized alphabetically below.
Binding MOAD (bindingmoad.org) gathers high-quality protein-ligand structures from the PDB (about 17,000 currently) and annotates as many as possible (about 5,600 currently) with measured binding affinity data collected from the scientific literature.70-72 BindingMOAD is thus particularly relevant to structure-based drug discovery. One may browse and search structure and affinity data via a protein classification, PDB ID, enzyme classification number, keyword or author.
ChemSpider (www.chemspider.com) is a freely accessible chemical database54 containing more than 26 million distinct molecules with links to information about properties and availability in over 400 data sources, such as compound catalogs and databases, including many of those listed in this article. The web interface uses a crowdsourcing approach to expand and improve the data set, by allowing users to enter or correct entries. One may query by, for example, compound name, structure, database identifier, and computed properties; available information for each compound includes names, properties, spectra, vendors, data sources, and patents.
DrugBank (drugbank.ca)3, 55, 73 is a smaller but richly annotated public database of approved and experimental drugs, including a total of about 6,700 small molecules and biopharmaceuticals. One may browse and query by, for example, structure, pathway, protein sequence, and drug interactions. The data set includes pharmacological and pharmacokinetic data, dosage forms, solubilities, drug-drug interactions, metabolism information, target and pathway data. An extensive set of downloads is provided.
GRAC and IUPHAR-DB (http://www.guidetopharmacology.org74, http://www.iuphar-db.org)75, 76 are two complementary and integrated databases which collect a range of pharmacological information on GPCRs, ion-channels and nuclear receptors from the primary literature. These data, which include small molecule activities and affinities, are reviewed by expert international subcommittees and consultants and are linked to related information in other online resources. These databases currently house data for about 1,800 small molecules and 600 different proteins spanning the targets of about half of all current licensed drugs.
PDBbind (pdbbind.org.cn and pdbbind.org)77-79, like BindingMOAD above, collects measured affinities for many types of complexes in the PDB, including protein-small molecule, protein-protein, and nucleic acid-small molecule systems. The current version at pdbbind-org.cn provides about 8,000 data, of which about 6,000 are for protein-small molecule complexes, and is free for academic and commercial use, on acceptance of a license agreement.
PDSP Ki (pdsp.med.unc.edu), the database of the Psychoactive Drug Screening Program at University of North Carolina19, contains about 55,000 binding measurements for 7,500 drugs and other compounds with 740 receptors, neurotransmitter transporters, ion channels, and enzymes. The query interface is based primarily on pulldown menus and Ki limits. At no cost to academics engaged in mental health research, the same group provides experimental compound screening services with a variety of assays, including bioavailability predictions (e.g., CaCo2) and cardiotoxicity (e.g., HERG).
SuperTarget (http://insilico.charite.de/supertarget/) provides various views of over 330,000 interactions involving about 6,000 targets and 200,000 compounds, along with annotated pathway diagrams and the ability to browse for targets categorically, such as by function and cellular location80, 81.
Therapeutic Targets Database (http://bidd.nus.edu.sg/group/cjttd/TTD_HOME.asp)82 focuses on proven and prospective drug targets and their associated drugs and candidate drugs, providing extensive links to related information, such as sequence and pathway data. Both text-based and chemical similarity searches are supported, and many datasets may be downloaded.
ZINC (zinc.docking.org), based at the University of California, San Francisco, is a free database of over 21 million commercially available compounds39. Compounds are organized into various subsets, such as target-focused, natural products, metabolites, lead-like and fragment-like, and are annotated with the time-frame for their availability. Small arbitrary subsets may also be assembled by the user. Compounds are downloadable in popular molecular docking formats with precomputed three-dimensional conformations, in order to facilitate virtual structure-based screening.
C. Trends in Medicinal Chemistry Data
The combined holdings of BindingDB, ChEMBL and PubChem enable a broad overview of trends in published medicinal chemistry data. Here, we examine rates of data production overall and by journal and institution, as well as statistical distributions of, for example, compound molecular weight and compounds per target protein. Clearly, many other analyses are also enabled by these resources.
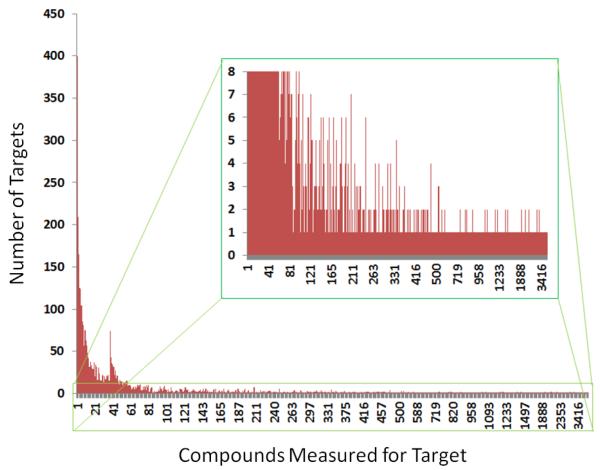
The number of unique small molecules published annually has increased year on year since 2008 (Figure 4), while the number of protein-small molecule binding measurements has followed a similar trend but at a higher level (Figure 5). (Note, however, that although ChEMBL has sought to exhaustively curate the core medicinal chemistry journals, it is not guaranteed that all articles in the targeted journals were captured every year.) The difference between these two quantities implies that multiple measurements are available for some compounds, and this relationship is depicted in Figure 6 for the data in BindingDB. Although nearly 180,000 compounds have only one measurement, about 80,000 have two, about 40,000 have three, and so on. In fact, as shown in the inset, there is a long tail in this distribution, due to a small number of compounds with tens or hundreds of measurements apiece. These outliers are mainly kinase inhibitors which have been tested against many mutants of many kinases, but several other classes are also represented there. The distribution of the number of compounds studied per target is depicted in Figure 7. Not surprisingly, there are many targets, such as neurotransmitter receptors, clotting factors and kinases, against which hundreds and even thousands of compounds have been tested. The bump in the distribution at about 40 compounds per target appears to result from the reuse of several compound panels in various assays. Further details of these distributions are available at the BindingDB web-site (http://bit.ly/uu6ZNn83, http://bit.ly/uz9HeV84). Finally, it is interesting to observe that, since 2004, the distribution of compound molecular weights has sharpened dramatically, with more between 200 and 600 Da, and most in the 200-400 Da range (Figure 8).
