Abstract
We have used a translational convergent functional genomics (CFG) approach to identify and prioritize genes involved in schizophrenia, by gene-level integration of genome-wide association study data with other genetic and gene expression studies in humans and animal models. Using this polyevidence scoring and pathway analyses, we identify top genes (DISC1, TCF4, MBP, MOBP, NCAM1, NRCAM, NDUFV2, RAB18, as well as ADCYAP1, BDNF, CNR1, COMT, DRD2, DTNBP1, GAD1, GRIA1, GRIN2B, HTR2A, NRG1, RELN, SNAP-25, TNIK), brain development, myelination, cell adhesion, glutamate receptor signaling, G-protein–coupled receptor signaling and cAMP-mediated signaling as key to pathophysiology and as targets for therapeutic intervention. Overall, the data are consistent with a model of disrupted connectivity in schizophrenia, resulting from the effects of neurodevelopmental environmental stress on a background of genetic vulnerability. In addition, we show how the top candidate genes identified by CFG can be used to generate a genetic risk prediction score (GRPS) to aid schizophrenia diagnostics, with predictive ability in independent cohorts. The GRPS also differentiates classic age of onset schizophrenia from early onset and late-onset disease. We also show, in three independent cohorts, two European American and one African American, increasing overlap, reproducibility and consistency of findings from single-nucleotide polymorphisms to genes, then genes prioritized by CFG, and ultimately at the level of biological pathways and mechanisms. Finally, we compared our top candidate genes for schizophrenia from this analysis with top candidate genes for bipolar disorder and anxiety disorders from previous CFG analyses conducted by us, as well as findings from the fields of autism and Alzheimer. Overall, our work maps the genomic and biological landscape for schizophrenia, providing leads towards a better understanding of illness, diagnostics and therapeutics. It also reveals the significant genetic overlap with other major psychiatric disorder domains, suggesting the need for improved nosology.
Keywords: biomarkers, convergent functional genomics, genetic risk prediction, pathways, schizophrenia
Introduction
‘Things fall apart; the center cannot hold'
– WB Yeats, The Second Coming
Schizophrenia is a devastating disorder affecting ∼1% of the population. While there is clear evidence for roles for both genes and environment, a comprehensive biological understanding of the disorder has been elusive so far. Most notably, there has been until recently a lack of concerted integration across functional and genetic studies, and across human and animal model studies, resulting in missed opportunities to see the whole picture.
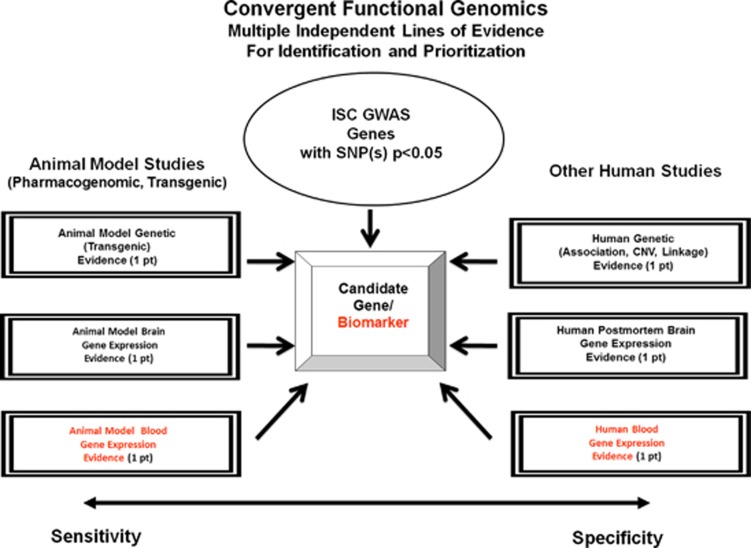
As part of a translational convergent functional genomics (CFG) approach, developed by us over the last decade,1, 2, 3, 4, 5 and expanding upon our earlier work on identifying genes for schizophrenia6 and biomarkers for psychosis,7 we set out to comprehensively identify candidate genes, pathways and mechanisms for schizophrenia, integrating the available evidence in the field to date. We have used data from published genome-wide association studies (GWAS) data sets for schizophrenia.8, 9 We integrated those data with gene expression data—human postmortem brain gene expression data, human induced pluripotent stem cell-derived neuronal cells10 and human blood gene expression data7 published by others and us, as well as with relevant animal model brain and blood gene expression data generated by our group6 and others. In addition, we have integrated as part of this comprehensive approach other genetic data—human genetic data (linkage, copy number variant (CNV) or association) for schizophrenia, as well as relevant mouse model genetic evidence (Figure 1, Table 1 and Figure 2). Animal model data provide sensitivity of detection, and human data provide specificity for the illness. Together, they help to identify and prioritize candidate genes for the illness, using a polyevidence CFG score, resulting in essence in a de facto field-wide integration putting together the best available evidence to date. Once that is done, biological pathway analyses can be conducted and mechanistic models can be constructed (Figure 3).
Figure 1.
Convergent functional genomics. GWAS, genome-wide association study; ISC, International Schizophrenia Consortium; SNP, single-nucleotide polymorphism.
Table 1. Top candidate genes for schizophrenia—CFG analysis of ISC GWAS data.
| Gene Symbol/name | ISC GWAS best P-value SNP | Animal model genetic evidence (TG ) | Animal model brain gene expression evidence | Animal model blood gene expression evidence6 | Human genetic evidence (association CNV or linkage) | Human postmortem brain gene expression evidence | Human blood/other peripheral tissue gene expression evidence | CFG score | GAIN-EA GWAS best P-value SNP | GAIN-AA GWAS best P-value SNP |
|---|---|---|---|---|---|---|---|---|---|---|
| DISC1, disrupted in schizophrenia 1 | 0.002934, rs10910616 | SZ18, 27, 78, 79, 80, 81, 82, 83, 84, 85 | (I) Antipsychotic treatment,86 mouse neurodevelopment87 | 1q42.2 (Assoc) SZ22, 67, 88, 89, 90, 91, 92, 93, 94 | (I) SZ HIP25 | (I) SZ lymphoblasts,25 PMBCs26 | 5.0 | 0.001562, rs12754490 | 0.0001308, rs11122318 | |
| HSPA1B, heat-shock 70-kDa protein 1B | 0.0009003, rs2763979 | (D) PCP HIP; (D) CLZ VT6 | (D) CLZ | 6p21.33 (Assoc) SZ44 | (I) SZ45 | (I) SZ, IPS-derived neurons10 | 5.0 | |||
| MBP, myelin basic protein | 0.01002, rs12959006 | (I) PCP and CLZ AMY; (D), PCP PFC,6 (I), SZ,95 (D) SZ96 | (I) PCP and CLZ | 18q23 (Assoc) SZ37 | (D) SZ38 | (D) SZ peripheral blood97 | 5.0 | 0.01345, rs9967028 | 0.03257, rs1629089 | |
| TCF4, transcription factor 4 | 0.0002902, rs17594665 | SZ98 | (I) PCP NAC6 | 18q21.2 (Assoc) SZ32, 33, 34, 35 | (I) SZ36 | (I) SZ, IPS-derived neurons;10 (D) delusions SZ7 | 5.0 | 0.01039, rs17594665 | 0.00126, rs1539951 | |
| MOBP, myelin-associated oligodendrocyte basic protein | 0.003529, rs1708044 | (I), PCP and CLZ AMY; (D), CLZ CP; (I), CLZ NAC; (D), PCP PFC; (D), CLZ VT,6 (I), Psychosis,99 (I), Response to antipsychotics,100 (D), SZ101 | (I) CLZ | 3p22.1 (Linkage) SZ102 | (D), SZ,38 (I), SZ and substance abuse103 | (D), SZ lymphocytes104 | 4.5 | 0.02583, rs1405798 | 0.004474, rs1538783 | |
| NCAM1, neural cell adhesion molecule 1 | 0.003917, rs11214441 | SZ105 | (I) SZ95 | 11q23.2 (Assoc) SZ106 | (I) SZ106, 107 | (D) SZ, IPS-derived neurons10 | 4.5 | 0.002043, rs1245133 | 0.001454, rs600964 | |
| NDUFV2, NADH dehydrogenase (ubiquinon) flavoprotein 2, 24kDa | 0.003243, rs8084822 | Response to antipsychotics108 | (D) PCP and CLZ | 18p11.22 (Assoc) SZ109 | (D), SZ Striatum,110 (I), SZ parito-occipital cortex1, 10 (I), SZ111 | (I), SZ lymphocytes110 | 4.5 | 0.004361, rs1893144 | 0.0009294, rs10468792 | |
| NRCAM, neuronal cell adhesion molecule | 0.006234, rs10250083 | SZ112 | (I) CLZ VT6 | 7q31.1 (Linkage) SZ113 | (D) SZ48 | (D) SZ serum49 | 4.5 | 0.004773, rs404287 | 0.002343, rs409797 | |
| RAB18, RAB18, member RAS oncogene family | 0.03817, rs12261690 | (I) PCP AMY; (D), PCP PFC; (D) CLZ VT6 | (I) PCP | 10p12.1 (Linkage) SZ114, 115, 116, 117, 118 | (D) SZ119 | (D) SZ whole blood120 | 4.5 | 0.01716, rs7476899 | 0.01231, rs11015796 | |
| ADCYAP1, adenylate cyclase-activating polypeptide 1 (pituitary) | 0.002876, rs9954574 | SZ121 | (I) CLZ NAC; (D) CLZ VT 6 | 18p11.32 (Assoc) SZ121, 122 | (D) SZ119 | 4.0 | 0.02558, rs1394890 | 0.005448, rs16953183 | ||
| ALDH1A1, aldehyde dehydrogenase 1 family, member A1 | 0.02526, rs11143438 | (I) PCP and CLZ AMY; (D), PCP and CLZ NAC,6 (I) Psychosis99 | (I) PCP | (D) SZ,111, 123 (I) SZ124 | (I) SZ, fibroblasts125 | 4.0 | 0.01389, rs7028573 | 0.01285, rs11999628 | ||
| ANK3, ankyrin 3, node of Ranvier (ankyrin G) | 0.001727, rs4948256 | (D) PCP AMY; (I), PCP CP;v(D), PCP NAC; (I), CLZ VT6 | 10q21.2 (Assoc) SZ48, 126 | (D) SZ48 | (I), SZ IPS-derived neurons10 | 4.0 | 0.006456, rs10509133 | 0.005837, rs7906905 | ||
| BDNF, brain-derived neurotrophic factor | 0.001666, rs10742178 | (I) SZ,127, 128, 129 (I) PCP in rats, 130 (I) MK-801 in rats131 | 11p14.1 (Assoc) SZ132, 133, 134, 135 | (D) SZ136, 137, 138 | (D) SZ serum,49, 139, 140, 141 (D) leukocytes psychosis142 | 4.0 | 0.005507, rs2131060 | |||
| CD9, CD9 molecule | 0.0455, rs3181291 | (I) PCP AMY; (D), PCP PFC; (I), PCP VT6 | (I) PCP and CLZ | (D) SZ143 | (D) SZ IPS-derived neurons10 | 4.0 | 0.01167, rs2268014 | 0.04739, rs7342306 | ||
| CNR1, cannabinoid receptor 1 (brain) | 0.002567, rs1324073 | SZ144 | (D) PCP and CLZ VT,6 (D) response to antipsychotics100 | 6q15 (Assoc), SZ50, 51 | (D) SZ52 | 4.0 | 0.001542, rs9451023 | 0.002128, rs873413 | ||
| COMT, catechol-O-methyltransferase | 0.04098, rs1544325 | SZ145, 146 | (I), CLZ VT6, (D), SZ147 | 22q11.21 (Assoc) SZ148 | (I), SZ149 | 4.0 | 0.01457, rs1544325 | |||
| CPLX2, complexin 2 | 0.04338, rs10213927 | SZ150 | (D), PCP and CLZ VT,6 (D), SZ127 | 5q35.2, (Assoc), SZ151 | (D), SZ136, 152, 153 | 4.0 | 0.01658, rs6887620 | |||
| DRD2, dopamine receptor D2 | 0.01151, rs12791990 | (D), PCP and CLZ PFC,6 (D), SZ154 | 11q23.2 (Assoc), SZ155, 156 | (D) SZ136, 157, 158 | (D) SZ delusions7 | 4.0 | 0.007265, rs4938021 | 0.01096, rs17529477 | ||
| DTNBP1, dystrobrevin binding protein 1 | 0.002634, rs12209943 | SZ159, 160, 161, 162 | 6p22.3 (Assoc) SZ163, 164, 165, 166, 167, 168 | (D), SZ169, 170, 171, 172 | (D) Lymphocytes173 | 4.0 | 0.009146, rs9477021 | 0.0001501, rs16876575 | ||
| FABP7, fatty acid–binding protein 7, brain | 0.01053, rs9490546 | SZ174 | (I), CLZ NAC; (I), PCP and CLZ PFC6 | 6q22.31 (Assoc), SZ174 | (I), SZ174 | 4.0 | ||||
| GABRB3, gamma-aminobutyric acid (GABA) A receptor, β3 | 0.004635, rs8037461 | (I), PCP AMY; (D), PCP HIP; (D), PCP PFC6 | 15q12 (Assoc) SZ175 | (I) SZ,176 (D) SZ177 | (I), SZ serum178 | 4.0 | 0.01579, rs12904865 | 0.0009769, rs4906835 | ||
| GAD1, glutamate decarboxylase 1 (brain, 67 kDa) | 0.03907, rs16859026 | (I), PCP AMY,6 (D), SZ,127, 128 (I), SZ176 | (I), CLZ | 2q31.1 (Assoc), SZ179 | (D), SZ,136, 176, 177, 179, 180, 181, 182, 183, 184, 185, 186, 187, 188, 189, 190, 191 (I), SZ192 | 4.0 | 0.008447, rs10191129 | 0.01776, rs2883888 | ||
| GNB1L, guanine nucleotide–binding protein (G-protein), β-polypeptide 1-like | 0.03659, rs17745302 | Impaired PPI193 | (I), Chronic haloperidol,194 (D), SZ147 | 22q11.21 (Assoc), SZ195 | (D), SZ 194 | 4.0 | ||||
| GRIA1, glutamate receptor, ionotropic, AMPA 1 | 0.0008031, rs2962816 | SZ196 | (D), PCP and CLZ AMY,6 (I), response to antipsychotics,100 (D), response to PCP197 | 5q33.2 (Assoc), SZ 198, 199 | (D), SZ200, 201, (I), SZ181, 202, 203 | 4.0 | 0.00659, rs10044974 | 0.006037, rs498660 | ||
| GRIA4, glutamate receptor, ionotrophic, AMPA 4 | 0.02792, rs649098 | Cognition (impaired PPI)204 | (I), PCP and CLZ AMY,6 (D), SZ205 | 11q22.3 (Assoc), SZ206 | (D), SZ,119, 207 (I), SZ202 | 4.0 | 0.001526, rs7116118 | 0.00343, rs2277280 | ||
| GRIN2B, glutamate receptor, ionotropic, N-methyl D-aspartate 2B | 0.001569, rs4363703 | SZ 208 | (D), CLZ AMY; (I), CLZ VT,6 MK-801-treated rats,209 (D), response to antipsychotics,100 (D), SZ,176 | 12p13.1 (Assoc), SZ155, 167 | (D), SZ,201 (I), SZ210 | 4.0 | 0.001427, rs1072388 | 0.003102, rs12826365 | ||
| GRM5, glutamate receptor, metabotropic 5 | 0.002559, rs992259 | SZ211 | (D), PCP AMY, (D), PCP VT,6 (D), SZ129 | 11q14.3 (Assoc), SZ212 | (D), SZ181 | 4.0 | 0.01842, rs170110 | 0.001263, rs1846475 | ||
| GSN, gelsolin | 0.04739, rs12376078 | (I), PCP AMY, (D), CLZ CP6 | (I) PCP and CLZ | 33.2 (Assoc), SZ213 | (D), SZ111, 192143, 214 | 4.0 | 0.002313, rs767770 | 0.0001564, rs4837835 | ||
| HINT1, histidine triad nucleotide–binding protein 1 | 0.00008672, rs11242025 | SZ215 | 5q23.3 (Assoc), SZ, 216, 217 | (D), SZ218 | (I), SZ whole blood120 | 4.0 | 0.008637, rs7734177 | |||
| HTR2A, 5-hydroxytryptamine (serotonin) receptor 2A | 0.02014, rs7985155 | (I), Response to antipsychotics,100 (D), SZ219 (I), SZ154 | 13q14.2 (Assoc), SZ155, 220 | (D), SZ136, 221, 222 | (D), SZ lymphocytes223 | 4.0 | 0.003461, rs17070879 | 0.002529, rs1886439 | ||
| KALRN, kalirin, RhoGEF kinase | 0.006285, rs3772756 | (I), CLZ VT6 | 3q21.1 (Assoc), SZ224 | (D), SZ225 | (D), SZ IPS-derived neurons10 | 4.0 | 0.01015, rs9832419 | 0.009074, rs1822791 | ||
| KIF2A, kinesin heavy chain member 2A | 0.005374, rs6864793 | (D), CLZ VT6 | (I) PCP and CLZ | 5q12.1 (Assoc), SZ226 | (D), SZ119 | 4.0 | 0.003396, rs153864 | 0.005207, rs10069830 | ||
| NR4A2, nuclear receptor subfamily 4, group A, member 2 | 0.0006887, rs12465886 | SZ227 | (I), PCP and CLZ HIP, (I), PCP and CLZ NAC;, (D), PCP PFC, (I), PCP and CLZ VT6 | 2q24.1 (Assoc), SZ,228 | (I), SZ IPS-derived neurons 10 | 4.0 | 0.001624, rs6743834 | 0.004081, rs16840214 | ||
| NRG1, neuregulin 1 | 0.001731, rs1158001 | SZ229, 230, 231 | 8p12 (Assoc), SZ94, 220, 232, 233, 234, 235 | (D), SZ38, 236, 237, 238 (I), SZ239, 240, 241 | (I), SZ IPS-derived neurons,10 (I), SZ, Leucocytes,242 (I), SZ lymphocyte, 243 (I), SZ delusions7 | 4.0 | 0.00104, rs2716960 | 0.000006564, rs6989777 | ||
| PDE4B, phosphodiesterase 4B, cAMP-specific | 0.003042, rs6588193 | SZ244 | (D), PCP,6 (I), psychosis,245 (D), response to antipsychotics246 | 1p31.3 (Assoc) SZ234, 247, 248, 249, 250 | (D) SZ247 | 4.0 | 0.02102, rs11805090 | 0.000103, rs17417507 | ||
| PRKCA, protein kinase C, alpha | 0.007991, rs6504428 | (I), CLZ PFC, (D), CLZ VT,6 (I), response to antipsychotics251 | 17q24.2, (Assoc), SZ252, 253 | (D), SZ136 | (D), SZ IPS-derived neurons10 | 4.0 | 0.01166, rs9908814 | 0.0004001, rs16959057 | ||
| RELN, reelin | 0.01368, rs2711865 | SZ254 | (I), CLZ PFC;, (D), PCP and CLZ VT,6 (D), SZ128, 255 | 7q22.1 (Assoc), SZ94, 155, 233, 256, 257, 258, 259 | (D), SZ136, 186, 187, 190, 260, 261 | 4.0 | 0.007165, rs10227303 | 0.004365, rs7794418 | ||
| RGS4, regulator of G-protein signaling 4 | 0.004835, rs4657235 | SZ262 | (I), PCP and CLZ AMY;, (I), CLZ HIP ;, (D), CLZ PFC;, (I), PCP and CLZ VT,6 (D), SZ263, 264 | 1q23.3 (Assoc), SZ167, 220, 265, 266 | (D), SZ,45, 210, 267, 268, 269, 270, 271 (I), SZ272 | 4.0 | 0.007928, rs12403644 | 0.007516, rs10917637 | ||
| SLC1A2, solute carrier family 1 (glial high-affinity glutamate transporter), member 2 | 0.02565, rs3794086 | (D), CLZ AMY, (D), CLZ VT6 | (I), CLZ | 11p13 (Assoc), SZ,273 | (D), SZ,153 (I), SZ274, 275 | 4.0 | 0.03109, rs3829280 | 0.002563, rs12270460 | ||
| SNAP25, synaptosomal-associated protein, 25 kDa | 0.01815, rs6032783 | SZ276 | (I), SZ127, 263 | 20p12.2 (Assoc), SZ277 | (D), SZ,181, 278, 279, 280, 281 (I), SZ181, 282, 283 | 4.0 | 0.005819, rs362616 | 0.01192, rs362560 | ||
| SYN2, synapsin II | 0.003144, rs2960421 | SZ284, 285 | (D), PCP and CLZ AMY, (D), PCP CP, (D), PCP and CLZ VT,6 (I), SZ,205 (D), SZ127, 263 | 3p25.2 (Assoc), SZ151, 286, 287 | (D), SZ,45, 288183 (I), SZ antipsychotic, treatment107 | 4.0 | 0.042, rs2618406 | 0.02541, rs17671592 | ||
| TNIK, TRAF2 and NCK interacting kinase | 0.001377, rs260769 | (I), CLZ VT6 | 3q26.31 (Assoc), SZ9, 89, 234, 289 | (I), SZ278 | (D), SZ lymphoblastoid cell lines290 | 4.0 | 0.006987, rs12639373 | 0.00007179, rs13065441 |
Abbreviations: AA, African American; AMY, amygdala; Assoc, association evidence; CFG, convergent functional genomics; CLZ, clozapine; CP, caudate putamen; D, decreased in expression; EA, European American; GWAS, genome-wide association study; I, increased in expression; IPS, pluripotent stem cell; ISC, International Schizophrenia Consortium; Linkage, linkage evidence; NAC, nucleus accumbens; PCP, phencyclidine; PFC, prefrontal cortex; PMBC, peripheral mononuclear blood cells; SNP, single-nucleotide polymorphism; SZ, schizophrenia; TG, transgenic; VT, ventral tegmentum.
