Abstract
Ollier disease and Maffucci syndrome are non-hereditary skeletal disorders characterized by multiple enchondromas (Ollier disease) combined with spindle cell hemangiomas (Maffucci syndrome). We report somatic heterozygous IDH1 (R132C and R132H) or IDH2 (R172S) mutations in 87% of enchondromas, benign cartilage tumors, and in 70% of spindle cell hemangiomas, benign vascular lesions. In total, 35 of 43 (81%) patients with Ollier disease and 10 of 13 (77%) patients with Maffucci syndrome carried IDH1 (98%) or IDH2 (2%) mutations in their tumors. Fourteen of sixteen patients displayed identical mutations in separate lesions. Immunohistochemistry for mutant R132H IDH1 protein suggested intraneoplastic and somatic mosaicism. IDH1 mutations in cartilage tumors are associated with hypermethylation and downregulation of expression of several genes. Mutations were also found in 40% of solitary central cartilaginous tumors and in four chondrosarcoma cell lines, enabling functional studies to assess the role of IDH1 and IDH2 mutations in tumor formation.
Enchondroma is a benign cartilage forming tumor within the medullary cavity of the bone 1-3. Patients with enchondromatosis syndrome, which encompasses seven major subtypes, develop multiple enchondromas. Most common are non-hereditary Ollier disease (subtype I) and Maffucci syndrome (subtype II), the latter distinguished by spindle cell hemangioma in addition to multiple enchondromas 1, 3. Malignant transformation of enchondromas towards chondrosarcomas occurs in >30% of the patients 3, 4.
Genome-wide screens have not identified a causative gene 5-9. These patients have an increased incidence of gliomas 3, 10 and juvenile granulosa cell tumors 3, 11-13. IDH1 and, more rarely IDH2 mutations in gliomas 14, 15, 16 and GNAS activating mutations in juvenile granulosa cell tumors 17 have been reported. Interestingly, IDH1 and IDH2 mutations were recently reported in solitary central and periosteal enchondromas and chondrosarcomas, including few tumors from patients with enchondromatosis 18. The possibility of GNAS mutations in enchondromas and chondrosarcomas has not been explored.
We therefore assessed whether IDH1, IDH2, or GNAS mutations may cause enchondroma and spindle cell hemangioma formation in Ollier disease and Maffucci syndrome. Sequence analysis of hotspot positions using lesional tissue from 43 patients with Ollier disease revealed that heterozygous R132C IDH1 (c.394C>T), R132H IDH1 (c.395G>A) or R172S IDH2 (c.516G>C) (Human Genome Variation Society) mutations were present in 33 patients (78%) (Supplementary Fig.1a-c). In Maffucci syndrome, 7 out of 13 patients (54%) carried R132C IDH1 mutations. Mutations were absent in DNA from patients' blood, muscle or saliva (Supplementary Fig.1b). Mutations in GNAS were absent.
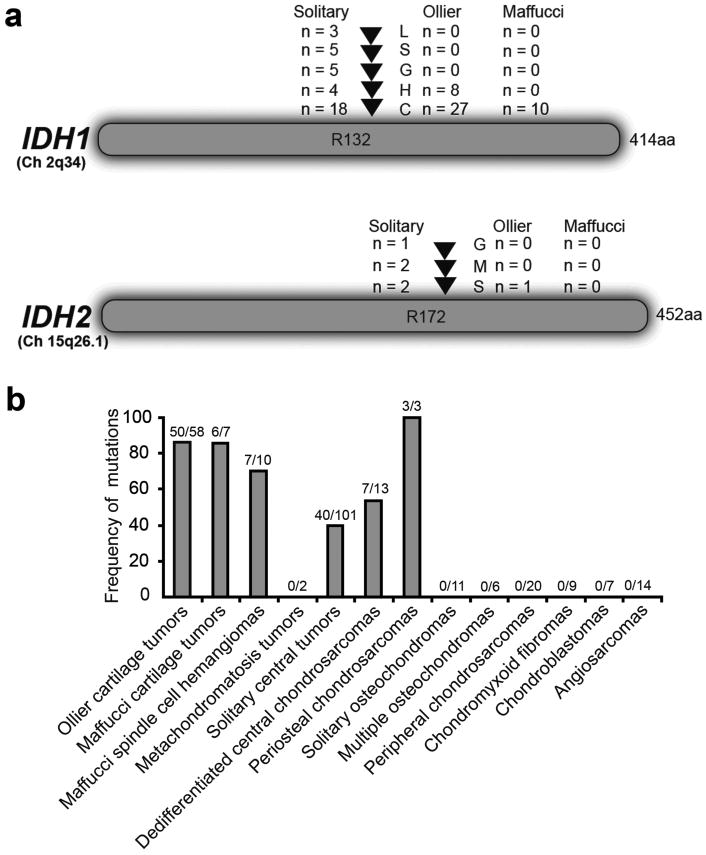
An additional 8 tumors demonstrated sub-threshold peaks at the position where R132C or R132H IDH1 mutations can be expected, suggesting that the mutant allele might be present in a small subpopulation of the tumor cells at the limits or below the detection level of Sanger sequencing. We therefore performed a hydrolysis probes assay, capable of detecting as low as 1% of mutant allele, for the detection of R132C and R132H IDH1 mutations 19, 20. Mutations were confirmed in 7 of 8 tumors (Supplementary Fig.1d-g), while from 1 tumor DNA was no longer available. Thus, in total 35 out of 43 (81%) and 10 of 13 (77%) patients with Ollier disease and Maffucci syndrome, respectively, showed IDH1 or IDH2 mutations (Fig.1a, Table 1, and Supplementary Table 1). Frequency of mutations in tumors is shown in Fig.1b.
Figure 1. Frequency of IDH1 and IDH2 mutations.
a) Distribution of the different R132 IDH1 and R172 IDH2 mutations among the patients with Ollier disease, Maffucci syndrome and solitary tumors. b) Frequency of somatic heterozygous IDH (IDH1 and IDH2) mutations in tumors of patients with Ollier disease and Maffucci syndrome, in comparison with different subtypes of solitary cartilaginous tumors and angiosarcomas.