Figure 4.
Trends in published unique small molecules by year. The data are the union (JChem 5.2 full structure search) of the holdings of BindingDB and ChEMBL for the four medicinal chemistry journals with the most data.
Figure 5.
Trends in published unique small molecules and associated binding data by year. The data are the union (JChem 5.2 full structure search) of the holdings of BindingDB and ChEMBL across 34 curated journals.
Figure 6.
Number of binding measurements per compound in BindingDB. For example, there are nearly 180,000 compounds with one binding measurement. Inset shows the long tail of the distribution, which contains a few compounds with hundreds of measurements each.
Figure 7.
Number of protein targets in BindingDB having a given number of compounds for which affinities were measured. For example, there are about 400 targets for which one compound’s affinity was tested. Inset shows the long tail of the distribution, which contains targets for which hundreds or thousands of compounds have been measured.
Figure 8.
Molecular weights of new compounds in ChEMBL, BindingDB and PubChem BioAssays, by year.
The number of new compounds in BindingDB and ChEMBL from the most highly represented journals each year is examined in Figure 4. (The analogous breakdown of new measurements per year parallels new compounds closely, though at a higher level, and is therefore not shown.) There is a rather consistent laddering of journals by the numbers of new compounds they publish, with most in Bioorg. Med. Chem. Lett., followed by J. Med. Chem and Bioorg. Med. Chem. Interestingly, although the total number of new compounds per year was rather level from 2004 to 2008 (Figure 4), this overall trend masked a drop in new compounds in J. Med. Chem. and a rise in compounds in Bioorg. Med. Chem. and Eur. J. Med. Chem. However, since 2008, the number of new compounds in all of these journals has risen together, and their relative shares have not changed appreciably.
Perhaps surprisingly, academia generates nearly half of the medicinal chemistry data in the combined holdings of BindingDB, ChEMBL and PubChem BioAssays (Figure 9). It is unlikely, however, that this distribution reflects the volume of data actually generated in these two sectors, as many corporate data are not published. Note, too, that about one third of the academic data derive from screening centers, such as The Scripps Research Institute Molecular Screening Center and the New Mexico Molecular Libraries Screening Center.
Figure 9.
Institutional sources of articles (A) and compound activity data (B) in BindingDB, PubChem BioAssay, and ChEMBL. These data include only measurements with a defined protein target. Each confirmatory PubChem BioAssay is counted as an article. Institutional sources were obtained based on keywords (e.g. “university”, “institute”) in the Affiliation information in PubMed article entries, and were spot-checked by hand.
D. Directions
1. Strengthening the databases
Coordination among databases
The existence of several publicly accessible medicinal chemistry databases provides substantial benefits, while defining a need for coordination to minimize duplication of effort and maximize the value to users. One current benefit is the availability of a diversity of user-interfaces and capabilities to support a range of applications and preferences. Another is a high level sustainability and stability in the face of potential data losses and the uncertainties of continued scientific funding for any single project. There is also a valuable opportunity to distribute the workload of journal curation across projects. Indeed, BindingDB, ChEMBL and PubChem are increasingly sharing data and curation efforts. For example, while ChEMBL’s outsourced curation focuses on core medicinal chemistry journals, such as J. Med. Chem. and Bioorg. Med. Chem. Lett., BindingDB is now engaged in curation of chemical biology journals and others not covered by ChEMBL, such as Chem. & Biol., Nature Chem. Biol., and ACS Chem. Biol. The protein-ligand datasets in the latter journals often are particularly interesting, because they involve proteins which are currently in the process of being identified as candidate drug-targets, or compounds that explore innovative chemistries. To further increase efficiencies, there is now a collaborative effort between BindingDB and ChEMBL to compare each other’s existing data holdings for discrepancies, and thus potentially errors. Ultimately, greatest efficiency and service to users may be achieved by following the models of other large database endeavors. For example, the worldwide Protein Data Bank (wwPDB, www.wwpdb.org)85 comprises four different projects, two in the United States2, 86, one in Japan87, and one in Europe88, which share a core dataset, as well as annotation and validation strategies, while presenting the data differently and with emphasis on different user communities.
Data quality
Data quality is of fundamental importance, and it is of interest to consider the origins and nature of errors in the public medicinal chemistry databases. Data errors may be separated into three classes: scientific errors, errors of transcription, and data handling errors. Scientific errors result from problems with an experiment or its technical analysis. Transcription errors arise during the writing and publication of the data or during the extraction of the data from the publication and its subsequent entry into the database. Data handling errors result from problems at the database itself, such as the introduction of a mismatch between a table of compounds and a table of targets during a database update. A systematic evaluation of the quality of data in these public databases would be of interest, and one could in principle use statistical sampling to characterize overall data quality without having to examine every entry. It would be even more valuable to identify and correct errors throughout these massive datasets, but this would be a much larger challenge. Some of the issues in error checking are now discussed.