Top genes with a CFG score of 4 and above (n=42) are shown. A more complete list of genes with CFG score of 3 and above (n=186) is available in the Supplementary Information. Gene symbols underlined are blood biomarker candidate genes. Best P-value SNP within the gene or flanking regions are depicted. P-values in bold are <0.001. The last two columns depict gene-level replication of findings, that is, best P-value SNPs in the same gene from two other independent cohorts (GAIN EA and GAIN AA). In total, 37 of our top 42 genes (88.1%) had at least a SNP with P<0.05, in both the GAIN-EA cohort and in the GAIN-AA cohort.
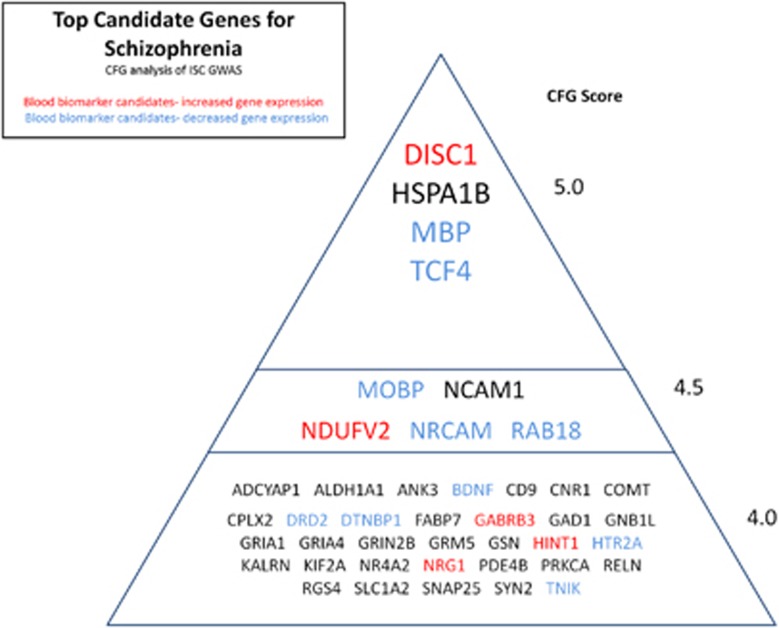
Figure 2.
Top candidate genes for schizophrenia. CFG, convergent functional genomics; GWAS, genome-wide association study; ISC, International Schizophrenia Consortium.
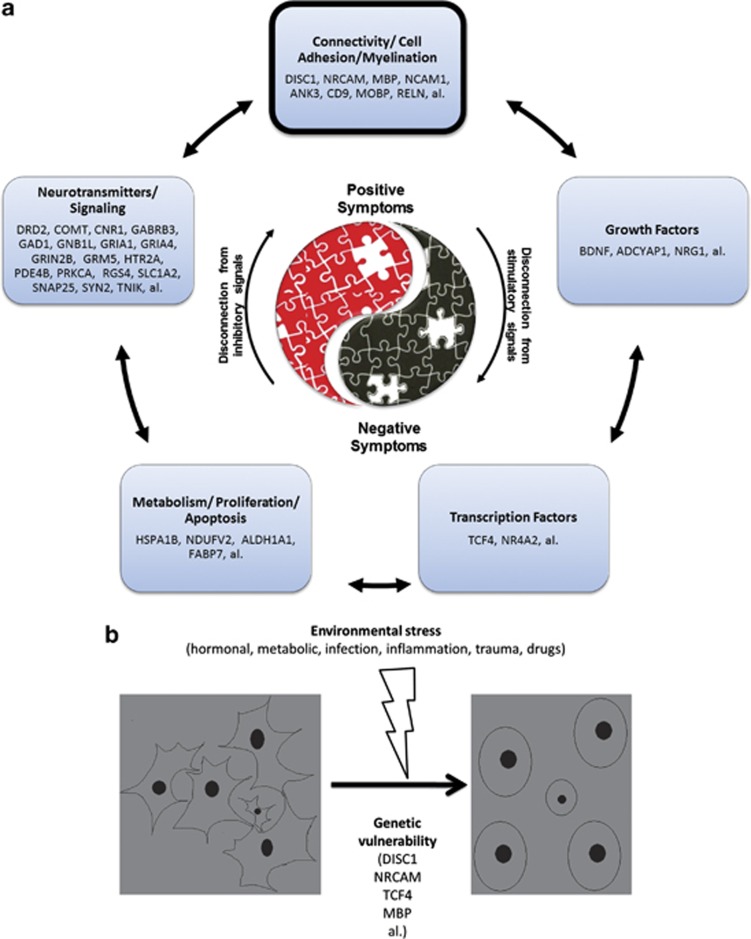
Figure 3.
Schizophrenia as a disease of disconnection. (a) Biology of schizophrenia, (b) gene–environment interplay.
An obvious next step is developing a way of applying that knowledge to genetic testing of individuals to determine risk for the disorder. On the basis of our comprehensive identification of top candidate genes described in this paper, we have chosen the nominally significant single-nucleotide polymorphisms (SNPs) inside those genes in the GWAS data set used for discovery (International Schizophrenia Consortium, ISC), and assembled a genetic risk prediction (GRP) panel out of those SNPs. We then developed a genetic risk prediction score (GRPS) for schizophrenia based on the presence or absence of the alleles of the SNPs associated with the illness in ISC, and tested the GRPS in independent cohorts (GAIN European Americans (EA), GAIN African Americans (AA), nonGAIN EA, nonGAIN AA)9 for which we had both genotypic and clinical data available, comparing the schizophrenia subjects to normal controls. Our results show that a panel of SNPs in top genes identified and prioritized by CFG analysis can differentiate between schizophrenia subjects and controls at a population level, although at an individual level the margin is minimal. The latter point suggests that, like for bipolar disorder,11 the contextual cumulative combinatorics of common variants and environment12 plays a major role in risk for illness. Moreover, the genetic risk component identified by us seems to be stronger for classic age at onset schizophrenia than for early onset and late-onset schizophrenia, suggesting that those subtypes may be different, either in having a larger environmental component or having a different genetic component.
We have also looked at genetic heterogeneity, overlap and reproducibility between independent GWAS for schizophrenia. We show that the overlap is minimal at a nominal P-value SNP level, but increases dramatically at a gene level, then at a CFG-prioritized gene level and finally at a pathway level. CFG provides a fit-to-disease prioritization of genes that leads to generalizability in independent cohorts, and counterbalances the fit-to-cohort prioritization inherent in classic SNP level genetic-only approaches, which have been plagued by poor reproducibility across cohorts. Finally, we have looked at overlap with candidate genes for other psychiatric disorders (bipolar disorder, anxiety disorders), as well as with other disorders affecting cognition (autism, Alzheimer disease (AD)), and provide evidence for shared genes.
Overall, this work sheds comprehensive light on the genetic architecture and pathophysiology of schizophrenia, provides mechanistic targets for therapeutic intervention and has implications for genetic testing to assess risk for illness before the illness manifests itself clinically.
Materials and methods
Genome-wide association studies data for schizophrenia
The GWAS data from the ISC was used for the discovery CFG work.8 This cohort consists of EA subjects (3322 schizophrenics and 3587 controls). SNPs with a nominal allelic P-value <0.05 were selected for our analysis. No Bonferroni correction was performed.
Four independent cohorts,9 two EA (GAIN EA 1170 schizophrenics and 1378 controls; nonGAIN EA 1149 schizophrenics and 1347 controls) and two AA (GAIN AA 915 schizophrenics and 949 controls; nonGAIN AA 78 schizophrenics and 20 controls), were used for testing the results of the discovery analyses. The GWAS GAIN and nonGAIN data used for analyses described in this paper were obtained from the database of Genotype and Phenotype (dbGaP) found at www.ncbi.nlm.nih.gov.
The software package PLINK (http://pngu.mgh.harvard.edu/~purcell) was used to extract individual genotype information for each subject from the GAIN GWAS data files. We analyzed EA, and separately, AA, schizophrenia subjects and controls.
Gene identification
To identify the genes that correspond to the selected SNPs, the lists of SNPs from the GWAS were uploaded to NetAFFX (Affymetrix, Santa Clara, CA, USA; http://www.affymetrix.com/analysis/index.affx). We used the Netaffx na32 Genotyping Annotation build. In the cases where a SNP mapped to multiple genes, we selected all the genes. SNPs for which no gene was identified were not included in our subsequent analyses.
Convergent functional genomics analyses
Databases
We have established in our laboratory (Laboratory of Neurophenomics, Indiana University School of Medicine; www.neurophenomics.info) manually curated databases of all the human gene expression (postmortem brain, blood, cell cultures), human genetic (association, CNVs, linkage) and animal model gene expression and genetic studies published to date on psychiatric disorders.12 Only the findings deemed significant in the primary publication, by the study authors, using their particular experimental design and thresholds, are included in our databases. Our databases include only primary literature data, and do not include review papers or other secondary data integration analyses, to avoid redundancy and circularity. These large and constantly updated databases have been used in our CFG cross-validation and prioritization (Figure 1).
Human postmortem brain gene expression evidence
Information about genes was obtained and imported in our databases by searching the primary literature with PubMed (http://ncbi.nlm.nih.gov/PubMed), using various combinations of keywords (for this work: schizophrenia, psychosis, human, brain, postmortem). Convergence was deemed to occur for a gene if there were published human postmortem brain data showing changes in expression of that gene in tissue from patients with schizophrenia.
Human blood and other peripheral tissue gene expression data
For human blood gene expression evidence, we have used previously generated data from our group,7 as well as published data from the literature. We also included recent data generated from induced pluripotent stem cell-derived neurons.10
Human genetic evidence (association, CNVs, linkage)
To designate convergence for a particular gene, the gene had to have independent published evidence of association, CNVs or linkage for schizophrenia. We sought to avoid using any association studies that included subjects that were also included in the ISC or GAIN GWAS. For CNVs, all the known genes on a CNV were taken. For linkage, the location of each gene was obtained through GeneCards (http://www.genecards.org), and the sex-averaged cM location of the start of the gene was then obtained through http://compgen.rutgers.edu/old/map-interpolator/. For linkage convergence, per our previously published criteria,2 the start of the gene had to map within 5 cM of the location of a marker linked to the disorder.
Animal model brain and blood gene expression evidence
For animal model brain and blood gene expression evidence, we have used our own comprehensive pharmacogenomic mouse model (phencyclidine and clozapine) data sets,6 as well as published reports from the literature curated in our databases.
Animal model genetic evidence (transgenic)
To search for mouse genetic evidence (transgenic) for our candidate genes, we utilized PubMed as well as the Mouse Genome Informatics (http://www.informatics.jax.org; Jackson Laboratory, Bar Harbor, ME, USA) database, and used the search ‘Genes and Markers' form to find transgenics for categories ‘Schizophrenia' as well as ‘abnormal nervous system physiology' (subcategory ‘abnormal sensorimotor gating').
Convergent functional genomics analysis scoring
We used a nominal P-value threshold for including genes from the ISC GWAS in the CFG analysis: having a SNP with P<0.05. All six cross-validating lines of evidence (other human data, animal model data) were weighted equally, receiving a maximum of 1 point each (for human genetic evidence: 0.5 points if it is linkage, 0.75 if it is from CNVs, 1 point if it is association). Thus, the maximum possible CFG score for each gene is 6. We have capped each line of evidence at 1 point, regardless of how many different studies support that line of evidence, to avoid potential ‘popularity' biases, where some genes are more studied than others.
The more lines of evidence, that is, the more times a gene shows up as a positive finding across independent studies, platforms, methodologies and species, the higher its CFG score (Figure 1). This is similar conceptually to the Google PageRank algorithm, in which the more links to a page, the higher it comes up on the search prioritization list.13 Human and animal model data, genetic and gene expression were integrated and tabulated, resulting in a polyevidence CFG score. It has not escaped our attention that other ways of weighing the lines of evidence may give slightly different results in terms of prioritization, if not in terms of the list of genes per se. Nevertheless, we feel this simple scoring system provides a good separation of genes, with sensitivity provided by animal model data and specificity provided by human data.
Pathway analyses
IPA 9.0. (Ingenuity Systems, Redwood City, CA, USA) was used to analyze the biological roles, including top canonical pathways, of the candidate genes resulting from our work (Table 2 and Supplementary Table S5), as well as used to identify genes in our data sets that are the target of existing drugs (Supplementary Table S2).
Table 2. Ingenuity pathway analyses of top candidate genes.
| Top canonical pathways CFG ⩾3 | P-value | Ratio |
|---|---|---|
| ISC (n=186 genes) | ||
| Glutamate receptor signaling | 9.25E−13 | 12/69 (0.174) |
| G-protein–coupled receptor signaling | 9.33E−13 | 27/530 (0.051) |
| CREB signaling in neurons | 1.76E−12 | 17/202 (0.084) |
| cAMP-mediated signaling | 3.55E−11 | 17/219 (0.078) |
| Neuropathic pain signaling in dorsal horn neurons | 3.64E−11 | 13/112 (0.116) |
| GAIN EA (n=173 genes) | ||
| Glutamate receptor signaling | 4.57E−16 | 14/69 (0.203) |
| CREB signaling in neurons | 4.72E−14 | 18/202 (0.089) |
| G-protein–coupled receptor signaling | 2E−13 | 27/530 (0.051) |
| cAMP-mediated signaling | 1.2E−12 | 18/219 (0.082) |
| Synaptic long-term potentiation | 1.58E−12 | 14/114 (0.123) |
| GAIN AA (n=201 genes) | ||
| cAMP-mediated signaling | 7.6E-17 | 23/219 (0.105) |
| Glutamate receptor signaling | 1.09E−16 | 15/69 (0.217) |
| Synaptic long-term potentiation | 2.24E−15 | 17/114 (0.149) |
| G-Protein–coupled receptor signaling | 2.43E−14 | 30/530 (0.057) |
| CREB signaling in neurons | 4.52E−14 | 19/202 (0.094) |
Abbreviations: AA, African American; CFG, convergent functional genomics; EA, European American; ISC, International Schizophrenia Consortium.
Discovery in ISC and reproducibility in two independent cohorts, GAIN EA and GAIN AA.
Intra-pathway epistasis testing
As an example,11 the ISC GWAS data were used to test for epistatic interactions among the best P-value SNPs in genes from our data set present in a top canonical biological pathway identified by Ingenuity pathway analysis (Supplementary Table S4). SNP × SNP allelic epistasis was tested for each distinct pair of SNPs between genes, using the PLINK software package.
Genetic risk prediction panel and scoring
As we had previously done for bipolar disorder,11 we developed a polygenic GRPS for schizophrenia based on the presence or absence of the alleles of the SNPs associated with illness, and tested the GRPS in independent cohorts for which we had both genotypic and clinical data available, comparing the schizophrenia subjects to normal controls. We tested two panels: a smaller one (GRPS-42) containing the single best P-value SNP in ISC in each of the top CFG prioritized genes (n=42), and a larger one (GRPS-542), containing all the nominally significant SNPs (n=542) in ISC in the top CFG prioritized genes (n=42; Tables 3, 4, Supplementary Table S3, and Figure 4).
Table 3. GRPS-42: non-differentiation between schizophrenics and controls in independent cohorts using a panel composed of the single best SNP from ISC in each of the top candidate genes (42 SNPs, in 42 genes).
| Description of panel | GAIN EA | GAIN AA |
|---|---|---|
| Single best P-value SNPs in each of the top 42 candidate genes from ISC GWAS, n=42 | P=0.10308, 39 out of the 42 ISC SNPs were present in GAIN EA | P=0.13567, 37 out of the 42 ISC SNPs were present in GAIN AA |
Abbreviations: AA, African American; EA, European American; GRPS, genetic risk prediction score; GWAS, genome-wide association study; ISC, International Schizophrenia Consortium; SNP, single-nucleotide polymorphism.
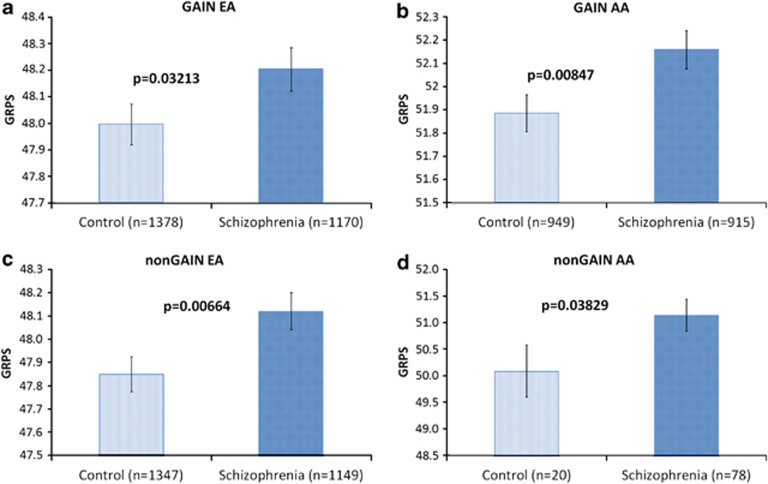
Table 4. GRPS-542: differentiation between schizophrenics and controls in four independent cohorts using a panel composed of all the nominally significant SNPs from ISC in the top candidate genes (542 SNPs in 42 genes).
| GAIN EA | GAIN AA |
|---|---|
| P=0.03213, 527 SNPs in 41 genes were present in GAIN EA | P=0.00847, 516 SNPs in 42 genes were present in GAIN AA |
| NonGAIN EA | NonGAIN AA |
| P=0.00664, 537 SNPs in 42 genes were present in nonGAIN EA | P=0.03829, 537 SNPs in 42 genes were present in nonGAIN AA |
Abbreviations: AA, African American; EA, European American; GRPS, genetic risk prediction score; ISC, International Schizophrenia Consortium; SNP, single-nucleotide polymorphism.
Figure 4.
Genetic risk prediction of schizophrenia in four independent cohorts. AA, African American; EA, European American; GRPS, genetic risk prediction score.
Of note, our SNP panels and choice of affected alleles were based solely on analysis of the ISC GWAS, which is our discovery cohort, completely independently from the test cohorts. Each SNP has two alleles (represented by base letters at that position). One of them is associated with the illness (affected), the other not (non-affected), based on the odds ratios from the discovery ISC GWAS. We assigned the affected allele a score of 1 and the non-affected allele a score of 0. A two-dimensional matrix of subjects by GRP panel alleles is generated, with the cells populated by 0 or 1. A SNP in a particular individual subject can have any permutation of 1 and 0 (1 and 1, 0 and 1, 0 and 0). By adding these numbers, the minimum score for a SNP in an individual subject is 0, and the maximum score is 2. By adding the scores for all the alleles in the panel, averaging that, and multiplying by 100, we generate for each subject an average score corresponding to a genetic loading for disease, which we call Genetic Risk Predictive Score (GRPS).