Table 1. Results of IDH1 and IDH2 mutation analysis.
| Total | Gender (M:F) (median age, years) | IDH1 mutation (%) | R132C IDH1 (CGT>TGT) | R132H IDH1 (CGT>CAT) | IDH2 mutation (%) | Total IDH1+IDH2 mutation | |
|---|---|---|---|---|---|---|---|
| Ollier Disease | |||||||
| Number of patients | 43 | 21:21*(24) | 34 (79%) | 1 (2%) | 35 (81%) | ||
| Enchondroma | 25 | 22 (88%) | 15 (68%) | 7 (32%) | 0 | 22 (88%) | |
| Chondrosarcoma grade I | 23 | 20 (87%) | 18 (90%) | 2 (10%) | 0 | 20 (87%) | |
| Chondrosarcoma grade II | 8 | 5 (63%) | 5 (100%) | 0 | 1 (12%) | 6 (75%) | |
| Chondrosarcoma grade III | 2 | 1 (50%) | 1 (100%) | 0 | 1 (50%) | 2 (100%) | |
| Total number of tumors | 58 | 48 (83%) | 39 (81%) | 9 (19%) | 2 (3%) | 50 (86%) | |
| Maffucci Syndrome | |||||||
| Number of patients | 13 | 5:8 (15) | 10 (77%) | 0 | |||
| Enchondroma | 5 | 4 (80%) | 4 (100%) | 0 | 0 | ||
| Chondrosarcoma grade I | 1 | 1 (100%) | 1 (100%) | 0 | 0 | ||
| Chondrosarcoma grade II | 1 | 1 (100%) | 1 (100%) | 0 | 0 | ||
| Spindle cell hemangioma | 10 | 7 (70%) | 7 (100%) | 0 | 0 | ||
| Total number of tumors | 17 | 13 (76%) | 13 (100%) | 0 | 0 | ||
| Solitary Tumors | |||||||
| Enchondroma | 9 | 3 (33%) | 2 (67%) | 1 (33%) | 2 (22%) | 5 (56%) | |
| Central chondrosarcoma grade I | 20 | 7** (35%) | 2 (29%) | 2 (29%) | 0 | 7 (35%) | |
| Central chondrosarcoma grade II | 57 | 18**(32%) | 9 (50%) | 1 (6%) | 3 (5%) | 21 (37%) | |
| Central chondrosarcoma grade III | 15 | 7** (47%) | 5 (71%) | 0 | 0 | 7 (47%) | |
| Dedifferentiated chondrosarcoma | 13 | 6** (46%) | 3 (50%) | 1 (17%) | 1 (8%) | 7 (54%) | |
| Periosteal chondrosarcoma | 3 | 3 (100%) | 3 | 0 | 0 | 3 (100%) |
unknown gender for one patient
also other types of mutations than R132C or R132H
Other subtypes of enchondromatosis syndrome are known to be caused by mutations in PTPN11 (metachondromatosis) 21, 22, ACP5 (spondyloenchondrodysplasia) 23, 24 and PTHLH duplication (symmetrical enchondromatosis) 25. Mutations in PTH1R, involved in enchondral bone formation, are found in ∼ 8% of patients with Ollier disease, but not in patients with Maffucci syndrome 5-7. Previously, our patients were reported negative for PTPN11 mutations 22. Here we did not detect PTH1R mutations in a screen of 35 patients. A custom-made Agilent tiling array (Supplementary Table 2) analysis did not find evidence of losses or gains of IDH1, IDH2, PTHLH, PTPN11, PTH1R, EXT1, EXT2 and ACP5. Thus, even though patients with enchondromatosis syndromes demonstrate overlapping clinical features, they appear to be genetically discrete entities, with the exception of Ollier disease and Maffucci syndrome, which we have now shown to contain IDH1 or IDH2 mutations.
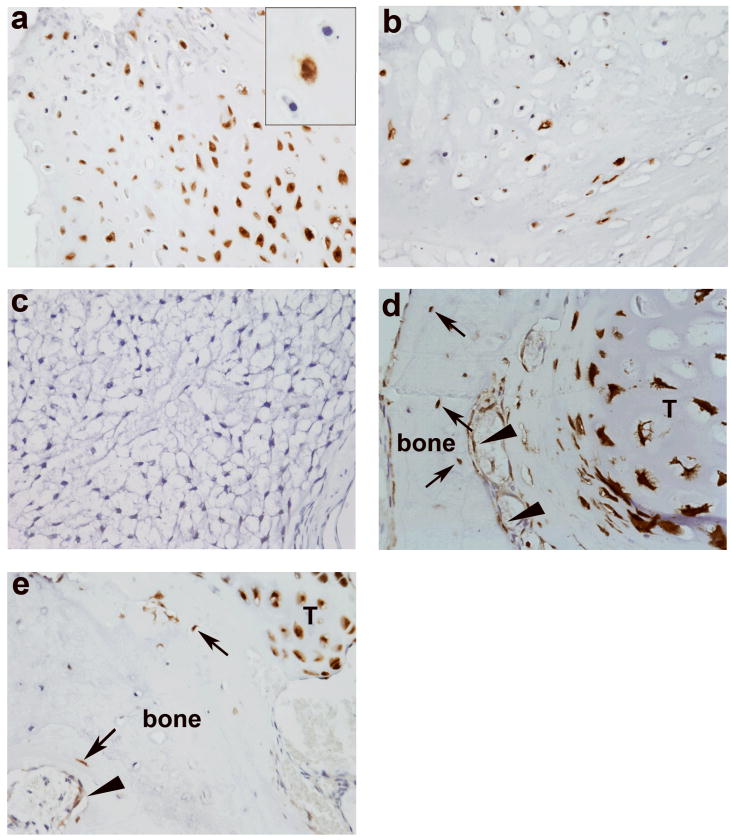
Since these disorders are not inherited and the enchondromas are often unilateral, we further hypothesized that mutations may occur in a somatic mosaic fashion. Fourteen of sixteen patients (88%) possessed identical mutations, including rare variants, in more than one tumor (Supplementary Table 1). We additionally used immunohistochemistry to determine the distribution of the R132H IDH1 mutant protein. Of 68 tumors from patients with Ollier disease, 17 tumors (25%) showed mutant protein expression while 51 (75%) tumors were negative (Table 2, Fig.2). We observed a mixture of cells without (wild-type) and with expression of R132H IDH1 mutant protein (of the same histologic type, i.e., not including entrapped elements and supporting elements), which we refer to as intraneoplastic mosaicism (Fig.2a and b). The percentage of positive tumor cells ranged from 50% to 95%. Intraneoplastic mosaicism is also described for other benign bone tumors. In fibrous dysplasia, experimental evidence showed that both normal and GNAS mutated cells were needed to develop fibrous dysplasia-like lesions 26. Also, in osteochondromas, benign cartilaginous tumors arising at the surface of the bone that are caused by mutations in EXT1 or EXT2, a mixture of EXT wild-type and EXT mutated cells was observed 27-30. EXT is involved in heparan sulphate biosynthesis, and it is hypothesized that EXT mutated cells that are deficient in heparan sulphate, need heparan sulphate from neighboring cells for cellular signaling and survival 31, 32.
