Abstract
Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (MS) offers the possibility of accurate, rapid, inexpensive identification of bacteria, fungi, and mycobacteria isolated in clinical microbiology laboratories. The procedures for preanalytic processing of organisms and analysis by MALDI-TOF MS are technically simple and reproducible, and commercial databases and interpretive algorithms are available for the identification of a wide spectrum of clinically significant organisms. Although only limited work has been reported on the use of this technique to identify molds, perform strain typing, or determine antibiotic susceptibility results, these are fruitful areas of promising research. As experience is gained with MALDI-TOF MS, it is expected that the databases will be expanded to resolve many of the current inadequate identifications (eg, no identification, genus-level identification) and algorithms for potential misidentification will be developed. The current lack of Food and Drug Administration approval of any MALDI-TOF MS system for organism identification limits widespread use in the United States.
It is rare that a technology can fundamentally alter well-established diagnostic testing methods, but that is precisely what the use of matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (MS) has done in Europe, where it is widely used for identification of bacteria, mycobacteria, and fungi. For >100 years, microbes were characterized by their biochemical properties, that is, the ability of organisms to use various substrates for growth and metabolic activity. As discovery of new organisms rapidly expanded the need for more complex identification algorithms, a proliferation of reference books1–3 documenting detailed identification tables for each family of organisms and the use of sophisticated commercial identification systems proliferated. In the last 20 years the use of gene sequencing techniques that targeted ribosomal RNA genes and various housekeeping genes permitted more precise identification of organisms but at significant technical and financial costs. These techniques are not practical for routine identification of organisms; however, they are useful for identification of uncommonly isolated bacteria and fungi. Indeed, the combination of gene sequencing and biochemical tests has proven valuable for the precise identification of organisms used for constructing databases for MALDI-TOF MS.
Mass Spectrometry in Biomarker Detection
In 1975, Anhalt and Fenselau4 described the use of biomarkers detected by mass spectrometry for the identification of bacteria. Despite the promise of this work and the development of significant advances in specimen preparation, mass spectrometry techniques, and bioinformatics during the next 25 years, adoption of this technology by clinical laboratories occurred slowly. Early studies targeted the analysis of polar fatty acids that comprised 5% to 8% of the dry cell weight of bacteria, whereas more recent studies focused on analysis of basic proteins, primarily in the range of 2000 to 20,000 Da (60% to 70% of the dry cell weight of bacteria). In 1994, Cain et al5 reported that MALDI-TOF MS could be used to differentiate selected bacteria by analysis of protein profiles from disrupted cells. Two years later, Claydon et al,6 Holland et al,7 and Krishnamurthy et al8 demonstrated the feasibility of processing intact bacterial cells. This work was extended to eukaryotic cells in 2000 and 2001, when investigators reported that whole fungal cells could also be identified using MALDI-TOF MS.9–11 Subsequent work, primarily from research laboratories, demonstrated that individual species of bacteria and fungi could be accurately identified by MALDI-TOF MS at the species or subspecies level; however, the versatility of the technology to identify the wide spectrum of bacteria, yeasts, molds, and mycobacteria that can be isolated in clinical specimens was only recently demonstrated.12–18
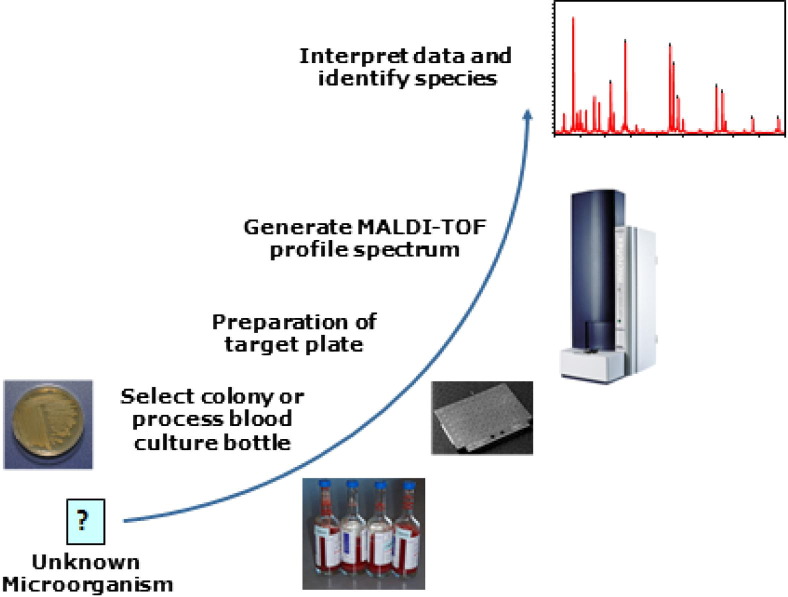
