Abstract
In this manuscript, we present the determination of glycation sites in synthetic neoglycoconjugates formed by conjugation of the antigenic monosaccharide hapten of Vibrio cholerae O1 serotype Ogawa to BSA using nano- liquid chromatography electrospray ionization quadrupole time-of-flight tandem mass spectroscopy (LC-ESI-QqTOF-MS/MS). The matrix-assisted laser desorption/ionization-TOF/TOF-MS/MS analyses of the tryptic digests of the glycoconjugates having a hapten:BSA ratio of 4.3:1, 6.6:1 and 13.2:1 revealed only three glycation sites, on the following lysine residues: Lys 235, Lys 437 and Lys 455. Digestion of the neoglycoconjugates with the proteases trypsin and GluC V8 gave complementary structural information and was shown to maximize the number of recognized glycation sites. Here, we report identification of 20, 27 and 33 glycation sites using LC-ESI-QqTOF-MS/MS analysis of a series of synthetic neoglycoconjugates with a hapten:BSA ratio of, respectively, 4.3:1, 6.6:1 and 13.2:1. We also tentatively propose that all the glycated lysine residues are located mainly near the outer surface of the protein.
Keywords: nano-LC-ESI-QqTOF-MS/MS, neoglycoconjugate vaccine models, hapten-BSA carrier ratio, Trypsin digestion, GluC V8 digestion
INTRODUCTION
Glycoconjugate vaccines are obtained by covalent attachment of synthetic or bacterial carbohydrate antigens to carrier proteins. Such conjugates provide T cell-dependent immunogenicity against the saccharide hapten. With the involvement of T cells, immunological memory is invoked, avidity maturation and isotypes switching occurs, to generate complement-activating protective antibodies.[1]
Different studies have been carried out on the analysis of glycoproteins using mass spectrometry.[2–4] Novel soft ionization techniques such as electrospray ionization (ESI) and matrix-assisted laser desorption ionization (MALDI) were applied successfully for the determination of the glycation sites of N- and O-glycoproteins, as well as for the general carbohydrate structure determination.[4,5] Generally, two different methods are applied. The first consists of hydrolysis of the glycans from the protein, followed by a purification step and mass spectrometry analysis of the intact carbohydrate portions.[6,7] Sometimes, it is necessary to derivatize the released polysaccharides in order to obtain a sufficient m/z signal of the released carbohydrate.[8] The second approach is based on the entire glycoprotein digestion with endoproteases, followed by mass spectral and tandem mass spectral analyses of the obtained digests.[9,10] The main advantage of this method is that the product ions formed correspond to the glycopeptide fragments, which contain the exact glycation site.[11] The determination of glycation sites is useful to verify structures of machine-synthesized large glycopeptides and/or glycoproteins where small-sized glycopeptide intermediates are used as building blocks. Thus, one of the main applications can be the quality control of neoglycoconjugate vaccines. In addition, it will permit the verification of site-specific glycation in future synthetic neoglycoconjugates.
In our previous work, we have reported the determination of the glycation sites of a series of neoglycoconjugate models by MALDI-time-of-flight tandem mass spectroscopy (TOF-MS/MS). These glycoconjugates were composed of the spacer-equipped terminal monosaccharide antigen of the O-specific polysaccharide (O-PS) of Vibrio cholerae O1, serotype Ogawa covalently linked to protein carrier BSA.[12] We were able to determine the hapten:BSA ratios of the different synthetic glycoconjugate preparations as being 4.3:1, 6.6:1 and 13.2:1. The obtained glycoconjugates were digested with trypsin, and the resulting digests were analyzed by MALDI-TOF/TOF-MS/MS in order to determine the conjugation sites between the carbohydrate and the carrier protein. In that study, we reported that three glycation sites on the Lys 235, Lys 437 and Lys 455 residues were identified on the three analyzed glycoconjugates. We postulated that perhaps selecting the protease trypsin may have not been the correct choice for the digestion of our hapten-BSA glycoconjugates. Additionally, ion suppression effects can occur during the MALDI-TOF-MS experiments and afford a low-quality MALDI-MS spectrum.[13,14]
We recently reported the determination of the glycation sites in Bacillus anthracis neoglycoconjugate vaccine by MALDI-TOF/TOF-collision-induced dissociation (CID)-MS/MS and liquid chromatography (LC)-ESI- quadrupole (Qq)TOF-MS/MS.[15] Among other things, it permitted us to evaluate the hapten-to-BSA ratio which was found to be 5.4:1. The carbohydrate-protein vaccine model was then digested using two different proteases: the trypsin and the GluC V8 endoproteinase. The different digests were subsequently subjected to MALDI-TOF/TOF-MS/MS and LC-ESI-QqTOF-MS/MS analysis. The study revealed that the LC-ESI-QqTOF-MS/MS analysis of the different digests allowed identification of a higher number of glycation sites (30 were discovered), when comparing to the MALDI-TOF/TOF-MS/MS analysis of the same digests (only five glycation sites identified). We thus concluded that the LC-ESI-QqTOF-MS/MS analysis of both tryptic and GluC V8 digests was more efficient to reveal the glycation sites on our synthetic carbohydrate-protein glycoconjugates.
In the present work, we applied the foregoing strategy.[15] Thus, we digested synthetic glycoconjugates with different hapten:BSA ratios (4.3:1, 6.6:1 and 13.2:1), formed by conjugation of the monosaccharide antigen Vibrio cholerae O1 serotype Ogawa to BSA, separately with trypsin and the GluC V8 endoproteinase. The different digests were then analyzed by LC-ESI-QqTOF-MS/MS, and the glycopeptides were sequenced manually in order to determine the glycation sites.
