Abstract
Infection with Clostridium difficile is a growing concern because of the increasing prevalence and spread of nosocomial infections. Emergence of the hypervirulent 027/NAP1/BI strain is also notable. Existing diagnostic methods have low sensitivity or are time-consuming. Therefore, establishing a rapid and accurate microbiological diagnostic assay is needed. We evaluated the Xpert C. difficile assay (Xpert CD assay; Cepheid, USA) to detect toxigenic C. difficile. This assay is a real-time multiplex PCR assay that can be used to detect toxigenic C. difficile strains and differentiate the C. difficile presumptive 027/NAP1/BI strain. A total of 253 loose stool specimens were collected and toxigenic cultures, VIDAS C. difficile A & B assays (VIDAS CDAB assay; bioMérieux, France), and the Xpert CD assay were performed. In comparison to toxigenic cultures, the sensitivity, specificity, and positive and negative predictive values were 100%, 94.6%, 83.1%, and 100%, respectively, for the Xpert CD assay and 40.8%, 98.0%, 100%, and 88.9%, respectively, for VIDAS CDAB assay. Because of the low prevalence of the PCR ribotype 027 in Korea, the evaluation of the usefulness of the Xpert CD assay for screening for the 027 strain was limited. The Xpert CD assay provides great sensitivity in diagnosing toxigenic C. difficile infection. In addition, this method has excellent usability because it is simple and fast.
Keywords: Clostridium difficile, Real-time PCR, Enzyme immunoassay
Clostridium difficile is the leading cause of antibiotic-associated diarrhea and pseudomembranous colitis. The increasing prevalence and severity of healthcare-associated C. difficile infections (CDI) is of great concern [1]. Moreover, emergence and spread of the hypervirulent 027/NAP1/BI strain of C. difficile have been reported in North America and Europe [2-4]. The characteristics of the PCR ribotype 027 strain are production of the C. difficile binary toxin (CDT), as well as toxin A/B, and a single nucleotide deletion at position 117 in the tcdC gene [4]. The diagnosis of CDI should be based on a combination of symptoms and a positive stool test result for C. difficile toxins or toxigenic C. difficile [5]. Enzyme immunoassays rapidly detect toxins A and B, but their sensitivity varies greatly among the various products [6]. Toxigenic cultures and cytotoxin assays are considered as gold standard methods for the detection of toxigenic C. difficile, but toxigenic cultures that combine anaerobic cultures and detection of toxin A and B production take at least 48 hr to complete. Cytotoxin assays using cultured cells are also time-consuming and costly, making them unsuitable for routine laboratory diagnosis. Therefore, a rapid and more accurate microbiological diagnostic assay is highly needed for providing optimal patient care and controlling the spread of infections in hospitals.
The Xpert C. difficile assay (Xpert CD assay; Cepheid, Sunnyvale, CA, USA) is a real-time multiplex PCR assay performed using the GeneXpert Dx system. The assay uses primers targeted to the cytotoxin gene (tcdB), binary toxin genes (cdtA and cdtB), and a single nucleotide deletion at position 117 in the tcdC gene. As a result, the Xpert CD assay can detect toxigenic C. difficile strains and differentiate C. difficile presumptive 027/NAP1/BI. We evaluated the Xpert CD assay for rapidity and accuracy in diagnosing CDI.
A total of 253 consecutive loose stool specimens were collected in a stool specimen container from suspected CDI patients from April to June 2011, in a tertiary hospital. For toxigenic cultures, alcohol-shocked stool specimens were inoculated on C. difficile selective agar (CDSA; Becton, Dickinson and Company, Sparks, MD, USA) and incubated at 37℃ in an anaerobic chamber (Forma scientific, Marietta, OH, USA) for 48 hr. Suspected C. difficile colonies were used to make a gram-stained smear to observe typical morphology. The species were identified by using the ATB 32A system (bioMérieux, Marcy l'Etoile, France). The identified C. difficile isolates were used to detect tcdA repetitive regions, tcdB as well as cdtA and cdtB genes, following the previously described PCR method [7] and using the PCR primers listed in Table 1.
Table 1.