The method as initially introduced is technically simple and rapid. Bacterial colonies are removed from agar culture plates, mixed with an excess of UV-absorbing matrix, and dried on steel target plates. The dried preparations are exposed to laser pulses, resulting in energy transfer from the matrix to the nonvolatile analyte molecules, with desorption (removal) of analyte into the gas phase. The ionized molecules are accelerated by electric potentials through a flight tube to the mass spectrometer, with separation of the biomarkers determined by their mass/charge ratio (m/z; z typically is 1). The profile of biomarkers is then compared with profiles of a collection of well-characterized organisms (Figure 1).
Figure 1.
MALDI-TOF MS for microorganism identification.
An Overview of MALDI-TOF MS
Critical for the successful use of MALDI-TOF MS in clinical laboratories is the demonstration that the method is reproducible and highly discriminatory. The preanalysis preparation of the sample for MALDI-TOF MS is critical for reproducible spectral profiles and test sensitivity.19 Bacteria or yeast can be selected from a culture plate or concentrated from broth cultures and either transferred directly to a target plate or pretreated with ethanol followed by protein extraction with formic acid and acetonitrile. Pretreatment is beneficial because it inactivates the organisms, enhances detection of biomarkers above 15 kDa, and improves sample stability.20 Although pretreatment can be omitted for analysis of most bacteria, it is necessary for the generation of yeast spectral profiles and yields higher spectral scores for all bacteria.18,21 Before analysis by MALDI-TOF MS, optimum sensitivity requires disruption of the cell wall structure by treating the cells with a strong organic acid (eg, formic, trifluoroacetic, and acetic) either before or concurrent with the addition of the matrix solution. Selection of the matrix influences the specific biomarkers that are detected (eg, proteins, phospholipids, and cyclic lipopeptides), with α-cyano-4-hydroxy-cinnamic acid used preferentially for detection of protein biomarkers.7 Thus, although a variety of sample preparation methods have been evaluated, a relatively uniform approach has evolved for the identification of bacteria and yeast from cultured specimens.22
Sample Preparation
Single colonies are selected and suspended in a small volume of 70% ethanol (microbial inactivation), briefly vortexed, and then concentrated by centrifugation. The supernatant is discarded and the cells are resuspended in 50 μL of 70% formic acid (cell wall disruption), an equal volume of acetonitrile is added (protein extraction), the sample is vortexed, and it is again concentrated by centrifugation. One microliter of the supernatant with the extracted proteins is spotted on the target plate, allowed to evaporate to near dryness, and then overlaid with the matrix consisting of a saturated solution of α-cyano-4-hydroxy-cinnamic acid in 50% acetonitrile and 2.5% trifluoroacetic acid. With this protocol, sufficient spectra are obtained for organism identification if the initial solution contains a minimum of 5 to 10 × 106 cells/mL.23,24 Most work in clinical laboratories is performed on isolated colonies grown on nutrient agar plates, so the presence of sufficient numbers of cells in a sample is not a limitation. Until recently, samples were spotted onto stainless steel plates, but Schuerenberg et al25 demonstrated that coating predetermined spots on steel plates (anchors) with a thin layer of polytetrafluoroethylene (Teflon) allowed the sample/matrix to concentrate on the anchor spot during solvent evaporation, resulting in more homogenous crystallization of the matrix and excellent biomarker spectra reproducibility. As a practical matter, using these plates makes sample spotting technically easier to perform manually and reduces the risk of cross-contamination of samples due to spreading of a sample into an adjacent spot. Commercially prepared, disposable sample platforms have also been developed for clinical laboratories, but there is little need for this because the target plates can be rapidly cleaned and reused for an indefinite period.
