Abstract
Protein molecules naturally emit streams of information-rich signals in the language of hydrogen exchange concerning the intimate details of their stability, dynamics, function, changes therein, and effects thereon, all resolved to the level of their individual amino acids. The effort to measure protein hydrogen exchange behavior, understand the underlying chemistry and structural physics of hydrogen exchange processes, and use this information to learn about protein properties and function has continued for 50 years. Recent work uses mass spectrometric analysis together with an earlier proteolytic fragmentation method to extend the hydrogen exchange capability to large biologically interesting proteins. This article briefly reviews the advances that have led us to this point and the understanding that has so far been achieved.
Protein hydrogen exchange (HX) studies focus principally on the main chain amide NH hydrogens. These hydrogens are distributed uniformly at every amino acid, except proline, in every protein molecule. They are just the hydrogens that get involved in the H-bonds that help to stabilize protein secondary and tertiary structure. Although they are covalently bound to the amide nitrogen and are often well-protected by their H-bonding interactions, they engage in continual exchange with the hydrogens of solvent water. In any given protein, HX rates are commonly spread over a very large dynamic range, many orders of magnitude wide. These rates encode detailed site-resolved information on protein structure, physical properties, and biochemical function. We need but to measure the signals and understand them in terms of the underlying chemistry and structural physics of protein HX processes.
The study of protein hydrogen exchange has a long history, dating back to the pioneering work of Kai U. Linderstrøm-Lang and his coworkers at the Carlsberg Laboratories in Copenhagen in the 1950s. In the excitement following Pauling's discovery of the α-helix and β-sheet, Linderstrøm-Lang realized that he might look for H-bonded structures by measuring protein hydrogen exchange. In typical style, Lang and his colleagues designed entirely new methods, built their own equipment, and pursued a series of ground-breaking HX studies on a variety of proteins and polypeptides [1]. In the early Carlsberg version of HX measurement, one dissolved the experimental protein in D2O, took samples as a function of H-D exchange time, transferred the protein-bound deuterium into pure H2O solvent, and quantified the deuterium by a sensitive mass technique that was able to measure the density of tiny water droplets to about one part in a million using the density gradient column pictured on the journal cover.
As it turns out, a good fraction of the Carlsberg data were in some way incorrect due to an artifact that has never been fully explained. Nevertheless, in a magical display of scientific insight, Lang inferred the basic dynamic mechanisms that underlie protein hydrogen exchange processes and wrote the equations that govern measurable HX, which we still use today. This was 10 years before the first protein structures were solved and 20 years before molecular dynamics came on the scene. Linderstrøm-Lang left us with the extraordinary potential that hydrogen exchange offers but also with challenges yet to be overcome. How can we measure HX behavior, ideally at amino acid resolution, understand how protein behavior and properties are reflected in measurable HX rates, and deploy the HX capability, experimentally and theoretically, to elicit that deeply hidden information?
Efforts to address these challenges have matured over the last half century. The development of multi-dimensional NMR methods now provides an efficient way to measure HX in rather small protein molecules. Larger, more interesting proteins require other approaches. The most recent advances implement the explosively developing capabilities of mass spectrometry together with an earlier proteolytic fragmentation-separation method. This article briefly recapitulates some of the important things that we have learned on the way to our present most promising vantage point.
HX Chemistry
We now know that amide hydrogen exchange is catalyzed only by the strong base, OH−-ion, and the strong acid, H3O+-ion. Also, there may be a small contribution due to catalysis by the H2O molecule (present at 55.5 M concentration) under conditions where chemical exchange is otherwise very slow (eq 1; see Figure 1). These restrictions stem from the extreme acid and base pKa values of the amide group as explained by the proton transfer theory of Manfred Eigen [2, 3]. The pKa value of the secondary amide (peptide) group is just over 18 for direct deprotonation, about −2 for protonation at the amide carbonyl, and apparently −11 for protonation at the amide nitrogen. The pKa values for hydroxide ion protonation and hydronium ion deprotonation are 15.7 and −2, respectively. These parameters come into play in the equilibrium proton transfer steps that determine HX rates, which involve direct deprotonation of the amide in the alkaline reaction and a pre-equilibrium amide protonation step in the acid reaction. Weaker acids and bases cannot effectively compete with catalysis by hydroxide ion and hydronium ion [2–4]. The same principles explain why hydrogens on the polar protein side chains exchange so much faster and make them less useful, or less interfering, depending on one's point of view.
Figure 1.
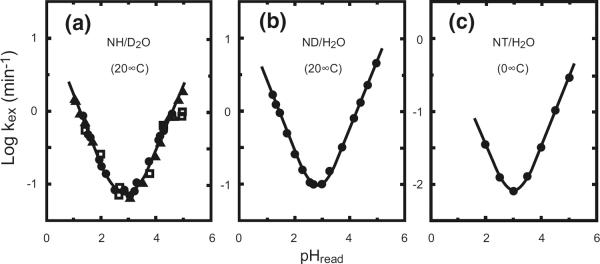
HX of the amide group in unstructured polypeptides. At the chemical level, amide HX rates vary linearly with hydronium and hydroxide ion concentration (eq 1). They depend also on temperature, neighboring side chains, and isotope effects [5, 6] (see URL HX2.Med.UPenn.edu/download). The HX halftime for unprotected amides is over 1 h at the pH minimum, between pH 2 and 3 at 0 °C, which makes possible the peptide separation/mass spectrometry technology.