Sequences of the PCR primers used in this study
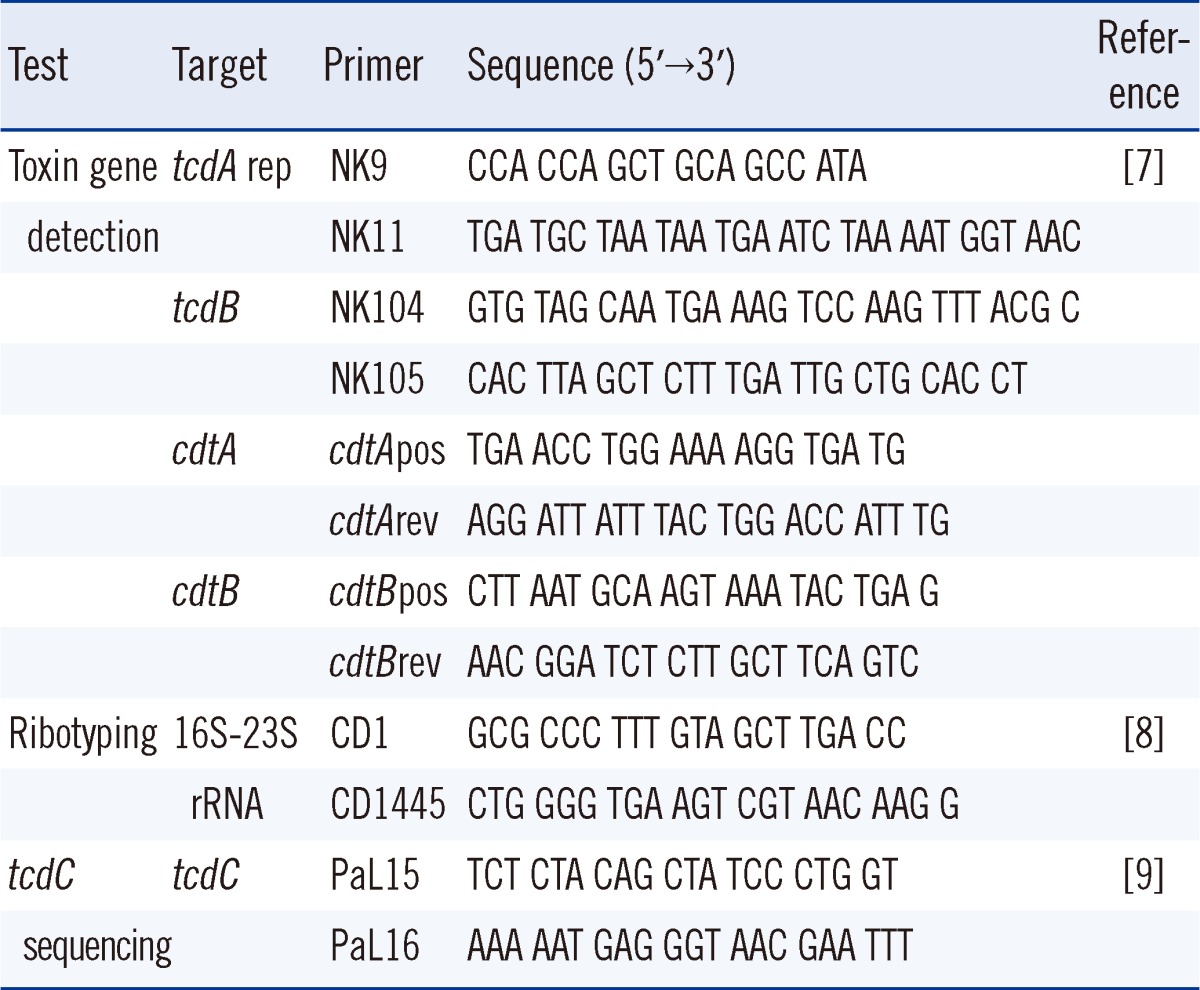
Abbreviations: tcdA rep, toxin A gene repetitive region; tcdB, toxin B gene; cdtA and cdtB, binary toxin genes.
Xpert CD assays were performed according to the manufacturer's instructions. A stool specimen was transferred to a vial containing a buffer solution by using a sterile swab. The vial was vortexed, and the solution was then transferred to a cartridge. The test was run on the GeneXpert DX module. The results were reported as C. difficile-positive 027/NAP1/BI presumptive negative, C. difficile-positive 027/NAP1/BI presumptive positive, C. difficile-negative, invalid, error, or no result. The test was repeated if the result was "invalid," "error," or "no result." Sequencing of the tcdC gene was performed on isolates that were positive for the presumptive 027/NAP1/BI strain. PCR ribotyping and tcdC sequencing were performed in accordance with previously described methods [8, 9] for isolates that tested positive for binary toxin genes in the Xpert CD assay in order to confirm the results.
VIDAS C. difficile A & B assays (VIDAS CDAB assay; bioMérieux) were performed according to the manufacturer's instructions. Test results are presented as positive, negative, or equivocal for toxins A and/or B. Specimens with equivocal results were retested once.
