Abstract
Heterografting and RNA transport experiments have demonstrated the long-distance mobility of StBEL5 RNA, its role in controlling tuber formation, and the function of the 503-nt 3′ untranslated region (UTR) of the RNA in mediating transport. Because the 3′ UTR of StBEL5 is a key element in regulating several aspects of RNA metabolism, a potato leaf cDNA library was screened using the 3′ UTR of StBEL5 as bait in the yeast three-hybrid (Y3H) system to identify putative partner RNA-binding proteins (RBPs). From this screen, 116 positive cDNA clones were isolated based on nutrient selection, HIS3 activation, and lacZ induction and were sequenced and classified. Thirty-five proteins that were predicted to function in either RNA- or DNA-binding were selected from this pool. Seven were monitored for their expression profiles and further evaluated for their capacity to bind to the 3′ UTR of StBEL5 using β-galactosidase assays in the Y3H system and RNA gel-shift assays. Among the final selections were two RBPs, a zinc finger protein, and one protein, StLSH10, from a family involved in light signaling. In this study, the Y3H system is presented as a valuable tool to screen and verify interactions between target RNAs and putative RBPs. These results can shed light on the dynamics and composition of plant RNA-protein complexes that function to regulate RNA metabolism.
Keywords: mobile RNA, potato, Solanum tuberosum, yeast three-hybrid, StBEL5, BEL1 family
Introduction
For plant development, phloem plays important roles in not only transporting nutrients, but also as a conduit for moving signal RNAs and proteins. Full-length, phloem-mobile mRNAs function to integrate environmental cues for plant development via this long-distance signaling pathway (Haywood et al., 2005; Banerjee et al., 2006). There are three types of mobile RNAs in plants; (1) pathogenic viral and viroid RNAs, (2) small RNAs including siRNAs and microRNAs, and (3) full-length cellular RNA transcripts (Kehr and Buhtz, 2008). Although many full-length transcripts have been identified in the phloem, only a few of these transcripts have been confirmed to be mobile through the phloem translocation stream. The best examples of mobile RNAs are StBEL5 (Banerjee et al., 2006), CmGAI (Haywood et al., 2005), and the Arabidopsis FLOWERING LOCUS T (Li et al., 2011; Lu et al., 2012). StBEL5 is a transcription factor that works in tandem with Knotted1-types to regulate plant growth (Chen et al., 2003, 2004). RNA detection methods and heterografting experiments demonstrated that StBEL5 transcripts are present in phloem cells and move across a graft union to localize in stolon tips, the site of tuber induction (Banerjee et al., 2006). This movement of RNA originates in leaf veins and petioles and is induced by a short-day photoperiod, regulated by the untranslated regions, and correlated with enhanced tuber production (Banerjee et al., 2006, 2009). Long-distance movement of the RNA of GA INSENSITIVE (GAI) has also been clearly established in both cucumber and pumpkin (Haywood et al., 2005; Ham et al., 2009). Recent results suggest that in addition to FT protein, FT RNA may also be moving to shoot apices to contribute to systemic floral signaling (Li et al., 2011; Lu et al., 2012).
In general, RNA molecules are associated with RNA-binding proteins (RBPs) in the cell, and a number of RNA-protein interactions have been established. RBPs function in splicing, nuclear export, RNA transport and localization, translation, and stability (Dreyfuss et al., 2002; Fedoroff, 2002). RBPs are involved in coordinating gene expression and also influence the localization of protein synthesis (Lunde et al., 2007). For example, a polypyrimidine-tract binding protein (PTB), designated as CmRBP50, was reported as the core protein of a phloem-mobile ribonucleoprotein complex consisting of six RNAs, including CmGAI RNA, and 16 proteins in pumpkin phloem sap (Ham et al., 2009). Commonly, it is the UTRs that function via protein interactions in facilitating the cellular localization of a transcript (Jansen, 2001), in mediating its stability (Lee and Jeong, 2006), or in regulating the efficiency of translation (Barreau et al., 2006). Binding motifs have been identified in the RNAs of animals that function in recognizing RBPs (for review, see Jansen, 2001). These motifs are most predominant in the 3′ UTR (Saunders and Cohen, 1999; Corral-Debrinski et al., 2000; Thio et al., 2000). There are numerous examples demonstrating the importance of the 3′ UTRs in recognizing RBPs that regulate metabolism and movement (Ferrandon et al., 1994; Padmanabhan and Richter, 2006; Irion and St. Johnston, 2007). As a prime example in plants, the 3′ UTR of StBEL5 plays a significant role in mediating its long-distance transport, controlling translation, and regulating stability (Banerjee et al., 2006, 2009), suggesting the presence of cis-elements in this UTR that are recognized by RNA-binding partners.
Although there are several useful biochemical approaches to analyze RNA-protein interactions, the yeast three-hybrid (Y3H) system (Sengupta et al., 1996; Hook et al., 2005) represents a simple but powerful tool for searching a large collection of cDNAs to identify proteins that bind a specific RNA of interest (Cassiday and Maher III, 2003; Gonsalvez et al., 2003; Maniataki et al., 2003; Moore et al., 2003; Campalans et al., 2004; Hwang et al., 2005). Not only does it allow the identification of RNA-protein binding partners but also the dissection of higher-order RNA-protein complexes (Bernstein et al., 2002). Using the 503-nt 3′ UTR of StBEL5 as bait, the Y3H system was used with a potato leaf cDNA library for screening binding partners that may be involved in the metabolism of the full-length, mobile RNA, StBEL5. Initially, more than 100 cDNA clones were isolated from the screening based on nutrient selection and HIS3 and β-galactosidase activation. Seven proteins were selected based on their putative RNA-binding properties for further analyses and RNA gel-shift assays. These results clearly demonstrate the utility of the Y3H system in identifying candidate RBPs.
Materials and Methods
Constructs for the Y3H system
The DNA fragments encoding full-length and truncated forms (D1, T2, and UA baits) of the 3′ UTR of StBEL5 were amplified with gene-specific primer sets (Table A5 in Appendix) and cloned into pIIIA/MS2-1. The plasmids containing the hybrid RNA fused to the full-length 3′ UTR and the truncated forms were transformed into the YBZ-1 yeast strain. For screening, an amplified leaf cDNA library from potato (Solanum tuberosum cv Désirée) similar in design to the stolon library described by Chen et al. (2003) was used. This library was directionally cloned into pAD-GAL4-2.1 (Stratagene, La Jolla, CA, USA) and was a generous gift from Salomé Prat, Madrid, Spain. The YBZ-1 strain and the pIIIA/MS2-1 plasmid were graciously provided Dr. Marvin Wickens, University of Wisconsin, Madison.
Screening of RNA-binding proteins
The YBZ-1 yeast strain containing the StBEL5 full-length 3′ UTR hybrid RNA was transformed with 60 μg of the potato cDNA library. The entire transformation mixture (about 10 mL) was spread onto plates containing SD/-his/-leu/-ura and 1.0 mM 3-aminotrizole (3-AT), a competitive inhibitor of the HIS3 gene product. To calculate the transformation efficiency, the transformation mixture was serially diluted (10−1, 10−2, 10−3) and grown on small SD/-leu/-ura plates. After 1st and 2nd rounds of screening on different concentration (0, 1, 5, 10, or 50 mM) of 3-AT-containing plates, positive colonies were applied to β-galactosidase assays in order to measure the induction of the lacZ reporter gene using a Yeast β-galactosidase Assay Kit (Pierce Biotechnology), according to the manufacturer’s protocol. From the assays, 116 colonies were selected. The 3′ UTR of StBEL5 with StPTB6-pAD (Mahajan et al., 2012) and an empty pAD were used as positive and negative controls, respectively.
Plasmid rescue, identification and categorization of the screened clones
In order to identify the positive clones from the screening, yeast plasmid rescue was performed with E.Z.N.A.® Yeast Plasmid Kit (OMEGA bio-tek) with slight modifications. Positive yeast colonies were picked from the plate and inoculated in 3.0 ml of SD/-leu. Overnight grown cells were pelleted and incubated at 30°C for at least 30 min after resuspension in 480 μl Buffer SE/β-mercaptoethanol and 40 μl lyticase solutions. After incubation, yeast plasmid DNA was isolated by following the kit protocols. The rescued yeast plasmids were transformed into E. coli HB101 competent cells, and the plasmids were isolated and sequenced using pGAD-specific primers (Table A5 in Appendix) at the DNA Facility, Iowa State University. For putative identities of the clones, sequences of the cDNAs were analyzed at the Dana–Farber Cancer Institute (DFCI) Gene Index1 potato database. Translated protein sequences were obtained from Translate2 and ORF Finder3 and analyzed using BLAST on the TAIR database4. Functional categorization of selected proteins was performed using the MIPS Arabidopsis thaliana database5. The domains of the B5RBPs (Figure 3) were analyzed using SMART6 and NCBI’s Conserved Domain Search. For Figure 4A, amino acid sequences of Arabidopsis and potato LSH proteins were organized into a phylogenetic tree with the MEGA 4.0.2 package and the neighbor-joining program. The numbers listed at the branching points are boot-strapping values that indicate the level of significance (percentage) for the separation of two branches.
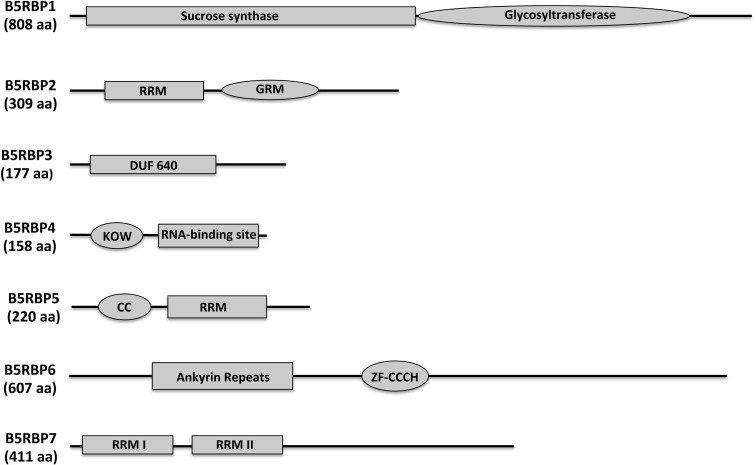
Figure 3.
Protein structure and prediction of conserved domains of the seven B5RBPs. The deduced amino acid sequences of the B5RBPs were analyzed using SMART (http://smart.embl-heidelberg.de/) and NCBI’s Conserved Domain Search. RRM, RNA recognition motif; DUF, domain of unknown function; GRM, glycine-rich motif; KOW, from Kyrpides et al. (1996); CC, coiled-coil motif; ZF-CCCH, zinc finger, CCCH-type.