Eq 1 expresses acid and base-catalyzed amide HX rates in these terms.
| (1) |
The terms in eq 1 are the unprotected chemical HX rate, kch, and the intrinsic second-order rate constants for the acid-catalyzed (A), the base-catalyzed (B), and the water-catalyzed (W) reactions. Chemical exchange rates go through a minimum between pH 2 and 3 and increase by a factor of 10 per pH unit at higher and lower pH (Figure 1). The amide chemical exchange rate is also very sensitive to temperature with activation energies close to 17 kcal/mol, which produces a factor of 10 in rate per 22 °C. Also, the rate is significantly influenced by neighboring side chains to the right and left and is sensitive to kinetic, equilibrium, and solvent isotope effects.
All of these factors have been well calibrated [5, 6] so that it is now possible to predict the unprotected chemical HX rate for all conditions. Calculations are based on reference values for the various possible isotopic conditions and the known effects thereon of pH, temperature, and neighboring side chains. Figure 1 illustrates the pH-dependent HX behavior for the unstructured polypeptide reference model, poly-D,L-alanine. An available spreadsheet handles all of the necessary calculations (URL is HX2.med.upenn.edu/download).
The kinetic properties of amide hydrogen exchange are exceedingly favorable for protein studies. Rates can be easily targeted onto the most useful time scale, simply by adjusting pH and temperature through the normal protein and laboratory range. HX isotope labeling experiments can be performed under conditions most pertinent for the system studied, and the analysis for carried label can be performed at any other chosen condition. The fairly slow HX rate for unprotected amide hydrogens at the pH minimum, over 1 h at pH ~2.5 and 0 °C (Figure 1), makes possible the proteolytic fragmentation-mass spectrometric analysis (see below).
HX and Protein Dynamics
The chemical HX rate in eq 1 refers to the freely exposed hydrogen. Protein structure imposes further slowing that builds on the chemical reference rates in Figure 1. We now know that hydrogens protected by H-bonding, whether in small organic molecules or large protein molecules, cannot exchange at all. Yet they do exchange. The modern view of HX rate determination follows closely the early insights of Linderstrøm-Lang. Lang's view was that the exchange process requires the transient separation of H-bond donor and acceptor groups in some unspecified dynamic exposure reaction, which clears the way for attack by solvent catalyst. In the absence of information on the dynamic exposure reaction, we refer to this behavior noncommittally (eqs 2–4) as structural opening (op) and reclosing (cl).
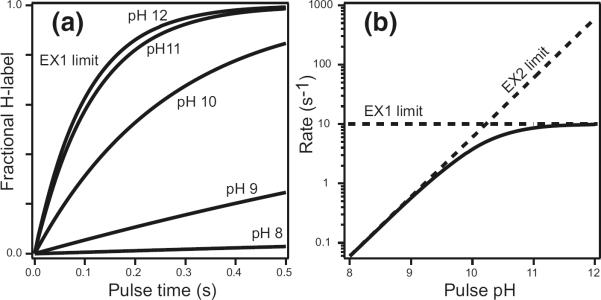
Lang pictured that when opening occurs, a kinetic competition ensues between the chemical exchange rate and structural reclosing. If reclosing is faster than chemical exchange (kcl > kch), then opening must occur many times before a successful exchange event. In this case, the opening reaction enters the kinetic expression as a pre-equilibrium step as in eq 2. The approximation shown in eq 2 holds for any significant degree of structural protection (kop/kcl = Kop < 1). The equilibrium constant (Kop) and the free-energy for the responsible opening reaction (eq 3) can then be directly obtained from the measured HX rate (kex) and the expected kch. In this so-called EX2 region (bimolecular exchange) HX rate depends sharply on pH, changing by a factor of 10 per pH unit as in eq 1. When the chemical rate is made progressively faster, for example by raising the pH (Figure 1), it may come to outpace the structural reclosing rate. HX rate will then ultimately limit at the structural opening rate (kop) (eq 4), known as the EX1 limit (monomolecular exchange) [1, 3]. This behavior is illustrated in Figure 2.
| (2) |
| (3) |
| (4) |
These equations apply to the usual steady-state situation where the protein is stable (kop< kcl) and in dynamic equilibrium with solvent and with its own higher energy transiently open states. The more general solution was given by Hvidt [7] (more accessible in [8]).
Figure 2.