Table 2. Immunohistochemistry for R132H mutant protein expression.
| Total nr of tumors | R132H positive | |
|---|---|---|
| Ollier Disease | ||
| Enchondroma | 46 | 14/43*(32%) |
| Chondrosarcoma grade I | 22 | 3/17* (18%) |
| Chondrosarcoma grade II | 10 | 0/8* |
| Maffucci syndrome | ||
| Enchondroma | 9 | 0/9 |
| Spindle cell hemangioma | 14 | 0/14 |
| Solitary tumors | ||
| Enchondroma | 19 | 4/19 (21%) |
| Central chondrosarcoma grade I | 42 | 4/38* (10%) |
| Central chondrosarcoma grade II | 36 | 1/32* (3%) |
| Central chondrosarcoma grade III | 14 | 0/11* |
| Central dedifferentiated chondrosarcoma | 26 | 1/24* (4%) |
| Periosteal chondrosarcoma | 6 | 1/6 (17%) |
| Solitary osteochondroma | 20 | 0/17* |
| Multiple osteochondroma | 7 | 0/7 |
| Peripheral chondrosarcoma | 45 | 0/35* |
| Peripheral dedifferentiated chondrosarcoma | 16 | 0/16 |
| Conventional hemangioma | 3 | 0/3 |
| Hemangioendothelioma | 2 | 0/2 |
| High grade angiosarcoma of bone | 44 | 0/44 |
| High grade angiosarcoma of soft tissue | 22 | 0/22 |
| Controls | ||
| Normal growth plate | 3 | 0/3 |
| Articular cartilage | 3 | 0/3 |
| Normal bone | 12 | 0/12 |
not all tumors included were evaluable due to tissue loss on tissue microarray
Figure 2. Immunohistochemistry for R132H IDH1 mutant protein.
a,b) Enchondroma (L1490) of patient with Ollier disease demonstrating strong cytoplasmic and nuclear staining of R132H IDH1 mutant protein. Note the mixture of wild-type and mutated cells indicating intraneoplastic mosaicism. Overall the percentage of positive tumor cells ranged from 50% to 95%. Insets show vitality of the negative cells at higher magnification. c) Grade II chondrosarcoma negative for R132H IDH1 mutant protein. d and e) Enchondromas from patients with Ollier disease demonstrating occasional positive cells in the surrounding normal bone. Some positive osteocytes (arrows) and osteoblasts (arrowheads) are seen. T: tumor tissue. (Magnification 400×)
We additionally studied the surrounding normal tissue of Ollier related and solitary mutated tumors and surprisingly, a very low frequency (on average <1%) of mutant protein was observed in osteoblasts, osteocytes, adipocytes and fibroblasts (Fig. 2d and e). Hydrolysis probes assay could be performed on DNA isolated from one normal bone of patient with Ollier disease, which was negative. Mutant R132H IDH1 protein was absent in 12 bones resected for reasons other than chondrosarcoma, normal growth plates and articular cartilage (Table 2). Therefore, our current data support somatic mosaicism, similar to somatic mosaic GNAS mutations causing polyostotic fibrous dysplasia 33, 34. Unfortunately, the nature of the material (decalcified paraffin-embedded bone tissue) and the occurrence of the mutation in single scattered cells do not allow verification using other techniques. However, the R132H IDH1 antibody was shown to be highly reliable in glioma diagnosis 35 and correlated well with sequence analysis in our series.
Twelve tumors were negative for IDH1 or IDH2 hotspot mutations. For 5 of these, all exons were sequenced and no mutations were identified. This was not surprising because only R132 IDH1 and R172 IDH2 mutations have been identified in other IDH-associated tumors. It is possible that because of intralesional mosaicism, only small sub-fraction of tumor cells contain the IDH1 or IDH2 hotspot mutations, which may be below the detection level of the techniques used. Alternatively, mutations in other genes such as TET2, which is mutually exclusive with IDH1 or IDH2 mutations in cases of acute myeloid leukemia 36, might be involved 18, 37.
Recently, point mutations in IDH1 or IDH2 were reported in 56% of solitary central and periosteal cartilaginous tumors 18, and the data within our control group are in concordance with these findings. In total 40 of 101 (40%) solitary central tumors, 7 of 13 (54%) dedifferentiated chondrosarcomas and 3 of 3 periosteal chondrosarcomas displayed IDH1 or IDH2 mutations (Fig.1b, Table 1). In 6 additional tumors, the mutant allele seemed to be present below the detection level of Sanger sequencing. IDH1 or IDH2 mutations were absent in other subtypes of cartilaginous tumors, in angiosarcomas (Fig.1b) and in patients' blood. Immunohistochemistry for the R132H IDH1 mutant protein on tissue microarrays (TMA) containing cartilaginous and vascular tumors confirmed that mutant protein expression was restricted to central, dedifferentiated and periosteal cartilage tumors, while all other tumors were negative (Table 2). Interestingly, four of eight solitary chondrosarcoma cell lines carry different types of mutations in IDH1 or IDH2 (Table 3). To the best of our knowledge, no cell lines with IDH1 or IDH2 mutations are currently available. IDH1 or IDH2 mutations were more frequently found in solitary central tumors located in hands and feet (11 out of 14) versus those located in long and flat bones (28 out of 84) (p=0.006, Pearson Chi-Square test), which was also reported previously 18. This correlation was absent in Ollier disease (20 out of 22 versus 28 out of 34, p=0.5, Pearson Chi-Square test). While in gliomas, mutations in IDH1 or IDH2 predict a favorable outcome 38, we found no significant prognostic value of these mutations in solitary central cartilaginous tumors using multivariate analysis (Cox Regression, p-value = 0.3).
Table 3. IDH1 or IDH2 mutations in solitary central chondrosarcoma cell lines and primary culture.
| Cell line | Tumor type | Tumor Grade | Passage | IDH1 | IDH2 | Reference |
|---|---|---|---|---|---|---|
| SW1353 | Solitary central | CSII | p12 | Wt | R172S | ATCC |
| JJ012 | Solitary central | CSII | p15 | R132G | Wt | 51 |
| CH2879 | Solitary central | CSIII | p16 | G105G | Wt | 52 |
| OUMS27 | Solitary central | CSIII | p18 | Wt | Wt | 53 |
| L835 | Solitary central | CSIII | p38 | R132C | Wt | Home made |
| C3842 | Ollier disease | CSII | p32 | Wt | Wt | 54 |
| L2975 | Dedifferentiated CS | p31 | Wt | R172W* | Home made | |
| NDCS1 | Dedifferentiated CS | p12 | Wt | Wt | 55 |
L2975 showed R172W IDH2 homozygous mutation.