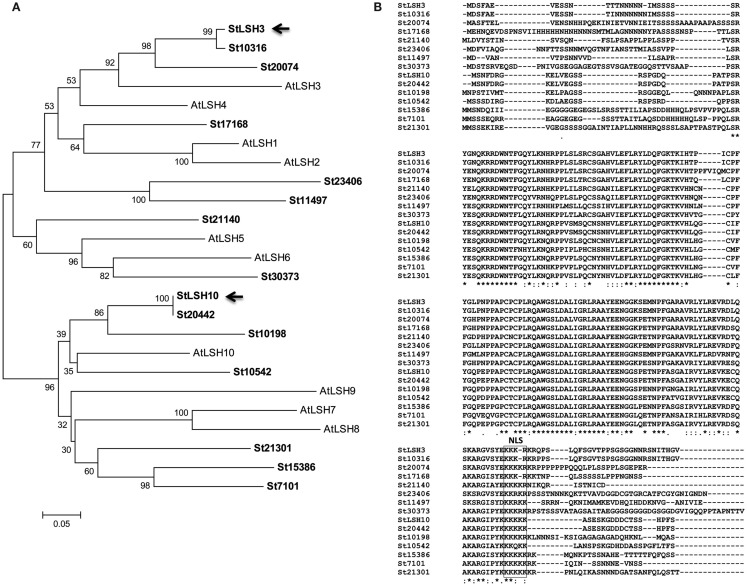
Figure 4.
Phylogenetic analysis (A) and alignment of the aa sequence (B) of the family of LSH proteins of potato (St). Included in the dendrogram (A) are the 10 LSH proteins of Arabidopsis (At). Arrows indicate the two StLSHs, LSH3 and -10, identified from the current yeast three-hybrid screening. The nuclear localization signal (NLS) is boxed (B). Asterisks below the aligned amino acids (B) indicate conserved residue identity.
RNA gel-shift assays
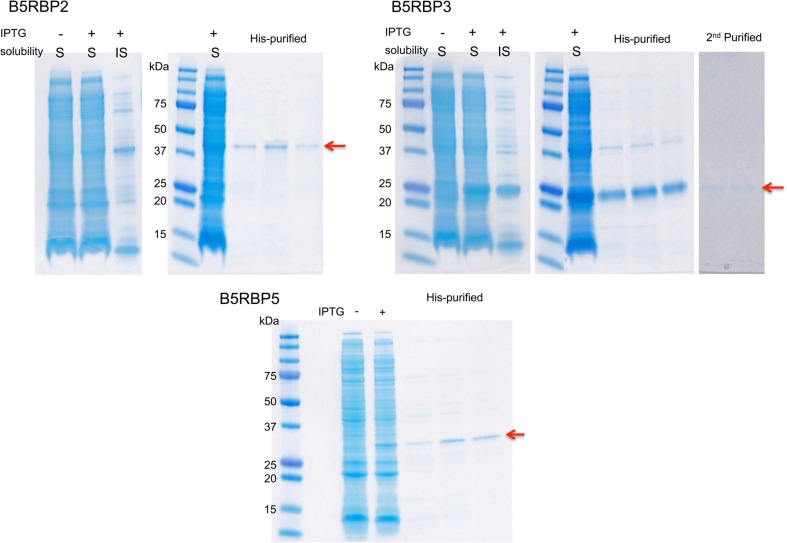
The PCR-amplified fragments with gene-specific primer sets (Table A5 in Appendix) were cloned into the pET-28a (+) plasmids after proper enzyme digestion to produce histidine tag (His)-fusion recombinant proteins (Figure A1 in Appendix). The constructs were transformed into E. coli BL21-Codon (DE3) cells (Stratagene). The recombinant proteins were induced with 0.4 mM IPTG, and purified using HisPur Cobalt Purification Kit (Pierce Biotechnology). For in vitro transcription to generate RNA probes, T3 promoter-containing sense primers were created by adding T3 sequences on the 5′ end of the sense primers of the target sequences (Table A5 in Appendix), and used for PCR amplification using Platinum Taq DNA Polymerase High Fidelity (Invitrogen). The gel-purified PCR product was transcribed using MEGAscript T3 (Ambion) incorporating biotin (biotin-11-UTP, Perkin Elmer) as described by the manufacturer’s manual. The biotin-labeled probe RNA was purified by gel purification using ZymocleanTM Gel RNA Recovery kit (ZymoResearch). Five femtomoles of biotin-labeled RNA probes were incubated with indicated amounts of purified recombinant proteins in the binding buffer provided by the Light Shift Chemiluminescent RNA EMSA kit (Pierce Biotechnology) on ice for 45 min. The RNA-protein complexes were separated in 2.5% agarose (for the full-length probe) or 5% polyacrylamide gel (for the IRE probe) and transferred onto BrightStar-Plus (Ambion) nylon membranes. The signal was detected using the EMSA kit according to the manufacturer’s manual.
Screening for RNA-binding proteins using the yeast three-hybrid system
As eloquently explained by Bernstein et al. (2002), the Y3H system is based on two expression vectors, one for the RNA bait and the other for the protein target, and three-hybrid components. When bait RNA interacts with the target protein, the reporter genes, HIS3 and lacZ, are activated and can be readily detected by simple biochemical assays (Hook et al., 2005; Seay et al., 2006).
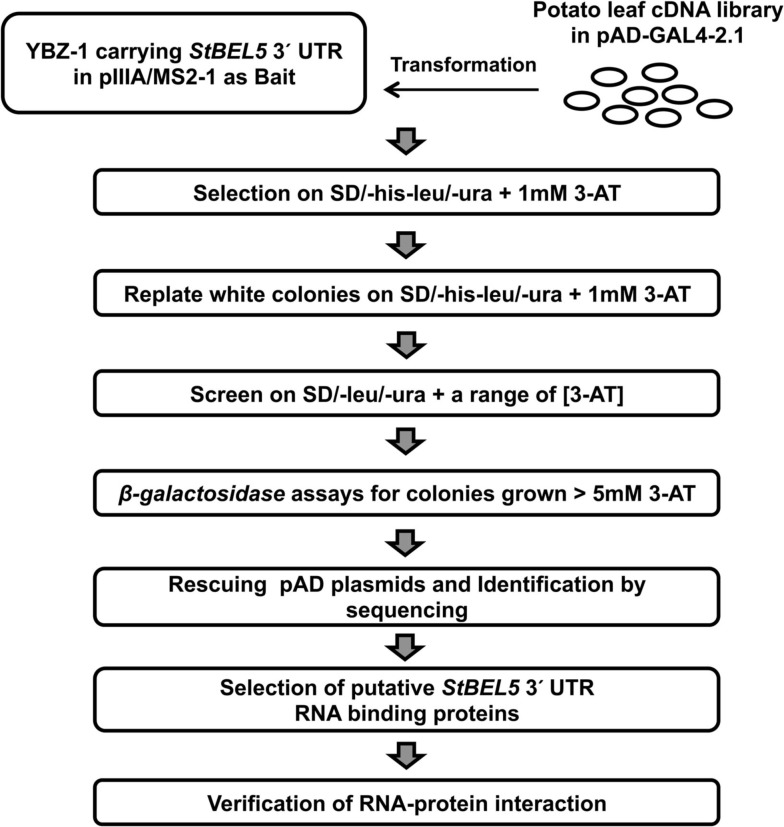
To isolate interacting proteins, the DNA fragment encoding 3′ UTR of StBEL5 as a bait was cloned into the MS2 portion of pIIIA/MS2-1 vector. The resulting plasmids carrying hybrid MS2-3′ UTR of StBEL5 were transformed into a yeast strain, YBZ-1, and the potato leaf cDNA library was sequentially transformed using conventional protocols with slight modifications (Bernstein et al., 2002; Seay et al., 2006). We analyzed approximately 6.5 × 105 yeast colonies, and the resulting transformed colonies were screened on SD/-his/-leu/-ura plates containing 1.0 mM 3-AT (Figure 1). From the first round, 448 colonies were selected as primary positives. Those selected positive colonies were replicated on SD/-his/-leu/-ura plates containing 1.0 mM 3-AT again to remove potential false positives. From these screenings, 281 colonies were chosen for further screening by using the two reporter genes, HIS3 and lacZ. SD/-his/-leu/-ura plates containing a series of 3-AT concentration (0, 1, 3, 5, 10, and 50 mM) were used for testing HIS3 expression. The 281 colonies were streaked on these plates, and 194 colonies were grown on SD/-his/-leu/-ura plates containing at least 5.0 mM 3-AT. Finally, 116 colonies were selected based on β-galactosidase and HIS3 activation and were sequenced and analyzed (Table A1 in Appendix). The overall strategy of the screening is summarized in Figure 1. As a comparison, using the yeast strain YBZ-1, 49 HIS3+ colonies were selected out of approximately 8.0 × 105 transformants according to Hook et al. (2005). The interactions of the 3′ UTR of StBEL5 with StPTB6 and the 3′ UTR of StBEL5 with empty pGAD vector were used as positive and negative controls for RNA-protein interaction, respectively. PTB proteins are multifunctional proteins that bind numerous mRNAs and are involved in a wide range of RNA metabolism, such as RNA stability, splicing, translational repression, and long-distance transport. There are six PTBs in the potato genome, and one, designated StPTB6, binds to untranslated regions of phloem-mobile mRNAs of potato (Mahajan et al., 2012).
Figure 1.
Schematic diagram of the Y3H system for screening interacting partner proteins from a leaf cDNA library using the 503-nt 3′ UTR of StBEL5 as bait.
Characterization of selected cDNA clones
For identification of the screened colonies, their sequences were analyzed and BLAST was performed using the Arabidopsis and potato databases, and a total of 89 clones (76.7%) exhibited significant matches to previously characterized or known genes (Table A2 in Appendix). Thirteen clones were identified as redundant (30.2% redundancy), and therefore, a total of 94 unique singletons were isolated from the Y3H screening (listed in Tables A1– A3 in Appendix). These 13 included LSH3 (Light-dependent Short Hypocotyls3), LSH10 (Light-dependent Short Hypocotyls10), C3H zinc finger transcription factor, sucrose synthase4, a Transducin/WD40 repeat-like superfamily protein, LTP12 (Lipid Transfer Protein 12), ELI3-1 (elicitor-activated gene 3-1), X-ray induced transcript 1, glutamate-1-semialdehyde-2, 1-aminomutase, and some ribosomal proteins and unknown proteins (Table A3 in Appendix). Interestingly, LSH10, a close sequence match to AtLSH10 (AT2G42610), was identified from 10 clones along with another LSH member, LSH3, which was isolated twice. Twenty-seven clones (23.3%) were categorized as undefined clones, i.e., “unknown” or “no hit” from the database search (Table A2 in Appendix). Functional classification revealed a total of eight cDNAs that encoded proteins with DNA/RNA-binding properties. Nineteen clones encoded proteins that are components of the machinery for protein synthesis (16.4%) at the initiation and/or elongation of translation, such as eIF5A, transducin/WD40 repeat-like superfamily, heat shock protein 70, and an alpha-tubulin protein (Doroshenk et al., 2009; Lin et al., 2009; Tables A1 and A4 in Appendix). Overall, 35 cDNAs (30.1% of total screened clones) encoding proteins with RNA-binding properties were identified (Table A4 in Appendix).