HX of protected amides. The additional protection afforded by H-bonded structure depends on the thermodynamic and kinetic parameters of transient opening reactions that separate the H-bond and expose the hydrogen to solvent. Exchange rate increases linearly with solvent catalyst concentration in the EX2 range but will ultimately limit at the structural opening rate (EX1 limit). In the EX2 region, the reference chemical rate (eq 1, Figure 1) is decreased by the fraction of time that the hydrogen is exposed, given by kop/(kop + kcl) ≈ Kop (from eq 2), therefore can indicate the free-energy of the responsible structural opening reaction (eq 3). In the EX1 region, the HX rate becomes equal to, and therefore provides a way to measure, the rate for the responsible structural opening (eq 4). EX1 behavior can be documented by its insensitivity to pH as shown here or by correlated MS behavior when multiple amides experience the same EX1 exchange rate so that MS results show a molecular species moving from the unlabeled mass to the fully labeled mass in one concerted step.
An alternative view of the physical basis for protein HX behavior gained prominence in the 1970s and 1980s, and has been convincingly disproved, but tends to re-emerge whenever a new cadre of workers enters the field. This happened in the 1990s with the development of 2D NMR-HX technologies and is repeating with the emergence of mass spectrometry-HX methods. The intuitively seductive picture is that slowly exchanging hydrogens are slow because they are buried within the protein and that the HX process requires penetration into the protein by water and/or solvent catalysts. One now knows that many highly protected hydrogens are in fact fully exposed to solvent at the surface of proteins and even in small, organic molecules, where burial is not an issue (e.g., [9, 10]. Mere contact with water is not enough. The hydrogens exchange slowly because they are involved in H-bonding interactions that sterically block normal HX chemistry. Pre-existing H-bonds must be separated to allow attack by and H-bonding to HX catalysts. EX1 HX behavior (Figure 2), seen for some well-buried amides, is inconsistent with a penetration mechanism, which would irrevocably produce EX2-like behavior. The dependence of HX rate on temperature and on degree of burial are against any penetration mechanism [3, 11, 12]. The enthalpy for entry of charged HX catalysts into the protein interior would be extremely large due to the desolvation penalty, and this is not seen. Partial desolvation would require the insertion of a large complex, still bearing a net charge. Direct studies of the ability of small molecules to penetrate into proteins and quench the fluorescence and phosphorescence of buried tryptophans show that small apolar diatomic and triatomic molecules can penetrate into proteins at some reasonable rate, but that charged species like hydroxide and hydronium ions cannot [13, 14].
To be sure, hydrogens that are buried within proteins must, in addition, be brought into contact with solvent. HX patterns shown by near-neighbor hydrogens, some buried and some not, show that this occurs by protein distortional motions that break H-bonds and bring the hydrogens out into solvent contact rather than by allowing charged HX catalyst to enter [11]. Structural openings that can bring protected hydrogens out into contact with solvent may involve segmental unfolding reactions, which can reach from small local unfoldings to whole molecule global unfolding. Recent work has tested the validity of protein stability parameters obtained by using eqs 2 to 4. The equilibrium and kinetic parameters of global and subglobal unfolding reactions appear to be accurately obtained [15, 16]. Alternatively, the dominant opening often depends on smaller distortional motions known noncommittally as local fluctuations [17], which can expose to exchange as little as one hydrogen at a time [11]. The relationships that connect HX rate with the parameters of local fluctuations are still unclear. A significant issue is whether the kint values obtained for unstructured peptides (eq 1) are pertinent in this case [18]. More generally, the factors that determine which dynamical mode will dominate (global unfolding, subglobal unfolding, local fluctuations) have not been well defined.
HX Measurement: Functional Labeling, Proteolytic Fragmentation, and HPLC Separation
Progress in HX measurement over the years has depended on technological innovation and commercialization at every step along the way. The advance of technology can be illustrated by the sequential panels in Figure 3.
Figure 3.
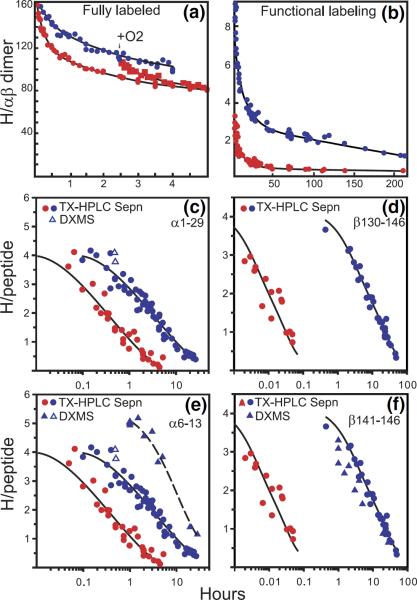
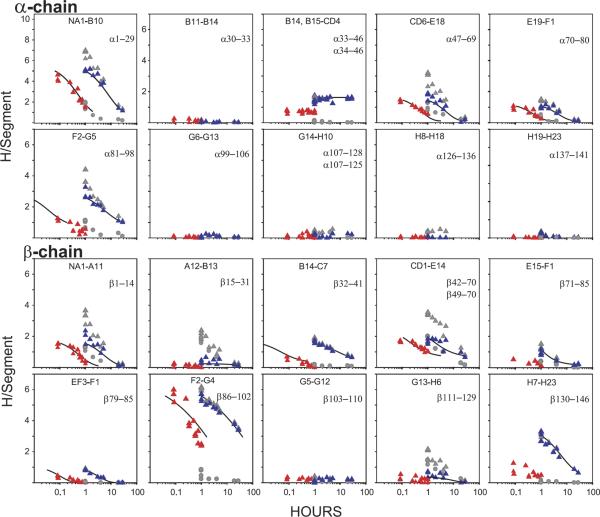