MATERIAL AND METHOD
Preparation of the hapten-BSA neoglycoconjugate
The synthesis of the hapten-BSA glycoconjugates has been described previously.[16] Briefly, the synthesis started by the preparation of the methyl 6-hydroxyhexanoyl α-glycoside form of the upstream terminal moiety of the O-PS of Vibrio cholerae serogroup O1 serotype Ogawa antigen (4-(3-deoxy-L-glycero)-2-O-methyl-α-D-perosamine). The spacer-equipped sugar was then reacted with 1,2-diaminoethane which converted the methyl ester group in the spacer into the corresponding amino amide. The latter was treated with diethyl squarate to produce squarate monoester. The hapten was finally conjugated with the BSA (Sigma Aldrich, Saint Louis, MO, USA) in 0.5 M borate buffer (pH 9.0) at the initial molar carbohydrate-protein ratio of 100:1. Isolation of the neoglycoconjugates was carried out by ultrafiltration using centrifugal filters with a molecular weight cut-off of 30,000 Da.
Neoglycoconjugates digestion with different proteases
The digestions of the hapten-BSA glycoconjugates were carried out with trypsin and GluC V8 protease (Sigma Aldrich, Saint Louis, MO, USA). Thus, 100 µg of the glycoconjugate was dissolved in a mixture of 0.1% RapiGest SF Surfactant (1 µg, Waters, USA) in 50 mM of NH4HCO3 (100 µL) at a pH of 8.0 and reduced by treatment with 2 µl of 10 mM dithiothreitol (Sigma Aldrich, Saint Louis, MO, USA) for 30 min at room temperature, followed by alkylation with 2 µl of a 50 mM iodoacetamide (Sigma Aldrich, Saint Louis, MO, USA) for 1 h at room temperature. A portion (50 µg) of the glycoconjugate was digested with trypsin using a 20 ng/ml of trypsin dissolved in NH4HCO3 (50 mM, 1 ml, pH 7.8) at a trypsin:-glycoprotein ratio of 1:25 (w/w) and incubated at 37 °C overnight with shaking. The other 50 µg of the glycoconjugate was digested using the GluC V8 endoprotease dissolved in NH4HCO3 (50 mM, 1 ml, pH 7.8) at a protease:glycoprotein ratio of 1:25 (w/w) and incubated at 37 °C overnight with shaking. The sample was then dried under vacuum, and the residue was dissolved in 20 µl of 1% acetic acid (Sigma-Aldrich, Oakville, ON, Canada). An aliquot of each sample (10 µl) was then cleaned up using ZipTip C18 (Millipore, Bedford, MA, USA) before mass spectral analysis.
LC-ESI-QqTOF-CID-MS/MS analysis
The peptides were separated on a DIONEX UltiMate3000 Nano LC System (Germering, Germany). 250 fmol of the digested glycoprotein was dissolved in 0.1% TFA and loaded onto a precolumn (300 µm i.d. × 5 mm, C18 PepMap100, 5 µm (LC Packing, Sunnyvale, CA)) in order to desalt and concentrate the sample. After their elution from the precolumn, the mixture of peptides and glycopeptides was separated on a nanoflow analytical column (75 µm i.d. × 15 cm, C18 PepMap 100, 3 µm, 100 A, (LC Packing, Sunnyvale, CA)) at a flow rate of 180 nl/min. The mobile phase eluents used were composed of 0.1% FA/0.01% TFA/2% ACN (A) and 0.08% FA/0.008% TFA/98% ACN (B). The elution started with 0% B for 10 min, followed by a gradient of 0 → 60% B in 55 min and 60 → 90% B in 3 min and was kept at 90% B for 3 min. The MS/MS analysis of the eluted peptides and glycopeptides was accomplished using an Applied Biosystems API-QSTAR XL QqTOF-MS/MS hybrid tandem mass spectrometer (Applied Biosystems International-MDS Sciex, Foster City, CA, USA) equipped with a nano-electrospray source (Protana XYZ manipulator) which produces the electrospray through a PicoTip needle (10 µm i.d., New Objectives, Wobum, MA, USA) carrying a voltage of 2400 V. The mass spectra were acquired from m/z 100 to m/z 2000.
RESULTS
As previously stated, the purpose of this study was to determine the glycation sites in synthetic hapten-BSA glycoconjugates with hapten-to-BSA ratios: 4.3:1, 6.6:1 and 13.2:1 prepared by conjugation of the antigenic monosaccharide hapten of Vibrio cholerae O1 serotype Ogawa to BSA.
Carbohydrate fragmentation nomenclature used throughout this manuscript to identify the various glycopeptides is that described by Domon and Costello[4] whereas the nomenclature used to identify the true peptide fragments is the one described by Roepstorff et al. and lately modified by Johnson and coworkers.[17,18]
Mass spectrometry determination of the glycation sites on the neoglycoconjugate possessing a hapten:BSA ratio of 4.3:1, 6.6:1 and 13.2:1
The following parameters were applied for the MASCOT library searching: carbamidomethyl (C) as fixed modifications, oxidation (M) as variable modifications, a peptide mass tolerance of ± 0.2 Da and a maximum of missed cleavages of 1.