Candidate protein partners for StBEL5 RNA
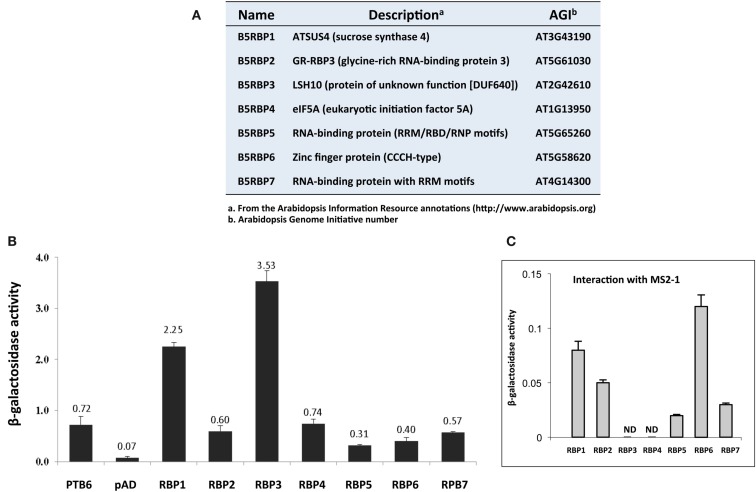
Based on activities of marker genes (HIS3 and lacZ), and their putative RNA-binding function, seven cDNA clones were selected for verification as protein partners of StBEL5, and designated as StBEL5 RNA-binding protein (B5RBP) one to seven (Figure 2A). Related proteins of three of the B5RBPs, B5RBP1, -4, and -6, were previously identified as RBPs in other species (Xu et al., 2004; Doroshenk et al., 2009; Ling et al., 2011), and three others, B5RBP2, -5, and -7, contain conserved RNA recognition motifs (RRMs; Figure 3). The most frequently identified clone from the Y3H screening, StLSH10 (B5RBP3), was also included (Table A3 in Appendix). Each of these proteins induced β-galactosidase activity in an interaction with the bait RNA to levels much higher than the negative controls and, in some cases, even higher than the positive control (Figures 2B,C).
Figure 2.
List of seven B5RBPs (A) and a quantitative analyses of their interactions with the 503-nt 3 ′ UTR of StBEL5 using a β-galactosidase assay (B). The 3′ UTR of StBEL5 with StPTB6-pAD was used as a positive control (B), and an empty pAD (B) or the pIIIA/MS2-1 bait vectors (C) were used as negative controls. The numbers above each bar (B) represent the means for β-galactosidase activity in triplicate, and standard errors are shown for each mean (B,C). The RNA produced by the pIIIA/MS2-1 vector in yeast, including the 60-nt MS2 sites, is approximately 272 nt in length (Bernstein et al., 2002).
B5RBP1 encodes a sucrose synthase (SUS4), containing a sucrose synthase motif for the sucrose metabolic pathway and a glycosyltransferase motif for biosynthetic processes (Figure 3). There were three reasons to include SUS4. First, it was identified as a cytoskeleton-associated RBP from developing rice seeds (Doroshenk et al., 2009). Second, in Arabidopsis, SUS4 was detected in the companion cells of the phloem (Fallahi et al., 2008) and third, it plays an important role in starch metabolism in potato tubers (Fu et al., 1995; Zrenner et al., 1995; Bieniawska et al., 2007). With regard to the role of SUS4 in potato tuber development, high-level increases in SUS4 promoter activity were observed during early tuber formation (Fu et al., 1995) and transgenic lines overexpressing SUS4 produced enhanced tuber yields (Baroja-Fernandez et al., 2009). SUS4 could be an example of a multifunctional RBP directly involved in potato tuber development.
B5RBP2 encodes a glycine-rich RNA-binding protein that contains a RRM for RNA-binding and a glycine-rich motif (GRM, Figure 3). Plant proteins that contain a GRM are grouped into five classes based on structure. B5RBP2 is considered a class IV member because it contains a RRM (Mangeon et al., 2010). Class IV GRM-proteins are subdivided based on their links to osmotic stress, cold stress, flower timing, development, and responsiveness to abscisic acid. Interestingly, B5RBP2 is orthologous to AtGRP7 (AT2G21660) a protein related to a RBP found in pumpkin phloem sap (Lin et al., 2009). B5RBP2 may function in potato phloem sap by interacting with StBEL5 RNA to facilitate long-distance movement. AtGRP7 is also involved in the regulation of alternative splicing, ribosome function, and RNA metabolism (Wachter et al., 2012).
B5RBP3 is alight-dependent short hypocotyl (LSH10, AT2G42610) protein with a Domain of Unknown Function (DUF640, Figure 3). In Arabidopsis, there are 10 LSH genes and in potato, 15 (Figure 4A). The function of most these proteins, ranging in size from 164 to 219 aa, are unknown except for AtLSH1, -3, and -4. AtLSH1 is a nuclear protein in Arabidopsis with a nuclear localization signal (NLS) in the C-terminal region. It is involved in light regulation of seedling development (Zhao et al., 2004). Both AtLSH3 and -4 appear to have a role in Arabidopsis shoot and floral organ differentiation since constitutive expression of these genes resulted in abnormal development (Takeda et al., 2011). As described earlier, LSH10 was the most frequently selected cDNA from the screen (Table A3 in Appendix). β-galactosidase activity of the B5RBP3/StBEL5 interaction was several-fold greater than the other B5RBPs (Figure 2B). All of the potato LSH proteins exhibit a highly conserved internal region representative of a domain from the DUF640 superfamily flanked by sequence of considerable variance at both the amino- and carboxy-termini (Figure 4B).
B5RBP4 encodes a eukaryotic initiation factor 5A (eIF5A, AT1G13950) containing both KOW (acronym of the authors surname, Kyrpides et al., 1996; Figure 3) and eIF5A motifs for ribosome binding, RNA-binding, and translation activity (Figure 3). Studies in bacteria suggest that the KOW motif mediates RNA and protein interactions (Steiner et al., 2002). For simplicity, the eIF5A motif is referred to as a RNA-binding site (Figure 3) since the motif is characterized as a S1-like RNA-binding domain (Peat et al., 1998). eIF5A is a multifunctional protein involved in RNA-binding, processing, turnover, and transport from the nucleus to cytoplasm and in transcription and translation (Burd and Dreyfuss, 1994; Cusack, 1999; Xu and Chen, 2001). Recently, eIF5A in yeast was shown to stimulate protein synthesis but was not required for the process (Henderson and Hershey, 2011). In pumpkin phloem sap, CmeIF5A was detected as a component of the RBP50-based ribonucleoprotein complex (Ham et al., 2009). Further characterization of CmelF5A revealed that hypusination (lysine residue modification) was necessary for RNA-binding and protein interaction and that both hypusinated and non-hypusinated CmelF5A existed in the phloem (Ma et al., 2010). B5RBP4 may function through a similar mechanism in potato phloem sap to mediate the formation of ribonucleoprotein complexes.
B5RBP5 encodes a RNA-binding (RRM/RNA-Binding Domain/Ribonucleoprotein → RRM/RBD/RNP) protein family member containing a coiled-coil motif (CC) and a RRM (Figure 3) that is orthologous to At5G65260, an Arabidopsis RNA-binding protein. At5G65260 is designated as a polyadenylation factor that can bind to the poly (A) tail and control its length (Hunt et al., 2008). The Arabidopsis transcription factor Long Hypocotyl5 (HY5) that is involved in photomorphogenesis was shown to mediate the expression of At5G65260 (Lee et al., 2007). At5G65260 expression was down-regulated in a loss-of-function HY5 mutant. It is conceivable that B5RBP5 levels in potato may also be regulated by light.
B5RBP6 encodes a zinc finger (CCCH-type) family protein containing two ankyrin repeats for protein-protein interactions, and two zinc finger-C3H1 domains (ZF-CCCH) for zinc ion binding, and nucleic acid binding (Figure 3). Unlike other zinc finger proteins that generally function as DNA-binding proteins (Laity et al., 2001), CCCH zinc finger proteins bind to AU-rich elements of RNAs (Brown, 2005). A study in mouse revealed a role for a CCCH zinc finger protein (tristetraprolin) in mRNA decay (Lai et al., 2006). In Trypanosoma brucei, the causative agent of sleeping sickness, the CCCH zinc finger protein (Tb2C3H20) functions in mRNA stability (Ling et al., 2011). In Arabidopsis, two CCCH-type zinc finger genes, designated AtSZF1, and AtSZF2, were salt-inducible and mediated responsiveness to salt (Sun et al., 2007). From the current screen, two CCCH-type zinc finger proteins were identified (Table A4 in Appendix) that shared sequence similarity with AT2G40140 (AtSZF2) and AT5G58620 from Arabidopsis.
B5RBP7 encodes another RNA-binding (RRM/RBD/RNP motifs) family protein that shares sequence similarity with Arabidopsis AtRNP1 (AT4G14300) containing two RRM domains (Figure 3). AtRNP1 is a target of Arabidopsis transportin 1 (AtTRN1) that is an ortholog of the human nuclear import receptor transportin1 protein (Ziemienowicz et al., 2003). AtRNP1 may function as a shuttle protein moving RNAs between the nucleus and cytoplasm. Interestingly, AtGRP7 also interacted with AtTRN1. As discussed above, B5RBP2 is orthologous to AtGRP7 suggesting that B5RBP7 and B5RBP2 may associate with StBEL5 RNA as a tandem complex.
Expression profiles
To assess transcript levels for select RBPs, expression values were obtained from the publicly available RNA-seq database from the RH genotype of the Potato Genome Sequencing Project (Xu et al., 2011). Abundance levels of StBEL5 and StHSP70 have been included as references. The potato HSP70 protein was selected during the screen (Table A4 in Appendix) and a HSP70-type was previously identified as a member of a phloem-mobile RNP complex in pumpkin (Ham et al., 2009). Relatively high and consistent levels of transcripts across all organs were observed for B5RBP2, -4, HSP70, and both RBPs, B5RBP5, and -7 (Table 1). The very high levels of eIF5A (B5RBP4) likely reflect its general, multifunctional role in several aspects of RNA metabolism (Zanelli and Valentini, 2007). B5RBP1 (sucrose synthase) scored abundant RNA levels in both stolons and young tubers indicative of its role in starch metabolism during tuber formation. The zinc finger CCCH protein (B5RBP6) was most abundant in petioles with a value of 334 FPKMs (fragments per kb per million mapped reads). A mobile RNA like StBEL5 is very abundant in petioles, an observation that is consistent with both its transcriptional source and its capacity to move long-distances through the phloem (Banerjee et al., 2006). Petioles serve two main functions: to provide support for the leaf lamina and to act as a protective sleeve for phloem cells that move sugar and signaling molecules (like RNA) from source leaves to sinks. RBPs are commonly detected in companion cells and sieve elements of leaf veins in position to chaperone mobile RNAs (Ham et al., 2009).
Table 1.
Expression profile of select B5RBPs mined using the RNA-seq data from the publically available Potato Genome Database (Xu et al., 2011).
| Gene | Flower | In vitro plant | Sprout | Leaf | Petiole | SAM | Stem | Stolon | Young tuber | Root |
|---|---|---|---|---|---|---|---|---|---|---|
| B5RBP1 | 43 | 170 | 120 | 10 | 39 | 46 | 62 | 175 | 407 | 151 |
| B5RBP2 | 91 | 144 | 191 | 129 | 152 | 201 | 113 | 252 | 257 | 165 |
| B5RBP3 | 0 | 14 | 33 | 0 | 77 | 23 | 46 | 149 | 590 | 53 |
| StLSH3 | 4 | 15 | 12 | 0 | 27 | 5 | 3 | 17 | 3 | 11 |
| B5RBP4 | 375 | 364 | 414 | 354 | 511 | 199 | 377 | 420 | 418 | 606 |
| B5RBP5 | 76 | 68 | 93 | 40 | 83 | 63 | 70 | 65 | 71 | 92 |
| B5RBP6 | 67 | 91 | 46 | 103 | 334 | 48 | 59 | 70 | 40 | 104 |
| B5RBP7 | 119 | 229 | 392 | 111 | 250 | 188 | 141 | 184 | 184 | 238 |
| StHSP70 | 65 | 64 | 108 | 44 | 100 | 122 | 140 | 236 | 192 | 160 |
| StBEL5 | 35 | 52 | 60 | 39 | 170 | 25 | 77 | 24 | 55 | 42 |
Nine organs and an in vitro plantlet are presented and abundance values are shown in FPKMs (fragments per kb per million mapped reads). StBEL5 is shown as a relative control. The potato HSP70 protein is included here because it was selected during the screen (Table A4 in Appendix) and previously was identified as a member of a phloem-mobile RNP complex in pumpkin (Ham et al., 2009).