HX results for hemoglobin (Hb) illustrating the progression of HX technology. (a) Whole Hb initially labeled to exchange equilibrium in tritiated water, then exchanged-out in oxy (red) and deoxy (blue) forms, measured by the early tritiumgel-filtration method [36]. Some allosterically sensitive sites are faster in oxyHb. When O2 is added to exchanging deoxy Hb, the sensitive sites are made faster and the separate curves merge. (b) Functional labeling results for whole Hb with exchange-in for only 1 min (pH 7.4, 0 °C [37]). The functional labeling method selectively labels some allosterically sensitive sites, making it possible to characterize their number and HX rates in both forms, and therefore the allosteric free-energy that they carry. (c) and (d) HX of functionally labeled Hb (35 min, 0 °C exchange-in) measured by the proteolytic fragmentation method with peptide separation by HPLC alone [38]. Two sets of allosterically sensitive amides are located at the segment level. They exchange by way of a local unfolding mechanism. (e) and (f) The integration of these approaches with mass spectrometric analysis which more closely defines some allosterically sensitive positions. (e) and (f) include MS data obtained with the same exchange-in as for the tritium experiments (open triangles), and more intensively (20 min, 20 °C, closed triangles) [35].
In the 1960s, shortly after the Linderstrøm-Lang era, the radioactive isotope, tritium, and its measurement by liquid scintillation counting, became easily available as a result of ongoing fusion and hydrogen bomb research. These advances, together with the development and commercialization of gel-filtration media, led to the accurate artifact-free tritium-gel-filtration HX technique [19], which dominated HX work for the next 20 years. A range of HX studies on proteins, nucleic acids, oligosaccharides, their synthetic analogs, and their multimolecular complexes followed.
Figure 3a shows some results for the archetypal regulatory protein, hemoglobin (Hb). When O2 is added to T-state deoxy Hb, causing it to switch to the R-state oxy Hb form, significant structure changes occur [20]. The exchange of some hydrogens is seen to be accelerated (Figure 3a), as might be expected from the more relaxed nature of oxy Hb. However, this undifferentiated global measurement cannot tell how many hydrogens are affected, what their rates are in either form, or where in the protein they might be. All straightforward whole molecule determinations share this limitation. Similar, although less extreme, uncertainties occur even when the experimental analysis can be focussed down onto submolecular peptides, as is possible by using the modern MS methods described below.
A functional labeling technique can provide more specific information by placing HX label selectively on just the sites that sense the allosteric transition [21]. For Figure 3b, tritium was initially exchanged-in to the faster exchanging oxy form for only a limited time, 1 min in this case. Allosterically sensitive and insensitive sites that exchange on this time scale are indiscriminately labeled. Hb was then switched to its slower exchanging deoxy form and exchanged out for a somewhat longer time. Label on allosterically insensitive sites is soon lost because, by definition, insensitive sites exchange at the same rate in both forms. But label on the allosterically sensitive sites is now much slower, and it survives the chase period. One is then left with a sample that has label selectively placed on just (some of) the sites that change in the function being studied. The number of sensitive sites and their rates in both forms can then be directly determined. Figure 3b kinetically distinguishes two sets of sensitive hydrogens that change in rate by almost 1000-fold and 10,000-fold between the T and R forms, indicating a change in stability of 3.7 and 5.0 kcal/mol in free-energy at these two positions (eq 2 and 3). Selectivity for the sensitive sites and accuracy of the HX determination increase with the rate difference, i.e., with the energetic importance of the structure change. Still, one cannot tell where in the protein these sensitive sites are.
The development of a proteolytic fragmentation/ HPLC separation method [22] was able to locate the positions of some of the allosterically sensitive sites at intermediate resolution [23]. The method is made possible by the fact that the HX halftime for unprotected amides is over 1 h at pH 2 to 3 and 0 °C (see Figure 1). One takes samples in time from the exchanging Hb experiment and plunges the protein into 0 °C solution at pH 2.5. Hb unfolds but HX of the now freely exposed amides is slow enough to allow, with only modest loss of HX label, a brief proteolysis of the protein into fragments and their separation by fast HPLC. Scintillation counting can then locate and quantify the tritium label at the fragment level. The expected loss of HX label can be accurately predicted from known rates [5] and can be checked in trial experiments.
Figure 3c and d show results for the peptide fragment 1–29 at the N-terminus of the Hb α-chain and 130–146 at the C-terminus of the β-chain. Four allosterically sensitive amino acids are found on each terminal fragment. The hydrogens in each set all exchange at about the same rate in oxy Hb and they all move in unison to a new rate in deoxy Hb slower by 9-fold (α-chain) and 750-fold (β-chain). The indication is that the two sets of sensitive hydrogens are exposed to exchange by the concerted unfolding of two different chain segments that lose interactions worth 1.2 and 3.6 kcal/mol in free-energy, respectively, in the allosteric T to R transition (eq 2 and 3).