The Nano-LC-ESI-QqTOF-MS/MS analysis of the tryptic digests of the neoglycoconjugates allowed the identification of two serum albumin protein isoforms from the Bos taurus species on the MASCOT library: serum albumin precursor (gi|1351907) with the following sequence coverage: 61% for the neoglycoconjugate possessing a hapten:BSA ratio of 4.3:1, 61% for the neoglycoconjugate with a hapten:BSA ratio of 6.6:1 and 47% for the neoglycoconjugate with a hapten: BSA ratio of 13.2:1. In addition, the MASCOT search also identified for the analysis of all the tryptic digests of the neoglycoconjugates, the serum albumin protein (gi|74267962).
The submission of the LC-ESI-QqTOF-MS/MS data of the GluC V8 digests to the MASCOT library allowed the identification of two serum albumin protein isoforms from the Bos taurus species: serum albumin precursor (gi|1351907, sequence coverage of 55% for the neoglycoconjugate with a hapten:BSA ratio 4.3:1, 45% for the neoglycoconjugate with a hapten:BSA ratio 6.6:1 and 40% for the neoglycoconjugate with a hapten:BSA ratio 13.2:1) and serum albumin (gi|74267962).
We have identified the molecular masses of the glycopeptides produced by the digested glycoconjugates by comparison of the m/z values of the obtained molecular ions which did not correspond to BSA digests in the MASCOT report, with the calculated m/z values of all the possible glycopeptides. Thus, the masses of the glycopeptides were identified by the addition of either one or two carbohydrate-hapten residues (513.2323 Da), corresponding to the m/z values of the formed glycopeptides. In other words, the m/z values of the peptides which matched to the theoretical mass of a ‘peptide + carbohydrate-linker’ were identified, extracted and subjected to low-energy CID-MS/MS for sequencing to identify the glycation sites.
Mass spectrometry determination of the glycation sites on the neoglycoconjugate possessing a hapten: BSA ratio of 4.3:1
The previously described strategy allowed us to identify 11 glycation sites through sequencing of the glycopeptides obtained from the tryptic digests of the neoglycoconjugate (Table 1).
Table 1.
Tryptic glycopeptides identified during LC-ESI-QqTOF-MS/MS analysis of the hapten-BSA glycoconjugate with a hapten:BSA ratio of 4.3:1, 6.6:1 and 13.2:1
| Peptide | Missed | Hapten:BSA ratio 4.3:1 | Hapten:BSA ratio 6.6:1 | Hapten:BSA ratio 13.2:1 | ||||
|---|---|---|---|---|---|---|---|---|