Despite the observation that B5RBP3 (LSH10) appeared 10 times from the Y3H screening (Table A3 in Appendix), its RNA levels were remarkably low in leaf RNA of the RH genotype (Table 1). Similar results for LSH10 RNA abundance levels were observed in RNA-seq data from the DM genotype (Xu et al., 2011). Among the proteins selected, this putative RBP exhibited the strongest induction of β-galactosidase activity (Figure 2). Whereas the second LSH protein selected in this screen, StLSH3, exhibited very low transcript values across all organs, transcript abundance values for B5RBP3 (Table 1) were extremely high in petioles (77 FPKMs), stolons (149 FPKMs), and young tubers (590 FPKMs). This latter transcript value was the second most abundant of any of the RNAs scored in this experiment.
Verification of RNA-protein interaction of the B5RBPs
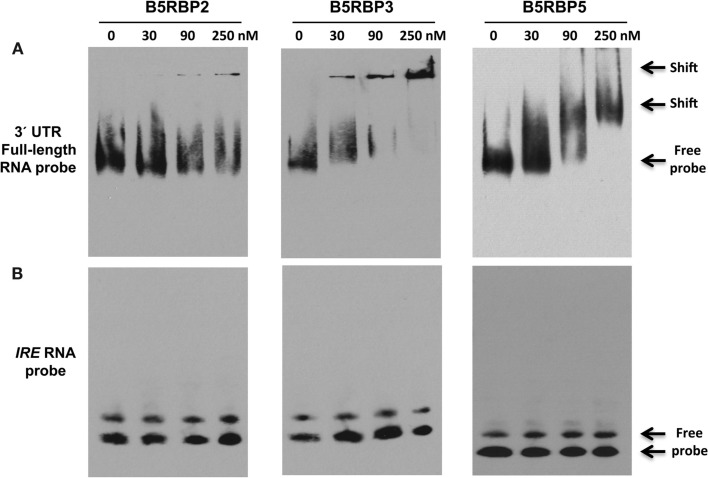
To validate the direct interaction of select proteins with the 3′ UTR of StBEL5, RNA gel-shift assays were performed with select B5RBPs. These assays were performed using biotin-incorporated RNA probes using the full-length 3′ UTR of StBEL5 and the purified recombinant B5RBP2, -3, and -5 proteins (Figure 5A). For showing specificity of the interaction, IRE RNA, which binds specifically with the iron responsive protein in the cell under iron-starved conditions, was used as a negative control (Figure 5B). Shifted bands were observed for all three interactions in a range of 30–250 nM of protein. B5RBP3 affected a shift with protein amounts as low as 30 nM, whereas shifted bands were clearly observed with the other two B5RBPs in the reactions containing 90 nM of protein. With comparable amounts of protein, no gel-shift was observed for the negative control, the iron responsive element (IRE, Figure 5B).
Figure 5.
Mobility shift assays for in the vitro interaction of the 3′ UTR of StBEL5 with select B5RBPs. (A) Full-length 3′ UTR of StBEL5 with B5RBP2, -3, and -5. The iron response element (IRE) is included as a negative control (B). Approximately 5 fmole of biotin-labeled bait RNA and protein concentrations ranging from 30 to 250 nM were used in each reaction.
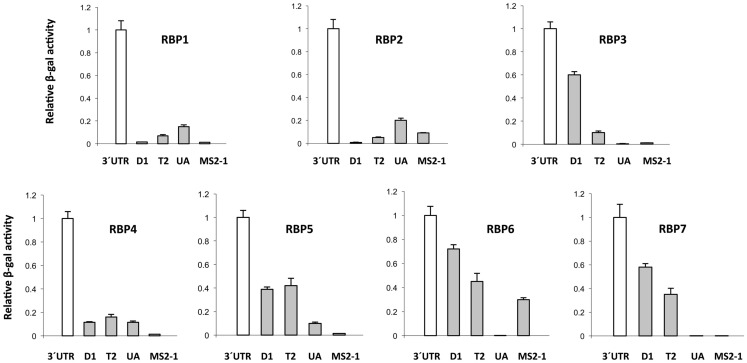
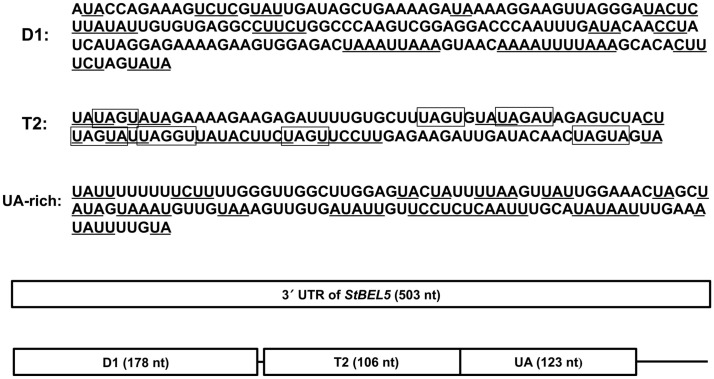
The 3′ UTR of StBEL5 is involved in several aspects of RNA metabolism and is replete with potential binding motifs (Banerjee et al., 2009). To identify shorter binding regions within the 3′ UTR that may be involved in protein/RNA interaction, truncated bait sequences were utilized in the β-galactosidase assay of the Y3H system (Figure 6). Three truncated sequences were used based on their conserved sequence motifs and their coverage of the UTR (Figure 7). The 5′ D1 sequence is enriched for CU motifs (underlined sequence, Figure 7). T2 contains several UAGU motifs (Figure 7, boxed), and the UA-bait sequence contains a number of uracil/adenine runs (underlined sequence, Figure 7). Overall, the greatest β-galactosidase activity was observed for the full-length 3′ UTR (Figure 6). Based on β-galactosidase activity, B5RBP3, -5, -6, and -7 exhibited the strongest interaction with sequence located toward the 5′ end of the UTR (D1 and T2 baits). B5RBP1 and -2 exhibited the strongest interaction with sequence located toward the 3′ end of the UTR (T2 and UA baits). B5RBP4, the potato ortholog of eIF5A, exhibited equivalent strength of activity with all three truncated baits suggesting a degree of non-specific binding. These results with the potato eIF5A are consistent with previous work showing that a pumpkin form of eIF5A exhibited RNA-binding that was non-sequence-specific in nature (Ma et al., 2010).
Figure 6.
Analysis of β-galactosidase activity for the interaction of select StBEL5 RNA-binding proteins (B5RBPs) with sequences within the 3′ UTR of StBEL5. The values in each graph are normalized to activity relative to the full-length UTR. 3′ UTR, full-length UTR; D1; a 178-nt sequence within the UTR starting from the stop codon; T2, a UAGU-rich region within the UTR; UA, UA-rich region toward the 3′ end of the UTR; MS2-1, RNA sequence from the bait vector serving as a negative control. See Figure 7 for details on these truncated StBEL5 bait sequences.
Figure 7.
Schematic diagram of the bait RNA sequences from the 3′ UTR of StBEL5 used in Figure 6. Included are three truncated sequences within the full-length 503-nt 3′ UTR: the 178-nt D1 sequence, the 106-nt T2 region, and the 123-nt UA-rich region (UA). Both UA-and CU-rich motifs are underlined. The UAGU motifs of the T2 sequence are boxed.
Conclusion
The Y3H system has been established as an efficient method for selecting protein partners of RNA from among thousands of putative partners, and for assaying binding affinity of specific RNA/protein interactions. With modifications, this system has been adapted for screening RNA/RNA interactions (Piganeau and Schroeder, 2006), for identifying protein/small signaling molecule complexes (Cottier et al., 2011), and for testing multi-component interactions (Bernstein et al., 2002). Although numerous false positives may arise during the screening process, there are several levels of selection that may be utilized to eliminate these. These include nutrient selection, HIS3 activation, addition of 3-aminotriazole, and 5-fluororotic acid to the media, and the induction of lacZ. As shown previously (Hook et al., 2005), both HIS3 and lacZ expression levels are directly related to binding affinity and may be used to assess the robustness of specific RNA/protein interactions (Mahajan et al., 2012) or to map specific motifs present in either bait or target sequences (Edwards et al., 2001; Mori et al., 2008; Stumpf et al., 2008).
Factors known to affect in vivo interactions include the intrinsic affinity of bait RNA and target protein, the length of the bait RNA, and accessibility of the insert to the target (Wurster and Maher III, 2010). RNA sequences less than 150 nt generally produce the most substantial and specific reporter activation. The inclusion of additional sequence can lead to a reduction in signal. In this study, the entire 503-nt 3′ UTR of StBEL5 was utilized to include a wide range of interactions. With this approach, subsequent analyses of shorter RNA sequences in RNA/protein interactions using either RNA gel-shift or Y3H assays may be used to identify the location of specific binding motifs. The source of the protein expression library will also play an important role in identification of protein partners. Here the use of a leaf cDNA library would provide wide coverage but could preclude the identification of important interactions with rare phloem-mobile proteins. Three of the RBPs compiled in the final list here (B5RBP1, -5, and -7) appear to function in the intracellular transport of RNAs. Two RBPs identified in the current study, eIF5A and HSP70, were isolated from a phloem-mobile RNP complex of pumpkin (Ham et al., 2009). A protein very close in sequence match to the glycine-rich RBP (B5RBP2) was also identified in pumpkin phloem sap (Lin et al., 2009). Both binding affinity and abundance of the target protein would impact the results of the Y3H screen. As demonstrated by StLSH10, despite its relatively low transcript level in leaves (Table 1), its affinity for the BEL5 UTR bait was demonstrated to be relatively high. The very high transcript levels observed for StLSH10 (B5RBP3) in stolons and young tubers (149 and 590 FPKMs, respectively) also suggest a possible role in tuber development. Screening a library of proteins from a sink organ like the tuberizing stolon would further expand our understanding of the RNP complex responsible for delivery of the mobile StBEL5 transcript to its functional site. Overall, these results demonstrate the utility of the Y3H system in screening and verifying interactions between target RNAs and putative RBPs.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Thanks to Kate Lueders for her valuable technical assistance and to Marv Wickens for graciously providing the Y3H vectors and yeast strains. Thanks also to Tian Lin and Pooja Sharma for their help in preparing the LSH phylogenetic tree and alignment. This research was supported by the NSF Plant Genome Research Program award no. DBI-0820659.
Appendix
Figure A1.