The ability to quantitatively evaluate stability and change in stability in terms of thermodynamically relevant structural free-energy is particularly valuable because regulatory proteins function as energy interconverting machines. The four subunits of Hb bind a total of four oxygens with positive cooperativity; initial oxygens are bound with reduced affinity and later ones more strongly. Qualitatively, the intra- and inter-subunit interactions that produce this behavior act through concerted structure changes. Quantitatively, the currency of these interactions is free-energy as codified in the equations of Wyman [24] and Monod et al. [25]. In Hb, the allosteric machinery is designed to interconvert oxygen binding free-energy and structure change free-energy (overall free-energy is conserved). When Hb binds its initial oxygens, the protein steals some of the binding energy (affinity is decreased), transduces it into structure change energy, carries this energy through the protein in the form of energetic structure changes, and transduces it back into enhanced binding energy at distant heme sites (affinity is increased). Thus, the initial ligands are bound with reduced energy, and the later ones with enhanced energy, generating the functionally important S-shaped binding curve. The HX experiment not only locates some of the operative segments but, uniquely, can measure how much allosteric free-energy is carried by each. Site-resolved changes in structural stabilization free-energy can be read out in terms of the stability of the allosterically sensitive segments against the unfolding reactions that govern their measurable HX rates (eq 3). Structure change can be measured by many methods but the ability to measure and localize changes in structural free-energy and thus track the pathways of energy-transfer through a protein has not been available before.
However, these methods are still limited. HPLC alone cannot rapidly resolve the large number of peptides produced by nonspecific acid proteases. Even for the few proteolytic fragments that could be obtained in relatively pure form, the results in Figure 3c and d do not closely specify the sensitive amino acid positions. For these purposes, it is necessary to obtain many more peptides.
HX Measurement: The MS Capability
The proteolytic fragmentation/HPLC separation method can be supplemented with a second dimension of peptide separation by mass spectrometry [26, 27]. This approach, when it is successful, can capture a large library of peptides that covers the entire length of an experimental protein. Further the need to use acid proteases at the initial fragmentation step, imposed by the requirement to perform the fragmentation-separation analysis at low pH, has a remarkable benefit. Acid proteases are notoriously non-specific. With sufficiently extensive proteolysis, many overlapping peptides can be obtained. The comparison of high accuracy data from overlapping peptides can make it possible to obtain HX structural resolution close to the amino acid level. Ultimately, one can look forward to the development of direct MS fragmentation methods that may efficiently accomplish this purpose, but the difficult H-D scrambling problem has so far frustrated this effort, although see [28].
In this method, the trace tritium label used before is replaced by labeling in D2O and the liquid scintillation analysis for tritium label is replaced by mass analysis for carried deuterium. The H-D exchange/MS capability can be deployed to measure exchange at the whole molecule level, analogous to the early work in Figure 3a or, more selectively, by functional labeling analogous to Figure 3b or, at higher structural resolution, by the proteolytic fragmentation method (Figure 3c–f).
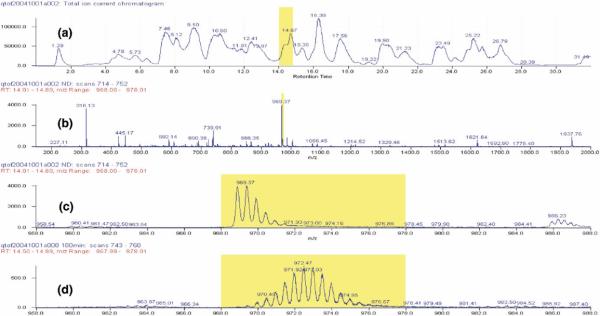
Figure 4 taken from recent experiments with α-synuclein [29] illustrates the method. The protein was placed into D2O. Aliquots of exchanging protein taken as a function of H-D exchange time were injected into an online flow system that carried them through two immobilized acid protease columns (pepsin and fungal protease XIII), then through an HPLC column to obtain a rough separation (Figure 4a), and then by electrospray into a QTOF mass spectrometer, which continually sweeps the mass range (2 s/sweep) as HPLC eluant enters (Figure 4b). The mass difference between undeuterated (Figure 4c) and partially deuterated (Figure 4d) peptide ions yields the number of D-labeled sites recovered for each peptide.
Figure 4.
H-D exchange/MS experiment. Amyloid (α-synuclein) HX labeled by timed exposure to D2O was dissociated with chemical denaturant at the condition of minimum HX rate (Figure 1), then passed through two immobilized acid protease columns, the proteolytic peptides were roughly resolved by HPLC, and then by on line ESI MS. (a) TIC trace of the HPLC eluant. (b) HPLC eluant was continually scanned by ESI MS, as shown for the eluant region marked in (a) MS results for the peptide ion marked in (b) are shown expanded for the protonated (c) and the partially deuterated (d) condition. Subtraction of the centroid mass of the unlabelled all-H peptide from the D-labeled peptide yields the number of D-labeled sites recovered on each peptide.