Biomarker Detection
Most biomarkers detected in MALDI spectra are intracellular proteins primarily in the range of 4000 to 15,000 Da.26 Lysis of organisms with organic solvents in acidic conditions favors extraction of basic cytoplasmic proteins, specifically ribosomal and mitochondrial proteins, cold shock proteins, heat shock proteins, DNA binding proteins, and RNA chaperone proteins.11,27 These are highly conserved housekeeping proteins and serve as ideal biomarkers for characterizing individual species.
Bacteria
A number of published studies have demonstrated the accuracy of MALDI-TOF MS for the identification of a broad spectrum of bacteria,12,14–16,28–33 including gram-positive cocci and rods, fermentative and nonfermentative gram-negative rods, and anaerobes. In general, >90% of all isolates are identified at the species level, 98% are identified at the genus level, and <1% are incorrectly identified. The most common reason an isolate is not identified is because it is not included in the database. For example, Justesen et al30 and Velloo et al33 reported <67% of anaerobes could be identified with the Bruker microflex MS systems (Billerica, MA). In contrast, Fedorko et al32 reported >86% of anaerobes were identified with the Bruker microflex system when an expanded database was used. Misidentifications are most commonly observed with taxonomically related bacteria, such as Shigella with Escherichia and Streptococcus pneumoniae with S. mitis. As experience is gained with MALDI-TOF MS, it is expected that the database will be expanded to resolve many of the current inadequate identifications (eg, no identification, genus-level identification) and algorithms for potential misidentification will be developed (eg, rapid bile solubility test to confirm a S. pneumoniae identification).
Yeast
Identification of yeasts isolated in culture can also be performed accurately,13,18,34,35 including differentiation of closely related species (eg, Candida albicans and C. dubliniensis; C. guilliermondii and C. kefyr; C. metapsilosis, C. orthopsilosis, and C. parapsilosis). More limited studies have been performed with filamentous fungi, primarily because routine identification is based on morphologic features; however, preliminary studies have demonstrated that Aspergillus, Fusarium, and Penicillium can be identified accurately at the species level.36–38
Organisms in Blood Culture Broths
A number of studies have demonstrated the usefulness of MALDI-TOF MS for identification of bacteria and yeasts isolated in blood culture broths.39–45 Processing these specimens is more complex because the nonmicrobial cells, serum proteins, and broth culture nutrients must be removed before the microbial cells are evaluated. However, definitive identification results from positive blood culture broths are generally available in <1 hour. Although, approximately 15% to 20% of the isolates are not initially identified, primarily because insufficient numbers of cells are available, modification of the extraction procedures has improved the test sensitivity.46,47 However, not all blood culture broth formulations produce adequate results, particularly media supplemented with charcoal.48–50
Mycobacteria
Identification of mycobacteria and other acid-fast organisms by MALDI-TOF MS poses a particular challenge because the organisms must be sacrificed before processing for safety reasons and cell lysis using methods developed for other microorganisms is inadequate. Saleeb et al51 resolved these problems by heating the bacterial suspension for 30 minutes at 95°C to sacrifice the mycobacteria, dispersing the bacteria using a micropestle, and lysing the organisms by vortexing the suspension with glass beads in the presence of formic acid and acetonitrile. They demonstrated that virtually all species of mycobacteria could be identified with a 90-minute turnaround time, significantly faster than the days to weeks that were required for identification by gene sequencing or biochemical tests.
Microbial Typing and Antimicrobial Susceptibility Testing
In addition to microbial identification, MALDI-TOF MS has been applied to two other areas: microbial typing and antimicrobial susceptibility testing. Because MALDI-TOF MS detects a large spectrum of proteins, the technique should be able to discriminate between closely related species and to classify organisms at the subspecies level. Work with virtually all genera of bacteria and many fungal isolates has demonstrated MALDI-TOF MS as a feasible approach for subspecies classification.52 However, only limited work has been performed demonstrating that the spectral profiles are reproducible and discriminating for strain typing and that results are comparable to conventional typing methods, such as pulsed-field gel electrophoresis. If future studies prove this is a viable technique, then MALDI-TOF MS could be used to both identify an organism and determine whether it is related to a previous clinical isolate (ie, perform prospective epidemiology).