Coomassie-stained gels of protein purification of three B5RBPs. Each protein was induced in E. coli, the soluble fraction was extracted, and the protein purified once or twice (B5RBP3) using the HisPur Cobalt purification kit (Pierce). The farthest right-hand lanes (arrows) represent from 15 to 100 ng of protein loaded and these samples were used for the RNA EMSAs. (+) indicates the addition of IPTG; (−) no IPTG; S, soluble fraction; IS, insoluble fraction.
Table A1.
Full list of the screened genes with their putative identities and concentration of 3-AT for screening.
| Clone no | AGIa | Descriptionb | E-value | Potatoc | 3-ATd |
|---|---|---|---|---|---|
| #1–1 | AT4G16260 | Glycosyl hydrolase superfamily protein | 2E−96 | TC194747 | 50 |
| #1–9 | AT4G17880 | Basic helix-loop-helix (bHLH) DNA-binding family protein | 3E−66 | TC207090 | 50 |
| #1–12 | AT2G37190 | Ribosomal protein L11 | 3E−71 | TC200521 | 10 |
| #1–21 | AT3G43190 | ATSUS4 | sucrose synthase 4 | 0 | TC194622 | 50 |
| #1–22 | AT3G43190 | ATSUS4 | sucrose synthase 4 | 0 | TC194622 | 10 |
| #1–25 | AT2G38040 | Acetyl co-enzyme a carboxylase carboxyltransferase alpha subunit | 2E−76 | TC202772 | 5 |
| #1–38 | AT2G40140 | ATSZF2 (salt-inducible zinc finger 2) | 8E−23 | TC208753 | 10 |
| #1–40 | AT4G05320 | Polyubiquitin 10 | 0 | TC204873 | 5 |
| #1–55 | AT1G58684 | Ribosomal protein S5 | E−111 | TC225313 | 5 |
| #1–57 | AT4G15470 | Bax inhibitor-1 | 1E−71 | TC200584 | 5 |
| #1–61 | AT3G14610 | Cytochrome P450, family 72, subfamily A, polypeptide 7 | E−124 | TC197461 | 5 |
| #2–5 | AT3G12120 | Fatty acid desaturase 2 | E−143 | TC197103 | 50 |
| #2–23 | AT5G57280 | S-Adenosyl-l-methionine-dependent methyltransferases | 9E−77 | AW906478 | 50 |
| #2–35 | AT1G26830 | ATCUL3A (cullin 3) | 0 | TC220451 | 5 |
| #2–36 | AT5G24530 | 2-Oxoglutarate (2OG) and Fe(II)-dependent oxygenase | 8E−37 | TC212020 | 50 |
| #2–51 | AT2G23940 | Protein of unknown function (DUF788) | 6E−60 | TC223843 | 5 |
| #2–52 | AT5G48720 | X-ray induced transcript 1 | 1E−26 | BG597043 | 50 |
| #2–57 | AT1G47260 | APFI/gamma carbonic anhydrase 2 | E−126 | TC195322 | 5 |
| #2–62 | AT5G48720 | XRI, XRI1 | X-ray induced transcript 1 | 6E−17 | BG597043 | 5 |
| #3–1 | AT3G18000 | S-Adenosyl-l-methionine-dependent methyltransferases | 0 | TC212421 | 10 |
| #3–4 | AT3G13340 | Transducin/WD40 repeat-like superfamily protein | 3E−92 | TC222486 | 10 |
| #3–7 | AT1G58684 | Ribosomal protein S5 | E−111 | TC225313 | 5 |
| #3–16 | AT1G53840 | ATPME1, PME1 | pectin methylesterase 1 | 9E−20 | EG012025 | 5 |
| #3–20 | AT3G05590 | Ribosomal protein L18 | 8E−83 | TC210667 | 5 |
| #3–33 | AT4G37980 | ELI3-1, ELI3, ATCAD7, CAD7 | elicitor-activated gene 3-1 | 4E−20 | TC223852 | 5 |
| #3–39 | AT5G61030 | Glycine-rich RNA-binding protein | 2E−30 | TC203116 | 50 |
| #3–43 | AT2G46500 | ATPI4K GAMMA 4 (phosphoinositide 4-kinase gamma 4) | 3E−54 | TC208976 | 5 |
| #3–53 | AT1G49040 | SCD1 | stomatal cytokinesis defective/SCD1 protein: WD40 containing | 2E−63 | TC206273 | 50 |
| #4–1 | AT5G19510 | Translation elongation factor EF1B/ribosomal protein S6 family protein | 2E−26 | TC197463 | 5 |
| #4–5 | AT2G33220 | GRIM-19 protein | 1E−63 | TC201317 | 5 |
| #4–13 | AT4G22140 | EBS | PHD finger family protein/bromo-adjacent homology (BAH) domain-containing | 6E−98 | TC216086 | 5 |
| #4–33 | AT5G52640 | Heat shock protein 90 | E−157 | TC200319 | 10 |
| #4–36 | AT1G22410 | Class-II DAHP synthetase family protein | 0 | TC194919 | 5 |
| #4–37 | AT5G39510 | Vesicle transport v-SNARE family protein | 6E−08 | TC196409 | 5 |
| #4–60 | AT3G43120 | SAUR-like auxin-responsive protein family | 7E−19 | TC219427 | 5 |
| #5–1 | AT2G42210 | ATOEP16-3, OEP16-3 | Mitochondrial import inner membrane translocase subunit | 2E−54 | TC204735 | 50 |
| #5–2 | AT3G01320 | SNL1 | SIN3-like 1 | 1E−42 | CV286681 | 10 |
| #5–4 | AT5G27700 | Ribosomal protein S21e | 4E−43 | TC214159 | 5 |
| #5–5 | AT5G25610 | RD22, ATRD22 | BURP domain-containing protein | 1E−32 | TC201553 | 10 |
| #5–6 | AT1G03860 | ATPHB2, PHB2 | prohibitin 2 | E−120 | TC196152 | 50 |
| #5–9 | AT4G09800 | S18 ribosomal protein | 5E−44 | TC207656 | 10 |
| #5–10 | AT2G42610 | LSH10 | Protein of unknown function (DUF640) | 1E−64 | TC202398 | 10 |
| #5–11 | AT2G42610 | LSH10 | Protein of unknown function (DUF640) | 1E−64 | TC202398 | 10 |
| #5–12 | AT1G49310 | Unknown protein | 0.002 | TC207198 | 5 |
| #5–13 | AT3G60520 | Unknown protein | 0.047 | TC196277 | 50 |
| #5–17 | AT1G27450 | APT1, ATAPT1 | adenine phosphoribosyl transferase 1 | 2E−78 | TC196278 | 5 |
| #5–21 | AT3G50500 | SNRK2.2 | SNF1-related protein kinase 2.2 | 3E−15 | EG012856 | 5 |
| #5–22 | AT1G60420 | DC1 domain-containing protein | E−100 | TC199573 | 50 |
| #5–23 | AT1G67785 | Unknown protein | 2E−16 | TC216692 | 5 |
| #5–24 | AT1G22100 | Inositol-pentakisphosphate 2-kinase family protein | 1E−67 | TC213605 | 5 |
| #5–26 | AT5G26220 | ChaC-like family protein | 1E−79 | TC195543 | 10 |
| #5–27 | AT2G42610 | LSH10 | Protein of unknown function (DUF640) | 2E−60 | TC205325 | 10 |
| #5–29 | AT2G42610 | LSH10 | Protein of unknown function (DUF640) | 5E−14 | TC205325 | 50 |
| #5–30 | AT1G19580 | GAMMA CA1 | gamma carbonic anhydrase 1 | E−129 | TC196517 | 5 |
| #5–31 | AT1G76670 | Nucleotide-sugar transporter family protein | E−147 | TC198970 | 10 |
| #5–32 | AT1G13950 | EIF-5A, ELF5A-1, ATELF5A-1, EIF5A | eukaryotic elongation factor 5A-1 | 3E−72 | TC202667 | 5 |
| #5–34 | AT2G42610 | LSH10 | Protein of unknown function (DUF640) | 2E−60 | TC202398 | 10 |
| #5–35 | AT2G23610 | ATMES3, MES3 | methyl esterase 3 | 5E−41 | TC221190 | 10 |
| #5–37 | AT3G25660 | Amidase family protein | 2E−73 | CV506455 | 50 |
| #5–38 | AT1G18080 | ATARCA (Transducin/WD40 repeat-like superfamily protein) | E−118 | TC197724 | 5 |
| #5–39 | AT2G31160 | LSH3 | Protein of unknown function (DUF640) | 1E−54 | CV502385 | 50 |
| #5–42 | AT2G42610 | LSH10 | Protein of unknown function (DUF640) | 1E−64 | TC202398 | 50 |
| #5–43 | AT4G28940 | Phosphorylase superfamily protein | E−113 | TC195965 | 5 |
| #5–44 | AT2G42610 | LSH10 | Protein of unknown function (DUF640) | 2E−70 | TC202398 | 10 |
| #5–46 | AT5G65020 | ANNAT2 | annexin 2 | E−88 | TC208485 | 5 |
| #5–48 | AT5G02380 | MT2B | metallothionein 2B | 0.007 | TC217979 | 50 |
| #6–1 | AT3G15610 | Transducin/WD40 repeat-like superfamily protein | 3E−34 | TC199143 | 5 |
| #6–2 | AT1G25520 | Uncharacterized protein family (UPF0016) | 3E−88 | TC215875 | 5 |
| #6–4 | AT2G19730 | Ribosomal L28e protein family | 4E−59 | TC200167 | 5 |
| #6–5 | AT3G16240 | DELTA-TIP, TIP2;1, DELTA-TIP1, AQP1, ATTIP2;1 | 3E−77 | TC195833 | 5 |
| #6–6 | AT2G42610 | LSH10 | Protein of unknown function (DUF640) | 1E−64 | TC202398 | 10 |
| #6–9 | AT2G31160 | LSH3 | Protein of unknown function (DUF640) | 2E−54 | DR034011 | 5 |
| #6–10 | AT1G58684 | Ribosomal protein S5 family protein | E−111 | TC225313 | 5 |
| #6–14 | AT2G42610 | LSH10 | Protein of unknown function (DUF640) | 1E−64 | TC202398 | 10 |
| #6–19 | AT1G36320 | Unknown protein | 2E−72 | TC198511 | 5 |
| #6–20 | AT5G59240 | Ribosomal protein S8e family protein | 4E−80 | TC205286 | 5 |
| #6–24 | AT5G63570 | GSA1 | glutamate-1-semialdehyde-2,1-aminomutase | E−115 | TC210671 | 5 |
| #6–25 | AT2G42610 | LSH10 | Protein of unknown function (DUF640) | 8E−65 | TC196879 | 10 |
| #6–26 | AT1G63000 | NRS/ER, UER1 | nucleotide-rhamnose synthase/epimerase-reductase | E−151 | TC194991 | 5 |
| #6–30 | AT1G43170 | RP1 | ribosomal protein 1 | 0 | TC196190 | 5 |
| #6–31 | AT5G65260 | RNA-binding (RRM/RBD/RNP motifs) family protein | E−60 | TC196348 | 5 |
| #6–35 | AT3G04400 | Ribosomal protein L14p/L23e family protein | 4E−69 | TC208592 | 5 |
| #6–37 | AT4G14960 | TUA6 | Alpha-tubulin/FtsZ family protein | 0 | TC196128 | 5 |