As before, the entire analysis for carried D-label is done in H2O solvent under conditions chosen to minimize the loss of label, namely at 0 °C or lower and at the pH minimum as shown in Figure 1. Unavoidably, some label will be lost during the time required for the analysis (“back exchange”). All D-label at exchangeable side-chain positions is totally lost with the possible exception of the NεH of argi-nine [5]. Also, label at the N-terminal amide, which is now a free amino group, is fully lost. Label on the penultimate amino acid amide is largely lost due to an end effect [5]. Through the length of each peptide, calculation using the chemical factors noted before leads one to expect a loss of only about 1% of the carried amide D-label per minute so that a final recovery of D-label in the range of 70 to 80% can be expected. However, experimental data so far available shows reproducible deviations from this value for any given peptide. Back exchange is generally dealt with empirically by calibrating and correcting for the deuterium loss on each peptide using the loss factor obtained from trial experiments with fully labeled samples.
Some Examples
The utility of these methods can be illustrated by results we have obtained in experiments with Hb (Figure 5) [35] and with α-synuclein amyloid (Parkinson's disease) (Figure 6) [29].
Figure 5.
H-D exchange/MS results for allosteric structure change in hemoglobin obtained as in Figure 4. Some allosterically sensitive amide sites were initially selectively labeled by the functional labeling method. Exchange-out results are compared for R-state oxy (red) and T-state deoxy (blue) Hb (obtained by subtracting the background from the raw data, both shown in gray; experimental details in [35]). HX was measured by the proteolytic fragmentation-separation method with analysis by MS. The peptides indicated span the entire length of the α- and the β-chains. The results show where changes occur and where they do not, and evaluate the number of amide sites affected within each sensitive segment. Several of the measured changes strongly suggest multi-residue segmental unfolding reactions (local unfolding) in which case the change in structural free-energy deposited at each allosterically sensitive position can be read out in terms of the change in HX rate between the two different allosteric forms (eq 3).
Figure 6.
H-D exchange/MS results for α-synuclein obtained as in Figure 4. Results for the soluble α-synuclein monomer (green) compare well with expected unprotected rates (dashed black curves). Results for the insoluble amyloid, shown in red, identify the well-protected chain segment that participates in the amyloid cross-β region and show the degree of protection in other regions.
The MS capability has provided peptides that scan the entire length of the Hb α- and β-subunits, as shown in Figure 5 [35]. Some of these results are compared with the earlier tritium exchange/HPLC results in Figure 3e and f. Good agreement is found. The much more complete results in Figure 5 show where structure change occurs and, importantly, where it does not. The comparison of data from overlapping peptides more closely places the positions of the allosteric changes. The allosterically sensitive positions found in this experiment, located at the α-chain N-terminus, the β-chain C-terminus, and the F helix and FG corners of both chains, are closely consistent with the allosteric model of Max Perutz [20]. In addition, these results support a local unfolding HX mechanism at the positions of some of the interesting allosteric changes, which makes it possible to measure the allosterically important free-energy that is carried at each of these sensitive positions.
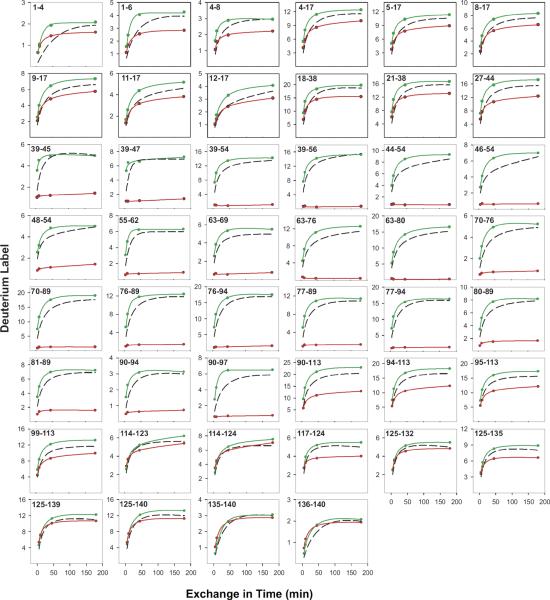
Results obtained for the soluble α-synuclein monomer and its amyloid form are shown in Figure 6. Amyloid is a massively aggregated and insoluble protein form that resists the usual structural studies, but not HX. Deuterium exchange-in was done at pDr 4 and 5 °C where exchange of even freely exposed amides proceeds on a ~10 min time scale (Figure 2) and so can be directly measured. Results for the α-synuclein monomer fairly closely match the expected unprotected rates, confirming its natively unfolded structure (compare the monomer data in green with the dashed black line predictions). Results for the insoluble amyloid (red in Figure 6) identify a 60 residue length in the systematically H-bonded cross-β amyloid region and place its position in the α-synuclein peptide. A 40 residue C-terminal length is wholly unprotected. An N-terminal length shows apparent heterogeneity with about 75% of the population freely exposed and 25% strongly protected. Overlapping peptides through the two boundary regions locate a somewhat ill-defined break from mostly unstructured to structured at residue position 38/39 ± 3 and a sharp discontinuity back to nonstructure at position 101/102 (±1). These results show the parts of the α-synuclein molecular length that participate in cross-β amyloid structure, give information on the condition of the segments that do not participate, imply how the chain is folded into the amyloid ribbon, and allow inferences concerning the general principles that govern amyloid formation and properties. For more extensive discussion see [29].