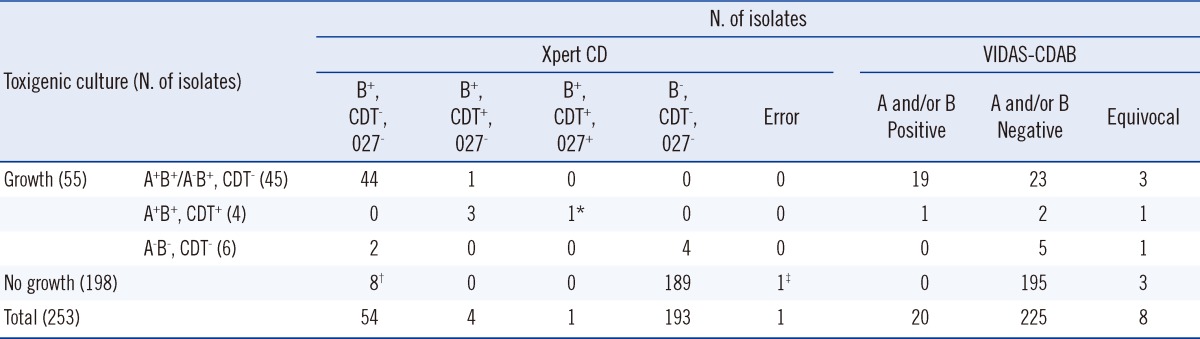
By anaerobic culture, 55 of 253 (21.7%) specimens yielded C. difficile isolates. Of these, 49 (19.4%) isolates were confirmed to be tcdB-positive (Table 2).
Table 2.
Evaluation of Xpert Clostridium difficile and VIDAS Clostridium difficile A & B assays for the detection of toxigenic Clostridium difficile isolates
*One presumptive 027/NAP1/BI strain identified as ribotype 078 on PCR ribotyping as well as a 39-base pair deletion and a point mutation at position 184 in tcdC; †Four specimens showed positive results by enrichment culture; ‡One "error" in the Xpert CD assay: no growth on anaerobic culture and negative on VIDAS-CDAB.
Abbreviations: Xpert CD, Cepheid Xpert Clostridium difficile assay; VIDAS-CDAB, VIDAS Clostridium difficile Toxin A&B assay; A, toxin A; B, toxin B; CDT, C. difficile binary toxin; 027, presumptive 027/NAP1/BI strain.
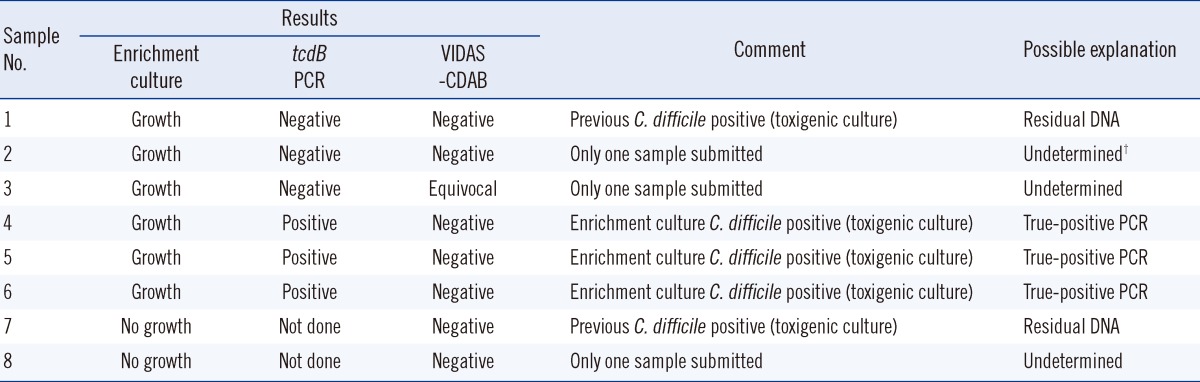
The Xpert CD assay detected tcdB in all 49 isolates identified as tcdB-positive by toxigenic culture (sensitivity 100%, Table 3). For 8 specimens that tested positive in the Xpert CD assay but were negative upon toxigenic culture, an enrichment culture was performed using cycloserine-cefoxitin fructose broth supplemented with 0.1% sodium taurocholate (TCCFB). Four of these eight specimens yielded a positive result for toxigenic C. difficile (Table 4). On the basis of analyses of other samples from the same patients, we suspect that at least 2 samples were contaminated with residual DNA [10]. In 3 undetermined cases, possible explanations for the discrepant results are residual DNA from prior CDI, false-positive PCR result, or true-positive PCR result.
Table 3.