| #6–38 | AT5G54770 | THI1, TZ, THI4 | thiazole biosynthetic enzyme, chloroplast (ARA6; THI1; THI4) | E−114 | TC194928 | 5 |
| #6–42 | AT5G01870 | Bifunctional inhibitor/lipid transfer protein/seed storage 2S albumin superfamily protein | 4E−13 | TC213303 | 5 |
| #6–44 | AT1G27400 | Ribosomal protein L22p/L17e family protein | 8E−70 | TC214142 | 5 |
| #7–1 | AT3G15610 | Transducin/WD40 repeat-like superfamily protein | E−120 | TC204693 | 5 |
| #7–2 | AT1G54290 | Translation initiation factor SUI1 family protein | 6E−51 | TC202798 | 5 |
| #7–3 | AT5G62220 | ATGT18, GT18 | glycosyltransferase 18 | E−131 | TC195956 | 5 |
| #7–7 | AT1G79920 | Heat shock protein 70 (Hsp 70) family protein | E−133 | TC204446 | 5 |
| #7–8 | AT1G18540 | Ribosomal protein L6 family protein | 3E−75 | TC198860 | 5 |
| #7–9 | AT1G18540 | Ribosomal protein L6 family protein | 2E−67 | TC198860 | 5 |
| #7–14 | No Hit | CK861625 | 5 | ||
| #7–15 | AT3G06030 | ANP3, MAPKKK12, NP3 | NPK1-related protein kinase 3 | E−39 | CK863139 | 5 |
| #7–18 | AT1G26355 | SP1L1 | SPIRAL1-like1 | 9E−22 | TC208981 | 5 |
| #7–19 | AT5G64300 | ATGCH, GCH, ATRIBA1, RFD1 | GTP cyclohydrolase II | 0 | TC207697 | 5 |
| #7–22 | AT5G58620 | Zinc finger (CCCH-type) family protein | E−144 | TC196771 | 5 |
| #7–23 | AT5G63570 | Glutamate-1-semialdehyde 2,1-aminomutase | E−115 | TC210671 | 5 |
| #7–28 | AT4G23895 | Pleckstrin homology (PH) domain-containing protein | 2E−97 | TC194970 | 5 |
| #7–29 | AT1G11680 | Putative obtusifoliol 14-alpha demethylase involved in sterol biosynthesis | E−169 | TC202830 | 5 |
| #7–32 | AT2G47730 | Glutathione transferase belonging to the phi class of GSTs | 4E−69 | TC218491 | 5 |
| #7–33 | AT3G25660 | Amidase family protein | 2E−73 | CV506455 | 5 |
| #7–35 | AT4G13350 | NIG | NSP (nuclear shuttle protein)-interacting GTPase | 4E−07 | TC200976 | 5 |
| #7–36 | AT1G07890 | Cytosolic ascorbate peroxidase APX1 | E−116 | TC195529 | 5 |
| #7–37 | AT4G14300 | RNA-binding (RRM/RBD/RNP motifs) family protein; AtRNP1 | 9.00E−73 | TC200460 | 5 |
| #7–40 | AT1G14685 | BPC2, BBR/BPC2, ATBPC2 | basic pentacysteine 2 | 3E−62 | TC202087 | 5 |
| #7–41 | AT4G38510 | ATPase, V1 complex, subunit B protein | 0 | TC197270 | 5 |
| #7–42 | AT4G37980 | Elicitor-activated gene 3-1 (ELI3-1) | 5E−20 | TC223852 | 5 |
| #7–46 | AT1G36320 | Unknown protein | 2E−76 | TC198511 | 5 |
| #8–4 | AT3G51590 | Lipid transfer protein 12 (LTP12) | 2E−14 | TC221371 | 5 |
| #8–17 | AT4G29410 | Ribosomal L28e protein family | 1E−46 | TC200167 | 5 |
| #8–26 | AT3G53120 | VPS37-1 | Modifier of rudimentary (Mod(r)) protein | 2E−47 | TC205847 | 5 |
| #8–30 | AT4G00100 | ATRPS13A, RPS13, PFL2, RPS13A | ribosomal protein S13A | 2E−78 | TC197315 | 5 |
| #8–44 | AT5G49650 | Xylulose kinase 2 | E−132 | TC211252 | 5 |
| #8–45 | AT3G51590 | Lipid transfer protein 12 (LTP12) | 2E−14 | TC221371 | 5 |
| #8–47 | AT1G67325 | Ran BP2/NZF zinc finger-like superfamily protein | 8E−98 | TC194820 | 5 |
aArabidopsis Genome Initiative number.
bFrom the Arabidopsis Information Resource annotations (http://www.arabidopsis.org).
cFrom DFCI Database.
dConcentration of 3-AT (mM) for screening the clones.
Table A2.
Summary of clones from the yeast three-hybrid screening.
| Number | Ratio (%) | |
|---|---|---|
| Total screened clones | 116 | |
| BLAST-match clones | 89 | 76.7 |
| BLAST-no match clones | 27 | 23.3 |
| Non-redundant clones | 81 | 69.8 |
| Redundant clones | 35 | 30.2 |
| Total singletons | 94 | |
| Functions | ||
| DNA/RNA-binding | 8 | 6.9 |
| Protein synthesis | 19 | 16.4 |
| Metabolism | 28 | 24.1 |
| Transcription activity | 7 | 6.0 |
| Structures | 18 | 15.5 |
| Others | 14 | 12.1 |
| Unknown/uncharacterized | 27 | 23.3 |
The number of the clones and their percentages are presented.
Table A3.
List of the redundant clones and their identities.
| AGIa | Descriptionb | Potatoc | No of clones |
|---|---|---|---|
| AT2G42610 | LSH10 | Protein of unknown function (DUF640) | TC202398 | 10 |
| AT1G58684 | Ribosomal protein S5 | TC225313 | 3 |
| AT1G18540 | Ribosomal protein L6 family protein | TC198860 | 2 |
| AT1G36320 | Unknown protein | TC198511 | 2 |
| AT2G31160 | LSH3 | Protein of unknown function (DUF640) | CV502385 | 2 |
| AT2G40140 | ATSZF2 (salt-inducible zinc finger 2) | TC208753 | 2 |
| AT3G15610 | Transducin/WD40 repeat-like superfamily protein | TC199143 | 2 |
| AT3G25660 | Amidase family protein | CV506455 | 2 |
| AT3G43190 | ATSUS4 | sucrose synthase 4 | TC194622 | 2 |
| AT3G51590 | Lipid transfer protein 12 (LTP12) | TC221371 | 2 |
| AT4G37980 | ELI3-1, ELI3, ATCAD7, CAD7 | elicitor-activated gene 3-1 | TC223852 | 2 |
| AT5G48720 | X-ray induced transcript 1 | BG597043 | 2 |
| AT5G63570 | Glutamate-1-semialdehyde 2,1-aminomutase | TC210671 | 2 |
aArabidopsis Genome Initiative number.
bFrom the Arabidopsis Information Resource annotations (http://www.arabidopsis.org).
cFrom DFCI Database.
Table A4.
List of the screened clones with RNA-binding properties.
| Clone No | AGIa | Descriptionb | Property |
|---|---|---|---|
| #1–9 | AT4G17880 | Basic helix-loop-helix (bHLH) DNA-binding family protein | DNA/RNA-binding |
| #1–38 | AT2G40140 | ATSZF2 (salt-inducible zinc finger 2) | DNA/RNA-binding |
| #3–39 | AT5G61030 | StGRP3 | glycine-rich RNA-binding protein 3 | DNA/RNA-binding |
| #4–13 | AT4G22140 | EBS | PHD finger family protein | DNA/RNA-binding |
| #6–31 | AT5G65260 | RNA-binding (RRM/RBD/RNP motifs) family protein | DNA/RNA-binding |
| #7–22 | AT5G58620 | Zinc finger (CCCH-type) family protein | DNA/RNA-binding |
| #7–37 | AT4G14300 | RNA-binding (RRM/RBD/RNP motifs) family protein; AtRNP1 | DNA/RNA-binding |
| #7–40 | AT1G14685 | BPC2, BBR/BPC2, ATBPC2 | basic pentacysteine 2 | DNA/RNA-binding |
| #1–21 | AT3G43190 | ATSUS4 | sucrose synthase 4 | RBP |
| #1–22 | AT3G43190 | ATSUS4 | sucrose synthase 4 | RBP |
| #3–4 | AT3G13340 | Transducin/WD40 repeat-like superfamily protein | RBP |
| #5–38 | AT1G18080 | ATARCA (Transducin/WD40 repeat-like superfamily protein) | RBP |
| #6–1 | AT3G15610 | Transducin/WD40 repeat-like superfamily protein | RBP |
| #6–37 | AT4G14960 | TUA6 | Alpha-tubulin/FtsZ family protein | RBP |
| #7–1 | AT3G15610 | Transducin/WD40 repeat-like superfamily protein | RBP |
| #7–7 | AT1G79920 | Heat shock protein 70 (Hsp 70) family protein | RBP |
| #1–12 | AT2G37190 | Ribosomal protein L11 | Translation |
| #1–55 | AT1G58684 | Ribosomal protein S5 | Translation |
| #3–7 | AT1G58684 | Ribosomal protein S5 | Translation |
| #3–20 | AT3G05590 | Ribosomal protein L18 | Translation |
| #4–1 | AT5G19510 | Translation elongation factor EF1B/ribosomal protein S6 family protein | Translation |
| #5–4 | AT5G27700 | Ribosomal protein S21e | Translation |
| #5–9 | AT4G09800 | S18 ribosomal protein | Translation |
| #6–4 | AT2G19730 | Ribosomal L28e protein family | Translation |
| #6–10 | AT1G58684 | Ribosomal protein S5 family protein | Translation |
| #6–20 | AT5G59240 | Ribosomal protein S8e family protein | Translation |
| #6–30 | AT1G43170 | RP1 | ribosomal protein 1 | Translation |
| #6–35 | AT3G04400 | Ribosomal protein L14p/L23e family protein | Translation |
| #6–44 | AT1G27400 | Ribosomal protein L22p/L17e family protein | Translation |
| #7–2 | AT1G54290 | Translation initiation factor SUI1 family protein | Translation |
| #7–8 | AT1G18540 | Ribosomal protein L6 family protein | Translation |
| #7–9 | AT1G18540 | Ribosomal protein L6 family protein | Translation |
| #8–17 | AT4G29410 | Ribosomal L28e protein family | Translation |
| #8–30 | AT4G00100 | ATRPS13A, RPS13, PFL2, RPS13A | ribosomal protein S13A | Translation |
| #5–32 | AT1G13950 | EIF-5A, ELF5A-1, ATELF5A-1, EIF5A | eukaryotic elongation factor 5A-1 | Translation/RBP |
aArabidopsis Genome Initiative number.
bFrom the Arabidopsis Information Resource annotations (http://www.arabidopsis.org).
Table A5.