Perspective
We find ourselves in an exciting time in the development of structural biology. Thanks to advanced struc tural methods, especially X-ray crystallography and multi-dimensional NMR, we now know a lot about what proteins look like. But that is just the beginning. There is a vast amount to learn about how the protein machines of biology actually work. In this effort it is fundamental to understand what the machines look like but one needs to know a lot more. Protein function depends on dynamics and energetics. This is the proper domain of hydrogen exchange experimentation. We now know a lot about hydrogen exchange, how to measure it and how to interpret it in terms of protein structure, dynamics, and energetics, and thus track how these fundamental properties produce protein function. The ability to obtain structurally resolved HX data and to interpret it based on firm knowledge of HX chemistry and the role of protein structural dynamics seems most promising for future studies. The coupling of mass spectrometry with hydrogen exchange provides a special entré into these previously hidden avenues. This article offers a brief overview of where we have been. H-D exchange/MS experimentation on many proteins is now being pursued in a rapidly expanding number of laboratories. One can look forward to continuing advances that will make routine the HX investigation of the large biomolecules that make biology work.
Acknowledgments
This work was supported by NIH research grant GM031847 and The Mathers Charitable Foundation.
References
- 1.Hvidt A, Nielsen SO. Hydrogen exchange in proteins. Adv. Protein Chem. 1966;21:287–386. doi: 10.1016/s0065-3233(08)60129-1. [DOI] [PubMed] [Google Scholar]
- 2.Eigen M. Proton transfer, acid-base catalysis, and enzymatic hydrolysis. Angew. Chem. Int. Ed. English. 1964;3:1–19. [Google Scholar]
- 3.Englander SW, Kallenbach NR. Hydrogen exchange and structural dynamics of proteins and nucleic-acids. Q. Rev. Biophys. 1984;16:521–655. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]
- 4.Perrin CL. Proton exchange in amides: Surprises from simple systems. Accts. Chem. Res. 1989;22:268–275. [Google Scholar]
- 5.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connelly GP, Bai Y, Jeng MF, Englander SW. Isotope effects in peptide group hydrogen exchange. Proteins. 1993;17:87–92. doi: 10.1002/prot.340170111. [DOI] [PubMed] [Google Scholar]
- 7.Hvidt A. A discussion of the pH dependence of the hydrogen-deuterium exchange of proteins. C. R. Trav. Lab. Carlsberg. 1964;34:299–317. [PubMed] [Google Scholar]
- 8.Krishna MMG, Hoang L, Lin Y, Englander SW. Hydrogen exchange methods to study protein folding. Methods. 2004;34:51–64. doi: 10.1016/j.ymeth.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 9.Rose MC, Stuehr J. Kinetics of proton transfer reactions in aqueous solution: rates of internally hydrogen bonded systems. J. Am. Chem. Soc. 1968;90:7205–7209. [Google Scholar]
- 10.Haslam JL, Eyring EM. Deuterium oxide solvent isotope effects on N—H…O, O—H…N, and N…H.N intramolecular hydrogen bonds. J. Phys. Chem. 1967;71:4470–4475. [Google Scholar]
- 11.Milne JS, Mayne L, Roder H, Wand AJ, Englander SW. Determinants of protein hydrogen exchange studied in equine cytochrome c. Protein Sci. 1998;7:739–745. doi: 10.1002/pro.5560070323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milne JS, Xu Y, Mayne LC, Englander SW. Experimental study of the protein folding landscape: Unfolding reactions in cytochrome c. J. Mol. Biol. 1999;290:811–822. doi: 10.1006/jmbi.1999.2924. [DOI] [PubMed] [Google Scholar]
- 13.Calhoun DB, Vanderkooi JM, Englander SW. Penetration of small molecules into proteins studied by quenching of phosphorescence and fluorescence. Biochemistry. 1983;22:1533–1539. doi: 10.1021/bi00276a003. [DOI] [PubMed] [Google Scholar]
- 14.Calhoun DB, Englander SW, Wright WW, Vanderkooi JM. Quenching of room temperature protein phosphorescence by added small molecules. Biochemistry. 1988;27:8466–8474. doi: 10.1021/bi00422a026. [DOI] [PubMed] [Google Scholar]
- 15.Bai Y, Milne JS, Mayne L, Englander SW. Protein stability parameters measured by hydrogen exchange. Proteins. 1994;20:4–14. doi: 10.1002/prot.340200103. [DOI] [PubMed] [Google Scholar]
- 16.Huyghues-Despointes BMP, Scholtz JM, Pace CN. Protein conformational stabilities can be determined from hydrogen exchange rates. Nat. Struct. Biol. 1999;6:910–912. doi: 10.1038/13273. [DOI] [PubMed] [Google Scholar]