The software package PLINK (http://pngu.mgh.harvard.edu/~purcell) was used to extract individual genotype information for each subject from the GAIN and nonGAIN GWAS data files. We analyzed separately EA and AA schizophrenia subjects and controls, to examine any potential ethnicity variability (Tables 3 and 4, and Supplementary Table S3). To test for significance, a one-tailed t-test was performed between the schizophrenia subjects and the control subjects, looking at differences in GRPS.
Results
Top candidate genes
To minimize false negatives, we initially cast a wide net, using as a filter a minimal requirement for a gene to have both some GWAS evidence and some additional independent evidence. We thus generated an initial list of 3194 unique genes with at least a SNP at P<0.05 in the discovery GWAS analyzed (ISC),8 that also had some additional evidence (human or animal model data), implicating them in schizophrenia (CFG score ⩾1; Table 5). This suggests, using these minimal thresholds and requirements, that the repertoire of genes potentially involved directly or indirectly in cognitive processes and schizophrenia may be quite large, similar to what we have previously seen for bipolar disorder.11
Table 5. Reproducibility between independent GWAS.
| Numbers and overlap across studies | ISC | GAIN EA | GAIN AA | ISC vs GAIN EA | ISC vs GAIN AA | GAIN EA vs GAIN AA | ISC vs. GAIN-EA vs. GAIN-AA (% of ISC) |
|---|---|---|---|---|---|---|---|
| SNPs P⩽0.05 | 45 972 | 42 336 | 57 118 | 2649 | 2986 | 2839 | 163 (0.4%) |
| Genes | 10 180 | 9002 | 11 260 | 6470 | 7583 | 6807 | 5518 (54.2%) |
| Genes CFG ⩾1 | 3194 | 2913 | 3524 | 2243 | 2564 | 2384 | 2012 (63.0%) |
| Genes CFG ⩾3 | 186 | 173 | 201 | 147 | 160 | 153 | 134 (72.0%) |
| Genes CFG ⩾4 | 42 | 41 | 45 | 37 | 37 | 38 | 35 (83.3%) |
| Pathways for genes with CFG ⩾1 | 217 | 210 | 205 | 194 | 188 | 180 | 176 (81.1%) |
| Pathways for genes with CFG ⩾3 | 79 | 85 | 108 | 72 | 76 | 81 | 72 (91.1%) |
| Pathways for genes with CFG ⩾4 | 34 | 50 | 75 | 33 | 34 | 48 | 33 (97.1%) |
Abbreviations: AA, African American; CFG, convergent functional genomics; EA, European American; GWAS, genome-wide association study; ISC, International Schizophrenia Consortium; SNP, single-nucleotide polymorphism.
Increasing consistency and overlap observed from nominally significant SNPs (0.4%) to genes, then to CFG prioritized genes, and finally to pathways of CFG prioritized genes (97.1%).
To minimize false positives, we then used the CFG analysis integrating multiple lines of evidence to further prioritize this list of genes, and focused our subsequent analyses on only the top CFG scoring candidate genes. Overall, 186 genes had a CFG score of 3 and above (⩾50% of maximum possible score of 6), and 42 had a CFG score of 4 and above (Tables 1 and 5, and Figure 2).
Our top findings from ISC (Table 1) were over-represented in two independent schizophrenia GWAS cohorts, the GAIN EA and GAIN AA. In total, 37 of the top 42 genes identified by our approach (88.1%) had at least a SNP with a P-value of <0.05 in those independent cohorts, an estimated twofold enrichment over what would be expected by chance alone at a genetic level (as there were 9002 genes at P<0.05 in the GAIN-EA GWAS, and the number of genes in the human genome is estimated at 20 500,14 the enrichment factor provided by our approach is (37/42)/(9002/20 500)≈2). Of note, there was no correlation between CFG prioritization and gene size, thus excluding a gene-size effect for the observed enrichment (Supplementary Figure S1).
Candidate blood biomarkers
Of the top candidate genes from Table 1 (see also Figure 2), 15 out of 42 have prior human blood evidence for change in schizophrenia, implicating them as potential blood biomarkers. The additional evidence provided by GWAS data suggests a genetic rather than purely environmental (medications, stress) basis for their alteration in disease, and their potential utility as trait rather than purely state markers.
Biological pathways
Pathway analyses were carried out on the top genes (Table 2), and on all the candidate genes (Supplementary Table S5). Notably, glutamate receptor signaling, G-protein–coupled receptor signaling and cAMP-mediated signaling were the top canonical pathways over-represented in schizophrenia, which may be informative for new drug discovery efforts by pharmaceutical companies.
Genetic risk prediction
Once the genes involved in a disorder are identified, and prioritized for likelihood of involvement, then an obvious next step is developing a way of applying that knowledge to genetic testing of individuals to determine risk for the disorder. Based on our identification of top candidate genes described above using CFG, we pursued a polygenic panel approach, with digitized binary scoring for presence or absence, similar to the one we have devised and used in the past for biomarkers testing5 and for genetic testing in bipolar disorder.11 Somewhat similar approaches but without CFG prioritization, attempted by other groups, have been either unsuccessful15 or have required very large panels of markers.8, 16
We first chose the single best P-value SNPs in each of our top CFG prioritized genes (n=42) in the ISC GWAS data set used for discovery, and assembled a GRP panel out of those SNPs (Table 3). We then developed a GRPS for schizophrenia based on the presence or absence of the alleles of the SNPs associated with the illness, and tested the GRPS in independent cohorts (GAIN EA and GAIN AA), comparing the schizophrenia subjects to normal controls (Table 3). The results were not significant. We concluded that genetic heterogeneity at a SNP level is a possible explanation for these negative results. We then sought to see if we get better separation with a larger panel, containing all the nominally significant SNPs (n=542) in the top CFG prioritized genes in ISC (n=42), on the premise that a larger panel may reduce the heterogeneity effects, as different SNPs might be more strongly associated with illness in different cohorts. We found that our larger panel of SNPs was indeed able to significantly distinguish schizophrenics from controls in both GAIN EA and GAIN AA, two independent cohorts of different ethnicities. To verify this unexpectedly strong result, we further tested our panel in two other independent cohorts, nonGAIN EA and nonGAIN AA, and obtained similarly significant results (Table 4 and Figure 4).
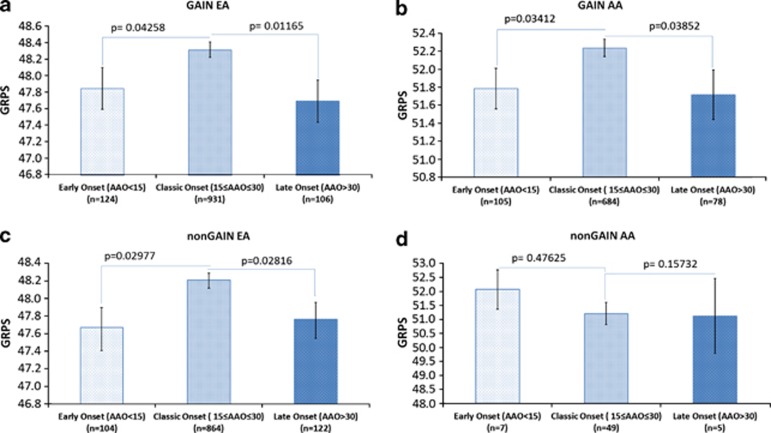
We also looked at whether our GRPS score distinguishes classic age of onset schizophrenia (defined by us as ages 15 to 30 years) from early onset (before 15 years) and late-onset (after 30 years) illness. Our results show that classic age of onset schizophrenia has a significantly higher GRPS than early or late-onset schizophrenia, in three out of the four independent cohorts of two different ethnicities (Figure 5).
Figure 5.
Genetic risk score and age at onset of schizophrenia. AA, African American; AAO, age at onset; EA, European American; GRPS, genetic risk prediction score.
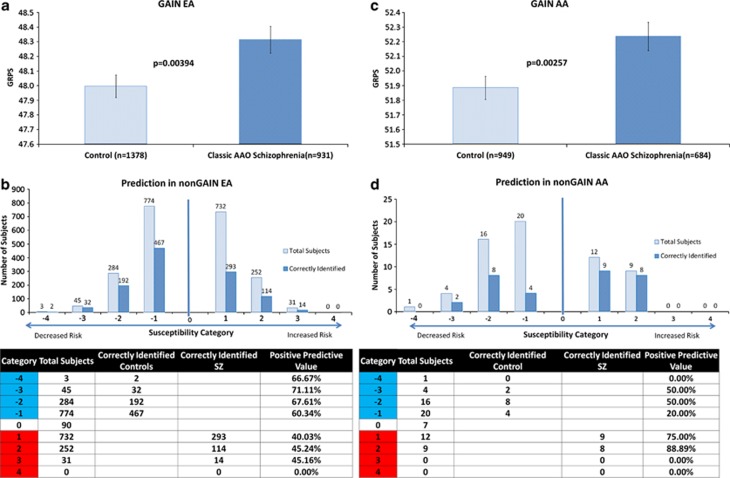
Finally, as we had done previously for bipolar disorder,11 we developed a prototype of how the GRPS score could be used in testing individuals to establish their category of risk for schizophrenia (Figure 6). The current iteration of the test, using the panel of 542 SNPs, seems to be able to distinguish in independent cohorts who is at lower risk for classic age of onset schizophrenia in two out of three EA subjects, and who is at higher risk for classic age of onset schizophrenia in three out of four AA subjects.
Figure 6.
Prototype of how genetic risk prediction score (GRPS) testing could be used at an individual rather than population level, to aid diagnostic and personalized medicine approaches. We used the average values and standard deviation values for GRPS from the GAIN samples from each ethnicity (European American (EA) and African American (AA)) as thresholds for predictive testing in the independent nonGAIN EA and nonGAIN AA cohorts. The average GRPS score for schizophrenics in the GAIN cohort is used as a cut-off for schizophrenics in the test cohort (that is, being above that threshold), and the average GRPS score for controls in the GAIN cohort is used as a cut-off for controls in the test nonGAIN cohort (that is, being below that threshold). The subjects who are in between these two thresholds are called undetermined. Furthermore, to stratify risk, we categorized subjects into risk categories (in red, increased risk; in blue, decreased risk): Category 1 if they fall within one standard deviation above the schizophrenics' threshold, and category −1 if they fall within one standard deviation below the controls threshold. Category 2 and −2, subjects are between one and two standard deviations from the thresholds, category 3 and −3, subjects are between two and three standard deviations, and category 4 and −4, subjects are those who fall beyond three standard deviations of the thresholds. The positive predictive value (PPV) of the tests increases in the higher categories, and the test is somewhat better at distinguishing controls in EA (that is, in a practical application, individuals that are lower risk of developing the illness), and schizophrenics in AA (that is, in a practical application, individuals that are higher risk of developing the illness).
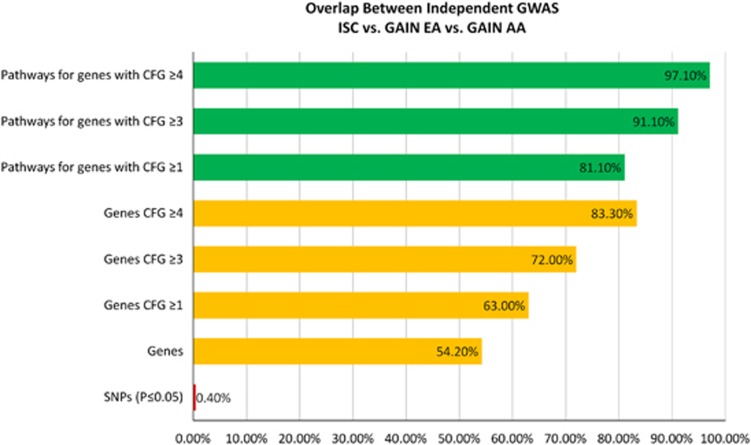
Overlap among studies
We examined the overlap at a nominally significant (P<0.05) SNP level between ISC, GAIN EA and GAIN AA, and found that a minority of these SNPs (0.4%) overlap (Table 5 and Figure 7). We then examined the overlap at a gene level, then CFG prioritized genes level and finally biological pathways level, and found increasing evidence of commonality and reproducibility of findings across studies.
Figure 7.
Overlap between independent genome-wide association study (GWAS). AA, African American; EA, European American; CFG, convergent functional genomics; ISC, International Schizophrenia Consortium; SNP, single-nucleotide polymorphism.
Discussion
Our CFG approach helped prioritize genes, such as DISC1 and MBP, with weaker evidence in the GWAS data but with strong independent evidence in terms of gene expression studies and other prior human or animal genetic work. Conversely, some of the top findings from GWAS, such as ZNF804A, have fewer different independent lines of evidence, and thus received a lower CFG prioritization score in our analysis (Supplementary Information-Table S1), although ZNF804A is clearly involved in schizophrenia-related cognitive processes.17 While we cannot exclude that more recently discovered genes have had less hypothesis-driven work done and thus might score lower on CFG, it is to be noted that the CFG approach integrates predominantly non-hypothesis driven, discovery-type data sets, such as gene expression, GWAS, CNV, linkage and quantitative traits loci. We also cap each line of evidence from an experimental approach (Figure 1) at a maximum score of 1, to minimize any ‘popularity' bias, whereas multiple studies of the same kind are conducted on better-established genes. In the end, it is gene-level reproducibility across multiple approaches and platforms that is built into the approach and gets prioritized most by CFG scoring during the discovery process. Our top results subsequently show good reproducibility and predictive ability in independent cohort testing, the litmus test for any such work.
At the very top of our list of candidate genes for schizophrenia, with a CFG score of 5, we have four genes: DISC1, TCF4, MBP and HSPA1B. An additional five genes have a CFG score of 4.5: MOBP, NRCAM, NCAM1, NDUFV2 and RAB18.
DISC1 (Disrupted-in Schizophrenia 1), encodes a scaffold protein that has an impact on neuronal development and function,18, 19, 20 including neuronal connectivity.21 DISC1 has been identified as a susceptibility gene for major mental disorders by multiple studies.22, 23, 24 DISC1 isoforms are upregulated in expression in blood cells in schizophrenia, thus serving as a potential peripheral biomarker as well.25, 26 Developmental stress interacts with DISC1 expression to produce neuropsychiatric phenotypes in mice.27 Notably, its interacting partners PDE4B,28 TNIK,29 FEZ1 (ref. 30) and DIXDC1 (ref. 31) are also present on our list of prioritized candidate genes, with CFG scores of 4, 4, 3.5 and 2.5, respectively (Table 1 and Supplementary Table S1).
TCF4 (transcription factor 4) encodes a basic helix-turn-helix transcription factor, expressed in immune system as well as neuronal cells. It is required for the differentiation of subsets of neurons in the developing brain. There are multiple alternatively spliced transcripts that encode different proteins, providing for biological diversity and heterogeneity. Defects in this gene are a cause of Pitt-Hopkins syndrome, characterized by mental retardation with or without associated facial dysmorphisms and intermittent hyperventilation. TCF4 has additional genetic evidence for association with schizophrenia-relevant phenotypes.32, 33, 34, 35 It is changed in expression in postmortem brain,36 induced pluripotent stem cell-derived neurons10 and blood from schizophrenia patients.7 Notably, it is a candidate blood biomarker for level of delusional symptoms (decreased in high delusional states) based on our previous work.7
MBP (myelin basic protein) is a major constituent of the myelin sheath of oligodendrocytes and Schwann cells in the nervous system. MBP-related transcripts are also present in the bone marrow and the immune system. MBP has additional genetic evidence for association with schizophrenia.37 It is decreased in expression in postmortem brain38 and blood39 from schizophrenia patients. MBP is also changed in expression in the brain and blood of a pharmacogenomics mouse model of schizophrenia, based on our previous work.6 It was also decreased in expression in a stress-reactive genetic mouse model of bipolar disorder,40 and treatment with the omega-3 fatty acid docosahexaenoic acid led to an increase in expression. Notably, MBP is a candidate blood biomarker for level of mood symptoms (increased in high mood states in bipolar subjects), based on our previous work.5 Overall, the data indicate that MBP and other myelin-related genes41, 42 may be involved in the effects of stress on psychosis and mood. Demyelinating disorders such as multiple sclerosis tend to be precipitated and exacerbated by stress, and have co-morbid psychiatric symptoms.43 Of note, other myelin-related genes are also present on our list of prioritized candidate genes: MOBP and MOG, with CFG scores of 4.5 and 3, respectively (Table 1 and Supplementary Table S1).
HSPA1B (heat-shock 70-kDa protein 1B), a chaperone involved in stress response, stabilizes existing proteins against aggregation and mediates the folding of newly translated proteins. HSPA1B has additional genetic evidence for association with schizophrenia.44 It is changed in expression in postmortem brain45 and induced pluripotent stem cell-derived neurons10 from schizophrenia patients. HSPA1B is also increased in expression in the brain and blood of a pharmacogenomics mouse model of schizophrenia, based on our previous work.6 It was also co-directionally changed in the brain and blood in a phramacogenomic mouse model of anxiety disorders, we have recently described,46 as well as in a stress-reactive genetic mouse model.40 Treatment with the omega-3 fatty acid docosahexaenoic acid reversed the increase in expression of HSPA1B in this stress-reactive genetic mouse model.47 Another closely related molecule, HSPA1A (heat-shock 70-kDa protein 1A), is also present on our list of prioritized candidate genes, with a CFG score of 3.5 (Supplementary Table S1). Heat-shock proteins may be involved in the biological and clinical overlap and interdependence between response to stress, anxiety and psychosis.
NRCAM (neuronal cell adhesion molecule) encodes a neuronal cell adhesion molecule. This ankyrin-binding protein is involved in neuron–neuron adhesion and promotes directional signaling during axonal cone growth. NRCAM is also expressed in non-neural tissues and may have a general role in cell–cell communication via signaling from its intracellular domain to the actin cytoskeleton during directional cell migration. It is decreased in expression in postmortem brain48 and peripherally in serum49 from schizophrenia patients. NRCAM is also changed in expression in the brain of a pharmacogenomics mouse model of schizophrenia, based on our previous work.6 It was also increased in the amygdala in a stress-reactive genetic mouse model studied by our group.40 Another closely related molecule, NCAM1 (neural cell adhesion molecule 1), is among our top candidate genes as well. These data support a central role for cell connectivity and cell adhesion in schizophrenia.
Another top candidate gene is CNR1 (cannabinoid receptor 1, brain). CNR1 is a member of the guanine-nucleotide-binding protein (G-protein) coupled receptor family, which inhibits adenylate cyclase activity in a dose-dependent manner. CNR1 has additional genetic evidence for association with schizophrenia.50, 51 It is decreased in expression in postmortem brain from schizophrenics.52 The other main cannabinoid receptor, CNR2 (cannabinoid receptor 2), is among our top candidate genes too (Supplementary Table S1), and is decreased in expression in postmortem brain from schizophrenics as well. These data support a role for the cannabinoid system in schizophrenia, perhaps through a deficiency of the endogenous cannabinoid signaling that leads to vulnerability to psychotogenic stress,53 and is accompanied by increased compensatory exogenous cannabinoid consumption that may have additional deleterious consequences.54
A number of glutamate receptor genes are present among our top candidate genes for schizophrenia (GRIA1, GRIA4, GRIN2B and GRM5), as well as GAD1, an enzyme involved in glutamate metabolism, and SLC1A2, a glutamate transporter (Table 1). Other genes involved in glutamate signaling present in our data, with a lower scores, are GRIN2A, SLC1A3, GRIA3, GRIK4, GRM1, GRM4 and GRM7 (Supplementary Table S1). Glutamate receptor signaling is one of the top canonical pathways over-represented in our analyses (Table 2), and that finding is reproduced in independent GWA data sets (Table 2). One has to be circumspect with interpreting such results, as glutamate signaling is quasi-ubiquitous in the brain, and a lot of prior hypothesis-driven work has focused on this area, potentially biasing the available evidence. Nevertheless, our results are striking, and contribute to the growing body of evidence that has emerged over the last few years implicating glutamate signaling as a point of convergence for findings in schizophrenia,55 as well as for autism56 and AD.57 Glutamate signaling is the target of active drug development efforts,58 which may be informed and encouraged by our current findings.