List of primers.
| Name | Primer sequence | Purpose |
|---|---|---|
| B5 3′ UTR F | atcccccgggATACCAGAAAGTCTCGTA | Cloning of 3′ UTR of StBEL5 |
| B5 3′ UTR R | cagcccgggGCTAATCTAATAATGATA | into pIIIA/MS2-1 for library screening |
| B5 3′ UTR F w T3 | AATTAACCCTCACTAAAGGGAGAATACCAGAAAGTCTCGTA | In vitro transcription of 3′ UTR of StBEL5 |
| B5 D1 F | atcccccgggATACCAGAAAGTCTCGTA | Cloning of physical dissection (D1) of 3′ UTR |
| B5 D1 R | cagcccgggTATACTAGAAAGTGTGCTTT | of StBEL5 into pIIIA/MS2-1 for validation |
| B5 T2 F | atcccccgggTATAGTATAGAAAAGAAGA | Cloning of physical dissection (T2) of 3′ UTR |
| B5 T2 R | cagcccgggTACTACTAGTTGTATCAATCT | of StBEL5 into pIIIA/MS2-1 for validation |
| B5 UA F | atcccccgggTATTTTTTTTCTTTTGGGTTG | Cloning of physical dissection (UA-rich) of 3′ UTR |
| B5 UA R | cagcccgggTACAAAATATTTCAAATTATA | of StBEL5 into pIIIA/MS2-1 for validation |
| B5 T1 F | atcccccgggACTCTTATATTGTG | Cloning of PT motif (T1) of 3′ UTR of StBEL5 |
| B5 T1 R | atcccccgggAAAGAAGTATATAT | into pIIIA/MS2-1 for validation |
| B5 D1 F w T3 | AATTAACCCTCACTAAAGGGAGAATACCAGAAAGTCTCGTA | In vitro transcription of D1 of StBEL5 |
| B5 D2 F w T3 | AATTAACCCTCACTAAAGGGAGATATACTTCTTTTTTTTATAG | In vitro transcription of D2 of StBEL5 |
| B5 D3 F w T3 | AATTAACCCTCACTAAAGGGAGATGGAAACTAGCTATAGTATA | In vitro transcription of D3 of StBEL5 |
| B5 T1 F w T3 | AATTAACCCTCACTAAAGGGAGAACTCTTATATTGTG | In vitro transcription of T1 of StBEL5 |
| B5RBP2 F | tacggatccATGGCTTTCTTTAACAAAGC | Cloning of B5RBP2 into pET28a for |
| B5RBP2 R | cgatgtcgacGGCTCTTCTGTCTGCATAAT | expression of His-fusion protein |
| B5RBP3 F | tacggatccATGTCAAATTTTGATAGAGG | Cloning of B5RBP3 into pET28a for |
| B5RBP3 R | cgatgtcgacAGAAAATGGGTGAGAAGAAG | expression of His-fusion protein |
| B5RBP5 F | tacggatccATGGAGGACGACGTTGAGAT | Cloning of B5RBP5 into pET28a for |
| B5RBP5 R | cgatgtcgacGAAGTAGGGGCTGTAACGCA | expression of His-fusion protein |
| pGAD-C1 Seq | CGATGATGAAGATACCCCAC | Sequencing of the screened clones |
Note: Lower case letters represent restriction enzyme sites used for cloning.
Footnotes
References
- Banerjee A. J., Lin T., Hannapel D. J. (2009). Untranslated regions of a mobile transcript mediate RNA metabolism. Plant Physiol. 151, 1831–1843 10.1104/pp.109.144428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A. K., Yu Y., Chatterjee M., Suh S. G., Miller W. A., Hannapel D. J. (2006). Dynamics of a mobile RNA of potato involved in a long-distance signaling pathway. Plant Cell 18, 3443–3457 10.1105/tpc.106.042473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroja-Fernandez E., Munoz F. J., Montero M., Etxeberria E., Sesma M. T., Ovecka M., Bahaji A., Ezquer I., Li J., Prat S., Pozueta-Romero J. (2009). Enhancing sucrose synthase activity in transgenic potato (Solanum tuberosum L.) tubers results in increased levels of starch, ADPglucose and UDPglucose and total yield. Plant Cell Physiol. 50, 1651–1662 10.1093/pcp/pcp108 [DOI] [PubMed] [Google Scholar]
- Barreau C., Paillard L., Osborne H. B. (2006). AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 33, 7138–7150 10.1093/nar/gki1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein D. S., Buter N., Stumpf C., Wickens M. (2002). Analyzing mRNA-protein complexes using a yeast three-hybrid system. Methods 26, 123–141 10.1016/S1046-2023(02)00015-4 [DOI] [PubMed] [Google Scholar]
- Bieniawska Z., Paul Barratt D. H., Garlick A. P., Thole V., Kruger N. J., Martin C., Zrenner R., Smith A. M. (2007). Analysis of the sucrose synthase gene family in Arabidopsis. Plant J. 49, 810–828 10.1111/j.1365-313X.2006.03011.x [DOI] [PubMed] [Google Scholar]
- Brown R. S. (2005). Zinc finger proteins: getting a grip on RNA. Curr. Opin. Struct. Biol. 15, 94–98 10.1016/j.sbi.2005.01.006 [DOI] [PubMed] [Google Scholar]
- Burd C. G., Dreyfuss G. (1994). RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 13, 1197–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campalans A., Kondorosi A., Crespi M. (2004). Enod40, a short open reading frame-containing mRNA, induces cytoplasmic localization of a nuclear RNA binding protein in Medicago truncatula. Plant Cell 16, 1047–1059 10.1105/tpc.019406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassiday L. A., Maher J. L., III. (2003). Yeast genetic selections to optimize RNA decoys for transcription factor NF-kappa B. Proc. Natl. Acad. Sci. U.S.A. 100, 3930–3935 10.1073/pnas.0736013100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Banerjee A. K., Hannapel D. J. (2004). The tandem complex of BEL and KNOX partners is required for transcriptional repression of ga20ox1. Plant J. 38, 276–284 10.1111/j.1365-313X.2004.02048.x [DOI] [PubMed] [Google Scholar]
- Chen H., Rosin F. M., Prat S., Hannapel D. J. (2003). Interacting transcription factors from the TALE superclass regulate tuber formation. Plant Physiol. 13, 1391–1404 10.1104/pp.103.022434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corral-Debrinski M., Blugeon C., Jacq C. (2000). In yeast, the 3′ untranslated region or the presequence of ATM1 is required for the exclusive localization of its mRNA to the vicinity of mitochondria. Mol. Cell. Biol. 20, 7881–7892 10.1128/MCB.20.21.7881-7892.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottier S., Mönig T., Wang Z., Svoboda J., Boland W., Kaiser M., Kombrink E. (2011). The yeast three-hybrid system as an experimental platform to identify proteins interacting with small signaling molecules in plant cells: potential and limitations. Front. Plant Sci. 2:101. 10.3389/fpls.2011.00101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusack S. (1999). RNA-protein complexes. Curr. Opin. Struct. Biol. 9, 66–73 10.1016/S0959-440X(99)80009-8 [DOI] [PubMed] [Google Scholar]
- Doroshenk K. A., Crofts A. J., Morris R. T., Wyrick J. J., Okita T. W. (2009). Proteomic analysis of cytoskeleton-associated RNA binding proteins in developing rice seed. J. Proteome Res. 8, 4641–4653 10.1021/pr900537p [DOI] [PubMed] [Google Scholar]
- Dreyfuss G., Kim V. N., Kataoka N. (2002). Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 3, 195–205 10.1038/nrm760 [DOI] [PubMed] [Google Scholar]
- Edwards T. A., Pyle S. E., Wharton R. P., Aggarwal A. K. (2001). Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell 105, 281–289 10.1016/S0092-8674(01)00318-X [DOI] [PubMed] [Google Scholar]
- Fallahi H., Scofield G. N., Badger M. R., Chow W. S., Furbank R. T., Ruan Y. L. (2008). Localization of sucrose synthase in developing seed and siliques of Arabidopsis thaliana reveals diverse roles for SUS during development. J. Exp. Bot. 59, 3283–3295 10.1093/jxb/ern180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff N. V. (2002). RNA-binding proteins in plants: the tip of an iceberg. Curr. Opin. Plant Biol. 5, 452–459 10.1016/S1369-5266(02)00280-7 [DOI] [PubMed] [Google Scholar]
- Ferrandon D., Elphick L., Nusslein-Volhard C., St Johnston D. (1994). Staufen protein associates with the 3′ UTR of bicoid mRNA to form particles that move in a microtubule-dependent manner. Cell 79, 1221–1223 10.1016/0092-8674(94)90013-2 [DOI] [PubMed] [Google Scholar]
- Fu H., Kim S. Y., Park W. D. (1995). High-level tuber expression and sucrose inducibility of a potato Sus4 sucrose synthase gene require 5′ and 3′ flanking sequences and the leader intron. Plant Cell 7, 1387–1394 10.2307/3870129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonsalvez G. B., Lehmann K. A., Ho D. K., Stanitsa E. S., Williamson J. R., Long R. M. (2003). RNA-protein interactions promote asymmetric sorting of the ASH1 mRNA ribonucleoprotein complex. RNA 9, 1383–1399 10.1261/rna.5120803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham B. K., Brandom J. L., Xoconostle-Cázares B., Ringgold V., Lough T. J., Lucas W. J. (2009). A polypyrimidine tract binding protein, pumpkin RBP50, forms the basis of a phloem-mobile ribonucleoprotein complex. Plant Cell 21, 197–215 10.1105/tpc.108.061317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haywood V., Yu T. S., Huang N. C., Lucas W. J. (2005). Phloem long-distance trafficking of GIBBERELLIC ACID-INSENSITIVE RNA regulates leaf development. Plant J. 42, 49–68 10.1111/j.1365-313X.2005.02351.x [DOI] [PubMed] [Google Scholar]
- Henderson A., Hershey J. W. (2011). Eukaryotic translation initiation factor (eIF) 5A stimulates protein synthesis in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 108, 6415–6419 10.1073/pnas.1008150108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook B., Bernstein D., Zhang B., Wickens M. (2005). RNA–protein interactions in the yeast three-hybrid system: affinity, sensitivity, and enhanced library screening. RNA 11, 227–233 10.1261/rna.7202705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt A. G., Xu R., Addepalli B., Rao S., Forbes K. P., Meeks L. R., Xing D., Mo M., Zhao H., Bandyopadhyay A., Dampanaboina L., Marion A., Von Lanken C., Li Q. Q. (2008). Arabidopsis mRNA polyadenylation machinery: comprehensive analysis of protein-protein interactions and gene expression profiling. BMC Genomics 9, 220. 10.1186/1471-2164-9-220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang M. S., Kim S. H., Lee J. H., Bae J. M., Paek K. H., Park Y. I. (2005). Evidence for interaction between the 2a polymerase protein and the 3a movement protein of cucumber mosaic virus. J. Gen. Virol. 86, 3171–3177 10.1099/vir.0.81139-0 [DOI] [PubMed] [Google Scholar]
- Irion U., St Johnston D. (2007). Bicoid RNA localization requires specific binding of an endosomal sorting complex. Nature 445, 554–558 10.1038/nature05503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R. P. (2001). mRNA localization: message on the move. Nat. Rev. Mol. Cell Biol. 2, 247–256 10.1038/35067016 [DOI] [PubMed] [Google Scholar]
- Kehr J., Buhtz A. (2008). Long distance transport and movement of RNA through the phloem. J. Exp. Bot. 59, 85–92 10.1093/jxb/erm176 [DOI] [PubMed] [Google Scholar]