| Sequence (star = glycation site) | Calculated | Observed | Deviation | Observed | Deviation | Observed | Deviation | |
| m/z (charge) | cleavage | m/z (charge) | (Da) | m/z (charge) | (Da) | m/z (charge) | (Da) | |
| SLGK*VGTR (Lys 455) | 665.8643 (+2) | 1 | 665.8810 (+2) | 0.0168 | 665.8694 (+2) | 0.0052 | 665.8699 (+2) | 0.0057 |
| EK*VLTSSAR (Lys 211) | 752.3987 (+2) | 1 | 752.4233 (+2) | 0.0246 | 752.4114 (+2) | 0.0127 | 752.3971 (+2) | −0.0016 |
| ALK*AWSVAR (Lys 235) | 757.9143 (+2) | 1 | 757.9261 (+2) | 0.0118 | 757.9189 (+2) | 0.0046 | 757.9248 (+2) | 0.0105 |
| K*QTALVELLK (Lys 548) | 828.4770 (+2) | 1 | 828.4992 (+2) | 0.0222 | 828.4872 (+2) | 0.0103 | 828.4857 (+2) | 0.0087 |
| CCTK*PESER (Lys 463) | 840.3662 (+2) | 1 | 840.3949 (+2) | 0.0287 | 840.3788 (+2) | 0.0126 | 840.3769 (+2) | 0.0107 |
| LK*PDPNTLCDEFKADEK (Lys 140) | 845.0720 (+3) | 3 | 845.0908 (+3) | 0.0188 | 845.0839 (+3) | 0.0119 | 845.0911 (+3) | 0.0191 |
| DTHK*SEIAHR (Lys 28) | 853.9209 (+2) | 1 | 853.9445 (+2) | 0.0236 | 853.9366 (+2) | 0.0158 | - | - |
| GACLLPK*IETMR (Lys 204) | 951.4892 (+2) | 1 | 951.5091 (+2) | 0.0199 | 951.5014 (+2) | 0.0122 | 951.5006 (+2) | 0.0114 |
| K*VPQVSTPTLVEVSR (Lys 437) | 1077.0887 (+2) | 0 | 1077.1129 (+2) | 0.0243 | 1077.0985 (+2) | 0.0099 | 1077.0975 (+2) | 0.0089 |
| LAK*EYEATLEECCAK (Lys 374) | 1164.5347 (+2) | 2 | 1164.5375 (+2) | 0.0028 | 1164.5613 (+2) | 0.0266 | 1164.5540 (+2) | 0.0193 |
| QNCDQFEK*LGEYGFQNALIVR (Lys 420) | 1014.8220 (+3) | 1 | 1014.8462 (+3) | 0.0242 | 1014.8390 (+3) | 0.0170 | 1014.8534 (+3) | 0.0314 |
| HKPK*ATEEQLK (Lys 561) | 607.9779 (+3) | 3 | - | - | 607.9966 (+3) | 0.0187 | 608.0033 (+3) | 0.0254 |
| LSQK*FPK (Lys 245) | 680.8716 (+2) | 3 | - | - | 680.8722 (+2) | 0.0007 | 680.8755 (+2) | 0.0040 |
| LCVLHEK*TPVSEK (Lys 489) | 685.0222 (+3) | 2 | - | - | 685.0256 (+3) | 0.0034 | 685.0310 (+3) | 0.0088 |
| CASIQK*FGER (Lys 228) | 854.9142 (+2) | 1 | - | - | 854.9284 (+2) | 0.0143 | 854.9290 (+2) | 0.0149 |
| VTK*CCTESLVNR (Lys 498) | 990.4743 (+2) | 1 | - | - | 990.4813 (+2) | 0.0071 | 990.4930 (+2) | 0.0188 |
| FK*DLGEEHFK (Lys 36) | 881.9304 (+2) | 2 | - | - | - | - | 881.9491 (+2) | 0.0188 |
| LKECCDK*PLLEK (Lys 304) | 1023.5103 (+2) | 3 | - | - | - | - | 1023.5348 (+2) | 0.0245 |
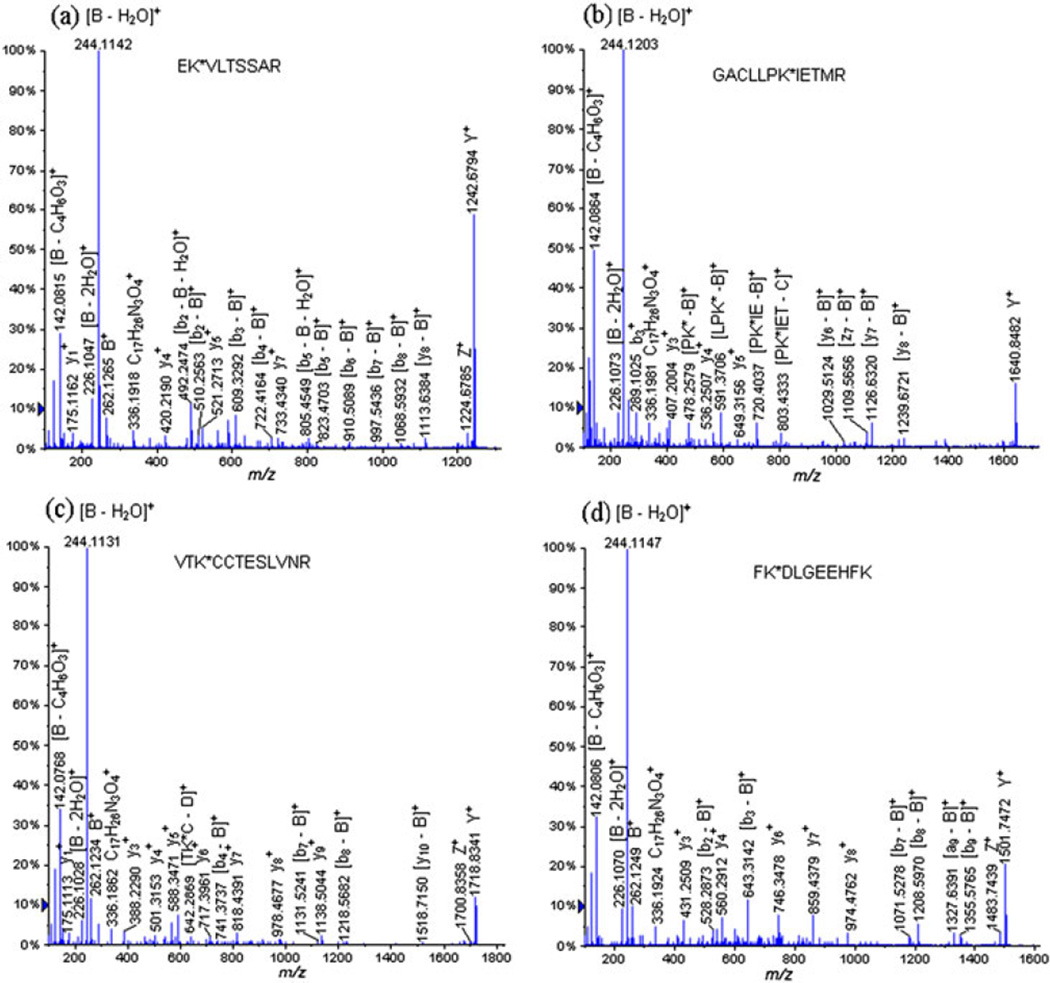
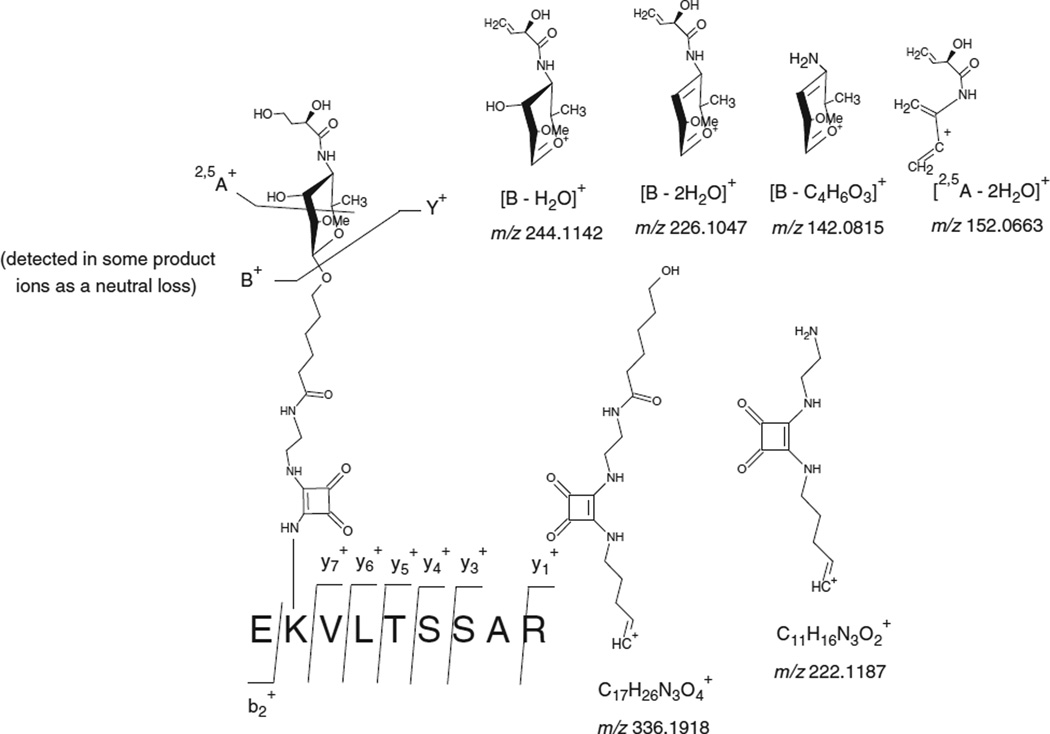