Our analysis also provides evidence for other genes that have long been of interest in schizophrenia, but have had previous variable evidence from genetic-only studies: BDNF, COMT, DRD2, DTNBP1 (dystrobrevin binding protein1/dysbindin; Table 1). In addition, our analysis provides evidence for genes that had previously not been widely implicated in schizophrenia, but do have relevant biological roles, demonstrating the value of empirical discovery-based approaches such as CFG (Table 1): ANK3,48 ALDH1A1 and ADCYAP1, which is a ligand for schizophrenia candidate gene VIPR2,59, 60 also present in our data set, albeit with a lower CFG score of 2. Other genes of interest in our full data set (Supplementary Table S1) include ADRBK2 (GRK3), first described by us as a candidate gene for psychosis,1 CHRNA7,61 and PDE10A,62 which are targets for drug development efforts.
Pathways and mechanisms
Our pathway analyses results are consistent with the accumulating evidence about the role of synaptic connections and glutamate signaling in schizophrenia, most recently from CNV studies63(Table 2, Supplementary Table S5, Figure 3). Very importantly, the same top pathways were consistent across independent GWA studies we analyzed (Tables 2, 5, and Supplementary Table S5). We also did a manual curation of the top candidate genes and their grouping into biological roles examining them one by one using PubMed and GeneCards, to come up with a heuristic model of schizophrenia (Figure 3). Overall, while multiple mechanistic entry points may contribute to schizophrenia pathogenesis (Figure 3a), it is likely at its core a disease of decreased cellular connectivity precipitated by environmental stress during brain development, on a background of genetic vulnerability (Figure 3b).
Genetic risk prediction
Of note, our SNP panels and choice of affected alleles were based solely on analysis of the discovery ISC GWAS, completely independently from the test GAIN EA, GAIN AA, nonGAIN EA and nonGAIN AA GWAS. Our results show that a relatively limited and well-defined panel of SNPs identified based on our CFG analysis could differentiate between schizophrenia subjects and controls in four independent cohorts of two different ethnicities, EA and AA. Moreover, the genetic risk component identified by us seems to be stronger for classic age of onset schizophrenia than for early or late-onset illness, suggesting that the latter two may be more environmentally driven or have a somewhat different genetic architecture. It is likely that such genetic testing will have to be optimized for different cohorts if done at a SNP level. Interestingly, at a gene and pathway level, the differences between studies seem much less pronounced than at a SNP level, if at all present (Table 5), suggesting that gene-level and pathway-level tests may have more universal applicability. In the end, such genetic data, combined with family history and other clinical information (phenomics),64 as well as with blood biomarker testing,5 may provide a comprehensive picture of risk of illness.65, 66
Reproducibility among studies
Our work provides striking evidence for the advantages, reproducibility and consistency of gene-level analyses of data, as opposed to SNP level analyses, pointing to the fundamental issue of genetic heterogeneity at a SNP level (Table 5 and Figure 7). In fact, it may be that the more biologically important a gene is for higher mental functions, the more heterogenity it has at a SNP level67 and the more evolutionary divergence,68 for adaptive reasons. On top of that, CFG provides a way to prioritize genes based on disease relevance, not study-specific effects (that is, fit-to-disease as opposed to fit-to-cohort). Reproducibility of findings across different studies, experimental paradigms and technical platforms is deemed more important (and scored as such by CFG) than the strength of finding in an individual study (for example, P-value in a GWAS). The CFG prioritized genes show even more reproducibility among independent GWAS cohorts (ISC, GAIN EA, GAIN AA) than the full list of unprioritized genes with nominal significant SNPs. The increasing overlap and reproducibility between studies of genes with a higher average CFG score points out to their biological relevance to disease architecture. Finally, at a pathway level, there is even more consistency across studies. Again, the pathways derived from the top CFG scoring genes show more consistency than the pathways derived from the lower CFG scoring genes. Overall, using our approach, we go from a reproducibilty between independent studies of 0.4% at the level of nominally significant SNPs to a reproducibility of 97.1% at the level of pathways derived from top CFG scoring genes.
Overlap with other psychiatric disorders
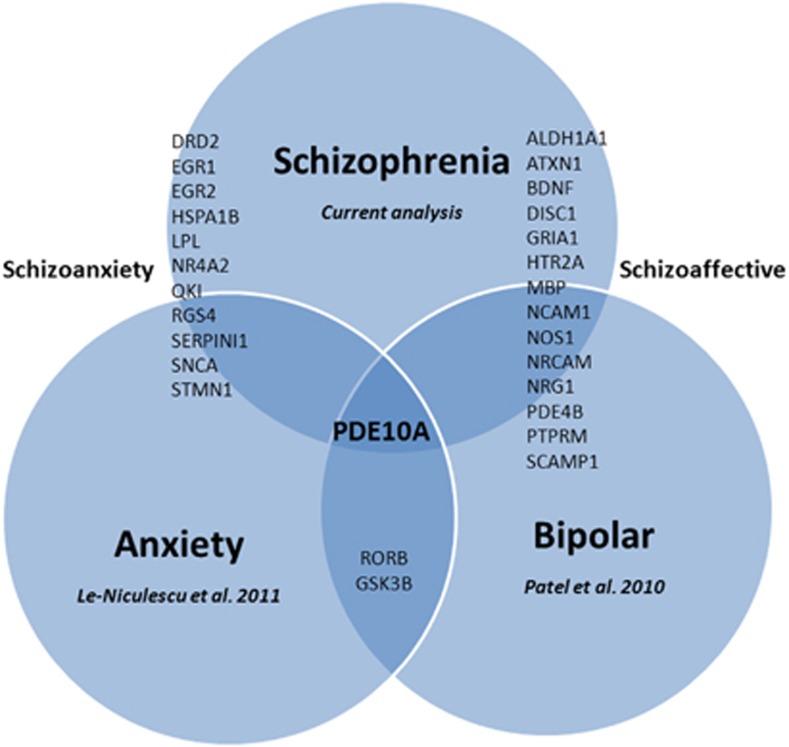
Despite using lines of evidence for our CFG approach that have to do only with schizophrenia, the list of genes identified has a notable overlap with other psychiatric disorders (Figure 8, Supplementary Table S1). This is a topic of major interest and debate in the field.12, 69 We demonstrate an overlap between top candidate genes for schizophrenia and candidate genes for anxiety and bipolar disorder, previously identified by us through CFG (Figure 8), thus providing a possible molecular basis for the frequently observed clinical co-morbidity and interdependence between schizophrenia and those other major psychiatric disorders, as well as cross-utility of pharmacological agents. In particular, PDE10A is at the overlap of all three major psychiatric domains, and may be of major interest for drug development.62 The overlap between schizophrenia and bipolar may have to do primarily with neurotrophicity and brain infrastructure (underlined by genes such as DISC1, NRG1, BDNF, MBP, NCAM1, NRCAM, PTPRM). The overlap between schizophrenia and anxiety may have to do primarily to do with reactivity and stress response (underlined by genes such as NR4A2, QKI, RGS4, HSPA1B, SNCA, STMN1, LPL). Notably, the overlap between schizophrenia and anxiety is of the same magnitude as the previously better appreciated overlap between schizophrenia and bipolar disorder,6, 70 supporting the consideration of a nosological domain of schizoanxiety disorder,46 by analogy to schizoaffective disorder. Clinically, while there are some reports of co-morbidity between schizophrenia and anxiety,71 it is an area that has possibly been under-appreciated and understudied. ‘Schizoanxiety disorder' may have heuristic value and pragmatic clinical utility.
Figure 8.
Genetic overlap among psychiatric disorders.
We also looked at the overlap with candidate genes for autism and AD from the literature (Supplementary Table S1), to elucidate whether schizophrenia, autism and AD might be on a spectrum, that is, whether autism might be a form of ‘schizophrenia praecox', similar to schizophrenia being referred to as ‘dementia praecox' (Kraepelin). We see significant overlap between the three disorders among the top genes with a CFG score of 4: a third of the genes overlap between schizophrenia and autism, and a quarter between schizophrenia and AD. Additional key genes of interest are lower on the list as well, with a CFG score of 3: CNTNAP2 for autism, MAPT and SNCA for AD (Supplementary Table S1).
Conclusions and future directions
First, in spite of its limitations, our analysis is arguably the most comprehensive integration of genetics and functional genomics to date in the field of schizophrenia, yielding a comprehensive view of genes, blood biomarkers, pathways and mechanisms that may underlie the disorder. From a pragmatic standpoint, we would like to suggest that our work provides new and/or more comprehensive insights on genes and biological pathways to target for new drug development by pharmaceutical companies, as well as potential new uses in schizophrenia for existing drugs, including omega-3 fatty acids (Supplementary Table S2).
Second, our current work and body of work over the years provides proof how a combined approach, integrating functional and genotypic data, can be used for complex disorders-psychiatric and non-psychiatric, as has been attempted by others as well.72, 73 What we are seeing across GWAS of complex disorders are not necessarily the same SNPs showing the strongest signal, but rather consistency at the level of genes and biological pathways. The distance from genotype to phenotype may be a bridge too far for genetic-only approaches, given genetic heterogeneity and the intervening complex layers of epigenetics and gene expression regulation.74 Consistency is much higher at a gene expression level (Table 5),75 and then at a biological pathway level. Using GWAS data in conjunction with gene expression data as part of CFG or integrative genomics76 approaches, followed by pathway-level analysis of the prioritized candidate genes, can lead to the unraveling of the genetic code of complex disorders such as schizophrenia.
Third, our work provides additional integrated evidence focusing attention and prioritizing a number of genes as candidate blood biomarkers for schizophrenia, with an inherited genetic basis (Table 1 and Figure 2). While prior evidence existed as to alterations in gene expression levels of those genes in whole-blood samples or lymphoblastoid cell lines from schizophrenia patients, it was unclear prior to our analysis whether those alterations were truly related to the disorder or were instead related only to medication effects and environmental factors.
Fourth, we have put together a panel of SNPs, based on the top candidate genes we identified. We developed a GRPS based on our panel, and demonstrate how in four independent cohorts of two different ethnicities, the GRPS differentiates between subjects with schizophrenia and normal controls. From a personalized medicine standpoint, genetic testing with highly prioritized panels of best SNP markers may have, upon further development (Figure 6) and calibration by ethnicity and gender, a role in informing decisions regarding early intervention and prevention efforts; for example, for classic age of onset schizophrenia before the illness fully manifests itself clinically, in young offspring from high-risk families. After the illness manifests itself, gene expression biomarkers and phenomic testing approaches, including clinical data, may have higher yield than genetic testing. A multi-modal integration of testing modalities would be the best approach to assess and track patients, as individual markers are likely to not be specific for a single disorder. The continuing re-evaluation in psychiatric nosology66, 77 brought about by recent advances will have to be taken into account as well for final interpretation of any such testing. The complexity, heterogeneity, overlap and interdependence of major psychiatric disorders as currently defined by DSM suggests that the development of tests for dimensional disease manifestations (psychosis, mood and anxiety)66 will ultimately be more useful and precise than developing tests for existing DSM diagnostic categories.
Finally, while we cannot exclude that rare genetic variants with major effects may exist in some individuals and families, we suggest a contextual cumulative combinatorics of common variants genetic model best explains our findings, and accounts for the thin genetic load margin between clinically ill subjects and normal controls, which leaves a major role to be played by gene expression (including epigenetic changes) and the environment. This is similar to our conclusions when studying bipolar disorder,11 and may hold true in general for complex medical disorders, psychiatric and non-psychiatric. Full-blown illness occurs when genetic and environmental factors converge, usually in young adulthood for schizophrenia. When they diverge, a stressful/hostile environment may lead to mild or transient illness even in normal genetic load individuals, whereas a favorable environment may lead to supra-normative functioning in certain life areas (such as creative endeavors) for individuals who carry a higher genetic load. The flexible interplay between genetic load, environment and phenotype may permit evolution to engender diversity, select and conserve alleles, and ultimately shape populations. Our emerging mechanistic understanding of psychosis as disconnectivity, mood as activity11 and anxiety as reactivity46 may guide such testing and understanding of population distribution as being on a multi-dimensional spectrum, from supra-normative to normal to clinical illness.
Acknowledgments
This work is, in essence, a field-wide collaboration. We would like to acknowledge our debt of gratitude for the efforts and results of the many other groups, cited in our paper, who have conducted and published empirical studies (human and animal model, genetic and gene expression) in schizophrenia. With their arduous and careful work, a convergent approach such as ours is possible. We would particularly like to thank the ISC and GAIN consortia. We would also like to thank the subjects who participated in these studies, their families and their caregivers. Without their contribution, such work to advance the understanding of mental illness would not be possible. Finally, we would like to acknowledge Elyn Saks for her insightful memoir, which inspired the Yeats quote at the beginning of the paper. This work was supported by an NIH Directors' New Innovator Award (1DP2OD007363) and a VA Merit Award (1I01CX000139-01) to ABN.
ABN is a founder of Mindscape Diagnostics. NJS is a founder of Cypher Genomics.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
Supplementary Material
References
- Niculescu AB, 3rd, Segal DS, Kuczenski R, Barrett T, Hauger RL, Kelsoe JR. Identifying a series of candidate genes for mania and psychosis: a convergent functional genomics approach. Physiol Genomics. 2000;4:83–91. doi: 10.1152/physiolgenomics.2000.4.1.83. [DOI] [PubMed] [Google Scholar]
- Ogden CA, Rich ME, Schork NJ, Paulus MP, Geyer MA, Lohr JB, et al. Candidate genes, pathways and mechanisms for bipolar (manic-depressive) and related disorders: an expanded convergent functional genomics approach. Mol Psychiatry. 2004;9:1007–1029. doi: 10.1038/sj.mp.4001547. [DOI] [PubMed] [Google Scholar]
- Bertsch B, Ogden CA, Sidhu K, Le-Niculescu H, Kuczenski R, Niculescu AB. Convergent functional genomics: a Bayesian candidate gene identification approach for complex disorders. Methods. 2005;37:274–279. doi: 10.1016/j.ymeth.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Rodd ZA, Bertsch BA, Strother WN, Le-Niculescu H, Balaraman Y, Hayden E, et al. Candidate genes, pathways and mechanisms for alcoholism: an expanded convergent functional genomics approach. Pharmacogenomics J. 2007;7:222–256. doi: 10.1038/sj.tpj.6500420. [DOI] [PubMed] [Google Scholar]
- Le-Niculescu H, Kurian SM, Yehyawi N, Dike C, Patel SD, Edenberg HJ, et al. Identifying blood biomarkers for mood disorders using convergent functional genomics. Mol Psychiatry. 2009;14:156–174. doi: 10.1038/mp.2008.11. [DOI] [PubMed] [Google Scholar]
- Le-Niculescu H, Balaraman Y, Patel S, Tan J, Sidhu K, Jerome RE, et al. Towards understanding the schizophrenia code: an expanded convergent functional genomics approach. Am J Med Genet B Neuropsychiatr Genet. 2007;144:129–158. doi: 10.1002/ajmg.b.30481. [DOI] [PubMed] [Google Scholar]
- Kurian SM, Le-Niculescu H, Patel SD, Bertram D, Davis J, Dike C, et al. Identification of blood biomarkers for psychosis using convergent functional genomics. Mol Psychiatry. 2011;16:37–58. doi: 10.1038/mp.2009.117. [DOI] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O′Donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Levinson DF, Duan J, Sanders AR, Zheng Y, Pe′er I, et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460:753–757. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SD, Le-Niculescu H, Koller DL, Green SD, Lahiri DK, McMahon FJ, et al. Coming to grips with complex disorders: genetic risk prediction in bipolar disorder using panels of genes identified through convergent functional genomics. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:850–877. doi: 10.1002/ajmg.b.31087. [DOI] [PubMed] [Google Scholar]
- Niculescu AB, Le-Niculescu H. Convergent Functional Genomics: what we have learned and can learn about genes, pathways, and mechanisms. Neuropsychopharmacology. 2010;35:355–356. doi: 10.1038/npp.2009.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le-Niculescu H, Patel SD, Niculescu AB. Convergent integration of animal model and human studies of bipolar disorder (manic-depressive illness) Curr Opin Pharmacol. 2010;10:594–600. doi: 10.1016/j.coph.2010.08.008. [DOI] [PubMed] [Google Scholar]
- Clamp M, Fry B, Kamal M, Xie X, Cuff J, Lin MF, et al. Distinguishing protein-coding and noncoding genes in the human genome. Proc Natl Acad Sci USA. 2007;104:19428–19433. doi: 10.1073/pnas.0709013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsumi A, Glatt SJ, Kanazawa T, Kawashige S, Uenishi H, Hokyo A, et al. The genetic validation of heterogeneity in schizophrenia. Behav Brain Funct. 2011;7:43. doi: 10.1186/1744-9081-7-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Aleksic B, Kinoshita Y, Okochi T, Kawashima K, Kushima I, et al. Genome-wide association study of schizophrenia in a Japanese population. Biol Psychiatry. 2011;69:472–478. doi: 10.1016/j.biopsych.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Rasetti R, Sambataro F, Chen Q, Callicott JH, Mattay VS, Weinberger DR. Altered cortical network dynamics: a potential intermediate phenotype for schizophrenia and association with ZNF804A. Arch Gen Psychiatry. 2011;68:1207–1217. doi: 10.1001/archgenpsychiatry.2011.103. [DOI] [PubMed] [Google Scholar]
- Ishizuka K, Kamiya A, Oh EC, Kanki H, Seshadri S, Robinson JF, et al. DISC1-dependent switch from progenitor proliferation to migration in the developing cortex. Nature. 2011;473:92–96. doi: 10.1038/nature09859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandon NJ, Sawa A. Linking neurodevelopmental and synaptic theories of mental illness through DISC1. Nat Rev Neurosci. 2011;12:707–722. doi: 10.1038/nrn3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porteous DJ, Millar JK, Brandon NJ, Sawa A. DISC1 at 10: connecting psychiatric genetics and neuroscience. Trends Mol Med. 2011;17:699–706. doi: 10.1016/j.molmed.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori T, Shimizu S, Koyama Y, Yamada K, Kuwahara R, Kumamoto N, et al. DISC1 regulates cell-cell adhesion, cell-matrix adhesion and neurite outgrowth Mol Psychiatry 20101577898–809. [DOI] [PubMed] [Google Scholar]