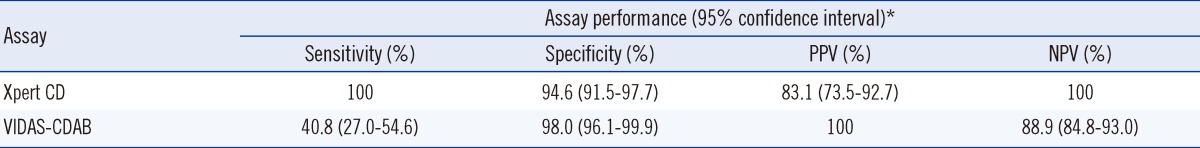
Assay performance of Xpert Clostridium difficile and VIDAS Clostridium difficile A & B assays for the detection of toxigenic Clostridium difficile isolates compared with toxigenic culture
*Sensitivity, specificity, PPV, and NPV are calculated as follows (×100): sensitivity, (number of true-positive assay results)/(sum of toxigenic culture-positive results); specificity, (number of true-negative assay results)/(sum of toxigenic culture-negative results); PPV, (number of true-positive assay results)/(sum of true-positive and false-positive assay results); NPV, (number of true-negative assay results)/(sum of true-negative and false-negative assay results).
Abbreviations: Xpert CD, Xpert Clostridium difficile assay; VIDAS-CDAB, VIDAS Clostridium difficile A & B assay; PPV, positive predictive value; NPV, negative predictive value.
Table 4.
Discordant results and further analysis of Xpert Clostridium difficile and toxigenic culture*
*All samples with initially no growth on anaerobic culture and Clostridium difficile-positive 027/NAP1/BI presumptive negative on Xpert CD assay; †False-positive PCR, residual DNA, or true-positive PCR.
Abbreviations: Xpert CD, Cepheid Xpert Clostridium difficile assay; VIDAS-CDAB, VIDAS Clostridium difficile Toxin A & B assay.
Compared to the toxigenic culture, the sensitivity, specificity, and positive and negative predictive values were 100%, 94.6%, 83.1%, and 100%, respectively, for the Xpert CD assay, and 40.8%, 98.0%, 100%, and 88.9%, respectively, for the VIDAS CDAB assay (Table 3). The overall agreement between the Xpert CD assay and toxigenic culture was 95.7%. Data from the enrichment culture were not included in the calculation of sensitivity, specificity, and positive and negative predictive values. One "error" case of the Xpert CD assay and 8 "equivocal" cases of the VIDAS CDAB assay were included in the calculation of assay performance (Table 2).
Binary toxin genes (cdtA and cdtB) were detected in 5 specimens by the Xpert CD assay, and 1 of them showed a 027/NAP1/BI presumptive positive result. The binary toxin genes were confirmed by toxin gene-specific PCR, PCR ribotyping, and tcdC sequencing. Four (including one 027/NAP1/BI presumptive positive isolate) of the 5 isolates revealed positive results for binary toxin genes. In addition, all 4 isolates showed an identical pattern to that of ribotype 078 and no deletion at position 117 of the tcdC gene. All ribotype 078 strains showed a 39-base pair deletion and a point mutation at position 184 in the tcdC gene [11].
Similar to a previous study, the evaluation of the usefulness of the Xpert CD assay for screening for the 027 strain was limited in this study due to the low prevalence of binary toxin-producing C. difficile strains (3.8% to 7.1%) and PCR ribotype 027 (0.6%) in Korea [12, 13]. A previously published study reported that the agreement between the Xpert CD assay and PCR-ribotyping was 93% [10]. Other studies reported discordant results for the presumptive 027/NAP1/BI strain between the Xpert CD assay and conventional typing and sequencing [14, 15]. These authors reported 1 ribotype 053 strain [15], 1 strain similar to the 078 strain [14], and 6 strains of unknown type. Ribotype 078 is the most frequent type present as a binary toxin-positive strain in Korea [12, 13]. Therefore, results of the presumptive 027/NAP1/BI strain must be interpreted with caution, particularly in Korea, where the prevalence of ribotype 027 is low.
The most significant advantage of the Xpert CD assay is its rapidity and simplicity. A loose stool specimen can be directly used, and the assay takes only 45 min.
In conclusion, the Xpert CD assay is a reliable method for detecting toxigenic C. difficile directly from stool specimens and provides greater sensitivity than an enzyme immunoassay.
Footnotes
No potential conflicts of interest relevant to this article were reported.