- 17.Bai Y, Englander SW. Future directions in folding: The multi-state nature of protein structure. Proteins. 1996;24:145–151. doi: 10.1002/(SICI)1097-0134(199602)24:2<145::AID-PROT1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 18.Maity H, Lim WK, Rumbley JN, Englander SW. Protein hydrogen exchange mechanism: Local fluctuations. Protein Sci. 2003;12:153–160. doi: 10.1110/ps.0225803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Englander SW. A hydrogen exchange method using tritium and Sephadex. Application to ribonuclease. Biochemistry. 1963;2:798–807. doi: 10.1021/bi00904a030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perutz MF. Mechanisms of cooperativity and allosteric regulation in proteins. Q Rev Biophys. 1989;22:139–237. doi: 10.1017/s0033583500003826. [DOI] [PubMed] [Google Scholar]
- 21.Englander JJ, Rogero JR, Englander SW. Protein- hydrogen exchange studied by a fragment separation method. Anal. Biochem. 1985;147:234–244. doi: 10.1016/0003-2697(85)90033-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rogero JR, Englander JJ, Englander SW. Individual breathing reactions measured by functional labeling and hydrogen exchange methods. Methods Enzymol. 1986;131:508–517. doi: 10.1016/0076-6879(86)31053-x. [DOI] [PubMed] [Google Scholar]
- 23.Englander SW, Englander JJ. Structure and energy change in hemoglobin by hydrogen exchange labeling. Methods Enzymol. 1994;232:26–42. doi: 10.1016/0076-6879(94)32041-1. [DOI] [PubMed] [Google Scholar]
- 24.Wyman J. Linked functions and reciprocal effects in hemoglobin: A second look. Adv. Protein Chem. 1964;19:223–286. doi: 10.1016/s0065-3233(08)60190-4. [DOI] [PubMed] [Google Scholar]
- 25.Monod J, Wyman J, Changeaux JP. On the nature of allosteric transitions: A plausible model. J. Mol. Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Z, Smith DL. Determination of amide-hydrogen exchange by mass spectrometry: A new tool for protein structure elucidation. Protein Sci. 1993;2:522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaltashov IA, Eyles SJ. Mass Spectrometry in Biophysics: Conformation and Dynamics of Biomolecules. John Wiley; Hoboken, NJ: 2005. p. 458. [Google Scholar]
- 28.Hoerner JK, Xiao H, Dobo A, Kaltashov IA. Is there hydrogen scrambling in the gas phase? Energetic and structural determinants of proton mobility within ions. J. Am. Chem. Soc. 2004;126:7709–7717. doi: 10.1021/ja049513m. [DOI] [PubMed] [Google Scholar]
- 29.Delmar C, Greenbaum EA, Mayne L, Englander SW, Woods VL., Jr. Structure and properties of α-synuclein and other amyloids determined at the amino acid level. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15477–15482. doi: 10.1073/pnas.0507405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hotchko M, Anand GS, Komives EA, Ten Eyck LF. Automated extraction of backbone deuteration levels from amide H/2H mass spectrometry experiments. Protein Sci. 2006;15:583–601. doi: 10.1110/ps.051774906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Engen JR, Smith DL. Investigating the higher order structure of proteins. Hydrogen exchange, proteolytic fragmentation, and mass spectrometry. Methods Mol. Biol. 2000;146:95–112. doi: 10.1385/1-59259-045-4:95. [DOI] [PubMed] [Google Scholar]
- 32.Eyles SJ, Kaltashov IA. Methods to study protein dynamics and folding by mass spectrometry. Methods. 2004;34:88–99. doi: 10.1016/j.ymeth.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 33.Hoofnagle AN, Resing KA, Ahn NG. Protein analysis by hydrogen exchange mass spectrometry. Annu. Rev. Biophys. Biomol. Struct. 2003;32:1–25. doi: 10.1146/annurev.biophys.32.110601.142417. [DOI] [PubMed] [Google Scholar]
- 34.Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom. Rev. 2006;25:158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 35.Englander JJ, Del Mar C, Li W, Englander SW, Kim JS, Stranz DD, Hamuro Y, Woods VC., Jr. Protein structure change studied by hydrogen-deuterium exchange, functional labeling, and mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 2003;100:7057–7062. doi: 10.1073/pnas.1232301100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Englander SW, Mauel C. Hydrogen exchange studies of respiratory proteins. Detection of discrete ligand induced changes in hemoglobin. J. Biol. Chem. 1972;247:2387–2394. [PubMed] [Google Scholar]
- 37.Liem RKH, Calhoun DB, Englander JJ, Englander SW. A high energy structure change in hemoglobin studied by difference hydrogen exchange. J. Biol. Chem. 1980;255:10687–10694. [PubMed] [Google Scholar]
- 38.Englander JJ, Rumbley JN, Englander SW. Signal transmission between subunits in the hemoglobin T-state. J. Mol. Biol. 1998;284:1707–1716. doi: 10.1006/jmbi.1998.2279. [DOI] [PubMed] [Google Scholar]