- Callicott JH, Straub RE, Pezawas L, Egan MF, Mattay VS, Hariri AR, et al. Variation in DISC1 affects hippocampal structure and function and increases risk for schizophrenia. Proc Natl Acad Sci USA. 2005;102:8627–8632. doi: 10.1073/pnas.0500515102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carless MA, Glahn DC, Johnson MP, Curran JE, Bozaoglu K, Dyer TD, et al. Impact of DISC1 variation on neuroanatomical and neurocognitive phenotypes. Mol Psychiatry. 2011;16:1096–1104. doi: 10.1038/mp.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KK, De Rienzo G, Drane L, Mao Y, Flood Z, Madison J, et al. Common DISC1 polymorphisms disrupt Wnt/GSK3beta signaling and brain development. Neuron. 2011;72:545–558. doi: 10.1016/j.neuron.2011.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata K, Lipska BK, Hyde TM, Ye T, Newburn EN, Morita Y, et al. DISC1 splice variants are upregulated in schizophrenia and associated with risk polymorphisms. Proc Natl Acad Sci USA. 2009;106:15873–15878. doi: 10.1073/pnas.0903413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olincy A, House R, Gao B, Recksiek P, Phang TL, Sullivan B, et al. Elevated DISC1 transcript levels in PBMCs during acute psychosis in patients with schizophrenia Transl Biomed 20112pii: 183. [PMC free article] [PubMed] [Google Scholar]
- Ayhan Y, Abazyan B, Nomura J, Kim R, Ladenheim B, Krasnova IN, et al. Differential effects of prenatal and postnatal expressions of mutant human DISC1 on neurobehavioral phenotypes in transgenic mice: evidence for neurodevelopmental origin of major psychiatric disorders. Mol Psychiatry. 2011;16:293–306. doi: 10.1038/mp.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar JK, Mackie S, Clapcote SJ, Murdoch H, Pickard BS, Christie S, et al. Disrupted in schizophrenia 1 and phosphodiesterase 4B: towards an understanding of psychiatric illness. J Physiol. 2007;584 (Part 2:401–405. doi: 10.1113/jphysiol.2007.140210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Charych EI, Pulito VL, Lee JB, Graziane NM, Crozier RA, et al. The psychiatric disease risk factors DISC1 and TNIK interact to regulate synapse composition and function. Mol Psychiatry. 2011;16:1006–1023. doi: 10.1038/mp.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang E, Burdick KE, Kim JY, Duan X, Guo JU, Sailor KA, et al. Interaction between FEZ1 and DISC1 in regulation of neuronal development and risk for schizophrenia. Neuron. 2011;72:559–571. doi: 10.1016/j.neuron.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KK, Ge X, Mao Y, Drane L, Meletis K, Samuels BA, et al. Dixdc1 is a critical regulator of DISC1 and embryonic cortical development. Neuron. 2010;67:33–48. doi: 10.1016/j.neuron.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennertz L, Rujescu D, Wagner M, Frommann I, Schulze-Rauschenbach S, Schuhmacher A, et al. Novel schizophrenia risk gene TCF4 influences verbal learning and memory functioning in schizophrenia patients. Neuropsychobiology. 2011;63:131–136. doi: 10.1159/000317844. [DOI] [PubMed] [Google Scholar]
- Li T, Li Z, Chen P, Zhao Q, Wang T, Huang K, et al. Common variants in major histocompatibility complex region and TCF4 gene are significantly associated with schizophrenia in Han Chinese. Biol Psychiatry. 2010;68:671–673. doi: 10.1016/j.biopsych.2010.06.014. [DOI] [PubMed] [Google Scholar]
- Quednow BB, Ettinger U, Mossner R, Rujescu D, Giegling I, Collier DA, et al. The schizophrenia risk allele C of the TCF4 rs9960767 polymorphism disrupts sensorimotor gating in schizophrenia spectrum and healthy volunteers. J Neurosci. 2011;31:6684–6691. doi: 10.1523/JNEUROSCI.0526-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudge J, Miller NA, Khrebtukova I, Lindquist IE, May GD, Huntley JJ, et al. Genomic convergence analysis of schizophrenia: mRNA sequencing reveals altered synaptic vesicular transport in post-mortem cerebellum. PLoS One. 2008;3:e3625. doi: 10.1371/journal.pone.0003625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruch K, Silberberg G, Aviv A, Shamir E, Bening-Abu-Shach U, Baruch Y, et al. Association between golli-MBP and schizophrenia in the Jewish Ashkenazi population: are regulatory regions involved. Int J Neuropsychopharmacol. 2009;12:885–894. doi: 10.1017/S1461145708009887. [DOI] [PubMed] [Google Scholar]
- Tkachev D, Mimmack ML, Ryan MM, Wayland M, Freeman T, Jones PB, et al. Oligodendrocyte dysfunction in schizophrenia and bipolar disorder. Lancet. 2003;362:798–805. doi: 10.1016/S0140-6736(03)14289-4. [DOI] [PubMed] [Google Scholar]
- Glatt SJ, Stone WS, Nossova N, Liew CC, Seidman LJ, Tsuang MT. Similarities and differences in peripheral blood gene-expression signatures of individuals with schizophrenia and their first-degree biological relatives. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:869–887. doi: 10.1002/ajmg.b.31239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le-Niculescu H, McFarland MJ, Ogden CA, Balaraman Y, Patel S, Tan J, et al. Phenomic, convergent functional genomic, and biomarker studies in a stress-reactive genetic animal model of bipolar disorder and co-morbid alcoholism. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:134–166. doi: 10.1002/ajmg.b.30707. [DOI] [PubMed] [Google Scholar]
- Haroutunian V, Davis KL. Introduction to the special section: myelin and oligodendrocyte abnormalities in schizophrenia. Int J Neuropsychopharmacol. 2007;10:499–502. doi: 10.1017/S1461145706007449. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Sakurai T, Davis KL, Buxbaum JD. Linking oligodendrocyte and myelin dysfunction to neurocircuitry abnormalities in schizophrenia. Prog Neurobiol. 2011;93:13–24. doi: 10.1016/j.pneurobio.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konradi C, Sillivan SE, Clay HB. Mitochondria, oligodendrocytes and inflammation in bipolar disorder: evidence from transcriptome studies points to intriguing parallels with multiple sclerosis. Neurobiol Dis. 2012;45:37–47. doi: 10.1016/j.nbd.2011.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pae CU, Drago A, Kim JJ, Mandelli L, De Ronchi D, Serretti A. The impact of heat shock protein 70 gene variations on clinical presentation and outcome in schizophrenic inpatients. Neuropsychobiology. 2009;59:135–141. doi: 10.1159/000218075. [DOI] [PubMed] [Google Scholar]
- Arion D, Unger T, Lewis DA, Levitt P, Mirnics K. Molecular evidence for increased expression of genes related to immune and chaperone function in the prefrontal cortex in schizophrenia. Biol Psychiatry. 2007;62:711–721. doi: 10.1016/j.biopsych.2006.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le-Niculescu H, Balaraman Y, Patel SD, Ayalew M, Gupta J, Kuczenski R, et al. Convergent functional genomics of anxiety disorders: translational identification of genes, biomarkers, pathways and mechanisms Translational Psychiatry 20111e9doi: 10.1038/tp.2011.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le-Niculescu H, Case NJ, Hulvershorn L, Patel SD, Bowker D, Gupta J, et al. Convergent functional genomic studies of omega-3 fatty acids in stress reactivity, bipolar disorder and alcoholism Translational Psychiatry 20111e4doi: 10.1038/tp.2011.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussos P, Katsel P, Davis KL, Bitsios P, Giakoumaki SG, Jogia J, et al. Molecular and genetic evidence for abnormalities in the nodes of ranvier in schizophrenia. Arch Gen Psychiatry. 2012;69:7–15. doi: 10.1001/archgenpsychiatry.2011.110. [DOI] [PubMed] [Google Scholar]
- Schwarz E, Guest PC, Rahmoune H, Harris LW, Wang L, Leweke FM, et al. Identification of a biological signature for schizophrenia in serum Mol Psychiatryadvance online publication, 12 April 2011 (e-pub ahead of print). [DOI] [PubMed]
- Ujike H, Takaki M, Nakata K, Tanaka Y, Takeda T, Kodama M, et al. CNR1, central cannabinoid receptor gene, associated with susceptibility to hebephrenic schizophrenia. Mol Psychiatry. 2002;7:515–518. doi: 10.1038/sj.mp.4001029. [DOI] [PubMed] [Google Scholar]
- Xu B, Woodroffe A, Rodriguez-Murillo L, Roos JL, van Rensburg EJ, Abecasis GR, et al. Elucidating the genetic architecture of familial schizophrenia using rare copy number variant and linkage scans. Proc Natl Acad Sci USA. 2009;106:16746–16751. doi: 10.1073/pnas.0908584106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggan SM, Hashimoto T, Lewis DA. Reduced cortical cannabinoid 1 receptor messenger RNA and protein expression in schizophrenia. Arch Gen Psychiatry. 2008;65:772–784. doi: 10.1001/archpsyc.65.7.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsom RJ, Osterlund C, Masini CV, Day HE, Spencer RL, Campeau S. Cannabinoid receptor type 1 antagonism significantly modulates basal and loud noise induced neural and hypothalamic-pituitary-adrenal axis responses in male Sprague-Dawley rats. Neuroscience. 2012;204:64–73. doi: 10.1016/j.neuroscience.2011.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray RM, Morrison PD, Henquet C, Di Forti M. Cannabis, the mind and society: the hash realities. Nat Rev Neurosci. 2007;8:885–895. doi: 10.1038/nrn2253. [DOI] [PubMed] [Google Scholar]
- Moghaddam B, Javitt D. From revolution to evolution: the glutamate hypothesis of schizophrenia and its implication for treatment. Neuropsychopharmacology. 2012;37:4–15. doi: 10.1038/npp.2011.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach BD, Osterweil EK, Bear MF. Mutations causing syndromic autism define an axis of synaptic pathophysiology. Nature. 2011;480:63–68. doi: 10.1038/nature10658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisse M, Halabisky B, Harris J, Devidze N, Dubal DB, Sun B, et al. Reversing EphB2 depletion rescues cognitive functions in Alzheimer model. Nature. 2011;469:47–52. doi: 10.1038/nature09635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javitt DC, Schoepp D, Kalivas PW, Volkow ND, Zarate C, Merchant K, et al. Translating glutamate: from pathophysiology to treatment. Sci Transl Med. 2011;3:102mr2. doi: 10.1126/scitranslmed.3002804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson DF, Duan J, Oh S, Wang K, Sanders AR, Shi J, et al. Copy number variants in schizophrenia: confirmation of five previous findings and new evidence for 3q29 microdeletions and VIPR2 duplications. Am J psychiatry. 2011;168:302–316. doi: 10.1176/appi.ajp.2010.10060876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacic V, McCarthy S, Malhotra D, Murray F, Chou HH, Peoples A, et al. Duplications of the neuropeptide receptor gene VIPR2 confer significant risk for schizophrenia. Nature. 2011;471:499–503. doi: 10.1038/nature09884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tregellas JR, Tanabe J, Rojas DC, Shatti S, Olincy A, Johnson L, et al. Effects of an alpha 7-nicotinic agonist on default network activity in schizophrenia. Biol psychiatry. 2011;69:7–11. doi: 10.1016/j.biopsych.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charych EI, Jiang LX, Lo F, Sullivan K, Brandon NJ. Interplay of palmitoylation and phosphorylation in the trafficking and localization of phosphodiesterase 10A: implications for the treatment of schizophrenia. J Neurosci. 2010;30:9027–9037. doi: 10.1523/JNEUROSCI.1635-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov G, Pocklington AJ, Holmans P, Ivanov D, Ikeda M, Ruderfer D, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry. 2012;17:142–153. doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niculescu AB, Lulow LL, Ogden CA, Le-Niculescu H, Salomon DR, Schork NJ, et al. PhenoChipping of psychotic disorders: a novel approach for deconstructing and quantitating psychiatric phenotypes. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:653–662. doi: 10.1002/ajmg.b.30404. [DOI] [PubMed] [Google Scholar]
- Niculescu AB., 3rd Polypharmacy in oligopopulations: what psychiatric genetics can teach biological psychiatry. Psychiatr Genet. 2006;16:241–244. doi: 10.1097/01.ypg.0000242195.74268.f9. [DOI] [PubMed] [Google Scholar]
- Niculescu AB, 3rd, Schork NJ, Salomon DR. Mindscape: A convergent perspective on life, mind, consciousness and happiness. J Affect Disord. 2010;123:1–8. doi: 10.1016/j.jad.2009.06.022. [DOI] [PubMed] [Google Scholar]
- Hennah W, Thomson P, McQuillin A, Bass N, Loukola A, Anjorin A, et al. DISC1 association, heterogeneity and interplay in schizophrenia and bipolar disorder. Mol Psychiatry. 2009;14:865–873. doi: 10.1038/mp.2008.22. [DOI] [PubMed] [Google Scholar]
- Bord L, Wheeler J, Paek M, Saleh M, Lyons-Warren A, Ross CA, et al. Primate disrupted-in-schizophrenia-1 (DISC1): high divergence of a gene for major mental illnesses in recent evolutionary history. Neurosci Res. 2006;56:286–293. doi: 10.1016/j.neures.2006.07.010. [DOI] [PubMed] [Google Scholar]
- Craddock N, Owen MJ. The Kraepelinian dichotomy—going, going... but still not gone. Br J Psychiatry. 2010;196:92–95. doi: 10.1192/bjp.bp.109.073429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams HJ, Craddock N, Russo G, Hamshere ML, Moskvina V, Dwyer S, et al. Most genome-wide significant susceptibility loci for schizophrenia and bipolar disorder reported to date cross-traditional diagnostic boundaries. Hum Mol Genet. 2011;20:387–391. doi: 10.1093/hmg/ddq471. [DOI] [PubMed] [Google Scholar]
- Buckley PF, Miller BJ, Lehrer DS, Castle DJ. Psychiatric comorbidities and schizophrenia. Schizophr Bull. 2009;35:383–402. doi: 10.1093/schbul/sbn135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen NC, Bagade S, McQueen MB, Ioannidis JP, Kavvoura FK, Khoury MJ, et al. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat Genet. 2008;40:827–834. doi: 10.1038/ng.171. [DOI] [PubMed] [Google Scholar]
- Jia P, Sun J, Guo AY, Zhao Z. SZGR: a comprehensive schizophrenia gene resource. Mol Psychiatry. 2010;15:453–462. doi: 10.1038/mp.2009.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan HY, Callicott JH, Weinberger DR. Intermediate phenotypes in schizophrenia genetics redux: is it a no brainer. Mol psychiatry. 2008;13:233–238. doi: 10.1038/sj.mp.4002145. [DOI] [PubMed] [Google Scholar]
- Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, et al. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478:519–523. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan JH, Lasky-Su J, Raby BA, Xu M, Molony C, Schadt EE, et al. Genomics and genome-wide association studies: an integrative approach to expression QTL mapping. Genomics. 2008;92:129–133. doi: 10.1016/j.ygeno.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O′Donovan MC, Craddock NJ, Owen MJ. Genetics of psychosis; insights from views across the genome. Hum Genet. 2009;126:3–12. doi: 10.1007/s00439-009-0703-0. [DOI] [PubMed] [Google Scholar]
- Clapcote SJ, Lipina TV, Millar JK, Mackie S, Christie S, Ogawa F, et al. Behavioral phenotypes of Disc1 missense mutations in mice. Neuron. 2007;54:387–402. doi: 10.1016/j.neuron.2007.04.015. [DOI] [PubMed] [Google Scholar]
- Hikida T, Jaaro-Peled H, Seshadri S, Oishi K, Hookway C, Kong S, et al. Dominant-negative DISC1 transgenic mice display schizophrenia-associated phenotypes detected by measures translatable to humans. Proc Natl Acad Sci USA. 2007;104:14501–14506. doi: 10.1073/pnas.0704774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Chang JH, Ge S, Faulkner RL, Kim JY, Kitabatake Y, et al. Disrupted-In-Schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell. 2007;130:1146–1158. doi: 10.1016/j.cell.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike H, Arguello PA, Kvajo M, Karayiorgou M, Gogos JA. Disc1 is mutated in the 129S6/SvEv strain and modulates working memory in mice. Proc Natl Acad Sci USA. 2006;103:3693–3697. doi: 10.1073/pnas.0511189103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvajo M, McKellar H, Arguello PA, Drew LJ, Moore H, MacDermott AB, et al. A mutation in mouse Disc1 that models a schizophrenia risk allele leads to specific alterations in neuronal architecture and cognition. Proc Natl Acad Sci USA. 2008;105:7076–7081. doi: 10.1073/pnas.0802615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Lang B, Nakamoto C, Zhang F, Pu J, Kuan SL, et al. Schizophrenia-related neural and behavioral phenotypes in transgenic mice expressing truncated Disc1. J Neurosci. 2008;28:10893–10904. doi: 10.1523/JNEUROSCI.3299-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletnikov MV, Ayhan Y, Xu Y, Nikolskaia O, Ovanesov M, Huang H, et al. Enlargement of the lateral ventricles in mutant DISC1 transgenic mice. Mol Psychiatry. 2008;13:115. doi: 10.1038/sj.mp.4002144. [DOI] [PubMed] [Google Scholar]
- Pletnikov MV, Ayhan Y, Nikolskaia O, Xu Y, Ovanesov MV, Huang H, et al. Inducible expression of mutant human DISC1 in mice is associated with brain and behavioral abnormalities reminiscent of schizophrenia Mol Psychiatry 200813173–186.15. [DOI] [PubMed] [Google Scholar]
- Chiba S, Hashimoto R, Hattori S, Yohda M, Lipska B, Weinberger DR, et al. Effect of antipsychotic drugs on DISC1 and dysbindin expression in mouse frontal cortex and hippocampus. J Neural Transm. 2006;113:1337–1346. doi: 10.1007/s00702-005-0414-1. [DOI] [PubMed] [Google Scholar]
- Schurov IL, Handford EJ, Brandon NJ, Whiting PJ. Expression of disrupted in schizophrenia 1 (DISC1) protein in the adult and developing mouse brain indicates its role in neurodevelopment. Mol Psychiatry. 2004;9:1100–1110. doi: 10.1038/sj.mp.4001574. [DOI] [PubMed] [Google Scholar]
- Palo OM, Antila M, Silander K, Hennah W, Kilpinen H, Soronen P, et al. Association of distinct allelic haplotypes of DISC1 with psychotic and bipolar spectrum disorders and with underlying cognitive impairments. Hum Mol Genet. 2007;16:2517–2528. doi: 10.1093/hmg/ddm207. [DOI] [PubMed] [Google Scholar]
- Potkin SG, Macciardi F, Guffanti G, Fallon JH, Wang Q, Turner JA, et al. Identifying gene regulatory networks in schizophrenia. Neuroimage. 2010;53:839–847. doi: 10.1016/j.neuroimage.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher J, Laje G, Abou Jamra R, Becker T, Muhleisen TW, Vasilescu C, et al. The DISC locus and schizophrenia: evidence from an association study in a central European sample and from a meta-analysis across different European populations. Hum Mol Genet. 2009;18:2719–2727. doi: 10.1093/hmg/ddp204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glessner JT, Reilly MP, Kim CE, Takahashi N, Albano A, Hou C, et al. Strong synaptic transmission impact by copy number variations in schizophrenia. Proc Natl Acad Sci USA. 2010;107:10584–10589. doi: 10.1073/pnas.1000274107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green EK, Grozeva D, Sims R, Raybould R, Forty L, Gordon-Smith K, et al. DISC1 exon 11 rare variants found more commonly in schizoaffective spectrum cases than controls. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:490–492. doi: 10.1002/ajmg.b.31187. [DOI] [PubMed] [Google Scholar]