CS : chondrosarcoma
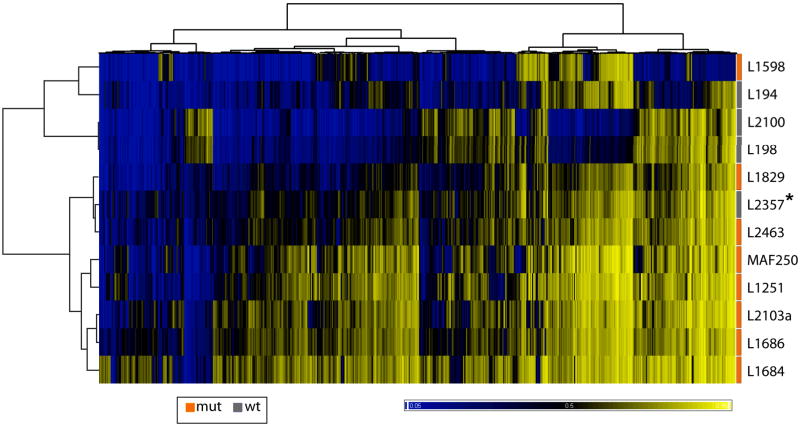
IDH1 or IDH2 mutations have also been reported at lower frequencies in various other tumors such as acute myeloid leukemia (AML) (8%) 39, 40, prostate cancer (2.7%) 40, 41, paragangliomas (0.7%) 40, 42 and thyroid carcinoma (16%) 43. The high mutation frequency in enchondromas and the fact that they are early events suggest a causal rather than a bystander role for IDH1 or IDH2 mutations in tumorigenesis in Ollier disease and Maffucci syndrome. In gliomas, mutant IDH1 or IDH2 leads to gain of function by producing 2-hydroxyglutarate (2HG), a structural analogue of α-KG, and by ultimately reducing α-KG production 44. In AML, it was demonstrated that mutant IDH protein results in DNA hypermethylation and impairment of hematopoietic differentiation 36, and in gliomas the presence of an IDH1 mutation is strongly associated with hypermethylation 45. Therefore, we used Illumina HumanMethylation27 BeadChip (Illumina Inc., CA) to assess a possible difference in methylation between enchondromas with (n = 8) and without (n = 4) IDH1 mutations detectable at Sanger sequencing. Unsupervised clustering based on the 2000 most variable CpG methylation sites resulted in 2 subgroups (Fig.3). One of these subgroups showed an overall higher methylation at the examined CpG sites and is therefore similar to the CpG island methylator phenotype (CIMP) as described in colon carcinoma and glioblastoma 45, 46. All but one enchondroma with an IDH1 mutation were CIMP+. Supervised clustering analysis indicated that 797 CpG sites are differentially methylated by more than 20% (at p<0.05) between enchondromas with and without IDH1 mutations. Interestingly 710 (89.1%) of these differentially methylated CpG sites were methylated in the enchondromas with IDH1 mutations (Supplementary Table 3). These results are in line with the hypothesis that IDH1 mutations induce methylation and thus contribute to the CIMP phenotype 36.
Figure 3. CpG island Methylator Phenotype in enchondromas with IDH1 mutations.
Heatmap depicting unsupervised clustering analysis based on the 2000 most variable CpG sites of enchondromas with IDH1 mutations (orange, n = 8) and without IDH1 mutation (gray, n=4). The level of DNA methylation (beta value) for each probe (columns) in each sample (rows) is represented by color scale as shown in the picture ranging from 0 (0% methylation, blue) to 1 (100% methylation, yellow). Asterisk indicates sample L2357 in which the R132G IDH1 mutant allele was detected in a subpopulation of cells. However, the mutation escaped detection at Sanger sequencing, and therefore the sample is labeled “wild-type”.
To assess the effect of IDH1 or IDH2 mutations on mRNA expression levels in cartilaginous tumors, we performed whole-genome gene expression analysis using Illumina Human-6 v3 array (Illumina Inc., CA). High quality mRNA was available for only three tumors in which mutation was negative (n=1) or below the threshold of Sanger sequencing (thus possibly carrying a low percentage of mutated cells)(n=2). Comparison with 18 tumors with clearly detectable IDH1 or IDH2 mutations using LIMMA analysis showed 36 differentially expressed probes encoding for 33 genes (Supplementary Table 4). 32 of 33 genes were down regulated in tumor samples with an IDH1 or IDH2 mutation. There was no overlap between the affected genes found in methylation and expression analysis.
One of the most differentially methylated genes was DLX5. There was a trend for downregulation of DLX5 but this was not significant in Ollier enchondromas versus controls (adj. p-value = 0.3, Supplementary Fig.2). The controls consisted of 2 growth plates and 4 articular/rib cartilage samples. The homeodomain transcription factor DLX5 is a cell autonomous positive regulator of chondrocyte maturation during endochondral ossification, promoting the conversion of immature proliferating chondrocytes into hypertrophic chondrocytes 47, 48 DLX5 also induces expression of Runx2 and osterix, promoting osteogenic differentiation 49, 50. Future studies should reveal whether down regulation of DLX5 through methylation as a consequence of IDH1 mutation delays hypertrophic differentiation of chondrocytes and inhibits subsequent osteogenic differentiation, thereby leaving clusters of proliferating chondrocytes behind.
In summary, we report a large multi-institutional series demonstrating somatic heterozygous IDH1 or, rarely, IDH2 point mutations in tumor tissues of 81% of patients with Ollier disease and 77% of patients with Maffucci syndrome, and provide evidence for intraneoplastic and somatic mosaicism. Future studies using deep sequencing approaches should reveal whether the percentage of patients carrying somatic mosaic IDH1 or IDH2 mutations is even higher than that detected in our series, or whether other genes are involved. We show the IDH1 mutation to be associated with hypermethylation and downregulation of several genes. Future studies should demonstrate a causal effect and it will be of great interest to assess how this dysregulation leads to enchondroma and spindle cell hemangioma formation. Finally, this is the first report of four chondrosarcoma cell lines carrying IDH1 or IDH2 mutations, providing good in vitro models for functional studies to dissect the role of IDH1 and IDH2 in Ollier disease and Maffucci syndrome, but also in tumorigenesis in general.
Data Deposition
MIAME-compliant data of tiling arrays, expression arrays and methylation arrays have been deposited in the GEO database (www.ncbi.nlm.nih.gov/geo/, accession number GSE30844).
Materials and Methods
Patients and Clinical Specimens
Fresh frozen tumor tissues (n = 60) of 44 patients with multiple cartilage tumors (36 patients with Ollier disease and 8 patients with Maffucci syndrome) (Table 1, Supplementary Table 1) were collected from EuroBoNet consortium (http://www.eurobonet.eu) 8 and the Laboratory of Human Molecular Genetics at the de Duve Institute, UCL (Brussels, Belgium). In addition, paraffin embedded tumor tissues (n = 15) from 12 patients were obtained from the files of the Children's Hospital (Boston, USA). Samples were handled according to the ethical guidelines of the host institution. All samples were coded and the ethical guidelines “Code for Proper Secondary Use of Human Tissue in The Netherlands” (Dutch Federation of Medical Scientific Societies) were followed in all procedures. Chondrosarcoma samples were graded according to Evans et al 56. Normal DNA derived from saliva, blood or muscle was available from 12 patients with Ollier disease. Patients' ages were documented at the time of operation. Demographic and survival data were obtained from the host institutions' patient records.