Although there are relatively few reports of MALDI-TOF MS used for antimicrobial susceptibility testing, this is a research area of exciting possibilities. Edwards-Jones et al53 and Du et al54 reported distinct spectral profiles that could be used to differentiate methicillin-susceptible and methicillin-resistant Staphylococcus aureus. If these results are validated, then expression of methicillin resistance could be confirmed significantly faster than with conventional test methods. Hrabak et al55 used MALDI-TOF MS to assess meropenem degradation by carbapenemases. Although this test method would not replace current methods, it could be used to confirm expression of carbapenemase activity, a procedure that now requires 1 to 2 days of additional testing. Rogers et al56 demonstrated that azole resistance in C. glabrata was associated with differential expression of 25 proteins, and Marinach et al57 compared the proteome of fluconazole-susceptible strains of C. albicans with that of fluconazole-resistant strains and demonstrated discrete profile changes that corresponded to minimum inhibitory concentration values determined by the Clinical Laboratory Standards Institute's microdilution reference method. If these results can be extended to additional clinical isolates and other classes of antifungal drugs, then the feasibility of rapid antifungal susceptibility testing is promising.
Conclusions
MALDI-TOF MS offers the possibility of accurate, rapid, and inexpensive identification of microorganisms. The procedures for preanalysis processing of organisms and analysis by MALDI-TOF MS are technically simple and reproducible, and commercial databases and interpretive algorithms are available for the identification of a wide spectrum of bacteria, yeast, and mycobacteria. Efforts by both users and manufacturers of commercial systems to include less commonly isolated organisms in the identification databases will further expand the utility of MALDI-TOF MS. Although only limited work has been reported on the use of mass spectrometry to identify molds, perform strain typing, or determine antibiotic susceptibility results, these are fruitful areas of promising research.
Footnotes
Supported by an intramural NIH research grant.
Disclosure: P.R.M. is employed by Becton Dickinson, which supported travel costs to the meeting in Troy, MI, but provided no additional support for preparation of the manuscript.
References
- 1.Versalovic J., Carroll K.C., Funke G., Jorgensen J.H., Landry M.L., Warnock D.W. ed 10. ASM Press; Washington, DC: 2011. Manual of Clinical Microbiology. [Google Scholar]
- 2.Borriello S.P., Murray P.R., Funke G. ed 10. Hodder Arnold; London: 2005. Topley and Wilson's Microbiology and Microbial Infections. [Google Scholar]
- 3.Garrity G.M. ed 2. Springer-Verlag; New York: 2001. Bergey's Manual of Systematic Bacteriology. [Google Scholar]
- 4.Anhalt J.P., Fenselau C. Identification of bacteria using mass-spectrometry. Anal Chem. 1975;47:219–225. [Google Scholar]
- 5.Cain T.C., Lubman D.M., Weber W.J. Differentiation of bacteria using protein profiles from matrix-assisted laser/desorption ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 1994;8:1026–1030. [Google Scholar]
- 6.Claydon M.A., Davey S.N., Edwards-Jones V., Gordon D.B. The rapid identification of intact microorganisms using mass spectrometry. Nature Biotechnol. 1996;14:1584–1586. doi: 10.1038/nbt1196-1584. [DOI] [PubMed] [Google Scholar]