- Liu YL, Fann CS, Liu CM, Chen WJ, Wu JY, Hung SI, et al. A single nucleotide polymorphism fine mapping study of chromosome 1q42.1 reveals the vulnerability genes for schizophrenia, GNPAT and DISC1: association with impairment of sustained attention. Biol Psychiatry. 2006;60:554–562. doi: 10.1016/j.biopsych.2006.04.024. [DOI] [PubMed] [Google Scholar]
- Greenwood TA, Lazzeroni LC, Murray SS, Cadenhead KS, Calkins ME, Dobie DJ, et al. Analysis of 94 candidate genes and 12 endophenotypes for schizophrenia from the Consortium on the Genetics of Schizophrenia. Am J Psychiatry. 2011;168:930–946. doi: 10.1176/appi.ajp.2011.10050723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouchi Y, Kubota Y, Kuramasu A, Watanabe T, Ito C. Gene expression profiling in whole cerebral cortices of phencyclidine- or methamphetamine-treated rats. Brain Res Mol Brain Res. 2005;140:142–149. doi: 10.1016/j.molbrainres.2005.07.011. [DOI] [PubMed] [Google Scholar]
- Fatemi SH, Pearce DA, Brooks AI, Sidwell RW. Prenatal viral infection in mouse causes differential expression of genes in brains of mouse progeny: a potential animal model for schizophrenia and autism. Synapse. 2005;57:91–99. doi: 10.1002/syn.20162. [DOI] [PubMed] [Google Scholar]
- Glatt SJ, Stone WS, Nossova N, Liew CC, Seidman LJ, Tsuang MT. Similarities and differences in peripheral blood gene-expression signatures of individuals with schizophrenia and their first-degree biological relatives. American journal of medical genetics Part B, Neuropsychiatric genetics: the official publication of the International Society of Psychiatric Genetics. 2011;156B:869–887. doi: 10.1002/ajmg.b.31239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzozka MM, Radyushkin K, Wichert SP, Ehrenreich H, Rossner MJ. Cognitive and sensorimotor gating impairments in transgenic mice overexpressing the schizophrenia susceptibility gene Tcf4 in the brain. Biol Psychiatry. 2010;68:33–40. doi: 10.1016/j.biopsych.2010.03.015. [DOI] [PubMed] [Google Scholar]
- Lowe XR, Lu X, Marchetti F, Wyrobek AJ. The expression of Troponin T1 gene is induced by ketamine in adult mouse brain. Brain Res. 2007;1174:7–17. doi: 10.1016/j.brainres.2007.07.039. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Kumanishi T, Hayashi S. Using a DNA microarray method to examine gene expression in brain from clozapine-injected mice. Ann N Y Acad Sci. 2004;1025:561–569. doi: 10.1196/annals.1316.069. [DOI] [PubMed] [Google Scholar]
- Matsuoka T, Tsunoda M, Sumiyoshi T, Takasaki I, Tabuchi Y, Seo T, et al. Effect of MK-801 on gene expressions in the amygdala of rats. Synapse. 2008;62:1–7. doi: 10.1002/syn.20455. [DOI] [PubMed] [Google Scholar]
- Lewis CM, Levinson DF, Wise LH, DeLisi LE, Straub RE, Hovatta I, et al. Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: Schizophrenia. Am J Hum Genet. 2003;73:34–48. doi: 10.1086/376549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitkus SN, Hyde TM, Vakkalanka R, Kolachana B, Weinberger DR, Kleinman JE, et al. Expression of oligodendrocyte-associated genes in dorsolateral prefrontal cortex of patients with schizophrenia. Schizophr Res. 2008;98:129–138. doi: 10.1016/j.schres.2007.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden NA, Weidenhofer J, Scott RJ, Schall U, Todd J, Michie PT, et al. Preliminary investigation of gene expression profiles in peripheral blood lymphocytes in schizophrenia. Schizophr Res. 2006;82:175–183. doi: 10.1016/j.schres.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Wood GK, Tomasiewicz H, Rutishauser U, Magnuson T, Quirion R, Rochford J, et al. NCAM-180 knockout mice display increased lateral ventricle size and reduced prepulse inhibition of startle. Neuroreport. 1998;9:461–466. doi: 10.1097/00001756-199802160-00019. [DOI] [PubMed] [Google Scholar]
- Atz ME, Rollins B, Vawter MP. NCAM1 association study of bipolar disorder and schizophrenia: polymorphisms and alternatively spliced isoforms lead to similarities and differences. Psychiatr Genet. 2007;17:55–67. doi: 10.1097/YPG.0b013e328012d850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan MK, Tsang TM, Harris LW, Guest PC, Holmes E, Bahn S. Evidence for disease and antipsychotic medication effects in post-mortem brain from schizophrenia patients. Mol Psychiatry. 2011;16:1189–1202. doi: 10.1038/mp.2010.100. [DOI] [PubMed] [Google Scholar]
- Ji B, La Y, Gao L, Zhu H, Tian N, Zhang M, et al. A comparative proteomics analysis of rat mitochondria from the cerebral cortex and hippocampus in response to antipsychotic medications. J Proteome Res. 2009;8:3633–3641. doi: 10.1021/pr800876z. [DOI] [PubMed] [Google Scholar]
- Washizuka S, Kametani M, Sasaki T, Tochigi M, Umekage T, Kohda K, et al. Association of mitochondrial complex I subunit gene NDUFV2 at 18p11 with schizophrenia in the Japanese population. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:301–304. doi: 10.1002/ajmg.b.30285. [DOI] [PubMed] [Google Scholar]
- Ben-Shachar D, Karry R. Sp1 expression is disrupted in schizophrenia; a possible mechanism for the abnormal expression of mitochondrial complex I genes, NDUFV1 and NDUFV2. PLoS One. 2007;2:e817. doi: 10.1371/journal.pone.0000817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabakaran S, Swatton JE, Ryan MM, Huffaker SJ, Huang JT, Griffin JL, et al. Mitochondrial dysfunction in schizophrenia: evidence for compromised brain metabolism and oxidative stress Mol Psychiatry 20049684–697.43. [DOI] [PubMed] [Google Scholar]
- Moy SS, Nonneman RJ, Young NB, Demyanenko GP, Maness PF. Impaired sociability and cognitive function in Nrcam-null mice. Behav Brain Res. 2009;205:123–131. doi: 10.1016/j.bbr.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekelund J, Lichtermann D, Hovatta I, Ellonen P, Suvisaari J, Terwilliger JD, et al. Genome-wide scan for schizophrenia in the Finnish population: evidence for a locus on chromosome 7q22. Hum Mol Genet. 2000;9:1049–1057. doi: 10.1093/hmg/9.7.1049. [DOI] [PubMed] [Google Scholar]
- Maziade M, Roy MA, Chagnon YC, Cliche D, Fournier JP, Montgrain N, et al. Shared and specific susceptibility loci for schizophrenia and bipolar disorder: a dense genome scan in Eastern Quebec families. Mol Psychiatry. 2005;10:486–499. doi: 10.1038/sj.mp.4001594. [DOI] [PubMed] [Google Scholar]
- Faraone SV, Matise T, Svrakic D, Pepple J, Malaspina D, Suarez B, et al. Genome scan of European-American schizophrenia pedigrees: results of the NIMH Genetics Initiative and Millennium Consortium. Am J Med Genet. 1998;81:290–295. [PubMed] [Google Scholar]
- Takahashi S, Faraone SV, Lasky-Su J, Tsuang MT. Genome-wide scan of homogeneous subtypes of NIMH genetics initiative schizophrenia families. Psychiatry Res. 2005;133:111–122. doi: 10.1016/j.psychres.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Freedman R, Leonard S, Olincy A, Kaufmann CA, Malaspina D, Cloninger CR, et al. Evidence for the multigenic inheritance of schizophrenia. Am J Med Genet. 2001;105:794–800. doi: 10.1002/ajmg.10100. [DOI] [PubMed] [Google Scholar]
- Straub RE, MacLean CJ, Ma Y, Webb BT, Myakishev MV, Harris-Kerr C, et al. Genome-wide scans of three independent sets of 90 Irish multiplex schizophrenia families and follow-up of selected regions in all families provides evidence for multiple susceptibility genes. Mol Psychiatry. 2002;7:542–559. doi: 10.1038/sj.mp.4001051. [DOI] [PubMed] [Google Scholar]
- Chu TT, Liu Y, Kemether E. Thalamic transcriptome screening in three psychiatric states. J Hum Genet. 2009;54:665–675. doi: 10.1038/jhg.2009.93. [DOI] [PubMed] [Google Scholar]
- Kuzman MR, Medved V, Terzic J, Krainc D. Genome-wide expression analysis of peripheral blood identifies candidate biomarkers for schizophrenia. J Psychiatr Res. 2009;43:1073–1077. doi: 10.1016/j.jpsychires.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Hashimoto R, Hashimoto H, Shintani N, Chiba S, Hattori S, Okada T, et al. Pituitary adenylate cyclase-activating polypeptide is associated with schizophrenia. Mol Psychiatry. 2007;12:1026–1032. doi: 10.1038/sj.mp.4001982. [DOI] [PubMed] [Google Scholar]
- Koga M, Ishiguro H, Horiuchi Y, Inada T, Ujike H, Itokawa M, et al. Replication study of association between ADCYAP1 gene polymorphisms and schizophrenia. Psychiatr Genet. 2010;20:123–125. doi: 10.1097/YPG.0b013e32833a1f52. [DOI] [PubMed] [Google Scholar]
- Galter D, Buervenich S, Carmine A, Anvret M, Olson L. ALDH1 mRNA: presence in human dopamine neurons and decreases in substantia nigra in Parkinson's disease and in the ventral tegmental area in schizophrenia. Neurobiol Dis. 2003;14:637–647. doi: 10.1016/j.nbd.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Barley K, Dracheva S, Byne W. Subcortical oligodendrocyte- and astrocyte-associated gene expression in subjects with schizophrenia, major depression and bipolar disorder. Schizophr Res. 2009;112:54–64. doi: 10.1016/j.schres.2009.04.019. [DOI] [PubMed] [Google Scholar]
- Wang L, Lockstone HE, Guest PC, Levin Y, Palotas A, Pietsch S, et al. Expression profiling of fibroblasts identifies cell cycle abnormalities in schizophrenia. J Proteome Res. 2010;9:521–527. doi: 10.1021/pr900867x. [DOI] [PubMed] [Google Scholar]
- Athanasiu L, Mattingsdal M, Kahler AK, Brown A, Gustafsson O, Agartz I, et al. Gene variants associated with schizophrenia in a Norwegian genome-wide study are replicated in a large European cohort. J Psychiatr Res. 2010;44:748–753. doi: 10.1016/j.jpsychires.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer JU, Schmitt A, Heck M, Schaeffer EL, Fendt M, Zink M, et al. Differential expression of presynaptic genes in a rat model of postnatal hypoxia: relevance to schizophrenia. Eur Arch Psychiatry Clin Neurosci. 2010;260 (Suppl 2:S81–S89. doi: 10.1007/s00406-010-0159-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai A, Mahadik SP. Increased truncated TrkB receptor expression and decreased BDNF/TrkB signaling in the frontal cortex of reeler mouse model of schizophrenia. Schizophr Res. 2008;100:325–333. doi: 10.1016/j.schres.2007.11.030. [DOI] [PubMed] [Google Scholar]
- Grottick AJ, Bagnol D, Phillips S, McDonald J, Behan DP, Chalmers DT, et al. Neurotransmission- and cellular stress-related gene expression associated with prepulse inhibition in mice. Brain Res Mol Brain Res. 2005;139:153–162. doi: 10.1016/j.molbrainres.2005.05.020. [DOI] [PubMed] [Google Scholar]
- Kaiser S, Foltz LA, George CA, Kirkwood SC, Bemis KG, Lin X, et al. Phencyclidine-induced changes in rat cortical gene expression identified by microarray analysis: implications for schizophrenia. Neurobiol Dis. 2004;16:220–235. doi: 10.1016/j.nbd.2004.01.011. [DOI] [PubMed] [Google Scholar]
- Guo C, Yang Y, Su Y, Si T. Postnatal BDNF expression profiles in prefrontal cortex and hippocampus of a rat schizophrenia model induced by MK-801 administration. J Biomed Biotechnol. 2010;2010:783297. doi: 10.1155/2010/783297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao HM, Kao HT, Porton B. BDNF Val66Met variant and age of onset in schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:505–506. doi: 10.1002/ajmg.b.30619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratacos M, Gonzalez JR, Mercader JM, de Cid R, Urretavizcaya M, Estivill X. Brain-derived neurotrophic factor Val66Met and psychiatric disorders: meta-analysis of case-control studies confirm association to substance-related disorders, eating disorders, and schizophrenia. Biol Psychiatry. 2007;61:911–922. doi: 10.1016/j.biopsych.2006.08.025. [DOI] [PubMed] [Google Scholar]
- Suchanek R, Owczarek A, Kowalski J. Association Study between BDNF C-281A polymorphism and paranoid schizophrenia in Polish population. J Mol Neurosci. 2012;46:217–222. doi: 10.1007/s12031-011-9582-7. [DOI] [PubMed] [Google Scholar]
- Decoster J, van Os J, Kenis G, Henquet C, Peuskens J, De Hert M, et al. Age at onset of psychotic disorder: cannabis, BDNF Val66Met, and sex-specific models of gene-environment interaction. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:363–369. doi: 10.1002/ajmg.b.31174. [DOI] [PubMed] [Google Scholar]
- Torrey EF, Barci BM, Webster MJ, Bartko JJ, Meador-Woodruff JH, Knable MB. Neurochemical markers for schizophrenia, bipolar disorder, and major depression in postmortem brains. Biol Psychiatry. 2005;57:252–260. doi: 10.1016/j.biopsych.2004.10.019. [DOI] [PubMed] [Google Scholar]
- Pillai A. Decreased expression of Sprouty2 in the dorsolateral prefrontal cortex in schizophrenia and bipolar disorder: a correlation with BDNF expression. PLoS One. 2008;3:e1784. doi: 10.1371/journal.pone.0001784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weickert CS, Hyde TM, Lipska BK, Herman MM, Weinberger DR, Kleinman JE. Reduced brain-derived neurotrophic factor in prefrontal cortex of patients with schizophrenia. Mol Psychiatry. 2003;8:592–610. doi: 10.1038/sj.mp.4001308. [DOI] [PubMed] [Google Scholar]
- Rizos EN, Papathanasiou M, Michalopoulou PG, Mazioti A, Douzenis A, Kastania A, et al. Association of serum BDNF levels with hippocampal volumes in first psychotic episode drug-naive schizophrenic patients. Schizophr Res. 2011;129:201–204. doi: 10.1016/j.schres.2011.03.011. [DOI] [PubMed] [Google Scholar]
- Grillo RW, Ottoni GL, Leke R, Souza DO, Portela LV, Lara DR. Reduced serum BDNF levels in schizophrenic patients on clozapine or typical antipsychotics. J Psychiatr Res. 2007;41:31–35. doi: 10.1016/j.jpsychires.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Miodownik C, Maayan R, Ratner Y, Lerner V, Pintov L, Mar M, et al. Serum levels of brain-derived neurotrophic factor and cortisol to sulfate of dehydroepiandrosterone molar ratio associated with clinical response to l-theanine as augmentation of antipsychotic therapy in schizophrenia and schizoaffective disorder patients. Clin Neuropharmacol. 2011;34:155–160. doi: 10.1097/WNF.0b013e318220d8c6. [DOI] [PubMed] [Google Scholar]
- Mondelli V, Cattaneo A, Murri MB, Di Forti M, Handley R, Hepgul N, et al. Stress and inflammation reduce brain-derived neurotrophic factor expression in first-episode psychosis: a pathway to smaller hippocampal volume. J Clin Psychiatry. 2011;72:1677–1684. doi: 10.4088/JCP.10m06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsel P, Davis KL, Haroutunian V. Variations in myelin and oligodendrocyte-related gene expression across multiple brain regions in schizophrenia: a gene ontology study. Schizophr Res. 2005;79:157–173. doi: 10.1016/j.schres.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Haller J, Szirmai M, Varga B, Ledent C, Freund TF. Cannabinoid CB1 receptor dependent effects of the NMDA antagonist phencyclidine in the social withdrawal model of schizophrenia. Behav Pharmacol. 2005;16:415–422. doi: 10.1097/00008877-200509000-00014. [DOI] [PubMed] [Google Scholar]
- Babovic D, O′Tuathaigh CM, O′Connor AM, O′Sullivan GJ, Tighe O, Croke DT, et al. Phenotypic characterization of cognition and social behavior in mice with heterozygous versus homozygous deletion of catechol-O-methyltransferase. Neuroscience. 2008;155:1021–1029. doi: 10.1016/j.neuroscience.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Huotari M, Garcia-Horsman JA, Karayiorgou M, Gogos JA, Mannisto PT. D-amphetamine responses in catechol-O-methyltransferase (COMT) disrupted mice. Psychopharmacology (Berl) 2004;172:1–10. doi: 10.1007/s00213-003-1627-3. [DOI] [PubMed] [Google Scholar]
- Jurata LW, Gallagher P, Lemire AL, Charles V, Brockman JA, Illingworth EL, et al. Altered expression of hippocampal dentate granule neuron genes in a mouse model of human 22q11 deletion syndrome. Schizophr Res. 2006;88:251–259. doi: 10.1016/j.schres.2006.07.017. [DOI] [PubMed] [Google Scholar]
- Shifman S, Bronstein M, Sternfeld M, Pisante-Shalom A, Lev-Lehman E, Weizman A, et al. A highly significant association between a COMT haplotype and schizophrenia. Am J Hum Genet. 2002;71:1296–1302. doi: 10.1086/344514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdolmaleky HM, Cheng KH, Faraone SV, Wilcox M, Glatt SJ, Gao F, et al. Hypomethylation of MB-COMT promoter is a major risk factor for schizophrenia and bipolar disorder. Hum Mol Genet. 2006;15:3132–3145. doi: 10.1093/hmg/ddl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y, Qin LH, Nishihara M, Sawada K, Kato K, Inoue S. Vulnerability of synaptic plasticity in the complexin II knockout mouse to maternal deprivation stress. Brain Res. 2005;1056:59–67. doi: 10.1016/j.brainres.2005.07.015. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Song JY, Kim JW, Jin SY, Hong MS, Park JK, et al. Association study of polymorphisms in synaptic vesicle-associated genes, SYN2 and CPLX2, with schizophrenia. Behav Brain Funct. 2005;1:15. doi: 10.1186/1744-9081-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastwood SL, Harrison PJ. Decreased expression of vesicular glutamate transporter 1 and complexin II mRNAs in schizophrenia: further evidence for a synaptic pathology affecting glutamate neurons. Schizophr Res. 2005;73:159–172. doi: 10.1016/j.schres.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Weidenhofer J, Bowden NA, Scott RJ, Tooney PA. Altered gene expression in the amygdala in schizophrenia: up-regulation of genes located in the cytomatrix active zone. Mol Cell Neurosci. 2006;31:243–250. doi: 10.1016/j.mcn.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Ortega-Alvaro A, Aracil-Fernandez A, Garcia-Gutierrez MS, Navarrete F, Manzanares J. Deletion of CB2 cannabinoid receptor induces schizophrenia-related behaviors in mice. Neuropsychopharmacology. 2011;36:1489–1504. doi: 10.1038/npp.2011.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Downing AC, Munsie LM, Chen P, Reed MR, Ruble CL, et al. Pharmacogenetic analysis of the mGlu2/3 agonist LY2140023 monohydrate in the treatment of schizophrenia Pharmacogenomics Jadvance online publication, 21 December 2010 (e-pub ahead of print). [DOI] [PubMed]
- McClay JL, Adkins DE, Aberg K, Bukszar J, Khachane AN, Keefe RS, et al. Genome-wide pharmacogenomic study of neurocognition as an indicator of antipsychotic treatment response in schizophrenia. Neuropsychopharmacology. 2011;36:616–626. doi: 10.1038/npp.2010.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan L, Kerr JR, Lafuente MJ, Maclean A, Chibalina MV, Liu B, et al. Altered expression and coregulation of dopamine signalling genes in schizophrenia and bipolar disorder. Neuropathol Appl Neurobiol. 2011;37:206–219. doi: 10.1111/j.1365-2990.2010.01128.x. [DOI] [PubMed] [Google Scholar]