For comparison with other cartilage tumors, we included DNA from solitary enchondromas (n =9), solitary central chondrosarcomas (n=92), central dedifferentiated chondrosarcomas (n=13), periosteal chondrosarcomas (n=3), 37 peripheral cartilaginous tumors [solitary osteochondroma (n = 11), peripheral chondrosarcomas (n=20), multiple osteochondromas (n=6)], as well as 9 chondromyxoid fibromas, 7 chondroblastomas, and 2 osteochondroma-like lesions of metachondromatosis. Matching blood-derived DNA was also available from 24 cases as controls. Additionally, we included DNA from angiosarcomas (n = 14) since patients with Maffucci syndrome have central cartilage tumors combined with vascular tumors. The angiosarcomas, chondromyxoid fibromas and chondroblastomas were analyzed for IDH1 mutations only. Thus, in total we analyzed 261 tumors from 242 patients.
DNA extraction and Mutation Analysis
Genomic DNA from frozen tumors containing at least 80% of tumors cells, as estimated on haematoxylin and eosin-stained frozen sections, from blood and from saliva was isolated as described earlier 8. DNA from paraffin embedded tissue was isolated after micro dissection as previously described 8. For cell lines and primary cultures, DNA was isolated from cell pellets using the Wizard Genomic DNA Purification Kit (Promega, Madison, WI), according to the manufacturer's instructions.
PCR amplification was performed on IDH1 exon 4 for all the samples. IDH2 exon 4 was amplified in samples without IDH1 mutation and GNAS exon 8 was studied in samples without IDH1 and IDH2 mutations. To correlate with possible PTH1R mutations we also amplified PTH1R exon 4 for G121E and A122T, exon 5 for R150C and exon 9 for R255H using DNA from 35 patients with Ollier disease and Maffucci syndrome. PCR was performed in 25μl reactions using 10ng DNA, 12.5 μl of iQ SYBR green Supermix (Bio-Rad, CA) and 10 pmol M13 tailed primers (Supplementary Table 5). The PCR was carried out in a CFX 96 ™ Real-Time PCR detection system (Bio-Rad, CA) at an initial denaturation step of 5 min 95°C followed by 40 cycles of 10 sec 95 °C, 10 sec 60 °C and 10 sec 72 °C. After a final elongation step of 10 min at 72 °C, a melt curve was obtained to check for the quality of the PCR products. PCR products were purified using the Qiagen MinElute ™ 96 UF PCR Purification kit (Qiagen) system and finally eluted in 25μl sterile water. PCR amplimers were sequenced by a commercial party using standard forward and reverse M13 primers (Macrogen Inc. Europe, Amsterdam). The sequence trace files were analyzed with Mutation Surveyor™ DNA variant software (version 3.97 SoftGenetics, PA).
To validate the R132C and R132H IDH1 mutations, we designed hydrolysis probes (Supplementary Table 6) assays using the Custom Taqman® Assay Design Tool (Applied Biosystems, Nieuwerkerk a/d Ijssel, NL). Assays were performed on 144 samples including tumors related to Ollier disease, Maffucci syndrome, solitary cartilaginous tumors, chondrosarcoma cell-lines, blood from Ollier patients as well as negative controls (healthy donor DNA) together with no template controls. qPCR was performed in 10 μL reactions as described earlier 57 in a CFX384™ Real-Time PCR Detection System (Bio-Rad, Veenendaal, NL) for 10 minutes at 95 °C and 40 cycles of 10 seconds at 92 °C and 30 seconds at 60 °C. The quantification cycle (Cq) was used for quality assessment and samples with Cq>35 for the wild-type allele were considered as DNA negative. The threshold for the mutant allele R132C IDH1 (c.394C>T) or R132H IDH1 (c.395G>A) was set after subtracting the highest background signal from the negative controls.
There was sufficient DNA left to perform sequence analysis of all exons of IDH1 and IDH2 from 5 of 12 tumors without mutation. One IDH1 mutated tumor was also sequenced. PCR was performed as mentioned above for exon 4 and primer sequences are listed in supplementary Table 5.
Tiling Resolution Targeted Oligonucleotide Arrays
Custom designed Agilent tiling oligonucleotide array CGH (Agilent, Amstelveen, The Netherlands) containing 15,000 probes with a tiling coverage of genes involved in the different types of enchondromatosis syndromes (IDH1, IDH2, ACP5, PTH1R, PTPN11, EXT1, EXT2 and PTHLH) (Supplementary Table 2) was performed to detect possible small, intragenic losses and gains in these genes. In total 16 enchondromas and chondrosarcomas of patients with Ollier disease and Maffucci syndrome, with (n=14) and without (n=2) IDH1 or IDH2 mutations were selected. Labeling and hybridization of genomic DNA from freshly frozen tumor and data processing were performed as described earlier 58.
Immunohistochemistry
To determine the protein expression of the R132H IDH1 mutant allele, immunohistochemistry was performed as described earlier 8 using R132H IDH1 antibody (1:200 dilution 5% in non-fat milk, citrate antigen retrieval, blocking for 30′ with 5% non-fat milk) from Dianova (Hamburg, Germany). We used 403 tumors (Table 2) on 19 tissue microarrays (TMA), for which details were published previously 8, 59-61. Additional cases from Ollier disease and Maffucci syndrome were collected through EMSOS, and clinical details for these patients are described separately 4. Glioma tissue with a known IDH1 mutation was used as a positive control and primary antibody was omitted as a negative control. Only strong cytoplasmic staining combined with nuclear staining was considered a positive result 35. To study possible mosaicism in the tumor and in surrounding normal tissues, we selected resection specimens from tumors expressing R132H IDH1 mutant protein (n = 7) and stained multiple tissue blocks from different areas. All except 9 tumors of patients with Ollier disease that were used for mutation analysis were also included in the TMA, and results were confirmed.
Statistical Analysis for Clinical Correlation
From 83 patients with solitary tumors, follow up data were available (range 2 to 335 months, mean 115.23). To investigate the relation of IDH1 or IDH2 mutations with patients' clinical features, multivariate survival analysis (Cox Regression) and cross-tabulations (Pearson Chi-Square) were performed using SPSS version 16.0 (Chicago, Illinois, USA). Statistical analysis was not performed for patients with Ollier disease because nearly all patients with available follow up data had IDH1 or IDH2 mutations. All the p-values reported are two-sided and p-values < 0.05 were considered to indicate statistical significance.