Although there is no perfect way to detect scientific errors, it is possible for an expert to judge the suitability of the method reported in the paper, as done, for example, in NIST’s evaluated database of the thermodynamics of enzyme-catalyzed reactions89. Concerns that might be identified in this way could include failure to ascertain the active enzyme concentration90, or reported enzyme inhibition by a compound that is a known aggregator91. Perhaps only the authors of an article can identify transcription errors that are enshrined in their publication, but errors introduced during the extraction of data from an article and their entry into a database can be detected by painstaking comparisons between database entries and associated articles. The same is true for data handling errors, but the latter, once detected, can often be corrected en masse by undoing the database manipulation that generated them. It is worth noting that meaningfully categorizing errors can also be challenging. For example, an error in stereochemistry may not be considered equally severe as an incorrect chemical structure. However, if these two types of error are put into different categories, rather than being lumped together, then more articles will need to be surveyed in order to gather meaningful statistics in both categories. There can also be ambiguities that are difficult to resolve, such as when a paper provides data for a protein target without specifying its subtype; e.g., beta-adrenergic receptor, as opposed to beta-1- or beta-2-adrenergic receptor. Other errors, such as in the name of an author, are significant but do not affect the scientific content of the database.
Evaluating and ultimately correcting the data extracted from tens of thousands of papers will be an enormous undertaking92, 93. Given the limited resources available to these projects, a community effort may be the only way to make inroads. It is in this spirit that the BindingDB project routinely emails article authors inviting them to correct any errors they may find in their BindingDB entries. Perhaps 1-2% of these messages receive a reply, and of these, about a third report an error. Users who notice errors in BindingDB, ChEMBL and PubChem are also invited to submit corrections at www.bindingdb.org/bind/sendmail.jsp, chembl-help@ebi.ac.uk, info@ncbi.nlm.nih.gov, respectively. However, a more systematic approach would be for experts to adopt specific protein targets, overseeing the crowdsourcing of corrections to the associated data10. Similar approaches are used already by Wikipedia, ChemSpider, and the IUPHAR databases.
Linking journals and databases
All of the literature data in these databases are entered by employees or contractors who read each article, extract the pertinent data, and enter it into one of the databases. This labor-intensive curation process is time-consuming and costly, and inevitably introduces errors. The magnitude of these parallel curation efforts is highlighted by the graphs in Figure 4 and Figure 5 and the data production rate will only grow in the coming years, as research in emerging economies accelerates, and technological advances yield a wealth of new candidate drug targets94. The challenge of keeping up with this data flow was the topic of a panel discussion at a recent database conference, which included the leaders of most of the largest databases discussed above, as well as many participants from industry, publishing and government95.
The consensus that emerged is that a new mechanism is needed, in which authors and/or journals make the data in their new articles available in a simple, machine-readable format. For example, authors might provide a file with a list of proten targets, SMILES strings, and affinity data. This could reside in the online supplementary information, or might be uploaded directly to a central web-portal from which any database team could draw those data which fall within the scope of their project. The field of structural biology offers two interesting models. In the case of macromolecular structures, authors routinely deposit their machine-readable structure data into one of the PDB portals so that they may be incorporated into the global wwPDB databases, and journals do not accept papers that report new structures without a PDBid. Small molecule structure data are typically published via Acta Crystallographica Section E, in which each online article is associated with a short crystallographic information file (cif), which users may freely download and use.
In the case of medicinal chemistry data, electronic submission should be quite straightforward, as most authors already have their data in machine-readable format when they are preparing their articles, in the form of spreadsheets and ChemDraw files, for example. The chief challenge for our community might be defining the precise set of data to be uploaded. For example, although it is clear that each compound should be defined, it may not be so clear how much information should be provided about the experimental method and conditions. Regardless of the details, it is clear that joining machine-readable data to every medicinal chemistry article will lead to a medicinal chemistry databases that are dramatically more sustainable, accurate and complete.
2. Next-generation capabilities
The public compound activity databases now provide an informatics foundation on which many new research capabilities can be built. For example, the fact that researchers increasingly read articles on computer screens rather than paper provides an opportunity for tighter integration between journals and databases96. Articles then become live, interactive media, which provide seamless access to a world of related information, while also serving as documentation for database entries (Phillip Bourne, personal communication). Building tighter, interactive connections between medicinal chemistry and pathway databases36-38, 63, 97-100 also has enormous potential to strengthen research. For example, the ability to display pathways while highlighting proteins already known to have small molecule binders will help medicinal chemists view their work in a broad biological context. It will also draw the attention of systems biologists to compounds which may be useful biological probes and to potential new avenues for drug discovery. The SuperTarget database80 is one significant effort along these lines, while the Reactome pathway database36, 66 and the Cytoscape software101 provide related network viewing capabilities by using the PSICQUIC web-service102, 103, which has links to multiple molecular interaction databases.
Data will also be more smoothly linked to computational analysis and prediction tools. For example, one might collect a set of active compounds at BindingDB, transfer them to another online resource which does machine-learning, and then use the result set of rules to computationally filter a compound catalog in search of new actives. The candidate actives could furthermore be piped through a database integrating pathway and medicinal chemistry data, in order to flag potentially unanticipated on- or off-target effects. Each step of such a process might be carried out on a different computer somewhere on the web, with the user directing the flow of data and collecting the output. Many other capabilities could be used in an online informatics network; for example, methods of predicting druggable protein binding sites104-108, or of estimating the physical properties of compounds. Software is already available that allows users to direct data flows involving multiple data and computational resources in a flexible manner109-115, and the continued development of such technologies will enable many new informatics tools to speed drug discovery.
E. Summary
Historically, medicinal chemistry data were not well connected to the informatics world, but this situation has now changed decisively. Here, we have focused on three prominent, publicly accessible chemical activity databases, BindingDB, ChEMBL, and PubChem, each with its own unique user-interface and scientific focus. These resources allow users to browse, query and download hundreds of thousands of data extracted from the medicinal chemistry literature, along with additional data from other sources, such as the NIH screening centers. We also more briefly reviewed seven complementary chemical databases also of interest to many medicinal chemists. Analysis of the holdings of BindingDB and ChEMBL indicate that the rate of publication of medicinal chemistry data has grown by about 50% since 2007 and appears to continue on an upward trend. This is exciting scientifically, but also means the work of extracting and managing the data is growing. We therefore discussed potential approaches to strengthening the database system, including further coordination among the various projects, community quality-control efforts, and the development of a simple mechanism for authors to make their data available in electronic format concurrently with publication. Finally, we discussed future research capabilities that will grow from integration of the medicinal chemistry databases with more biologically oriented databases, as well as with web-based tools for computational analysis and prediction. In sum, the emerging system of publicly accessible medicinal chemistry databases is rapidly becoming a critical infrastructure component for drug-discovery efforts world-wide, and is opening doors to valuable, new applications at the interfaces of chemistry and biology.