- Dean B, Pavey G, Scarr E, Goeringer K, Copolov DL. Measurement of dopamine D2-like receptors in postmortem CNS and pituitary: differential regional changes in schizophrenia. Life Sci. 2004;74:3115–3131. doi: 10.1016/j.lfs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Chen XW, Feng YQ, Hao CJ, Guo XL, He X, Zhou ZY, et al. DTNBP1, a schizophrenia susceptibility gene, affects kinetics of transmitter release. J Cell Biol. 2008;181:791–801. doi: 10.1083/jcb.200711021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng YQ, Zhou ZY, He X, Wang H, Guo XL, Hao CJ, et al. Dysbindin deficiency in sandy mice causes reduction of snapin and displays behaviors related to schizophrenia. Schizophr Res. 2008;106:218–228. doi: 10.1016/j.schres.2008.07.018. [DOI] [PubMed] [Google Scholar]
- Hattori S, Murotani T, Matsuzaki S, Ishizuka T, Kumamoto N, Takeda M, et al. Behavioral abnormalities and dopamine reductions in sdy mutant mice with a deletion in Dtnbp1, a susceptibility gene for schizophrenia. Biochem Biophys Res Commun. 2008;373:298–302. doi: 10.1016/j.bbrc.2008.06.016. [DOI] [PubMed] [Google Scholar]
- Takao K, Toyama K, Nakanishi K, Hattori S, Takamura H, Takeda M, et al. Impaired long-term memory retention and working memory in sdy mutant mice with a deletion in Dtnbp1, a susceptibility gene for schizophrenia. Mol Brain. 2008;1:11. doi: 10.1186/1756-6606-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voisey J, Swagell CD, Hughes IP, Lawford BR, Young RM, Morris CP. Analysis of HapMap tag-SNPs in dysbindin (DTNBP1) reveals evidence of consistent association with schizophrenia. Eur Psychiatry. 2010;25:314–319. doi: 10.1016/j.eurpsy.2009.11.011. [DOI] [PubMed] [Google Scholar]
- Rethelyi JM, Bakker SC, Polgar P, Czobor P, Strengman E, Pasztor PI, et al. Association study of NRG1, DTNBP1, RGS4, G72/G30, and PIP5K2A with schizophrenia and symptom severity in a Hungarian sample. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:792–801. doi: 10.1002/ajmg.b.31049. [DOI] [PubMed] [Google Scholar]
- Schwab SG, Knapp M, Mondabon S, Hallmayer J, Borrmann-Hassenbach M, Albus M, et al. Support for association of schizophrenia with genetic variation in the 6p22.3 gene, dysbindin, in sib-pair families with linkage and in an additional sample of triad families. Am J Hum Genet. 2003;72:185–190. doi: 10.1086/345463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatjo-Vilas M, Papiol S, Estrada G, Bombin I, Peralta V, Rosa A, et al. Dysbindin-1 gene contributes differentially to early- and adult-onset forms of functional psychosis. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:322–333. doi: 10.1002/ajmg.b.31166. [DOI] [PubMed] [Google Scholar]
- Fallin MD, Lasseter VK, Avramopoulos D, Nicodemus KK, Wolyniec PS, McGrath JA, et al. Bipolar I disorder and schizophrenia: a 440-single-nucleotide polymorphism screen of 64 candidate genes among Ashkenazi Jewish case-parent trios. Am J Hum Genet. 2005;77:918–936. doi: 10.1086/497703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov G, Ivanov D, Williams NM, Preece A, Nikolov I, Milev R, et al. Strong evidence for association between the dystrobrevin binding protein 1 gene (DTNBP1) and schizophrenia in 488 parent-offspring trios from Bulgaria. Biol Psychiatry. 2004;55:971–975. doi: 10.1016/j.biopsych.2004.01.025. [DOI] [PubMed] [Google Scholar]
- Weickert CS, Straub RE, McClintock BW, Matsumoto M, Hashimoto R, Hyde TM, et al. Human dysbindin (DTNBP1) gene expression in normal brain and in schizophrenic prefrontal cortex and midbrain. Arch Gen Psychiatry. 2004;61:544–555. doi: 10.1001/archpsyc.61.6.544. [DOI] [PubMed] [Google Scholar]
- Talbot K, Eidem WL, Tinsley CL, Benson MA, Thompson EW, Smith RJ, et al. Dysbindin-1 is reduced in intrinsic, glutamatergic terminals of the hippocampal formation in schizophrenia. J Clin Invest. 2004;113:1353–1363. doi: 10.1172/JCI20425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weickert CS, Rothmond DA, Hyde TM, Kleinman JE, Straub RE. Reduced DTNBP1 (dysbindin-1) mRNA in the hippocampal formation of schizophrenia patients. Schizophr Res. 2008;98:105–110. doi: 10.1016/j.schres.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot K, Louneva N, Cohen JW, Kazi H, Blake DJ, Arnold SE. Synaptic dysbindin-1 reductions in schizophrenia occur in an isoform-specific manner indicating their subsynaptic location. PLoS One. 2011;6:e16886. doi: 10.1371/journal.pone.0016886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagnon YC, Roy MA, Bureau A, Merette C, Maziade M. Differential RNA expression between schizophrenic patients and controls of the dystrobrevin binding protein 1 and neuregulin 1 genes in immortalized lymphocytes. Schizophr Res. 2008;100:281–290. doi: 10.1016/j.schres.2007.12.471. [DOI] [PubMed] [Google Scholar]
- Watanabe A, Toyota T, Owada Y, Hayashi T, Iwayama Y, Matsumata M, et al. Fabp7 maps to a quantitative trait locus for a schizophrenia endophenotype. PLoS Biol. 2007;5:e297. doi: 10.1371/journal.pbio.0050297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergen SE, Fanous AH, Walsh D, O′Neill FA, Kendler KS. Polymorphisms in SLC6A4, PAH, GABRB3, and MAOB and modification of psychotic disorder features. Schizophr Res. 2009;109:94–97. doi: 10.1016/j.schres.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock WM, Cardon K, Bustillo J, Roberts RC, Perrone-Bizzozero NI. Altered expression of genes involved in GABAergic transmission and neuromodulation of granule cell activity in the cerebellum of schizophrenia patients. Am J Psychiatry. 2008;165:1594–1603. doi: 10.1176/appi.ajp.2008.07121845. [DOI] [PubMed] [Google Scholar]
- Hashimoto T, Arion D, Unger T, Maldonado-Aviles JG, Morris HM, Volk DW, et al. Alterations in GABA-related transcriptome in the dorsolateral prefrontal cortex of subjects with schizophrenia. Mol Psychiatry. 2008;13:147–161. doi: 10.1038/sj.mp.4002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver H, Susser E, Danovich L, Bilker W, Youdim M, Goldin V, et al. SSRI augmentation of antipsychotic alters expression of GABA(A) receptor and related genes in PMC of schizophrenia patients. Int J Neuropsychopharmacol. 2011;14:573–584. doi: 10.1017/S1461145710001471. [DOI] [PubMed] [Google Scholar]
- Straub RE, Lipska BK, Egan MF, Goldberg TE, Callicott JH, Mayhew MB, et al. Allelic variation in GAD1 (GAD67) is associated with schizophrenia and influences cortical function and gene expression. Mol Psychiatry. 2007;12:854–869. doi: 10.1038/sj.mp.4001988. [DOI] [PubMed] [Google Scholar]
- Hashimoto T, Bazmi HH, Mirnics K, Wu Q, Sampson AR, Lewis DA. Conserved regional patterns of GABA-related transcript expression in the neocortex of subjects with schizophrenia. Am J Psychiatry. 2008;165:479–489. doi: 10.1176/appi.ajp.2007.07081223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benes FM, Lim B, Matzilevich D, Subburaju S, Walsh JP. Circuitry-based gene expression profiles in GABA cells of the trisynaptic pathway in schizophrenics versus bipolars. Proc Natl Acad Sci USA. 2008;105:20935–20940. doi: 10.1073/pnas.0810153105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T, Volk DW, Eggan SM, Mirnics K, Pierri JN, Sun Z, et al. Gene expression deficits in a subclass of GABA neurons in the prefrontal cortex of subjects with schizophrenia. J Neurosci. 2003;23:6315–6326. doi: 10.1523/JNEUROSCI.23-15-06315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirnics K, Middleton FA, Marquez A, Lewis DA, Levitt P. Molecular characterization of schizophrenia viewed by microarray analysis of gene expression in prefrontal cortex. Neuron. 2000;28:53–67. doi: 10.1016/s0896-6273(00)00085-4. [DOI] [PubMed] [Google Scholar]
- Akbarian S, Huang HS. Molecular and cellular mechanisms of altered GAD1/GAD67 expression in schizophrenia and related disorders. Brain Res Rev. 2006;52:293–304. doi: 10.1016/j.brainresrev.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Heckers S, Stone D, Walsh J, Shick J, Koul P, Benes FM. Differential hippocampal expression of glutamic acid decarboxylase 65 and 67 messenger RNA in bipolar disorder and schizophrenia. Arch Gen Psychiatry. 2002;59:521–529. doi: 10.1001/archpsyc.59.6.521. [DOI] [PubMed] [Google Scholar]
- Guidotti A, Auta J, Davis JM, Di-Giorgi-Gerevini V, Dwivedi Y, Grayson DR, et al. Decrease in reelin and glutamic acid decarboxylase67 (GAD67) expression in schizophrenia and bipolar disorder: a postmortem brain study. Arch Gen Psychiatry. 2000;57:1061–1069. doi: 10.1001/archpsyc.57.11.1061. [DOI] [PubMed] [Google Scholar]
- Impagnatiello F, Guidotti AR, Pesold C, Dwivedi Y, Caruncho H, Pisu MG, et al. A decrease of reelin expression as a putative vulnerability factor in schizophrenia. Proc Natl Acad Sci USA. 1998;95:15718–15723. doi: 10.1073/pnas.95.26.15718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa E, Davis J, Grayson DR, Guidotti A, Pappas GD, Pesold C. Dendritic spine hypoplasticity and downregulation of reelin and GABAergic tone in schizophrenia vulnerability. Neurobiol Dis. 2001;8:723–742. doi: 10.1006/nbdi.2001.0436. [DOI] [PubMed] [Google Scholar]
- Woo TU, Walsh JP, Benes FM. Density of glutamic acid decarboxylase 67 messenger RNA-containing neurons that express the N-methyl-D-aspartate receptor subunit NR2A in the anterior cingulate cortex in schizophrenia and bipolar disorder. Arch Gen Psychiatry. 2004;61:649–657. doi: 10.1001/archpsyc.61.7.649. [DOI] [PubMed] [Google Scholar]
- Veldic M, Caruncho HJ, Liu WS, Davis J, Satta R, Grayson DR, et al. DNA-methyltransferase 1 mRNA is selectively overexpressed in telencephalic GABAergic interneurons of schizophrenia brains. Proc Natl Acad Sci USA. 2004;101:348–353. doi: 10.1073/pnas.2637013100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldic M, Guidotti A, Maloku E, Davis JM, Costa E. In psychosis, cortical interneurons overexpress DNA-methyltransferase 1. Proc Natl Acad Sci USA. 2005;102:2152–2157. doi: 10.1073/pnas.0409665102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakak Y, Walker JR, Li C, Wong WH, Davis KL, Buxbaum JD, et al. Genome-wide expression analysis reveals dysregulation of myelination-related genes in chronic schizophrenia. Proc Natl Acad Sci USA. 2001;98:4746–4751. doi: 10.1073/pnas.081071198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paylor R, Glaser B, Mupo A, Ataliotis P, Spencer C, Sobotka A, et al. Tbx1 haploinsufficiency is linked to behavioral disorders in mice and humans: implications for 22q11 deletion syndrome. Proc Natl Acad Sci USA. 2006;103:7729–7734. doi: 10.1073/pnas.0600206103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro H, Koga M, Horiuchi Y, Noguchi E, Morikawa M, Suzuki Y, et al. Supportive evidence for reduced expression of GNB1L in schizophrenia. Schizophr Bull. 2010;36:756–765. doi: 10.1093/schbul/sbn160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams NM, Glaser B, Norton N, Williams H, Pierce T, Moskvina V, et al. Strong evidence that GNB1L is associated with schizophrenia. Hum Mol Genet. 2008;17:555–566. doi: 10.1093/hmg/ddm330. [DOI] [PubMed] [Google Scholar]
- Wiedholz LM, Owens WA, Horton RE, Feyder M, Karlsson RM, Hefner K, et al. Mice lacking the AMPA GluR1 receptor exhibit striatal hyperdopaminergia and ‘schizophrenia-related' behaviors. Mol Psychiatry. 2008;13:631–640. doi: 10.1038/sj.mp.4002056. [DOI] [PubMed] [Google Scholar]
- Toyooka K, Usui M, Washiyama K, Kumanishi T, Takahashi Y. Gene expression profiles in the brain from phencyclidine-treated mouse by using DNA microarray. Ann N Y Acad Sci. 2002;965:10–20. doi: 10.1111/j.1749-6632.2002.tb04147.x. [DOI] [PubMed] [Google Scholar]
- Magri C, Gardella R, Barlati SD, Podavini D, Iatropoulos P, Bonomi S, et al. Glutamate AMPA receptor subunit 1 gene (GRIA1) and DSM-IV-TR schizophrenia: a pilot case-control association study in an Italian sample. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:287–293. doi: 10.1002/ajmg.b.30294. [DOI] [PubMed] [Google Scholar]
- Leon CA, Schumacher J, Kluck N, Herold C, Schulze TG, Propping P, et al. Association study of the GRIA1 and CLINT1 (Epsin 4) genes in a German schizophrenia sample. Psychiatr Genet. 2011;21:114. doi: 10.1097/YPG.0b013e328341a334. [DOI] [PubMed] [Google Scholar]
- Sokolov BP. Expression of NMDAR1, GluR1, GluR7, and KA1 glutamate receptor mRNAs is decreased in frontal cortex of ‘‘neuroleptic-free'' schizophrenics: evidence on reversible up-regulation by typical neuroleptics. J Neurochem. 1998;71:2454–2464. doi: 10.1046/j.1471-4159.1998.71062454.x. [DOI] [PubMed] [Google Scholar]
- Ibrahim HM, Hogg AJ, Jr, Healy DJ, Haroutunian V, Davis KL, Meador-Woodruff JH. Ionotropic glutamate receptor binding and subunit mRNA expression in thalamic nuclei in schizophrenia. Am J Psychiatry. 2000;157:1811–1823. doi: 10.1176/appi.ajp.157.11.1811. [DOI] [PubMed] [Google Scholar]
- Dracheva S, McGurk SR, Haroutunian V. mRNA expression of AMPA receptors and AMPA receptor binding proteins in the cerebral cortex of elderly schizophrenics. J Neurosci Res. 2005;79:868–878. doi: 10.1002/jnr.20423. [DOI] [PubMed] [Google Scholar]
- Stadler F, Kolb G, Rubusch L, Baker SP, Jones EG, Akbarian S. Histone methylation at gene promoters is associated with developmental regulation and region-specific expression of ionotropic and metabotropic glutamate receptors in human brain. J Neurochem. 2005;94:324–336. doi: 10.1111/j.1471-4159.2005.03190.x. [DOI] [PubMed] [Google Scholar]
- Sagata N, Iwaki A, Aramaki T, Takao K, Kura S, Tsuzuki T, et al. Comprehensive behavioural study of GluR4 knockout mice: implication in cognitive function. Genes Brain Behav. 2010;9:899–909. doi: 10.1111/j.1601-183X.2010.00629.x. [DOI] [PubMed] [Google Scholar]
- Chong VZ, Young LT, Mishra RK. cDNA array reveals differential gene expression following chronic neuroleptic administration: implications of synapsin II in haloperidol treatment. J Neurochem. 2002;82:1533–1539. doi: 10.1046/j.1471-4159.2002.01104.x. [DOI] [PubMed] [Google Scholar]
- Makino C, Fujii Y, Kikuta R, Hirata N, Tani A, Shibata A, et al. Positive association of the AMPA receptor subunit GluR4 gene (GRIA4) haplotype with schizophrenia: linkage disequilibrium mapping using SNPs evenly distributed across the gene region. Am J Med Genet B Neuropsychiatr Genet. 2003;116B:17–22. doi: 10.1002/ajmg.b.10041. [DOI] [PubMed] [Google Scholar]
- Beneyto M, Meador-Woodruff JH. Lamina-specific abnormalities of NMDA receptor-associated postsynaptic protein transcripts in the prefrontal cortex in schizophrenia and bipolar disorder. Neuropsychopharmacology. 2008;33:2175–2186. doi: 10.1038/sj.npp.1301604. [DOI] [PubMed] [Google Scholar]
- Kochlamazashvili G, Senkov O, Grebenyuk S, Robinson C, Xiao MF, Stummeyer K, et al. Neural cell adhesion molecule-associated polysialic acid regulates synaptic plasticity and learning by restraining the signaling through GluN2B-containing NMDA receptors. J Neurosci. 2010;30:4171–4183. doi: 10.1523/JNEUROSCI.5806-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvanova M, Lakso M, Wong G. Identification of genes regulated by memantine and MK-801 in adult rat brain by cDNA microarray analysis. Neuropsychopharmacology. 2004;29:1070–1079. doi: 10.1038/sj.npp.1300398. [DOI] [PubMed] [Google Scholar]
- Bowden NA, Scott RJ, Tooney PA. Altered gene expression in the superior temporal gyrus in schizophrenia. BMC Genomics. 2008;9:199. doi: 10.1186/1471-2164-9-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney GG, Burno M, Campbell UC, Hernandez LM, Rodriguez D, Bristow LJ, et al. Metabotropic glutamate subtype 5 receptors modulate locomotor activity and sensorimotor gating in rodents. J Pharmacol Exp Ther. 2003;306:116–123. doi: 10.1124/jpet.103.048702. [DOI] [PubMed] [Google Scholar]
- Devon RS, Anderson S, Teague PW, Muir WJ, Murray V, Pelosi AJ, et al. The genomic organisation of the metabotropic glutamate receptor subtype 5 gene, and its association with schizophrenia. Mol Psychiatry. 2001;6:311–314. doi: 10.1038/sj.mp.4000848. [DOI] [PubMed] [Google Scholar]
- Xi ZR, Qin W, Yang YF, He G, Gao SH, Ren MS, et al. Transmission disequilibrium analysis of the GSN gene in a cohort of family trios with schizophrenia. Neurosci Lett. 2004;372:200–203. doi: 10.1016/j.neulet.2004.09.041. [DOI] [PubMed] [Google Scholar]
- Davis KL, Haroutunian V. Global expression-profiling studies and oligodendrocyte dysfunction in schizophrenia and bipolar disorder. Lancet. 2003;362:758. doi: 10.1016/S0140-6736(03)14297-3. [DOI] [PubMed] [Google Scholar]
- Barbier E, Zapata A, Oh E, Liu Q, Zhu F, Undie A, et al. Supersensitivity to amphetamine in protein kinase-C interacting protein/HINT1 knockout mice. Neuropsychopharmacology. 2007;32:1774–1782. doi: 10.1038/sj.npp.1301301. [DOI] [PubMed] [Google Scholar]
- Chen Q, Wang X, O′Neill FA, Walsh D, Kendler KS, Chen X. Is the histidine triad nucleotide-binding protein 1 (HINT1) gene a candidate for schizophrenia. Schizophr Res. 2008;106:200–207. doi: 10.1016/j.schres.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurotaki N, Tasaki S, Mishima H, Ono S, Imamura A, Kikuchi T, et al. Identification of novel schizophrenia loci by homozygosity mapping using DNA microarray analysis. PLoS One. 2011;6:e20589. doi: 10.1371/journal.pone.0020589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vawter MP, Crook JM, Hyde TM, Kleinman JE, Weinberger DR, Becker KG, et al. Microarray analysis of gene expression in the prefrontal cortex in schizophrenia: a preliminary study. Schizophr Res. 2002;58:11–20. doi: 10.1016/s0920-9964(01)00377-2. [DOI] [PubMed] [Google Scholar]
- Steward LJ, Kennedy MD, Morris BJ, Pratt JA. The atypical antipsychotic drug clozapine enhances chronic PCP-induced regulation of prefrontal cortex 5-HT2A receptors. Neuropharmacology. 2004;47:527–537. doi: 10.1016/j.neuropharm.2004.04.020. [DOI] [PubMed] [Google Scholar]
- McClay JL, Adkins DE, Aberg K, Stroup S, Perkins DO, Vladimirov VI, et al. Genome-wide pharmacogenomic analysis of response to treatment with antipsychotics. Mol Psychiatry. 2011;16:76–85. doi: 10.1038/mp.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polesskaya OO, Sokolov BP. Differential expression of the ‘‘C'' and ‘‘T'' alleles of the 5-HT2A receptor gene in the temporal cortex of normal individuals and schizophrenics. J Neurosci Res. 2002;67:812–822. doi: 10.1002/jnr.10173. [DOI] [PubMed] [Google Scholar]
- Garbett K, Gal-Chis R, Gaszner G, Lewis DA, Mirnics K. Transcriptome alterations in the prefrontal cortex of subjects with schizophrenia who committed suicide. Neuropsychopharmacol Hung. 2008;10:9–14. [PubMed] [Google Scholar]