Figure 1(a) displays the mass spectra obtained form the extraction of the precursor ion at m/z 752.4233 (+2) corresponding to the glycated peptide EK*VLTSSAR (Lys 211). The CID-MS/MS analysis of this glycated peptide reveals a signature of the carbohydrate moiety through the following product ions: B+ at m/z 262.1265, [B − H2O]+ at m/z 244.1142, [B − 2H2O]+ at m/z 226.1047, [2,5A − 2H2O]+ at m/z 152.0663, [B − C4H6O3]+ at m/z 142.0815 and [2,5A − CH4O3]+ at m/z 124.0708 (Fig. 2). Moreover, we were also able to detect the entire peptide attached to the linker portion: Y+ at m/z 1242.6794 and Z+ at m/z 1224.6785. It has also to be noticed that the product ion at m/z 336.1918 can be assigned to two different structures: (Fig. 2). However, the product at m/z 336.1918 was also detected in our previous work on Bacillus anthracis neoglycoconjugate vaccine model and assigned to the structure: [C17H26N3O4]+ which corresponds to a linker portion attached to a lysine residue (Fig. 2).[15] Similarly, the product ion [C11H16N3O2]+ at m/z 222.1187 was produced from a linker portion attached to a lysine residue. This latter product ion was also detected in our previous work on the glycation sites determination of Bacillus anthracis neoglycoconjugate vaccine model.[15]
Figure 1.
LC-ESI-QqTOF-MS/MS spectra of the tryptic glycopeptides (a) EK*VLTSSAR (Lys 211) at m/z 752.4233 (+2), (b) GACLLPK*IETMR (Lys 204) at m/z 951.5091 (+2), (c) VTK*CCTESLVNR (Lys 498) at m/z 990.4813 (+2) and FK*DLGEEHFK (Lys 36) at m/z 881.9491 (+2).
Figure 2.
Different product ions involving the fragmentation of the carbohydrate hapten observed during the LC-ESI-QqTOF-MS/MS analysis of the tryptic glycopeptide EK*VLTSSAR (Lys 211) at m/z 752.4233 (+2).
In addition, the CID-MS/MS analysis of this glycated peptide EK*VLTSSAR (Lys 211), at m/z 752.4233 (+2), allowed us to observe the formation of product ions corresponding to glycopeptide fragments obtained by the loss of their carbohydrate portion: [y8 − B]+ at m/z 1113.6384, [b8 − B]+ at m/z 1068.5932, [b7 − B]+ at m/z 997.5436, [b6 − B]+ at m/z 910.5089, [b5 − B]+ at m/z 823.4703, [b5 − B − H2O]+ at m/z 805.4549, [b4 − B]+ at m/z 722.4164, [b4 − B − H2O]+ at m/z 704.4081, [b3 − B]+ at m/z 609.3292, [b3 − B − H2O]+ at m/z 591.3171, [b2 − B]+ at m/z 510.2563 and [b2 − B − H2O]+ at m/z 492.2474. The series of detected y- and b-ions, which covered the sequence of the glycopeptides, are listed in Table 2.
Table 2.