F. Acknowledgements
This publication was made possible by grants no. GM61300 from the National Institutes of Health and FP7-HEALTH-2007-223411 from the European Commission FP7 programme. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or the European Commission.
Abbreviations Used
- BKD
Binary Kernel Discrimination
- cif
crystallographic information file
- CSV
comma-separated values
- Da
daltons
- GSK
GlaxoSmithKline
- HERG
human ether related gene
- MLPCN
Molecular Libraries Probe Production Centers Network
- MLSCN
Molecular Libraries Screening Centers Network
- MMDB
Molecular Modeling Database
- NCBI
National Center for Biotechnology Information
- PDB
Protein Data Bank
- Rin1
CDD
- Conserved Domain Database
SAR
- structure-activity relationship
SVM
- Support Vector Machine
UK, United Kingdom
- wwPDB
worldwide Protein Data Bank
G. Biographies
George Nicola, Ph.D., M.B.A. George Nicola attended the University of Massachusetts-Amherst, graduating with a BS in Biomedical Computing in 2000. He went on to graduate school at the Medical University of South Carolina in Charleston where he earned a Ph.D. in Biochemistry in 2005, and subsequently completed an MBA from the Rady School of Management, University of California San Diego in 2009. Dr. Nicola has been a postdoctoral associate at The Scripps Research Institute and was awarded a Research Scholar Fellowship from the American Cancer Society in 2007. He has held an appointment as Adjunct Professor of Chemistry at the University of San Diego, and is currently a Project Scientist at the Skaggs School of Pharmacy at the University of California San Diego.
Tiqing Liu, Ph.D. Tiqing Liu received his B.S. in Chemistry from East China Normal University in 1986, M.S. in Physical Chemistry in Peking University in 1989, and Ph.D. in Chemical Physics at University of Minnesota at Twin Cities in 2000. After two terms of postdoctoral studies in the fields of computational chemistry, he became an Application Developer and Programmer for BindingDB.
Michael K. Gilson, M.D., Ph.D. Mike Gilson received his A.B. in Bioengineering from Harvard College in 1981. He then joined the M.D.-Ph.D. program at Columbia University, where his graduate work with Barry Honig led to the Ph.D. in 1988; his M.D. was completed in 1989. After a medicine residency at Stanford University Hospital, Mike joined the laboratory of J. Andrew McCammon as an HHMI Physician Research Fellow. In 1994, he joined the faculty of the Center for Advanced Research in Biotechnology in Rockville, Maryland, and he is currently a professor in the Skaggs School of Pharmacy and Pharmaceutical Sciences at the University of California, San Diego. His research focuses on theoretical, computational and informatic aspects of molecular recognition, and the computer-aided design of drugs and other molecules.
References
- 1.Portoghese PS. My farewell to the Journal of Medicinal Chemistry. J Med Chem. 2011;54:8235. doi: 10.1021/jm201370n. [DOI] [PubMed] [Google Scholar]
- 2.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, Chang Z, Woolsey J. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bader G, Donaldson S. [last accessed: May 16, 2012];Pathguide: the pathway resource list. http://www.pathguide.org/
- 5.Bader GD, Cary MP, Sander C. Pathguide: a pathway resource list. Nucleic Acids Res. 2006;34:D504–D506. doi: 10.1093/nar/gkj126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Apodaca R. [last accessed: May 29, 2012];Depth-First. http://depth-first.com/articles/2011/10/12/sixty-four-free-chemistry-databases/
- 7.Gaulton A, Overington JP. Role of open chemical data in aiding drug discovery and design. Future Med Chem. 2010;2:903–907. doi: 10.4155/fmc.10.191. [DOI] [PubMed] [Google Scholar]
- 8.Wassermann AM, Bajorath J. BindingDB and ChEMBL: online compound databases for drug discovery. Expert Opin Drug Discov. 2011;6:683–687. doi: 10.1517/17460441.2011.579100. [DOI] [PubMed] [Google Scholar]
- 9.Li Q, Cheng T, Wang Y, Bryant SH. PubChem as a public resource for drug discovery. Drug Discov Today. 2010;15:1052–1057. doi: 10.1016/j.drudis.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams AJ. Public chemical compound databases. Curr Opin Drug Discov Devel. 2008;11:393–404. [PubMed] [Google Scholar]
- 11.Williams AJ. Internet-based tools for communication and collaboration in chemistry. Drug Discov Today. 2008;13:502–506. doi: 10.1016/j.drudis.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 12.Gozalbes R, Pineda-Lucena A. Small molecule databases and chemical descriptors useful in chemoinformatics: an overview. Comb Chem High Throughput Screen. 2011;14:548–558. doi: 10.2174/138620711795767857. [DOI] [PubMed] [Google Scholar]
- 13.Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, Hersey A, Light Y, McGlinchey S, Michalovich D, Al-Lazikani B, Overington JP. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012;40:D1100–D1107. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. [last accessed: May 16, 2012];ChEMBL. https://www.ebi.ac.uk/chembl/
- 15.Bryant S. PubChem: An information resource linking chemistry and biology. Abstracts of Papers of the American Chemical Society. 2006:231. [Google Scholar]
- 16.Bolton EE, Wang Y, Thiessen PA, Bryant SH. PubChem: Integrated Platform of Small Molecules and Biological Activities. In: Ralph AW, David CS, editors. Annual Reports in Computational Chemistry. Vol. 4. Elsevier; Bethesda, MD: 2008. pp. 217–241. [Google Scholar]
- 17.Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Bryant SH. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009;37:W623–W633. doi: 10.1093/nar/gkp456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evans JM, Lopez E, Roth BL. The NIMH Psychoactive Drug Screening Program’s Ki database: An on-line, searchable database of receptor-ligand affinity values. Society for Neuroscience Abstracts. 2001;27:2076. [Google Scholar]
- 19.Roth B. [last accessed: May 16, 2012];PDSP Ki. http://pdsp.med.unc.edu/indexR.html.