- Fukuda Y, Koga M, Arai M, Noguchi E, Ohtsuki T, Horiuchi Y, et al. Monoallelic and unequal allelic expression of the HTR2A gene in human brain and peripheral lymphocytes. Biol Psychiatry. 2006;60:1331–1335. doi: 10.1016/j.biopsych.2006.06.024. [DOI] [PubMed] [Google Scholar]
- Kushima I, Nakamura Y, Aleksic B, Ikeda M, Ito Y, Shiino T, et al. Resequencing and association analysis of the KALRN and EPHB1 genes and their contribution to schizophrenia susceptibility Schizophr Bulladvance online publication, 1 November 2010 (e-pub ahead of print). [DOI] [PMC free article] [PubMed]
- Hill JJ, Hashimoto T, Lewis DA. Molecular mechanisms contributing to dendritic spine alterations in the prefrontal cortex of subjects with schizophrenia. Mol Psychiatry. 2006;11:557–566. doi: 10.1038/sj.mp.4001792. [DOI] [PubMed] [Google Scholar]
- Li C, Zheng Y, Qin W, Tao R, Pan Y, Xu Y, et al. A family-based association study of kinesin heavy chain member 2 gene (KIF2) and schizophrenia. Neurosci Lett. 2006;407:151–155. doi: 10.1016/j.neulet.2006.08.025. [DOI] [PubMed] [Google Scholar]
- Rojas P, Joodmardi E, Hong Y, Perlmann T, Ogren SO. Adult mice with reduced Nurr1 expression: an animal model for schizophrenia. Mol Psychiatry. 2007;12:756–766. doi: 10.1038/sj.mp.4001993. [DOI] [PubMed] [Google Scholar]
- Huang J, Perlis RH, Lee PH, Rush AJ, Fava M, Sachs GS, et al. Cross-disorder genomewide analysis of schizophrenia, bipolar disorder, and depression. Am J Psychiatry. 2010;167:1254–1263. doi: 10.1176/appi.ajp.2010.09091335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O′Tuathaigh CM, Babovic D, O′Sullivan GJ, Clifford JJ, Tighe O, Croke DT, et al. Phenotypic characterization of spatial cognition and social behavior in mice with ‘knockout' of the schizophrenia risk gene neuregulin 1. Neuroscience. 2007;147:18–27. doi: 10.1016/j.neuroscience.2007.03.051. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Sigurdsson E, Steinthorsdottir V, Bjornsdottir S, Sigmundsson T, Ghosh S, et al. Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet. 2002;71:877–892. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YJ, Johnson MA, Lieberman MD, Goodchild RE, Schobel S, Lewandowski N, et al. Type III neuregulin-1 is required for normal sensorimotor gating, memory-related behaviors, and corticostriatal circuit components. J Neurosci. 2008;28:6872–6883. doi: 10.1523/JNEUROSCI.1815-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker RM, Christoforou A, Thomson PA, McGhee KA, Maclean A, Muhleisen TW, et al. Association analysis of Neuregulin 1 candidate regions in schizophrenia and bipolar disorder. Neurosci Lett. 2010;478:9–13. doi: 10.1016/j.neulet.2010.04.056. [DOI] [PubMed] [Google Scholar]
- van Schijndel JE, van Loo KM, van Zweeden M, Djurovic S, Andreassen OA, Hansen T, et al. Three-cohort targeted gene screening reveals a non-synonymous TRKA polymorphism associated with schizophrenia. J Psychiatr Res. 2009;43:1195–1199. doi: 10.1016/j.jpsychires.2009.04.006. [DOI] [PubMed] [Google Scholar]
- Mozhui XW, Chen J, Mulligan MK, Li Z, Ingles J, Chen X, et al. Genetic regulation of Nrxn1 expression: an integrative cross-species analysis of schizophrenia candidate genes. Translational Psychiatry. 2011;1:e38. doi: 10.1038/tp.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Sarginson J, Kong A, Yates P, Steinthorsdottir V, Gudfinnsson E, et al. Association of neuregulin 1 with schizophrenia confirmed in a Scottish population. Am J Hum Genet. 2003;72:83–87. doi: 10.1086/345442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris LW, Lockstone HE, Khaitovich P, Weickert CS, Webster MJ, Bahn S. Gene expression in the prefrontal cortex during adolescence: implications for the onset of schizophrenia. BMC Med Genomics. 2009;2:28. doi: 10.1186/1755-8794-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colantuoni C, Hyde TM, Mitkus S, Joseph A, Sartorius L, Aguirre C, et al. Age-related changes in the expression of schizophrenia susceptibility genes in the human prefrontal cortex. Brain Struct Funct. 2008;213:255–271. doi: 10.1007/s00429-008-0181-5. [DOI] [PubMed] [Google Scholar]
- Law AJ, Shannon Weickert C, Hyde TM, Kleinman JE, Harrison PJ. Neuregulin-1 (NRG-1) mRNA and protein in the adult human brain. Neuroscience. 2004;127:125–136. doi: 10.1016/j.neuroscience.2004.04.026. [DOI] [PubMed] [Google Scholar]
- Hahn CG, Wang HY, Cho DS, Talbot K, Gur RE, Berrettini WH, et al. Altered neuregulin 1-erbB4 signaling contributes to NMDA receptor hypofunction in schizophrenia. Nat Med. 2006;12:824–828. doi: 10.1038/nm1418. [DOI] [PubMed] [Google Scholar]
- Law AJ, Lipska BK, Weickert CS, Hyde TM, Straub RE, Hashimoto R, et al. Neuregulin 1 transcripts are differentially expressed in schizophrenia and regulated by 5′ SNPs associated with the disease. Proc Natl Acad Sci USA. 2006;103:6747–6752. doi: 10.1073/pnas.0602002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto R, Straub RE, Weickert CS, Hyde TM, Kleinman JE, Weinberger DR. Expression analysis of neuregulin-1 in the dorsolateral prefrontal cortex in schizophrenia. Mol Psychiatry. 2004;9:299–307. doi: 10.1038/sj.mp.4001434. [DOI] [PubMed] [Google Scholar]
- Petryshen TL, Middleton FA, Kirby A, Aldinger KA, Purcell S, Tahl AR, et al. Support for involvement of neuregulin 1 in schizophrenia pathophysiology Mol Psychiatry 200510366–374.28. [DOI] [PubMed] [Google Scholar]
- Middleton FA, Pato CN, Gentile KL, McGann L, Brown AM, Trauzzi M, et al. Gene expression analysis of peripheral blood leukocytes from discordant sib-pairs with schizophrenia and bipolar disorder reveals points of convergence between genetic and functional genomic approaches. Am J Med Genet B Neuropsychiatr Genet. 2005;136:12–25. doi: 10.1002/ajmg.b.30171. [DOI] [PubMed] [Google Scholar]
- Siuciak JA, McCarthy SA, Chapin DS, Martin AN. Behavioral and neurochemical characterization of mice deficient in the phosphodiesterase-4B (PDE4B) enzyme. Psychopharmacology (Berl) 2008;197:115–126. doi: 10.1007/s00213-007-1014-6. [DOI] [PubMed] [Google Scholar]
- Dlaboga D, Hajjhussein H, O′Donnell JM. Chronic haloperidol and clozapine produce different patterns of effects on phosphodiesterase-1B, -4B, and -10A expression in rat striatum. Neuropharmacology. 2008;54:745–754. doi: 10.1016/j.neuropharm.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Fatemi SH, Folsom TD, Reutiman TJ, Braun NN, Lavergne LG. Levels of phosphodiesterase 4A and 4B are altered by chronic treatment with psychotropic medications in rat frontal cortex. Synapse. 2010;64:550–555. doi: 10.1002/syn.20762. [DOI] [PubMed] [Google Scholar]
- Fatemi SH, King DP, Reutiman TJ, Folsom TD, Laurence JA, Lee S, et al. PDE4B polymorphisms and decreased PDE4B expression are associated with schizophrenia. Schizophr Res. 2008;101:36–49. doi: 10.1016/j.schres.2008.01.029. [DOI] [PubMed] [Google Scholar]
- Kahler AK, Otnaess MK, Wirgenes KV, Hansen T, Jonsson EG, Agartz I, et al. Association study of PDE4B gene variants in Scandinavian schizophrenia and bipolar disorder multicenter case-control samples. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:86–96. doi: 10.1002/ajmg.b.30958. [DOI] [PubMed] [Google Scholar]
- Pickard BS, Thomson PA, Christoforou A, Evans KL, Morris SW, Porteous DJ, et al. The PDE4B gene confers sex-specific protection against schizophrenia. Psychiatr Genet. 2007;17:129–133. doi: 10.1097/YPG.0b013e328014492b. [DOI] [PubMed] [Google Scholar]
- Tomppo L, Hennah W, Lahermo P, Loukola A, Tuulio-Henriksson A, Suvisaari J, et al. Association between genes of disrupted in schizophrenia 1 (DISC1) interactors and schizophrenia supports the role of the DISC1 pathway in the etiology of major mental illnesses. Biol Psychiatry. 2009;65:1055–1062. doi: 10.1016/j.biopsych.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ML, Chen CH. Microarray analysis of differentially expressed genes in rat frontal cortex under chronic risperidone treatment. Neuropsychopharmacology. 2005;30:268–277. doi: 10.1038/sj.npp.1300612. [DOI] [PubMed] [Google Scholar]
- Carroll LS, Williams NM, Moskvina V, Russell E, Norton N, Williams HJ, et al. Evidence for rare and common genetic risk variants for schizophrenia at protein kinase C, alpha. Mol Psychiatry. 2010;15:1101–1111. doi: 10.1038/mp.2009.96. [DOI] [PubMed] [Google Scholar]
- Jablensky A, Morar B, Wiltshire S, Carter K, Dragovic M, Badcock JC, et al. Polymorphisms associated with normal memory variation also affect memory impairment in schizophrenia. Genes Brain Behav. 2011;10:410–417. doi: 10.1111/j.1601-183X.2011.00679.x. [DOI] [PubMed] [Google Scholar]
- Ballmaier M, Zoli M, Leo G, Agnati LF, Spano P. Preferential alterations in the mesolimbic dopamine pathway of heterozygous reeler mice: an emerging animal-based model of schizophrenia. Eur J Neurosci. 2002;15:1197–1205. doi: 10.1046/j.1460-9568.2002.01952.x. [DOI] [PubMed] [Google Scholar]
- Fatemi SH, Reutiman TJ, Folsom TD. Chronic psychotropic drug treatment causes differential expression of Reelin signaling system in frontal cortex of rats. Schizophr Res. 2009;111:138–152. doi: 10.1016/j.schres.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Alkelai A, Lupoli S, Greenbaum L, Kohn Y, Kanyas-Sarner K, Ben-Asher E, et al. DOCK4 and CEACAM21 as novel schizophrenia candidate genes in the Jewish population. Int J Neuropsychopharmacol. 2011. pp. 20:1–2011. [DOI] [PubMed]
- Kahler AK, Djurovic S, Kulle B, Jonsson EG, Agartz I, Hall H, et al. Association analysis of schizophrenia on 18 genes involved in neuronal migration: MDGA1 as a new susceptibility gene. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1089–1100. doi: 10.1002/ajmg.b.30726. [DOI] [PubMed] [Google Scholar]
- Ovadia G, Shifman S. The genetic variation of RELN expression in schizophrenia and bipolar disorder. PLoS One. 2011;6:e19955. doi: 10.1371/journal.pone.0019955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shifman S, Johannesson M, Bronstein M, Chen SX, Collier DA, Craddock NJ, et al. Genome-wide association identifies a common variant in the reelin gene that increases the risk of schizophrenia only in women. PLoS Genet. 2008;4:e28. doi: 10.1371/journal.pgen.0040028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knable MB, Barci BM, Webster MJ, Meador-Woodruff J, Torrey EF.Molecular abnormalities of the hippocampus in severe psychiatric illness: postmortem findings from the Stanley Neuropathology Consortium Mol Psychiatry 20049609–620.544. [DOI] [PubMed] [Google Scholar]
- Grayson DR, Jia X, Chen Y, Sharma RP, Mitchell CP, Guidotti A, et al. Reelin promoter hypermethylation in schizophrenia. Proc Natl Acad Sci USA. 2005;102:9341–9346. doi: 10.1073/pnas.0503736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillet N, Pattyn A, Contet C, Kieffer BL, Goridis C, Brunet JF. Generation and characterization of Rgs4 mutant mice. Mol Cell Biol. 2005;25:4221–4228. doi: 10.1128/MCB.25.10.4221-4228.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald ML, Eaton ME, Dudman JT, Konradi C. Antipsychotic drugs elevate mRNA levels of presynaptic proteins in the frontal cortex of the rat. Biol Psychiatry. 2005;57:1041–1051. doi: 10.1016/j.biopsych.2005.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McOmish CE, Burrows EL, Howard M, Hannan AJ. PLC-beta1 knockout mice as a model of disrupted cortical development and plasticity: behavioral endophenotypes and dysregulation of RGS4 gene expression. Hippocampus. 2008;18:824–834. doi: 10.1002/hipo.20443. [DOI] [PubMed] [Google Scholar]
- So HC, Chen RY, Chen EY, Cheung EF, Li T, Sham PC. An association study of RGS4 polymorphisms with clinical phenotypes of schizophrenia in a Chinese population. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:77–85. doi: 10.1002/ajmg.b.30577. [DOI] [PubMed] [Google Scholar]
- Prasad KM, Almasy L, Gur RC, Gur RE, Pogue-Geile M, Chowdari KV, et al. RGS4 polymorphisms associated with variability of cognitive performance in a family-based schizophrenia sample. Schizophr Bull. 2010;36:983–990. doi: 10.1093/schbul/sbp002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden NA, Scott RJ, Tooney PA. Altered expression of regulator of G-protein signalling 4 (RGS4) mRNA in the superior temporal gyrus in schizophrenia. Schizophr Res. 2007;89:165–168. doi: 10.1016/j.schres.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Lipska BK, Mitkus S, Caruso M, Hyde TM, Chen J, Vakkalanka R, et al. RGS4 mRNA expression in postmortem human cortex is associated with COMT Val158Met genotype and COMT enzyme activity. Hum Mol Genet. 2006;15:2804–2812. doi: 10.1093/hmg/ddl222. [DOI] [PubMed] [Google Scholar]
- Mirnics K, Middleton FA, Stanwood GD, Lewis DA, Levitt P. Disease-specific changes in regulator of G-protein signaling 4 (RGS4) expression in schizophrenia. Mol Psychiatry. 2001;6:293–301. doi: 10.1038/sj.mp.4000866. [DOI] [PubMed] [Google Scholar]
- Prasad KM, Chowdari KV, Nimgaonkar VL, Talkowski ME, Lewis DA, Keshavan MS. Genetic polymorphisms of the RGS4 and dorsolateral prefrontal cortex morphometry among first episode schizophrenia patients. Mol Psychiatry. 2005;10:213–219. doi: 10.1038/sj.mp.4001562. [DOI] [PubMed] [Google Scholar]
- Erdely HA, Tamminga CA, Roberts RC, Vogel MW. Regional alterations in RGS4 protein in schizophrenia. Synapse. 2006;59:472–479. doi: 10.1002/syn.20265. [DOI] [PubMed] [Google Scholar]
- Volk DW, Eggan SM, Lewis DA. Alterations in metabotropic glutamate receptor 1alpha and regulator of G protein signaling 4 in the prefrontal cortex in schizophrenia. Am J Psychiatry. 2010;167:1489–1498. doi: 10.1176/appi.ajp.2010.10030318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Shibata H, Ninomiya H, Tashiro N, Iwata N, Ozaki N, et al. Association study of polymorphisms in the excitatory amino acid transporter 2 gene (SLC1A2) with schizophrenia. BMC Psychiatry. 2004;4:21. doi: 10.1186/1471-244X-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L, Vawter MP. Shared gene expression alterations in schizophrenia and bipolar disorder. Biol Psychiatry. 2008;64:89–97. doi: 10.1016/j.biopsych.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matute C, Melone M, Vallejo-Illarramendi A, Conti F. Increased expression of the astrocytic glutamate transporter GLT-1 in the prefrontal cortex of schizophrenics. Glia. 2005;49:451–455. doi: 10.1002/glia.20119. [DOI] [PubMed] [Google Scholar]
- Jeans AF, Oliver PL, Johnson R, Capogna M, Vikman J, Molnar Z, et al. A dominant mutation in Snap25 causes impaired vesicle trafficking, sensorimotor gating, and ataxia in the blind-drunk mouse. Proc Natl Acad Sci USA. 2007;104:2431–2436. doi: 10.1073/pnas.0610222104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanous AH, Zhao Z, van den Oord EJ, Maher BS, Thiselton DL, Bergen SE, et al. Association study of SNAP25 and schizophrenia in Irish family and case-control samples. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:663–674. doi: 10.1002/ajmg.b.31037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatt SJ, Everall IP, Kremen WS, Corbeil J, Sasik R, Khanlou N, et al. Comparative gene expression analysis of blood and brain provides concurrent validation of SELENBP1 up-regulation in schizophrenia. Proc Natl Acad Sci USA. 2005;102:15533–15538. doi: 10.1073/pnas.0507666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Egbufoama S, Vawter MP. SNAP-25 reduction in the hippocampus of patients with schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry. 2003;27:411–417. doi: 10.1016/S0278-5846(03)00027-7. [DOI] [PubMed] [Google Scholar]
- Karson CN, Mrak RE, Schluterman KO, Sturner WQ, Sheng JG, Griffin WS. Alterations in synaptic proteins and their encoding mRNAs in prefrontal cortex in schizophrenia: a possible neurochemical basis for ′hypofrontality′. Mol Psychiatry. 1999;4:39–45. doi: 10.1038/sj.mp.4000459. [DOI] [PubMed] [Google Scholar]
- Hemby SE, Ginsberg SD, Brunk B, Arnold SE, Trojanowski JQ, Eberwine JH. Gene expression profile for schizophrenia: discrete neuron transcription patterns in the entorhinal cortex. Arch Gen Psychiatry. 2002;59:631–640. doi: 10.1001/archpsyc.59.7.631. [DOI] [PubMed] [Google Scholar]
- Gabriel SM, Haroutunian V, Powchik P, Honer WG, Davidson M, Davies P, et al. Increased concentrations of presynaptic proteins in the cingulate cortex of subjects with schizophrenia. Arch Gen Psychiatry. 1997;54:559–566. doi: 10.1001/archpsyc.1997.01830180077010. [DOI] [PubMed] [Google Scholar]
- Clark D, Dedova I, Cordwell S, Matsumoto I.A proteome analysis of the anterior cingulate cortex gray matter in schizophrenia Mol Psychiatry 200611459–470.23. [DOI] [PubMed] [Google Scholar]
- Dyck BA, Skoblenick KJ, Castellano JM, Ki K, Thomas N, Mishra RK. Synapsin II knockout mice show sensorimotor gating and behavioural abnormalities similar to those in the phencyclidine-induced preclinical animal model of schizophrenia. Schizophr Res. 2007;97:292–293. doi: 10.1016/j.schres.2007.08.026. [DOI] [PubMed] [Google Scholar]
- Dyck BA, Skoblenick KJ, Castellano JM, Ki K, Thomas N, Mishra RK. Behavioral abnormalities in synapsin II knockout mice implicate a causal factor in schizophrenia. Synapse. 2009;63:662–672. doi: 10.1002/syn.20643. [DOI] [PubMed] [Google Scholar]
- Chen Q, He G, Qin W, Chen QY, Zhao XZ, Duan SW, et al. Family-based association study of synapsin II and schizophrenia. Am J Hum Genet. 2004;75:873–877. doi: 10.1086/425588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saviouk V, Moreau MP, Tereshchenko IV, Brzustowicz LM. Association of synapsin 2 with schizophrenia in families of Northern European ancestry. Schizophr Res. 2007;96:100–111. doi: 10.1016/j.schres.2007.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vawter MP, Thatcher L, Usen N, Hyde TM, Kleinman JE, Freed WJ. Reduction of synapsin in the hippocampus of patients with bipolar disorder and schizophrenia. Mol Psychiatry. 2002;7:571–578. doi: 10.1038/sj.mp.4001158. [DOI] [PubMed] [Google Scholar]
- Potkin SG, Turner JA, Guffanti G, Lakatos A, Fallon JH, Nguyen DD, et al. A genome-wide association study of schizophrenia using brain activation as a quantitative phenotype. Schizophr Bull. 2009;35:96–108. doi: 10.1093/schbul/sbn155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MV, Rollins B, Sequeira PA, Mesen A, Byerley W, Stein R, et al. Exon expression in lymphoblastoid cell lines from subjects with schizophrenia before and after glucose deprivation. BMC Med Genomics. 2009;2:62. doi: 10.1186/1755-8794-2-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.