LC-ESI-QqTOF-MS/MS analysis of the glycated peptide EK*VLTSSAR (Lys 211) at m/z 752.4233 (+2)
| Molecular ion | Calculated | Experimental | Deviation | |
|---|---|---|---|---|
| m/z | m/z | Da | ||
| Y+ | 1242.6683 | 1242.6794 | 0.0111 | |
| Z+ | 1224.6577 | 1224.6785 | 0.0208 | |
| [y8 − B]+ | 1113.6252 | 1113.6384 | 0.0132 | |
| [b8 − B]+ | 1068.5572 | 1068.5932 | 0.0360 | |
| [b7 − B]+ | 997.5201 | 997.5436 | 0.0235 | |
| [b6 − B]+ | 910.4880 | 910.5089 | 0.0209 | |
| [b5 − B]+ | 823.4560 | 823.4703 | 0.0143 | |
| [b5 − B − H2O]+ | 805.4454 | 805.4549 | 0.0095 | |
|
|
733.4197 | 733.4340 | 0.0143 | |
| [b4 − B]+ | 722.4083 | 722.4164 | 0.0081 | |
| [b4 − B − H2O]+ | 704.3977 | 704.4081 | 0.0104 | |
|
|
634.3513 | 634.3576 | 0.0063 | |
| [b3 − B]+ | 609.3243 | 609.3292 | 0.0049 | |
| [b3 − B − H2O]+ | 591.3137 | 591.3171 | 0.0034 | |
|
|
521.2673 | 521.2713 | 0.0040 | |
| [b2 − B]+ | 510.2558 | 510.2563 | 0.0005 | |
| [b2 − B − H2O]+ | 492.2452 | 492.2474 | 0.0022 | |
|
|
420.2196 | 420.2190 | −0.0006 | |
|
|
336.1918 | 336.1918 | 0.0000 | |
|
|
333.1875 | 333.1851 | −0.0024 | |
| B+ | 262.1290 | 262.1265 | −0.0025 | |
| [B − H2O]+ | 244.1179 | 244.1142 | −0.0037 | |
| [B − 2H2O]+ | 226.1079 | 226.1047 | −0.0032 | |
|
|
222.1237 | 222.1187 | −0.0050 | |
|
|
175.1184 | 175.1162 | −0.0022 | |
| [2,5A − 2H2O]+ | 152.0706 | 152.0663 | −0.0043 | |
| [B − C4H6O3]+ | 142.0863 | 142.0815 | −0.0048 | |
| [2,5A − CH4O3]+ | 124.0757 | 124.0708 | −0.0049 | |
Figure 1(b) displays another example CID-MS/MS analysis of a glycated peptide detected during the analysis of the tryptic digests of the hapten-BSA neoglycoconjugate with a hapten:BSA ratio of 4.3:1: GACLLPK*IETMR (Lys 204) at m/z 951.5091 (+2).
The LC-ESI-QqTOF-MS/MS analysis of the GluC V8 digests of the neoglycoconjugate with a hapten:BSA ratio of 4.3:1 permitted the localization of ten glycation sites through the sequencing of the observed glycopeptides listed in Table 3.
Table 3.
GluC V8 glycopeptide digests identified during LC-ESI-QqTOF-MS/MS analysis of the hapten-BSA glycoconjugate with a hapten:BSA ratio of 4.3:1, 6.6:1 and 13.2:1
| Peptide | Missed | Hapten:BSA ratio 4.3:1 | Hapten:BSA ratio 6.6:1 | Hapten:BSA ratio 13.2:1 | ||||
|---|---|---|---|---|---|---|---|---|
| Sequence (star = glycation site) | Calculated | Observed | Deviation | Observed | Deviation | Observed | Deviation | |
| m/z (charge) | cleavage | m/z (charge) | (Da) | m/z (charge) | (Da) | m/z (charge) | (Da) | |
| K*SHCIAE (Lys 309) | 679.3189 (+2) | 0 | 679.3320 (+2) | 0.0131 | 679.3343 (+2) | 0.0154 | 679.3284 (+2) | 0.0095 |
| LTKVHK*E (Lys 266) | 684.3745 (+2) | 0 | 684.3867 (+2) | 0.0122 | 684.3851 (+2) | 0.0106 | 684.3795 (+2) | 0.0050 |
| K*VTKCCTE (Lys 495) | 769.8575 (+2) | 0 | 769.8677 (+2) | 0.0102 | 769.8720 (+2) | 0.0145 | 769.8668 (+2) | 0.0093 |
| K*QEPERNE (Lys 117) | 771.8678 (+2) | 2 | 771.8852 (+2) | 0.0174 | 771.8795 (+2) | 0.0117 | 771.8883 (+2) | 0.0205 |