- 20. [last accessed: May 16, 2012]; http://bit.ly/ws4vLt.http://www.bindingdb.org/jsp/dbsearch/PrimarySearch_pubmed.jsp?pubmed=50006488&pub med_submit=TBD.
- 21. [last accessed: May 16, 2012]; http://bit.ly/AyOWyq. http://www.bindingdb.org/bind/ByKI.jsp?specified=IC50.
- 22. [last accessed: May 16, 2012]; http://bit.ly/AAUiVz. http://www.bindingdb.org/bind/ByMolWeight.jsp.
- 23.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 24. [last accessed: May 16, 2012]; http://bit.ly/xuN2IY. http://www.bindingdb.org/bind/BySequence.jsp.
- 25. [last accessed: May 16, 2012]; http://bit.ly/zL842y. http://www.bindingdb.org/bind/chemsearch/marvin/index.jsp.
- 26. [last accessed: May 16, 2012]; http://bit.ly/w0A1G5. http://www.bindingdb.org/bind/BatchStructures.jsp.
- 27. [last accessed: May 16, 2012];Drugs@FDA. http://www.accessdata.fda.gov/scripts/cder/drugsatfda/index.cfm.
- 28. [last accessed: May 16, 2012]; http://bit.ly/wXIziD. http://www.bindingdb.org/bind/ByFDAdrugs.jsp.
- 29. [last accessed: May 16, 2012]; http://bit.ly/wHLXDl. http://www.bindingdb.org/bind/ByAuthor.jsp.
- 30. [last accessed: May 16, 2012]; http://bit.ly/ydpChT. http://www.bindingdb.org/bind/ByJournal.jsp.
- 31. [last accessed: May 16, 2012]; http://bit.ly/yMYvx2. http://www.bindingdb.org/bind/ByInstitution.jsp.
- 32. [last accessed: May 16, 2012]; http://bit.ly/zVd6z2. http://www.bindingdb.org/bind/ByPDBids.jsp.
- 33. [last accessed: May 16, 2012]; http://bit.ly/x9f9Yd. http://www.bindingdb.org/bind/ByPDBids_100.jsp.
- 34.Creative Commons: Attribution-ShareAlike 3.0 Unported. Mountain View, CA: 2011. http://creativecommons.org/licenses/by-sa/3.0/ [Google Scholar]
- 35. [last accessed: May 16, 2012]; http://bit.ly/zq9oW3. http://www.bindingdb.org/jsp/dbsearch/PrimarySearch_ki.jsp?polymerid=50000007&target=ABL1&tag=polkd&column=Kd&energyterm=kcal/mole&startPg=0&Increment=50&submit=Search.
- 36.Joshi-Tope G, Gillespie M, Vastrik I, D’Eustachio P, Schmidt E, de Bono B, Jassal B, Gopinath GR, Wu GR, Matthews L, Lewis S, Birney E, Stein L. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33:D428–D432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogata H, Goto S, Fujibuchi W, Kanehisa M. Computation with the KEGG pathway database. Biosystems. 1998;47:119–128. doi: 10.1016/s0303-2647(98)00017-3. [DOI] [PubMed] [Google Scholar]
- 38.Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, Buetow KH. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Irwin JJ, Shoichet BK. ZINC--a free database of commercially available compounds for virtual screening. J Chem Inf Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. [last accessed: May 16, 2012]; http://bit.ly/zX0SfQ. http://www.bindingdb.org/bind/chemsearch/marvin/BatchStructures.jsp.
- 41.Csizmadia F. JChem: Java applets and modules supporting chemical database handling from web browsers. J Chem Inf Comput Sci. 2000;40:323–324. doi: 10.1021/ci9902696. [DOI] [PubMed] [Google Scholar]
- 42.JChem. Vol. 5.6. ChemAxon; Budapest, Hungary: 2011. [Google Scholar]
- 43.Harper G, Bradshaw J, Gittins JC, Green DVS, Leach AR. Prediction of biological activity for high-throughput screening using binary kernel discrimination. Journal of Chemical Information and Computer Sciences. 2001;41:1295–1300. doi: 10.1021/ci000397q. [DOI] [PubMed] [Google Scholar]
- 44.Jorissen RN, Gilson MK. Virtual screening of molecular databases using a support vector machine. J Chem Inf Model. 2005;45:549–561. doi: 10.1021/ci049641u. [DOI] [PubMed] [Google Scholar]
- 45.Cortes C, Vapnik V. Support-vector networks. Machine Learning. 1995;20:273–297. [Google Scholar]
- 46. [last accessed: May 16, 2012]; http://bit.ly/yTctqN. http://bindingdb.org/validation_sets/index.jsp.