DNA Methylation Profiling
Total 12 samples which includes 8 enchondromas with IDH1 mutation (4 Ollier enchondromas, 2 Maffucci enchondromas and 2 solitary enchondromas) and 4 enchondromas (1 Ollier enchondroma, 3 solitary enchondromas) without IDH1 or IDH2 mutations were used. Of these 4 enchondromas without IDH1 mutation, one had R132G IDH1 mutated cells present in the subpopulation, below the threshold level of Sanger sequencing. Bisulfite treatment was performed using EZ DNA Methylation™ Kit (Zymo Research, Orange, CA). Bisulfite converted DNA was then hybridized to Illumina HumanMethylation27 BeadChip (Illumina Inc., San Diego, CA) by following manufacturer's instructions. Infinium Unsupervised clustering analysis was performed using the Ward's clustering algorithm based on Euclidian distance. The 2000 most variable CpG sites (excluding those on the X and Y chromosomes) were used in the clustering analysis.
Genome-wide gene expression analysis
A total of 21 tumors including 6 enchondromas and 10 chondrosarcomas (6 grade I, 4 grade II) of Ollier disease and Maffucci syndrome as well as 1 solitary enchondroma and 4 solitary chondrosarcomas grade II and 6 controls (2 growth plates, 4 normal cartilage) were used. We determined differential expression between tumors with IDH1 or IDH2 mutations (n = 18) versus tumors without detectable IDH1or IDH2 mutation using Sanger sequencing (n=3). Two of these demonstrated subthreshold peaks for R132G and R132C IDH1 mutation suggesting a mutation in a minor subpopulation of tumor cells. Experimental procedures using the Illumina Human-6 v3.0 Expression BeadChips were performed as described previously 8, 62, 63. LIMMA analysis 64 was used to determine differential expression between the groups. Probes with Benjamini and Hochberg false discovery rate-adjusted P-values (adjP) < 0.05 and a log fold change (logFC) > 0.1 were considered to be significantly differentially expressed.
Supplementary Material
Acknowledgments
We are grateful to all of the patients and their families for their participation. The study is financially supported by The Netherlands Organization for Scientific Research (917-76-315 to JVMGB and TCP), the Liddy Shriver Sarcoma Initiative (to JVMGB and JO), the Interuniversity Attraction Poles initiated by the Belgian Federal Science Policy, network 6/05; the National Institute of Health, Program Project P01 AR048564; and the F.R.S.-FNRS (Fonds de la Recherche Scientifique)(all to MV), the Manton Center for Orphan Disease Research at Children's Hospital Boston (to KK) and is performed within EuroBoNet, a European Commission granted Network of Excellence for studying the pathology and genetics of bone tumors (018814). We would like to thank S. Romeo and C.M.A. Reijnders for providing DNA from cartilage tumors. We are grateful to A. B. Mohseny for help with statistics, D. van der Geest and T. Krenács for constructing TMAs, P. Wijers-Koster, D. de Jong, B. van den Akker, R. Duim, M. Winter, I. H. Briaire-de Bruijn, and M.E. Bowen for expert technical assistance. C.J.F. Waaijer, P.C.W. Hogendoorn and C.E. de Andrea are acknowledged for fruitful discussion. We would like to acknowledge F. Bertoni, E. L. Staals and P. Bacchini (Rizzoli Institute, Italy) for kindly providing peripheral dedifferentiated chondrosarcomas and vascular tumors, T. Kalinski (Otto-von-Guericke-University, Magdeburg, Germany) for C3842, M. Namba (Okayama University Medical School, Shikata, Japan) for OUMS27, Dr. T. Ariizumi (Niigata University Graduate School of Medical and Dental Sciences, Niigata, Japan) for NDCS1, and A. Llombart Bosch for CH2879 cell lines. Drs. J. Mulliken, J. Upton, and S. Fishman of the Vascular Anomalies Center at Children's Hospital Boston kindly provided spindle cell hemangiomas. S. Verdegaal, A.H.M. Taminiau and M.A.J. van de Sande are acknowledged for contributing patient data. We are thankful to S. Boeuf (Heidelberg University, Germany) R. Forsyth (Ghent University, Belgium) P. Mainil-Varlet (Bern University, Switzerland) W. Wuyts (University of Antwerp, Belgium) for providing a single Ollier or Maffucci case, or control samples. The continuous support of the Netherlands Committee on Bone Tumors is highly acknowledged.
Footnotes
Authors' contribution: The study was designed, written and reviewed by TCP and JVMGB. Mutation analysis was designed and performed by TCP, MAJHvR, JVMGB, KS, TvW and RvE. Immunohistochemistry was conducted and evaluated by TCP, MAJHvR and JVMGB. TCP, SV, JGvO and DM contributed tissue microarrays. Expression profiling was designed and performed by AMCJ, TCP, JVMGB, JO and analyzed by JO and MK. Methylation profiling was designed by AMCJ, JVMGB and LS, performed by PdA, and PdA and PF analyzed results. KHN, SD, LS, BK, BL, MS, RS, NL, LK, CG, PP, MV, LMB, KCK each contributed frozen or paraffin embedded tissues for multiple patients with Ollier disease or Maffucci syndrome and acquired patient data. The manuscript was approved by all coauthors.
Competing Financial Interests: Authors have no competing financial interests.