References
- 1.Kelly CP, LaMont JT. Clostridium difficile--more difficult than ever. N Engl J Med. 2008;359:1932–1940. doi: 10.1056/NEJMra0707500. [DOI] [PubMed] [Google Scholar]
- 2.McDonald LC, Killgore GE, Thompson A, Owens RC, Jr, Kazakova SV, Sambol SP, et al. An epidemic, toxin gene-variant strain of Clostridium difficile. N Engl J Med. 2005;353:2433–2441. doi: 10.1056/NEJMoa051590. [DOI] [PubMed] [Google Scholar]
- 3.Barbut F, Mastrantonio P, Delmée M, Brazier J, Kuijper E, Poxton I. Prospective study of Clostridium difficile infections in Europe with phenotypic and genotypic characterisation of the isolates. Clin Microbiol Infect. 2007;13:1048–1057. doi: 10.1111/j.1469-0691.2007.01824.x. [DOI] [PubMed] [Google Scholar]
- 4.Warny M, Pepin J, Fang A, Killgore G, Thompson A, Brazier J, et al. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet. 2005;366:1079–1084. doi: 10.1016/S0140-6736(05)67420-X. [DOI] [PubMed] [Google Scholar]
- 5.Cohen SH, Gerding DN, Johnson S, Kelly CP, Loo VG, McDonald LC, et al. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the society for healthcare epidemiology of America (SHEA) and the infectious diseases society of America (IDSA) Infect Control Hosp Epidemiol. 2010;31:431–455. doi: 10.1086/651706. [DOI] [PubMed] [Google Scholar]
- 6.Planche T, Aghaizu A, Holliman R, Riley P, Poloniecki J, Breathnach A, et al. Diagnosis of Clostridium difficile infection by toxin detection kits: a systematic review. Lancet Infect Dis. 2008;8:777–784. doi: 10.1016/S1473-3099(08)70233-0. [DOI] [PubMed] [Google Scholar]
- 7.Terhes G, Urbán E, Sóki J, Hamid KA, Nagy E. Community-acquired Clostridium difficile diarrhea caused by binary toxin, toxin A, and toxin B gene-positive isolates in Hungary. J Clin Microbiol. 2004;42:4316–4318. doi: 10.1128/JCM.42.9.4316-4318.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Neill GL, Ogunsola FT, Brazier JS, Duerden BI. Modification of a PCR ribotyping method for application as a routine typing scheme for Clostridium difficile. Anaerobe. 1996;2:205–209. [Google Scholar]
- 9.Spigaglia P, Mastrantonio P. Comparative analysis of Clostridium difficile clinical isolates belonging to different genetic lineages and time periods. J Med Microbiol. 2004;53:1129–1136. doi: 10.1099/jmm.0.45682-0. [DOI] [PubMed] [Google Scholar]
- 10.Babady NE, Stiles J, Ruggiero P, Khosa P, Huang D, Shuptar S, et al. Evaluation of the Cepheid Xpert Clostridium difficile Epi assay for diagnosis of Clostridium difficile infection and typing of the NAP1 strain at a cancer hospital. J Clin Microbiol. 2010;48:4519–4524. doi: 10.1128/JCM.01648-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goorhuis A, Bakker D, Corver J, Debast SB, Harmanus C, Notermans DW, et al. Emergence of Clostridium difficile infection due to a new hypervirulent strain, polymerase chain reaction ribotype 078. Clin Infect Dis. 2008;47:1162–1170. doi: 10.1086/592257. [DOI] [PubMed] [Google Scholar]
- 12.Kim H, Lee Y, Moon HW, Lim CS, Lee K, Chong Y. Emergence of Clostridium difficile ribotype 027 in Korea. Korean J Lab Med. 2011;31:191–196. doi: 10.3343/kjlm.2011.31.3.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim H, Jeong SH, Roh KH, Hong SG, Kim JW, Shin MG, et al. Investigation of toxin gene diversity, molecular epidemiology, and antimicrobial resistance of Clostridium difficile isolated from 12 hospitals in South Korea. Korean J Lab Med. 2010;30:491–497. doi: 10.3343/kjlm.2010.30.5.491. [DOI] [PubMed] [Google Scholar]
- 14.Huang H, Weintraub A, Fang H, Nord CE. Comparison of a commercial multiplex real-time PCR to the cell cytotoxicity neutralization assay for diagnosis of Clostridium difficile infections. J Clin Microbiol. 2009;47:3729–3731. doi: 10.1128/JCM.01280-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tenover FC, Akerlund T, Gerding DN, Goering RV, Boström T, Jonsson AM, et al. Comparison of strain typing results for Clostridium difficile isolates from North America. J Clin Microbiol. 2011;49:1831–1837. doi: 10.1128/JCM.02446-10. [DOI] [PMC free article] [PubMed] [Google Scholar]