- 47.Ertl P. JME Molecular Editor. Novartis; 2012. [Google Scholar]
- 48.ChemAxon. Marvin Sketch. 2012;5.7 [Google Scholar]
- 49.JDraw. Accelrys; 2012. [Google Scholar]
- 50.Weininger D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J Chem Inf Model. 1988;28:31–36. [Google Scholar]
- 51.Velankar S, Best C, Beuth B, Boutselakis CH, Cobley N, Sousa Da Silva AW, Dimitropoulos D, Golovin A, Hirshberg M, John M, Krissinel EB, Newman R, Oldfield T, Pajon A, Penkett CJ, Pineda-Castillo J, Sahni G, Sen S, Slowley R, Suarez-Uruena A, Swaminathan J, van Ginkel G, Vranken WF, Henrick K, Kleywegt GJ. PDBe: Protein Data Bank in Europe. Nucleic Acids Res. 2010;38:D308–D317. doi: 10.1093/nar/gkp916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Chen Y, Clapham P, Coates G, Fairley S, Fitzgerald S, Gordon L, Hendrix M, Hourlier T, Johnson N, Kahari A, Keefe D, Keenan S, Kinsella R, Kokocinski F, Kulesha E, Larsson P, Longden I, McLaren W, Overduin B, Pritchard B, Riat HS, Rios D, Ritchie GR, Ruffier M, Schuster M, Sobral D, Spudich G, Tang YA, Trevanion S, Vandrovcova J, Vilella AJ, White S, Wilder SP, Zadissa A, Zamora J, Aken BL, Birney E, Cunningham F, Dunham I, Durbin R, Fernandez-Suarez XM, Herrero J, Hubbard TJ, Parker A, Proctor G, Vogel J, Searle SM. Ensembl 2011. Nucleic Acids Res. 2011;39:D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bairoch A, Apweiler R, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Natale DA, O’Donovan C, Redaschi N, Yeh LS. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. [last accessed: May 16, 2012];ChemSpider: The free chemical database. http://www.chemspider.com/
- 55.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Degtyarenko K, de Matos P, Ennis M, Hastings J, Zbinden M, McNaught A, Alcantara R, Darsow M, Guedj M, Ashburner M. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res. 2008;36:D344–D350. doi: 10.1093/nar/gkm791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gamo FJ, Sanz LM, Vidal J, de Cozar C, Alvarez E, Lavandera JL, Vanderwall DE, Green DV, Kumar V, Hasan S, Brown JR, Peishoff CE, Cardon LR, Garcia-Bustos JF. Thousands of chemical starting points for antimalarial lead identification. Nature. 2010;465:305–310. doi: 10.1038/nature09107. [DOI] [PubMed] [Google Scholar]
- 58.Meister S, Plouffe DM, Kuhen KL, Bonamy GM, Wu T, Barnes SW, Bopp SE, Borboa R, Bright AT, Che J, Cohen S, Dharia NV, Gagaring K, Gettayacamin M, Gordon P, Groessl T, Kato N, Lee MC, McNamara CW, Fidock DA, Nagle A, Nam TG, Richmond W, Roland J, Rottmann M, Zhou B, Froissard P, Glynne RJ, Mazier D, Sattabongkot J, Schultz PG, Tuntland T, Walker JR, Zhou Y, Chatterjee A, Diagana TT, Winzeler EA. Imaging of Plasmodium liver stages to drive next-generation antimalarial drug discovery. Science. 2011;334:1372–1377. doi: 10.1126/science.1211936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guiguemde WA, Shelat AA, Bouck D, Duffy S, Crowther GJ, Davis PH, Smithson DC, Connelly M, Clark J, Zhu F, Jimenez-Diaz MB, Martinez MS, Wilson EB, Tripathi AK, Gut J, Sharlow ER, Bathurst I, El Mazouni F, Fowble JW, Forquer I, McGinley PL, Castro S, Angulo-Barturen I, Ferrer S, Rosenthal PJ, Derisi JL, Sullivan DJ, Lazo JS, Roos DS, Riscoe MK, Phillips MA, Rathod PK, Van Voorhis WC, Avery VM, Guy RK. Chemical genetics of Plasmodium falciparum. Nature. 2010;465:311–315. doi: 10.1038/nature09099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Zhou Z, Han L, Karapetyan K, Dracheva S, Shoemaker BA, Bolton E, Gindulyte A, Bryant SH. PubChem’s BioAssay Database. Nucleic Acids Res. 2012;40:D400–D412. doi: 10.1093/nar/gkr1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. [last accessed: May 16, 2012]; http://1.usa.gov/x3eREX. http://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?cid=184691.
- 62. [last accessed: May 16, 2012]; http://1.usa.gov/x2nfRP. http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?aid=33734&loc=ea_ras.
- 63.Geer LY, Marchler-Bauer A, Geer RC, Han L, He J, He S, Liu C, Shi W, Bryant SH. The NCBI BioSystems database. Nucleic Acids Res. 2010;38:D492–D496. doi: 10.1093/nar/gkp858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Minoru K. A database for post-genome analysis. Trends in Genetics. 1997;13:375–376. doi: 10.1016/s0168-9525(97)01223-7. [DOI] [PubMed] [Google Scholar]
- 65.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. [last accessed: May 16, 2012];Reactome. http://www.reactome.org/ReactomeGWT/entrypoint.html.