- 7.Holland R.D., Wilkes J.G., Rafii F., Sutherland J.B., Persons C.C., Voorhees K.J., Lay J.O., Jr Rapid identification of intact whole bacteria based on spectral patterns using matrix-assisted laser desorption/ionization with time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 1996;14:911–917. doi: 10.1002/(SICI)1097-0231(19960731)10:10<1227::AID-RCM659>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 8.Krishnamurthy T., Ross P.L., Rajamani U. Detection of pathogenic and non-pathogenic bacteria by matrix assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 1996;10:883–888. doi: 10.1002/(SICI)1097-0231(19960610)10:8<883::AID-RCM594>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 9.Li T.Y., Liu B.H., Chen Y.C. Characterization of Aspergillus spores by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2000;14:2393–2400. doi: 10.1002/1097-0231(20001230)14:24<2393::AID-RCM178>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 10.Welham K.J., Domin M.A., Johnson K., Jones L., Ashton D.S. Characterization of fungal spores by laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2000;14:307–310. doi: 10.1002/(SICI)1097-0231(20000315)14:5<307::AID-RCM823>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 11.Amiri-Eliasi B., Fenselau C. Characterization of protein biomarkers desorbed by MALDI from whole fungal cells. Anal Chem. 2001;73:5228–5231. doi: 10.1021/ac010651t. [DOI] [PubMed] [Google Scholar]
- 12.Degand N., Carbonnelle E., Dauphin B., Beretti J.L., Le Bourgeois M., Sermet-Gaudelus I., Segonds C., Berche P., Nassif X., Ferroni A. Matrix-assisted laser desorption ionization–time of flight mass spectrometry for identification of nonfermenting gram-negative bacilli isolated from cystic fibrosis patients. J Clin Microbiol. 2008;46:3361–3367. doi: 10.1128/JCM.00569-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marklein G., Josten M., Klanke U., Muller E., Horre R., Maier T., Wenzel T., Kostrzewa M., Bierbaum G., Hoerauf A., Sahl H.G. Matrix-assisted laser desorption ionization–time of flight mass spectrometry for fast and reliable identification of clinical yeast isolates. J Clin Microbiol. 2009;47:2912–2917. doi: 10.1128/JCM.00389-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mellmann A., Cloud J., Maier T., Keckevoet U., Ramminger I., Iwen P., Dunn J., Hall G., Wilson D., Lasala P., Kostrzewa M., Harmsen D. Evaluation of matrix-assisted laser desorption ionization time-of-flight mass spectrometry in comparison to 16 S rRNA gene sequencing for species identification of nonfermenting bacteria. J Clin Microbiol. 2008;46:1946–1954. doi: 10.1128/JCM.00157-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mellmann A., Bimet F., Bizet C., Borovskaya A.D., Drake R.R., Eigner U., Fahr A.M., He Y., Ilina E.N., Kostrzewa M., Maier T., Mancinelli L., Moussaoui W., Prevost G., Putignani L., Seachord C.L., Tang Y.W., Harmsen D. High interlaboratory reproducibility of matrix-assisted laser desorption ionization time of flight mass spectrometry-based species identification of non-fermenting bacteria. J Clin Microbiol. 2009;47:3732–3734. doi: 10.1128/JCM.00921-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seng P., Drancourt M., Gouriet F., La Scola B., Fournier P.E., Rolain J.M., Raoult D. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009;49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 17.Parisi D., Magliulo M., Nanni P., Casale M., Forina M., Roda A. Analysis and classification of bacteria by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and a chemometric approach. Anal Bioanal Chem. 2008;391:2127–2134. doi: 10.1007/s00216-008-2161-2. [DOI] [PubMed] [Google Scholar]
- 18.van Veen S.Q., Claas E.C.J., Kuijper E.J. High throughput identification of bacteria and yeast by matrix-assisted laser desorption ionization–time of flight mass spectrometry in conventional medical microbiology laboratories. J Clin Microbiol. 2010;48:900–907. doi: 10.1128/JCM.02071-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Findeisen P., Sismanidis D., Riedl M., Costina V., Neumaier M. Preanalytical impact of sample handling on proteome profiling experiments with matrix- assisted laser desorption/ionization time-of-flight mass spectrometry. Clin Chem. 2005;51:2409–2411. doi: 10.1373/clinchem.2005.054585. [DOI] [PubMed] [Google Scholar]