| K*SLHTLFGDE (Lys 88) | 830.4093 (+2) | 1 | 830.4932 (+2) | 0.0839 | 830.4246 (+2) | 0.0153 | 830.4196 (+2) | 0.0103 |
| DKGACLLPK*IE (Lys 204) | 878.9556 (+2) | 0 | 878.9783 (+2) | 0.0227 | 878.9669 (+2) | 0.0113 | 878.9643 (+2) | 0.0087 |
| AK*DAFLGSFLYE (Lys 346) | 937.4590 (+2) | 1 | 937.4871 (+2) | 0.0281 | 937.4759 (+2) | 0.0169 | 937.4798 (+2) | 0.0208 |
| KK*FWGKYLYE (Lys 156) | 937.9824 (+2) | 0 | 937.9877 (+2) | 0.0053 | 937.9862 (+2) | 0.0038 | 937.9648 (+2) | −0.0176 |
| LLYYANK*YNGVFQE (Lys 183) | 1118.0465 (+2) | 0 | 1118.0711 (+2) | 0.0246 | 1118.0587 (+2) | 0.0122 | 1118.0511 (+2) | 0.0046 |
| VSRSLGK*VGTRCCTKPE (Lys 455) | 816.7456 (+3) | 0 | 816.7646 (+3) | 0.0190 | 816.7583 (+3) | 0.0127 | 816.7483 (+3) | 0.0027 |
| K*TPVSE (Lys 489) | 587.2979 (+2) | 0 | - | - | 587.3103 (+2) | 0.0124 | 587.3041 (+2) | 0.0062 |
| GPK*LVVSTQTALA (Lys 597) | 899.4959 (+2) | 0 | - | - | 899.5080 (+2) | 0.0121 | 899.5092 (+2) | 0.0133 |
| LLKHKPK*ATEE (Lys 561) | 903.9960 (+2) | 1 | - | - | 904.0190 (+2) | 0.0230 | 903.9916 (+2) | −0.0044 |
| K*QIKKQTALVE (Lys 544) | 900.0117 (+2) | 0 | - | - | 900.0234 (+2) | 0.0117 | 900.0165 (+2) | 0.0048 |
| VTK*LVTD (Lys 256) | 644.8478 (+2) | 0 | - | - | - | - | 644.8498 (+2) | 0.0020 |
| HVK*LVNE (Lys 65) | 676.3588 (+2) | 0 | - | - | - | - | 676.3647 (+2) | 0.0059 |
| VEK*DAIPE (Lys 318) | 707.3534 (+2) | 2 | - | - | - | - | 707.3578 (+2) | 0.0044 |
| CCDK*PLLE (Lys 304) | 774.3520 (+2) | 1 | - | - | - | - | 774.3662 (+2) | 0.0142 |
| LCK*VASLRE (Lys 100) | 794.9162 (+2) | 0 | - | - | - | - | 794.9163 (+2) | 0.0001 |
| VSRSLGK*VGTRCCTK*PE (Lys 455, Lys 463) | 987.8230 (+3) | 0 | - | - | - | - | 987.8313 (+3) | 0.0083 |
| KK*FWGK*YLYE (Lys 156, 160) | 1194.5985 (+2) | 0 | - | - | - | - | 1194.5626 (+2) | −0.0359 |
| NFVAFVDK*CCAADDKE (Lys 580) | 1201.5300 (+2) | 3 | - | - | - | - | 1201.5350 (+2) | 0.0050 |
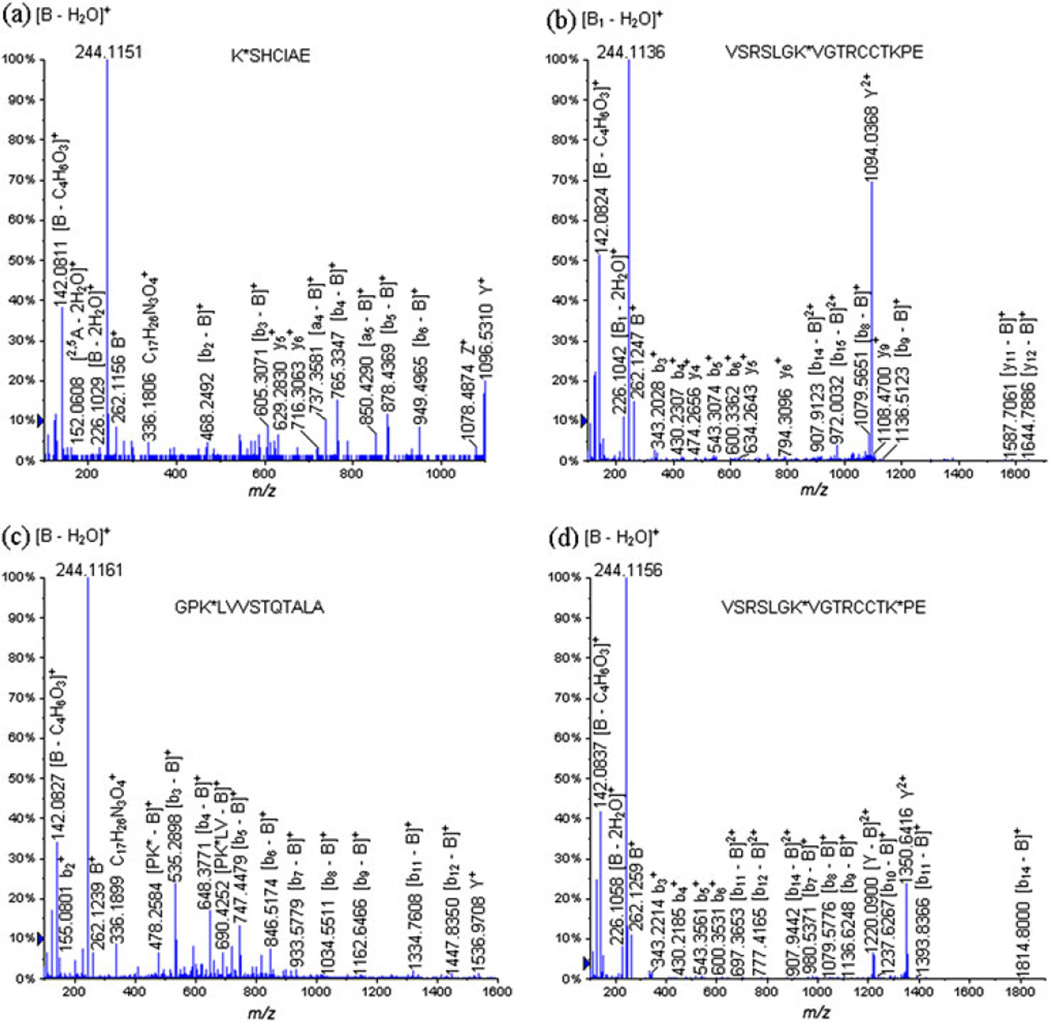
Figure 3 displays two examples of glycopeptides identified during the LC-ESI-QqTOF-MS/MS analysis of the GluC V8 digests of the neoglycoconjugate with a hapten-BSA ratio of 4.3: (a) K*SHCIAE (Lys 309) at m/z 679.3320 (+2) and (b) VSRSLGK*VGTRCCTKPE (Lys 455) at m/z 816.7646 (+3). The fragmentation pathway followed by the glycopeptide VSRSLGK*VGTRCCTKPE (Lys 455) at m/z 816.7646 (+3) allowed us to identify the glycation site exclusively on the Lys 455 residue (Fig. 3(b), Table 4). The precursor ion VSRSLGK*VGTRCCTKPE (Lys 455) at m/z 816.7646 (+3) produced characteristic carbohydrate moiety product ions, identified as follows: B+ at m/z 262.1247, [B − H2O]+ at m/z 244.1136, [B − 2H2O]+ at m/z 226.1042, [2,5A − 2H2O]+ at m/z 152.0669, [B − C4H6O3]+ at m/z 142.0824 and [2,5A1 − CH4O3]+ at m/z 124.0714. Furthermore, we detected the following product ion Y2+ at m/z 1094.0368 corresponding to the entire glycopeptide minus the loss of the entire carbohydrate