Reference List
- 1.Spranger J, Kemperdieck H, Bakowski H, Opitz JM. Two peculiar types of enchondromatosis. Pediatr Radiol. 1978;7:215–219. doi: 10.1007/BF02386711. [DOI] [PubMed] [Google Scholar]
- 2.Lucas DR, Bridge JA. Chondromas: enchondroma,periosteal chondroma,and enchondromatosis in World Health Organization classification of tumours. In: Fletcher CDM, Unni KK, Mertens F, editors. Pathology and genetics of tumours of soft tissue and bone. IARC Press; Lyon: 2002. pp. 237–240. [Google Scholar]
- 3.Pansuriya TC, Kroon HM, Bovee JVMG. Enchondromatosis: insights on the different subtypes. Int J Clin Exp Pathol. 2010;3:557–569. [PMC free article] [PubMed] [Google Scholar]
- 4.Verdegaal SHM, et al. Incidence, predictive factors and prognosis of chondrosarcoma in patients with Ollier disease and Maffucci syndrome; an international multicenter study of 161 patients. Manuscript submitted. 2011 doi: 10.1634/theoncologist.2011-0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hopyan S, et al. A mutant PTH/PTHrP type I receptor in enchondromatosis. Nat Genet. 2002;30:306–310. doi: 10.1038/ng844. [DOI] [PubMed] [Google Scholar]
- 6.Rozeman LB, et al. Enchondromatosis (Ollier disease, Maffucci syndrome) is not caused by the PTHR1 mutation p.R150C. Hum Mutat. 2004;24:466–473. doi: 10.1002/humu.20095. [DOI] [PubMed] [Google Scholar]
- 7.Couvineau A. PTHR1 mutations associated with Ollier disease result in receptor loss of function. Hum Mol Genet. 2008;17:2766–2775. doi: 10.1093/hmg/ddn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pansuriya TC, et al. Genome-wide analysis of Ollier disease: Is it all in the genes? Orphanet J Rare Dis. 2011;6:2. doi: 10.1186/1750-1172-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pansuriya TC, et al. Maffucci syndrome: A genome-wide analysis using high resolution single nucleotide polymorphism and expression arrays on four cases. Genes Chromosomes Cancer. 2011;50:673–679. doi: 10.1002/gcc.20889. [DOI] [PubMed] [Google Scholar]
- 10.Ranger A, Szymczak A. Do intracranial neoplasms differ in Ollier disease and maffucci syndrome? An in-depth analysis of the literature. Neurosurgery. 2009;65:1106–1113. doi: 10.1227/01.NEU.0000356984.92242.D5. [DOI] [PubMed] [Google Scholar]
- 11.Schwartz HS, et al. The malignant potential of enchondromatosis. J Bone Joint Surg Am. 1987;69:269–274. [PubMed] [Google Scholar]
- 12.Rietveld L, et al. First case of juvenile granulosa cell tumor in an adult with Ollier disease. Int J Gynecol Pathol. 2009;28:464–467. doi: 10.1097/PGP.0b013e3181a05af4. [DOI] [PubMed] [Google Scholar]
- 13.Leyva-Carmona M, Vazquez-Lopez MA, Lendinez-Molinos F. Ovarian juvenile granulosa cell tumors in infants. J Pediatr Hematol Oncol. 2009;31:304–306. doi: 10.1097/MPH.0b013e318196a70e. [DOI] [PubMed] [Google Scholar]
- 14.Yan H, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartmann C, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118:469–474. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 16.Dang L, Jin S, Su SM. IDH mutations in glioma acute myeloid leukemia. Trends Mol Med. 2010;16:387–397. doi: 10.1016/j.molmed.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 17.Kalfa N, et al. Activating mutations of the stimulatory g protein in juvenile ovarian granulosa cell tumors: a new prognostic factor? J Clin Endocrinol Metab. 2006;91:1842–1847. doi: 10.1210/jc.2005-2710. [DOI] [PubMed] [Google Scholar]
- 18.Amary MF, et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol. 2011;224:334–343. doi: 10.1002/path.2913. [DOI] [PubMed] [Google Scholar]
- 19.van Krieken JH, et al. KRAS mutation testing for predicting response to anti-EGFR therapy for colorectal carcinoma: proposal for an European quality assurance program. Virchows Arch. 2008;453:417–431. doi: 10.1007/s00428-008-0665-y. [DOI] [PubMed] [Google Scholar]
- 20.Wolff JN, Gemmell NJ. Combining allele-specific fluorescent probes restriction assay in real-time PCR to achieve SNP scoring beyond allele ratios of 1:1000. BioTechniques. 2008;44:193–4. 196, 199. doi: 10.2144/000112719. [DOI] [PubMed] [Google Scholar]
- 21.Sobreira NL, et al. Whole-genome sequencing of a single proband together with linkage analysis identifies a Mendelian disease gene. PLoS Genet. 2010;6:e1000991. doi: 10.1371/journal.pgen.1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bowen ME, et al. Loss-of-Function Mutations in PTPN11 Cause Metachondromatosis, but Not Ollier Disease or Maffucci Syndrome. PLoS Genet. 2011;7:e1002050. doi: 10.1371/journal.pgen.1002050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lausch E, et al. Genetic deficiency of tartrate-resistant acid phosphatase associated with skeletal dysplasia, cerebral calcifications and autoimmunity. Nat Genet. 2011;43:132–137. doi: 10.1038/ng.749. [DOI] [PubMed] [Google Scholar]
- 24.Briggs TA, et al. Tartrate-resistant acid phosphatase deficiency causes a bone dysplasia with autoimmunity and a type I interferon expression signature. Nat Genet. 2011;43:127–131. doi: 10.1038/ng.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collinson M, et al. Symmetrical enchondromatosis is associated with duplication of 12p11.23 to 12p11.22 including PTHLH. Am J Med Genet A. 2010;152A:3124–3128. doi: 10.1002/ajmg.a.33567. [DOI] [PubMed] [Google Scholar]
- 26.Bianco P, et al. Reproduction of human fibrous dysplasia of bone in immunocompromised mice by transplanted mosaics of normal and Gsalpha-mutated skeletal progenitor cells. J Clin Invest. 1998;101:1737–1744. doi: 10.1172/JCI2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones KB, et al. A mouse model of osteochondromagenesis from clonal inactivation of Ext1 in chondrocytes. Proc Natl Acad Sci U S A. 2010;107:2054–2059. doi: 10.1073/pnas.0910875107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Andrea CE, Prins FA, Wiweger MI, Hogendoorn PCW. Growth Plate Regulation and Osteochondroma Formation: Insights from Tracing Proteoglycans in Zebrafish Models and Human Cartilage. J Pathol. 2011;224:160–168. doi: 10.1002/path.2886. [DOI] [PubMed] [Google Scholar]
- 29.de Andrea CE, et al. Secondary Peripheral Chondrosarcoma Evolving from Osteochondroma as a Result of the Outgrowth of Cells with Functional. EXT Oncogene. 2011 doi: 10.1038/onc.2011.311. [DOI] [PubMed] [Google Scholar]