- 20.Winkler M.A., Uher J., Cepa S. Direct analysis and identification of Helicobacter and Campylobacter species by MALDI-TOF mass spectrometry. Anal Chem. 1999;71:3416–3419. doi: 10.1021/ac990135r. [DOI] [PubMed] [Google Scholar]
- 21.Qian J., Cutler J.E., Cole R.B., Cai Y. MALDI-TOF mass signatures for differentiation of yeast species, strain grouping and monitoring of morphogenesis markers. Anal Bioanal Chem. 2008;392:439–449. doi: 10.1007/s00216-008-2288-1. [DOI] [PubMed] [Google Scholar]
- 22.Freiwald A., Sauer S. Phylogenetic classification and identification of bacteria by mass spectrometry. Nat Protoc. 2009;4:732–742. doi: 10.1038/nprot.2009.37. [DOI] [PubMed] [Google Scholar]
- 23.Evason D.J., Claydon M.A., Gordon D.B. Exploring the limits of bacterial identification by intact cell mass spectrometry. J Am Soc Mass Spectrom. 2001;12:49–54. doi: 10.1016/S1044-0305(00)00192-6. [DOI] [PubMed] [Google Scholar]
- 24.Hathout Y., Demirev P.A., Ho Y.P., bundy J.L., Ryzhov V., Sapp L., Stutler J., Jackman J., Fenselau C. Identification of Bacillus spores by matrix assisted laser desorption ionization mass spectrometry. Appl Environ Microbiol. 1999;65:4313–4319. doi: 10.1128/aem.65.10.4313-4319.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuerenberg M., Luebbert C., Eickhoff H., Kalkum M., Lehrach H., Nordhoff E. Prestructured MALDI-MS sample supports. Anal Chem. 2000;72:3436–3442. doi: 10.1021/ac000092a. [DOI] [PubMed] [Google Scholar]
- 26.Fenselau C., Demirev P.A. Characterization of intact microorganisms by MALDI mass spectrometry. Mass Spectrom Rev. 2001;20:157–171. doi: 10.1002/mas.10004. [DOI] [PubMed] [Google Scholar]
- 27.Ryzhov V., Fenselau C. Characterization of the protein subset desorbed by MALDI from whole bacterial cells. Anal Chem. 2001;73:746–750. doi: 10.1021/ac0008791. [DOI] [PubMed] [Google Scholar]
- 28.Bizzini A., Durussel C., Bille J., Greub G., Prod'hom G. Performance of matrix-assist end laser desorption ionization time of flight mass spectrometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. J Clin Microbiol. 2010;48:1549–1554. doi: 10.1128/JCM.01794-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cherkaoui A., Hibbs J., Emonet S., Tangomo M., Girard M., Francois P., Schrenzel J. Comparison of two matrix assisted laser desorption ionication time of flight mass spectrometry methods with conventional phenotypic identification for routine identification of bacteria to the species level. J Clin Microbiol. 2010;48:1169–1175. doi: 10.1128/JCM.01881-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Justesen U., Holm A., Knudsen E., Andersen L., Jensen T., Kemp M., Skov M., Gahrn-Hansen B., Moller J. Species identification of clinical isolates of anaerobic bacteria: a comparison of two matrix assisted laser desorption ionization time-of-flight mass spectrometry systems. J Clin Microbiol. 2011;49:4314–4318. doi: 10.1128/JCM.05788-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martiny D., Busson L., Wybo I., Ait El Haj R., Dediste A., Vandenbeerg O. Comparison of the Microflex LT and Vitek MS systems for the routine identification of bacteria by matrix-assisted laser desorption-ionization time-of-flight mass spectrometry. J Clin Microbiol. 2012 doi: 10.1128/JCM.05971-11. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fedorko D., Drake S.K., Stock F., Murray P. Identification of clinical isolates of anaerobic bacteria using matrix assisted laser desorption ionization time-of-flight mass spectrometry. Eur J Clin Microbiol Infect Dis. 2012 doi: 10.1007/s10096-012-1563-4. (in press) [DOI] [PubMed] [Google Scholar]
- 33.Veloo A., Knoester M., Degener J., Kuiper E. Comparison of two matrix assisted laser desorption ionization time of flight mass spectrometry methods for the identification of clinically relevant anaerobic bacteria. Clin Microbiol Infect. 2011;17:1501–1506. doi: 10.1111/j.1469-0691.2011.03467.x. [DOI] [PubMed] [Google Scholar]