- 67.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Madej T, Addess KJ, Fong JH, Geer LY, Geer RC, Lanczycki CJ, Liu C, Lu S, Marchler-Bauer A, Panchenko AR, Chen J, Thiessen PA, Wang Y, Zhang D, Bryant SH. MMDB: 3D structures and macromolecular interactions. Nucleic Acids Res. 2012;40:D461–D464. doi: 10.1093/nar/gkr1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Molecular Libraries Probe Production Centers Network (MLPCN) [last accessed: May 16, 2012]; http://mli.nih.gov/mli/mlpcn/
- 70.Carlson HA. [last accessed: May 16, 2012];Binding MOAD. http://bindingmoad.org/
- 71.Hu L, Benson ML, Smith RD, Lerner MG, Carlson HA. Binding MOAD (Mother Of All Databases) Proteins. 2005;60:333–340. doi: 10.1002/prot.20512. [DOI] [PubMed] [Google Scholar]
- 72.Benson ML, Smith RD, Khazanov NA, Dimcheff B, Beaver J, Dresslar P, Nerothin J, Carlson HA. Binding MOAD, a high-quality protein-ligand database. Nucleic Acids Research. 2008;36:D674–D678. doi: 10.1093/nar/gkm911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Knox C, Law V, Jewison T, Liu P, Ly S, Frolkis A, Pon A, Banco K, Mak C, Neveu V, Djoumbou Y, Eisner R, Guo AC, Wishart DS. DrugBank 3.0: a comprehensive resource for ’omics’ research on drugs. Nucleic Acids Res. 2011;39:D1035–D1041. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Alexander SP, Mathie A, Peters JA. Guide to Receptors and Channels (GRAC) Br J Pharmacol. (5th edition) 2011;164(Suppl 1):S1–S324. doi: 10.1111/j.1476-5381.2011.01649_1.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sharman JL, Mpamhanga CP, Spedding M, Germain P, Staels B, Dacquet C, Laudet V, Harmar AJ, Nc I. IUPHAR-DB: new receptors and tools for easy searching and visualization of pharmacological data. Nucleic Acids Res. 2011;39:D534–D538. doi: 10.1093/nar/gkq1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Harmar AJ, Hills RA, Rosser EM, Jones M, Buneman OP, Dunbar DR, Greenhill SD, Hale VA, Sharman JL, Bonner TI, Catterall WA, Davenport AP, Delagrange P, Dollery CT, Foord SM, Gutman GA, Laudet V, Neubig RR, Ohlstein EH, Olsen RW, Peters J, Pin JP, Ruffolo RR, Searls DB, Wright MW, Spedding M. IUPHAR-DB: the IUPHAR database of G protein-coupled receptors and ion channels. Nucleic Acids Res. 2009;37:D680–D685. doi: 10.1093/nar/gkn728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang R, Fang X, Lu Y, Wang S. The PDBbind database: collection of binding affinities for protein-ligand complexes with known three-dimensional structures. J Med Chem. 2004;47:2977–2980. doi: 10.1021/jm030580l. [DOI] [PubMed] [Google Scholar]
- 78.Cheng T, Li X, Li Y, Liu Z, Wang R. Comparative assessment of scoring functions on a diverse test set. J Chem Inf Model. 2009;49:1079–1093. doi: 10.1021/ci9000053. [DOI] [PubMed] [Google Scholar]
- 79.Wang R, Fang X, Lu Y, Yang CY, Wang S. The PDBbind database: methodologies and updates. J Med Chem. 2005;48:4111–4119. doi: 10.1021/jm048957q. [DOI] [PubMed] [Google Scholar]
- 80.Gunther S, Kuhn M, Dunkel M, Campillos M, Senger C, Petsalaki E, Ahmed J, Urdiales EG, Gewiess A, Jensen LJ, Schneider R, Skoblo R, Russell RB, Bourne PE, Bork P, Preissner R. SuperTarget and Matador: resources for exploring drug-target relationships. Nucleic Acids Res. 2008;36:D919–D922. doi: 10.1093/nar/gkm862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hecker N, Ahmed J, von Eichborn J, Dunkel M, Macha K, Eckert A, Gilson MK, Bourne PE, Preissner R. SuperTarget goes quantitative: update on drug-target interactions. Nucleic Acids Res. 2012;40:D1113–D1117. doi: 10.1093/nar/gkr912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhu F, Shi Z, Qin C, Tao L, Liu X, Xu F, Zhang L, Song Y, Liu X, Zhang J, Han B, Zhang P, Chen Y. Therapeutic target database update 2012: a resource for facilitating target-oriented drug discovery. Nucleic Acids Res. 2012;40:D1128–D1136. doi: 10.1093/nar/gkr797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. [last accessed: May 16, 2012]; http://bit.ly/uu6ZNn. http://www.bindingdb.org/bind/ByDataLigand.jsp.
- 84. [last accessed: May 16, 2012]; http://bit.ly/uz9HeV. http://www.bindingdb.org/bind/ByMonomersTarget.jsp.
- 85.Berman H, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nature Structural Biology. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 86.Battle for BMRB Biological Magnetic Resonance Data Bank. Nature Structural Biology. 1995;2:811–812. [PubMed] [Google Scholar]
- 87.Kinjo AR, Suzuki H, Yamashita R, Ikegawa Y, Kudou T, Igarashi R, Kengaku Y, Cho H, Standley DM, Nakagawa A, Nakamura H. Protein Data Bank Japan (PDBj): maintaining a structural data archive and resource description framework format. Nucleic Acids Res. 2012;40:D453–D460. doi: 10.1093/nar/gkr811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Velankar S, Alhroub Y, Best C, Caboche S, Conroy MJ, Dana JM, Fernandez Montecelo MA, van Ginkel G, Golovin A, Gore SP, Gutmanas A, Haslam P, Hendrickx PM, Heuson E, Hirshberg M, John M, Lagerstedt I, Mir S, Newman LE, Oldfield TJ, Patwardhan A, Rinaldi L, Sahni G, Sanz-Garcia E, Sen S, Slowley R, Suarez-Uruena A, Swaminathan GJ, Symmons MF, Vranken WF, Wainwright M, Kleywegt GJ. PDBe: Protein Data Bank in Europe. Nucleic Acids Res. 2012;40:D445–D452. doi: 10.1093/nar/gkr998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Goldberg RN, Tewari YB, Bhat TN. Thermodynamics of enzyme-catalyzed reactions--a database for quantitative biochemistry. Bioinformatics. 2004;20:2874–2877. doi: 10.1093/bioinformatics/bth314. [DOI] [PubMed] [Google Scholar]
- 90.Kuzmic P, Elrod KC, Cregar LM, Sideris S, Rai R, Janc JW. High-throughput screening of enzyme inhibitors: simultaneous determination of tight-binding inhibition constants and enzyme concentration. Anal Biochem. 2000;286:45–50. doi: 10.1006/abio.2000.4685. [DOI] [PubMed] [Google Scholar]
- 91.McGovern SL, Caselli E, Grigorieff N, Shoichet BK. A common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening. J Med Chem. 2002;45:1712–1722. doi: 10.1021/jm010533y. [DOI] [PubMed] [Google Scholar]
- 92.Williams AJ, Ekins S. A quality alert and call for improved curation of public chemistry databases. Drug Discov Today. 2011;16:747–750. doi: 10.1016/j.drudis.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 93.Williams AJ, Ekins S, Tkachenko V. Towards a gold standard: regarding quality in public domain chemistry databases and approaches to improving the situation. Drug Discov Today. 2012 doi: 10.1016/j.drudis.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 94.Wang S, Georg GI. Transition in leadership: opportunities and challenges. J Med Chem. 2012;55:1. doi: 10.1021/jm201695q. [DOI] [PubMed] [Google Scholar]
- 95.In U.S. Government Chemical Databases and Open Chemistry, Frederick, MD. Aug, 2011. pp. 25–26.