- 30.Reijnders CM, et al. No haploinsufficiency but loss of heterozygosity for EXT in multiple osteochondromas. Am J Pathol. 2010;177:1946–1957. doi: 10.2353/ajpath.2010.100296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bovee JVMG. EXTra hit for mouse osteochondroma. Proc Natl Acad Sci U S A. 2010;107:1813–1814. doi: 10.1073/pnas.0914431107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clément A, et al. Regulation of zebrafish skeletogenesis by ext2/dackel and papst1/pinscher. PLoS Genet. 2008;4:e1000136. doi: 10.1371/journal.pgen.1000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cohen MM., Jr Fibrous dysplasia is a neoplasm. Am J Med Genet. 2001;98:290–293. doi: 10.1002/1096-8628(20010201)98:4<290::aid-ajmg1112>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 34.Lietman SA, Ding C, Levine MA. A highly sensitive polymerase chain reaction method detects activating mutations of the GNAS gene in peripheral blood cells in McCune-Albright syndrome or isolated fibrous dysplasia. J Bone Joint Surg Am. 2005;87:2489–2494. doi: 10.2106/JBJS.E.00160. [DOI] [PubMed] [Google Scholar]
- 35.Ikota H, Nobusawa S, Tanaka Y, Yokoo H, Nakazato Y. High-throughput immunohistochemical profiling of primary brain tumors and non-neoplastic systemic organs with a specific antibody against the mutant isocitrate dehydrogenase 1 R132H protein. Brain Tumor Pathol. 2011;28:107–114. doi: 10.1007/s10014-010-0016-y. [DOI] [PubMed] [Google Scholar]
- 36.Figueroa ME, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas DM. Lessons from the deep study of rare tumours. J Pathol. 2011;224:306–308. doi: 10.1002/path.2928. [DOI] [PubMed] [Google Scholar]
- 38.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807, 1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mardis ER, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yen KE, Bittinger MA, Su SM, Fantin VR. Cancer-associated IDH mutations: biomarker and therapeutic opportunities. Oncogene. 2010;29:6409–6417. doi: 10.1038/onc.2010.444. [DOI] [PubMed] [Google Scholar]
- 41.Kang MR, et al. Mutational analysis of IDH1 codon 132 in glioblastomas and other common cancers. Int J Cancer. 2009;125:353–355. doi: 10.1002/ijc.24379. [DOI] [PubMed] [Google Scholar]
- 42.Gaal J, et al. Isocitrate dehydrogenase mutations are rare in pheochromocytomas and paragangliomas. J Clin Endocrinol Metab. 2010;95:1274–1278. doi: 10.1210/jc.2009-2170. [DOI] [PubMed] [Google Scholar]
- 43.Hemerly JP, Bastos AU, Cerutti JM. Identification of several novel non-p.R132 IDH1 variants in thyroid carcinomas. Eur J Endocrinol. 2010;163:747–755. doi: 10.1530/EJE-10-0473. [DOI] [PubMed] [Google Scholar]
- 44.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noushmehr H, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toyota M, et al. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chin HJ, et al. Studies on the role of Dlx5 in regulation of chondrocyte differentiation during endochondral ossification in the developing mouse limb. Dev Growth Differ. 2007;49:515–521. doi: 10.1111/j.1440-169X.2007.00940.x. [DOI] [PubMed] [Google Scholar]
- 48.Zhu H, Bendall AJ. Dlx5 Is a cell autonomous regulator of chondrocyte hypertrophy in mice and functionally substitutes for Dlx6 during endochondral ossification. PLoS ONE. 2009;4:e8097. doi: 10.1371/journal.pone.0008097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee MH, Kwon TG, Park HS, Wozney JM, Ryoo HM. BMP-2-induced Osterix expression is mediated by Dlx5 but is independent of Runx2. Biochem Biophys Res Commun. 2003;309:689–694. doi: 10.1016/j.bbrc.2003.08.058. [DOI] [PubMed] [Google Scholar]
- 50.Ulsamer A, et al. BMP-2 induces Osterix expression through up-regulation of Dlx5 and its phosphorylation by p38. J Biol Chem. 2008;283:3816–3826. doi: 10.1074/jbc.M704724200. [DOI] [PubMed] [Google Scholar]
- 51.Scully SP, et al. Marshall Urist Award. Interstitial collagenase gene expression correlates with in vitro invasion in human chondrosarcoma. Clin Orthop Relat Res. 2000:291–303. doi: 10.1097/00003086-200007000-00038. [DOI] [PubMed] [Google Scholar]
- 52.Gil-Benso R, et al. Establishment and characterization of a continuous human chondrosarcoma cell line, ch-2879: comparative histologic and genetic studies with its tumor of origin. Lab Invest. 2003;83:877–887. doi: 10.1097/01.lab.0000073131.34648.ea. [DOI] [PubMed] [Google Scholar]
- 53.Kunisada T, et al. A new human chondrosarcoma cell line (OUMS-27) that maintains chondrocytic differentiation. Int J Cancer. 1998;77:854–859. doi: 10.1002/(sici)1097-0215(19980911)77:6<854::aid-ijc10>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 54.Kalinski T, et al. Establishment and characterization of the permanent human cell line C3842 derived from a secondary chondrosarcoma in Ollier's disease. Virchows Arch. 2005;446:287–299. doi: 10.1007/s00428-004-1194-y. [DOI] [PubMed] [Google Scholar]
- 55.Kudo N, et al. Establishment of novel human dedifferentiated chondrosarcoma cell line with osteoblastic differentiation. Virchows Arch. 2007;451:691–699. doi: 10.1007/s00428-007-0426-3. [DOI] [PubMed] [Google Scholar]
- 56.Evans HL, Ayala AG, Romsdahl MM. Prognostic factors in chondrosarcoma of bone. A clinicopathologic analysis with emphasis on histologic grading. Cancer. 1977;40:818–831. doi: 10.1002/1097-0142(197708)40:2<818::aid-cncr2820400234>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 57.van Eijk R, et al. Rapid KRAS, EGFR, BRAF and PIK3CA mutation analysis of fine needle aspirates from non-small-cell lung cancer using allele-specific qPCR. PLoS ONE. 2011;6:e17791. doi: 10.1371/journal.pone.0017791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Szuhai K, et al. Tiling resolution array-CGH shows that somatic mosaic deletion of the EXT gene is causative in EXT gene mutation negative multiple osteochondromas patients. Hum Mutat. 2011;32:2036–49. doi: 10.1002/humu.21423. [DOI] [PubMed] [Google Scholar]
- 59.Verbeke SL, et al. Distinct histological features characterize primary angiosarcoma of bone. Histopathology. 2011;58:254–264. doi: 10.1111/j.1365-2559.2011.03750.x. [DOI] [PubMed] [Google Scholar]
- 60.Meijer D, et al. Expression of aromatase and estrogen receptor alpha in chondrosarcoma, but no beneficial effect of inhibiting estrogen signaling both in vitro and in vivo. Clinical Sarcoma Research. 2011;1 doi: 10.1186/2045-3329-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rozeman LB, et al. Dedifferentiated peripheral chondrosarcomas: regulation of EXT-downstream molecules and differentiation-related genes. Mod Pathol. 2009;22:1489–1498. doi: 10.1038/modpathol.2009.120. [DOI] [PubMed] [Google Scholar]
- 62.Hallor KH, et al. Genomic Profiling of Chondrosarcoma: Chromosomal Patterns in Central and Peripheral Tumors. Clin Cancer Res. 2009;15:2685–2694. doi: 10.1158/1078-0432.CCR-08-2330. [DOI] [PubMed] [Google Scholar]
- 63.Buddingh' EP, et al. Tumor-infiltrating macrophages are associated with metastasis suppression in high-grade osteosarcoma: a rationale for treatment with macrophage-activating agents. Clin Cancer Res. 2011 doi: 10.1158/1078-0432.CCR-10-2047. [DOI] [PubMed] [Google Scholar]
- 64.Smyth GK, et al. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.