- 34.Stevenson L.G., Drake S.K., Shea Y.R., Zelazny A.M., Murray P.R. Evalutation of matrix-assisted laser desorption ionization–time of flight mass spectrometry for identification of clinically important yeast species. J Clin Microbiol. 2010;48:3482–3486. doi: 10.1128/JCM.00687-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dhiman N., Hall L., Wohlfiel S.L., Buckwalter S.P., Wengenack N.L. Performance and cost analysis of matrix assisted laser desorption ionization time of flight mass spectrometry for routine identification of yeast. J Clin Microbiol. 2011;49:1614–1616. doi: 10.1128/JCM.02381-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seyfarth F., Ziemer M., Sayer H.G., Burmester A., Erhard M., Welker M., Schliemann S., Straube E., Hipler U. The use of ITS DNA sequence analysis and MALDI-TOF mass spectrometry in diagnosing an infection with Fusarium proliferatum. Exp Dermat. 2008;17:965–971. doi: 10.1111/j.1600-0625.2008.00726.x. [DOI] [PubMed] [Google Scholar]
- 37.Hettick J.M., Green B.J., Buskirk A.D., Kashon M.L., Slaven J.E., Janotka E., Blachere F.M., Schmechel D., Beezhold D.H. Discrimination of Aspergillus isolates at the species and strain level by matrix assisted laser desorption/ionization time of flight mass spectrometry fingerprinting. Anal Biochem. 2008;380:276–281. doi: 10.1016/j.ab.2008.05.051. [DOI] [PubMed] [Google Scholar]
- 38.Hettick J.M., Green B.J., Buskirk A.D., Kashon M.L., Slaven J.E., Janotka E., Blachere F.M., Schmechel D., Beezhold D.H. Discrimination of Penicillium isolates by matrix assisted laser desorption ionization time of flight mass spectrometry fingerprinting. Rapid Commun Mass Spectrom. 2008;22:2555–2560. doi: 10.1002/rcm.3649. [DOI] [PubMed] [Google Scholar]
- 39.La Scola B., Raoult D. Direct identification of bacteria in positive blood culture bottles by matrix assisted laser desorption ionization time of flight mass spectrometry. PLoS One. 2009;4:e8041. doi: 10.1371/journal.pone.0008041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stevenson L.G., Drake S.K., Murray P.R. Rapid identification of bacteria in positive blood culture broths by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol. 2010;48:444–447. doi: 10.1128/JCM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferroni A., Suarez S., Beretti J.L., Dauphin B., Bille E., Meyer J. Real time identification of bacteria and Candida species in positive blood culture broths by matrix assisted laser desorption ionization time of flight mass spectrometry. J Clin Microbiol. 2010;48:1542–1548. doi: 10.1128/JCM.02485-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Christner M., Rohde H., Wolters M., Sobottka I., Wegscheider K., Aepfelbacher M. Rapid identification of bacteria from positive blood culture bottles by use of matrix assisted laser desorption ionization time of flight mass spectrometry fingerprinting. J Clin Microbiol. 2010;48:1584–1591. doi: 10.1128/JCM.01831-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prod'hom G., Bizzinii A., Durussel C., Bille J., Greub G. Matrix assisted laser desorption ionization time of flight mass spectrometry for direct bacterial identification from positive blood culture pellets. J Clin Microbiol. 2010;48:1481–1483. doi: 10.1128/JCM.01780-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ferreira L., Sanchez-Juanes F., Porras-Guerra I., Garcia-Garcia M.I., Garcia-Sanchez J.E., Gonzalez-Buitrago J.M. Microorganism direct identification from blood culture by matrix assisted laser desorption ionization time-of-flight mass spectrometry. Clin Microbiol Infect. 2011;17:546–551. doi: 10.1111/j.1469-0691.2010.03257.x. [DOI] [PubMed] [Google Scholar]