- 96.Bourne P. Will a Biological Database Be Different from a Biological Journal? PLoS Comput Biol. 2005;1:e34. doi: 10.1371/journal.pcbi.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kamburov A, Wierling C, Lehrach H, Herwig R. ConsensusPathDB—a database for integrating human functional interaction networks. Nucleic Acids Research. 2009;37:D623–D628. doi: 10.1093/nar/gkn698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pico A, Kelder T, van Iersel M, Hanspers K, Conklin B, Evelo C. WikiPathways: pathway editing for the people. PLoS Biol. 2008;6:e184. doi: 10.1371/journal.pbio.0060184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Frolkis A, Knox C, Lim E, Jewison T, Law V, Hau DD, Liu P, Gautam B, Ly S, Guo AC, Xia J, Liang Y, Shrivastava S, Wishart DS. SMPDB: The Small Molecule Pathway Database. Nucleic Acids Research. 2010;38:D480–D487. doi: 10.1093/nar/gkp1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sreenivasaiah PK, Rani S, Cayetano J, Arul N, Kim DH. IPAVS: Integrated Pathway Resources, Analysis and Visualization System. Nucleic Acids Research. 2012;40:D803–D808. doi: 10.1093/nar/gkr1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kerrien S, Aranda B. [last accessed: May 16, 2012];PSICQUIC View. http://www.ebi.ac.uk/Tools/webservices/psicquic/view/main.xhtml.
- 103.Aranda B, Blankenburg H, Kerrien S, Brinkman FS, Ceol A, Chautard E, Dana JM, De Las Rivas J, Dumousseau M, Galeota E, Gaulton A, Goll J, Hancock RE, Isserlin R, Jimenez RC, Kerssemakers J, Khadake J, Lynn DJ, Michaut M, O’Kelly G, Ono K, Orchard S, Prieto C, Razick S, Rigina O, Salwinski L, Simonovic M, Velankar S, Winter A, Wu G, Bader GD, Cesareni G, Donaldson IM, Eisenberg D, Kleywegt GJ, Overington J, Ricard-Blum S, Tyers M, Albrecht M, Hermjakob H. PSICQUIC and PSISCORE: accessing and scoring molecular interactions. Nat Methods. 2011;8:528–529. doi: 10.1038/nmeth.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nicola G, Smith CA, Abagyan R. New method for the assessment of all drug-like pockets across a structural genome. J Comput Biol. 2008;15:231–240. doi: 10.1089/cmb.2007.0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.An J, Totrov M, Abagyan R. Comprehensive identification of “druggable” protein ligand binding sites. Genome Inform. 2004;15:31–41. [PubMed] [Google Scholar]
- 106.Keller TH, Pichota A, Yin Z. A practical view of ’druggability’. Curr Opin Chem Biol. 2006;10:357–361. doi: 10.1016/j.cbpa.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 107.Halgren T. New method for fast and accurate binding-site identification and analysis. Chem Biol Drug Des. 2007;69:146–148. doi: 10.1111/j.1747-0285.2007.00483.x. [DOI] [PubMed] [Google Scholar]
- 108.Henrich S, Salo-Ahen OM, Huang B, Rippmann FF, Cruciani G, Wade RC. Computational approaches to identifying and characterizing protein binding sites for ligand design. J Mol Recognit. 2010;23:209–219. doi: 10.1002/jmr.984. [DOI] [PubMed] [Google Scholar]
- 109.Stevenson JM, Mulready PD. Pipeline pilot 2.1. Journal of the American Chemical Society. 2003;125:1437–1438. [Google Scholar]
- 110.Pipeline Pilot. Vol. 8.5. Accelrys; 2012. [Google Scholar]
- 111.Berthold MR, Cebron N, Dill F, Gabriel TR, Kotter T, Meinl T, Ohl P, Thiel K, Wiswedel B. KNIME – The Konstanz Information Miner. SIGKDD Explorations. 2009;11:26–31. [Google Scholar]
- 112.Berthold MH, Cebron N, Dill F, Di Fatta G, Gabriel TR, Georg F, Moinl T, Ohl P, Sieb C, Wiswedol B. KNIME: the Konstanz Information Miner. Industrial Simulation Conference; Palermo, Italy: University of Palermo; Jun 5-7, 2006. [Google Scholar]
- 113.Hull D, Wolstencroft K, Stevens R, Goble C, Pocock MR, Li P, Oinn T. Taverna: a tool for building and running workflows of services. Nucleic Acids Res. 2006;34:W729–W732. doi: 10.1093/nar/gkl320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Oinn T, Greenwood M, Addis M, Alpdemir MN, Ferris J, Glover K, Goble C, Goderis A, Hull D, Marvin D, Li P, Lord P, Pocock MR, Senger M, Stevens R, Wipat A, Wroe C. Taverna: lessons in creating a workflow environment for the life sciences. Concurrency and Computation: Practice and Experience. 2006;18:1067–1100. [Google Scholar]
- 115.Altintas I, Berkley C, Jaeger E, Jones M, Ludascher B, Mock S. Kepler: an extensible system for design and execution of scientific workflows. 16th International Conference on Scientific and Statistical Database Management; Santorini Island, Greece: Petros Nomikos Conference Center; Jun 21-23, 2004. [Google Scholar]