- Kyrpides N. C., Woese C. R., Ouzounis C. A. (1996). KOW: a novel motif linking a bacterial transcription factor with ribosomal proteins. Trends Biochem. Sci. 21, 425–426 10.1016/S0968-0004(96)30036-4 [DOI] [PubMed] [Google Scholar]
- Lai W. S., Parker J. S., Grissom S. F., Stumpo D. J., Blackshear P. J. (2006). Novel mRNA targets for tristetraprolin (TTP) identified by global analysis of stabilized transcripts in TTP-deficient fibroblast. Mol. Cell Biol. 26, 9196–9208 10.1128/MCB.00945-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laity J. H., Lee B. M., Wright P. E. (2001). Zinc finger proteins: new insights into structural and functional diversity. Curr. Opin. Struct. Biol. 11, 39–46 10.1016/S0959-440X(00)00167-6 [DOI] [PubMed] [Google Scholar]
- Lee H. K., Jeong S. (2006). Beta-catenin stabilizes cyclooxygenase-2 mRNA by interacting with AU-rich elements of 3′ UTR. Nucleic Acids Res. 34, 5705–5714 10.1093/nar/gkj083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., He K., Stolc V., Lee H., Figueroa P., Gao Y., Tongprasit W., Zhao H., Lee I., Deng X. (2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19, 731–749 10.1105/tpc.106.047688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Gu M., Shi N., Zhang H., Yang X., Osman T., Liu Y., Wang H., Vatish M., Jackson S., Hong Y. (2011). Mobile FTmRNA contributes to the systemic florigen signalling in floral induction. Sci. Rep. 1, 73. 10.1038/srep00088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M. K., Lee Y. J., Lough T. J., Phinney B. S., Lucas W. J. (2009). Analysis of the pumpkin phloem proteome provides insights into angiosperm sieve tube function. Mol. Cell Proteomics 8, 343–356 [DOI] [PubMed] [Google Scholar]
- Ling A. S., Trotter J. R., Hendriks E. F. (2011). A zinc finger protein, TbZC3H20, stabilizes two developmentally regulated mRNAs in Trypanosomes. J. Biol. Chem. 286, 20152–20162 10.1074/jbc.M110.139261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K. J., Huang N. C., Liu Y. S., Lu C. A., Yu T. S. (2012). Long-distance movement of Arabidopsis FLOWERING LOCUS T RNA participates in systemic floral regulation. RNA Biol. 1, 9. [DOI] [PubMed] [Google Scholar]
- Lunde B. M., Moore C., Varani G. (2007). RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 8, 479–490 10.1038/nrm2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Miura E., Ham B. K., Cheng H. W., Lee Y. J., Lucas W. J. (2010). Pumpkin eIF5A isoforms interact with components of the translational machinery in the cucurbit sieve tube system. Plant J. 64, 536–550 10.1111/j.1365-313X.2010.04347.x [DOI] [PubMed] [Google Scholar]
- Mahajan A., Bhogale S., Kang I. H., Hannapel D. J., Banerjee A. K. (2012). The mRNA of a Knotted1-like transcription factor of potato is phloem mobile. Plant Mol. Biol. 79, 595–608 10.1007/s11103-012-9931-0 [DOI] [PubMed] [Google Scholar]
- Mangeon A., Junqueira R. M., Sachetto-Martins G. (2010). Functional diversity of the plant glycine-rich proteins superfamily. Plant Signal. Behav. 5, 99–104 10.4161/psb.5.2.10336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniataki E., Martinez de Alba E., Sägesser R., Tabler M., Tsagris M. (2003). Viroid RNA systemic spread may depend on the interaction of a 71-nucleotide bulged hair-pin with the host protein VirP1. RNA 9, 346–354 10.1261/rna.2162203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore F. L., Jaruzelska J., Fox M. S., Urano J., Firpo M. T., Turek P. J., Dorfman D. M., Pera R. A. (2003). HumanPumilio-2 is expressed in embryonic stemcells and germcells and interacts with DAZ (Deleted in AZoospermia) and DAZ-like proteins. Proc. Natl. Acad. Sci. U.S.A. 100, 538–543 10.1073/pnas.1835831100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori D., Sasagawa N., Kino Y., Ishiura S. (2008). Quantitative analysis of CUG-BP1 binding to RNA repeats. J. Biochem. 143, 377–383 10.1093/jb/mvm230 [DOI] [PubMed] [Google Scholar]
- Padmanabhan K., Richter J. D. (2006). Regulated Pumilio-2 binding controls RINGO/Spy mRNA translation and CPEB activation. Genes Dev. 20, 199–209 10.1101/gad.1383106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peat T. S., Newman J., Waldo G. S., Berendzen J., Terwilliger T. C. (1998). Structure of translation initiation factor 5A from Pyrobaculum aerophilum at 1.75 A resolution. Structure 6, 1207–1214 10.1016/S0969-2126(98)00120-8 [DOI] [PubMed] [Google Scholar]
- Piganeau N., Schroeder R. (2006). Identification and detection of RNA-RNA interactions using the yeast RNA hybrid system. Nat. Protoc. 1, 689–694 10.1038/nprot.2006.111 [DOI] [PubMed] [Google Scholar]
- Xu X., Pan S., Cheng S., Zhang B., Mu D., Ni P., Zhang G., Yang S., Li R., Wang J., Orjeda G., Guzman F., Torres M., Lozano R., Ponce O., Martinez D., De la Cruz G., Chakrabarti S. K., Patil V. U., Skryabin K. G., Kuznetsov B. B., Ravin N. V., Kolganova T. V., Beletsky A. V., Mardanov A. V., Di Genova A., Bolser D. M., Martin D. M., Li G., Yang Y., Kuang H., Hu Q., Xiong X., Bishop G. J., Sagredo B., Mejía N., Zagorski W., Gromadka R., Gawor J., Szczesny P., Huang S., Zhang Z., Liang C., He J., Li Y., He Y., Xu J., Zhang Y., Xie B., Du Y., Qu D., Bonierbale M., Ghislain M., Herrera Mdel R., Giuliano G., Pietrella M., Perrotta G., Facella P., O’Brien K., Feingold S. E., Barreiro L. E., Massa G. A., Diambra L., Whitty B. R., Vaillancourt B., Lin H., Massa A. N., Geoffroy M., Lundback S., DellaPenna D., Buell C. R., Sharma S. K., Marshall D. F., Waugh R., Bryan G. J., Destefanis M., Nagy I., Milbourne D., Thomson S. J., Fiers M., Jacobs J. M., Nielsen K. L., Sønderkær M., Iovene M., Torres G. A., Jiang J., Veilleux R. E., Bachem C. W., de Boer J., Borm T., Kloosterman B., van Eck H., Datema E., Hekkert B. L., Goverse A., van Ham R. C., Visser R. G., Potato Genome Sequencing Consortium (2011). Genome sequence and analysis of the tuber crop potato. Nature 475, 189–195 10.1038/nature10158 [DOI] [PubMed] [Google Scholar]
- Saunders C., Cohen R. S. (1999). The role of oocyte transcription, the 5′ UTR, and translation repression and derepression in Drosophila gurken mRNAand protein localization. Mol. Cell 3, 43–45 10.1016/S1097-2765(00)80173-2 [DOI] [PubMed] [Google Scholar]
- Seay D., Hook B., Evans K., Wickens M. (2006). A three-hybrid screen identifies mRNAs controlled by a regulatory protein. RNA 12, 1594–1600 10.1261/rna.145306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta D. J., Zhang B., Kraemer B., Pochart P., Fields S., Wickens M. (1996). A three-hybrid system to detect RNA-protein interactions in vivo. Proc. Natl. Acad. Sci.U.S.A. 93, 8496–8501 10.1073/pnas.93.16.8496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner T., Kaiser J. T., Marinkovic S., Huber R., Wahl M. C. (2002). Crystal structures of transcription factor NusG in light of its nucleic acid- and protein-binding activities. EMBO J. 21, 4641–4653 10.1093/emboj/cdf455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf C. R., Kimble J., Wickens M. (2008). A Caenorhabditis elegans PUF protein family with distinct RNA binding specificity. RNA 14, 1550–1557 10.1261/rna.1095908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Jiang H., Xu Y., Li H., Wu X., Xie Q., Li C. (2007). The CCCH-type zinc finger proteins AtSZF1 and AtSZF2 regulate salt stress responses in Arabidopsis. Plant Cell Physiol. 48, 1148–1158 10.1093/pcp/pcm088 [DOI] [PubMed] [Google Scholar]
- Takeda S., Hanano K., Kariya A., Shimizu S., Zhao L., Matsui M., Tasaka M., Aida M. (2011). Cup-shaped cotyledon1 transcription factor activates the expression of LSH4 and LSH3, two members of the ALOG gene family, in shoot organ boundary cells. Plant J. 66, 1066–1077 10.1111/j.1365-313X.2011.04571.x [DOI] [PubMed] [Google Scholar]
- Thio G. L., Ray R. P., Barcelo G., Schupbach T. (2000). Localization of gurken RNA in Drosophila oogenesis requires elements in the 5′ and 3′ regions of the transcript. Dev. Biol. 15, 435–446 10.1006/dbio.2000.9690 [DOI] [PubMed] [Google Scholar]
- Wachter A., Rühl C., Stauffer E. (2012). The role of polypyrimidine tract-binding proteins and other hnRNP proteins in plant splicing regulation. Front. Plant Sci. 3:81. 10.3389/fpls.2012.00081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurster S. E., Maher J., III. (2010). Selections that optimize RNA display in the yeast three-hybrid system. RNA 16, 253–258 10.1261/rna.1880410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu A., Chen K. Y. (2001). Hypusine is required for a sequence-specific interaction of eukaryotic initiation factor 5A with post systematic evolution of ligands by exponential enrichment RNA. J. Biol. Chem. 276, 2555–2561 10.1074/jbc.M008973200 [DOI] [PubMed] [Google Scholar]
- Xu A., Jao D. L.-E., Chen K. Y. (2004). Identification of mRNA that binds to eukaryotic initiation factor 5A by affinity co-purification and differential display. Biochem. J. 384, 585–590 10.1042/BJ20041232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanelli C. F., Valentini S. R. (2007). Is there a role for eIF5A in translation? Amino Acids 33, 351–358 10.1007/s00726-007-0533-0 [DOI] [PubMed] [Google Scholar]
- Zhao L., Nakazawa M., Takase T., Manabe K., Kobayashi M., Seki M., Shinozaki K., Matsui M. (2004). Overexpression of LSH1, a member of an uncharacterised gene family, causes enhanced light regulation of seedling development. Plant J. 37, 694–706 10.1111/j.1365-313X.2003.01993.x [DOI] [PubMed] [Google Scholar]
- Ziemienowicz A., Haasen D., Staiger D., Merkle T. (2003). Arabidopsis transportin1 is the nuclear import receptor for the circadian clock-regulated RNA-binding protein AtGRP7. Plant Mol. Biol. 53, 201–212 10.1023/B:PLAN.0000009288.46713.1f [DOI] [PubMed] [Google Scholar]
- Zrenner R., Salanoubat M., Willmitzer L., Sonnewald U. (1995). Evidence of the crucial role of sucrose synthase for sink strength using transgenic potato plants (Solanum tuberosum L.). Plant J. 7, 97–107 10.1046/j.1365-313X.1995.07010097.x [DOI] [PubMed] [Google Scholar]