- 45.Kok J., Thomas L.C., Olma T., Chen S.C.A., Iredell J.R. Identification of bacteria in blood culture broths using matrix assisted laser desorption ionization Sepsityper and time of flight mass spectrometry. PLoS One. 2011;6:e23285. doi: 10.1371/journal.pone.0023285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schubert S., Weinert K., Wagner C., Gunzi B., Wieser A., Maier T., Kostrzewa M. Novel, improved sample preparation for rapid, direct identification from positive blood cultures using matrix assisted laser desorption ionization time of flight mass spectrometry. J Mol Diag. 2011;13:701–706. doi: 10.1016/j.jmoldx.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loonen A., Jansz A., Stalpers J., Wolffs P., van den Brule A. An evaluation of three processing methods and the effect of reduced culture times for faster direct identification of pathogens from BacT/Alert blood cultures by MALDI-TOF MS. Eur J Clin Microbiol Infect Dis. 2011 doi: 10.1007/s10096-011-1480-y. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmidt V., Jarosch A., Marz P., Sander C., Vacata V., Kalka-Moll W. Rapid identification of bacteria in positive blood culture by matrix assisted laser desorption ionization time of flight mass spectrometry. Eur J Clin Microbiol Infect Dis. 2011 doi: 10.1007/s10096-011-1312-0. (in press) [DOI] [PubMed] [Google Scholar]
- 49.Szabados F., Michels M., Kaase M., Gatermann S. The sensitivity of direct identification from positive Bac/ALERT (bioMerieux) blood culture bottles by matrix-assisted laser desorption ionization time-of-flight mass spectrometry is low. Clin Microbiol Infect. 2011;17:192–195. doi: 10.1111/j.1469-0691.2010.03229.x. [DOI] [PubMed] [Google Scholar]
- 50.Romero-Gomez M.P., Mingorance J. The effect of blood culture bottle type in the rate of direct identification from positive cultures by matrix assisted laser desorption/ionization time-of-flight mass spectrometry. J Infect. 2011;62:251–253. doi: 10.1016/j.jinf.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 51.Saleeb P.G., Drake S.K., Murray P.R., Zelazny A.M. Identification of mycobacteria in solid culture media by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. J Clin Microbiol. 2011;49:1790–1794. doi: 10.1128/JCM.02135-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murray P.R. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry: usefulness for taxonomy and epidemiology. Clin Microbiol Infect. 2010;16:1626–1630. doi: 10.1111/j.1469-0691.2010.03364.x. [DOI] [PubMed] [Google Scholar]
- 53.Edwards-Jones V., Claydon M., Evason D., Walker J., Fox A., Gordon D. Rapid discrimination between methicillin-sensitive and methicillin-resistant Staphylococcus aureus by intact cell mass spectrometry. J Med Microbiol. 2000;49:295–300. doi: 10.1099/0022-1317-49-3-295. [DOI] [PubMed] [Google Scholar]
- 54.Du Z., Yahng A.R., Guo Z., Song Y., Wang J. Identification of Staphylococcus aureus and determination of its methicillin resistance by matrix assisted laser desorption ionization time-of-flight mass spectrometry. Anal Chem. 2002;74:5487–5491. doi: 10.1021/ac020109k. [DOI] [PubMed] [Google Scholar]
- 55.Hrabak J., Walkova R., Studentova V., Chudackova E., Bergerova T. Carbapenemase activity detection by matrix assisted laser desorption ionization time-of-flight mass spectrometry. J Clin Microbiol. 2011;49:3222–3227. doi: 10.1128/JCM.00984-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rogers P.D., Vermitsky J.P., Edlind T.D., Hilliard G.M. Proteomic analysis of experimentally induced azole resistance in Candida glabrata. J Antimicrob Chemother. 2006;58:434–438. doi: 10.1093/jac/dkl221. [DOI] [PubMed] [Google Scholar]
- 57.Marinach C., Alanio A., Palous M., Kwasek S., Fekkar A., Brossas J.Y., Brun S., Snounou G., Hennequin C., Sanglard D., Datry A., Golmard J.L., Mazier D. MALDI-TOF MS based drug susceptibility testing of pathogens: the example of Candida albicans and fluconzaole. Proteomics. 2009;9:4627–4631. doi: 10.1002/pmic.200900152. [DOI] [PubMed] [Google Scholar]