portion. In addition, we also observed product ions corresponding to distinct fragmentation of the peptide backbone minus the loss of the entire carbohydrate moiety; needless to say, these product ions included the previously derivatized lysine: [y12 − B]+ at m/z 1644.7886, [y11 − B]+ at m/z 1587.7061, [b9 − B]+ at m/z 1136.5123, [b8 − B]+ at m/z 1079.5651, [b16 − B]2+ at m/z 1020.5277, [b7 − B]+ at m/z 980.5699, [b15 − B]2+ at m/z 972.0032, [b14 − B]2+ at m/z 907.9123 and [GK* − B]+ at m/z 438.2288. Therefore, these product ions allowed us to sequence and characterize this glycopeptide correctly.
Figure 3.
LC-ESI-QqTOF-MS/MS spectra of the glycopeptides obtained during the digestion of the neoglycoconjugates with the GluC V8 endoproteinase: (a) K*SHCIAE (Lys 309) at m/z 679.3320 (+2), (b) VSRSLGK*VGTRCCTKPE (Lys 455) at m/z 816.7646 (+3), (c) GPK*LVVSTQTALA (Lys 597) at m/z 899.5080 (+2) and VSRSLGK*VGTRCCTK*PE (Lys 455, Lys 463) at m/z 987.8313 (+3).
Table 4.
LC-ESI-QqTOF-MS/MS analysis of the glycated peptide VSRSLGK*VGTRCCTKPE (Lys 455) at m/z 816.7646 (+3)
| Molecular ion | Calculated | Experimental | Deviation | |
|---|---|---|---|---|
| m/z | m/z | Da | ||
| [y12 − B]+ | 1644.7821 | 1644.7886 | −0.0065 | |
| [y11 − B]+ | 1587.7607 | 1587.7061 | 0.0546 | |
| [b9 − B]+ | 1136.5422 | 1136.5123 | 0.0299 | |
|
|
1108.4868 | 1108.4700 | 0.0168 | |
| Y2+ | 1094.0541 | 1094.0368 | 0.0172 | |
| [b8 − B]+ | 1079.6208 | 1079.5651 | 0.0557 | |
|
|
1051.4654 | 1051.4701 | −0.0047 | |
| [b16 − B]2+ | 1020.5278 | 1020.5277 | 0.0001 | |
| [b7 − B]+ | 980.5524 | 980.5699 | −0.0175 | |
| [b15 − B]2+ | 972.0014 | 972.0032 | −0.0018 | |
| 950.4177 | 950.4802 | −0.0625 | ||
| [b14 − B]2+ | 907.9539 | 907.9123 | 0.0416 | |
| 794.3166 | 794.3096 | 0.0070 | ||
| 634.2859 | 634.2643 | 0.0216 | ||
|
|
600.3469 | 600.3362 | 0.0107 | |
|
|
543.3255 | 543.3074 | 0.0181 | |
|
|
474.2553 | 474.2656 | −0.0103 | |
| [GK* − B]+ | 438.2342 | 438.2288 | 0.0054 | |
|
|
430.2414 | 430.2307 | 0.0107 | |
|
|
373.2076 | 373.2188 | −0.0112 | |
|
|
343.2094 | 343.2028 | 0.0066 | |
| 336.1918 | 336.1787 | 0.0131 | ||
| B+ | 262.1290 | 262.1247 | 0.0043 | |
| 245.1126 | 245.1127 | −0.0001 | ||
| [B − H2O]+ | 244.1179 | 244.1136 | 0.0043 | |
| [B − 2H2O]+ | 226.1079 | 226.1042 | 0.0037 | |
|
|
222.1237 | 222.1242 | −0.0005 | |
| 187.1083 | 187.1204 | −0.0121 | ||
| [2,5A − 2H2O]+ | 152.0706 | 152.0669 | 0.0037 | |
| [y1 − H2]+ | 146.0453 | 146.0706 | −0.0253 | |
| [B − C4H6O3]+ | 142.0863 | 142.0824 | 0.0039 | |
| [2,5A1 − CH4O3]+ | 124.0757 | 124.0714 | 0.0043 | |
Finally, the following product ions, characteristic to a spacer portion attached to a lysine residue were also identified as follows: [C17H26N3O4]+ at m/z 336.1787 and [C11H16N3O2]+ at m/z 222.1242. The bidirectional sequence of the glycated peptide was also determined through the identification of the y- and b-ions (Fig. 3(b), Table 4).
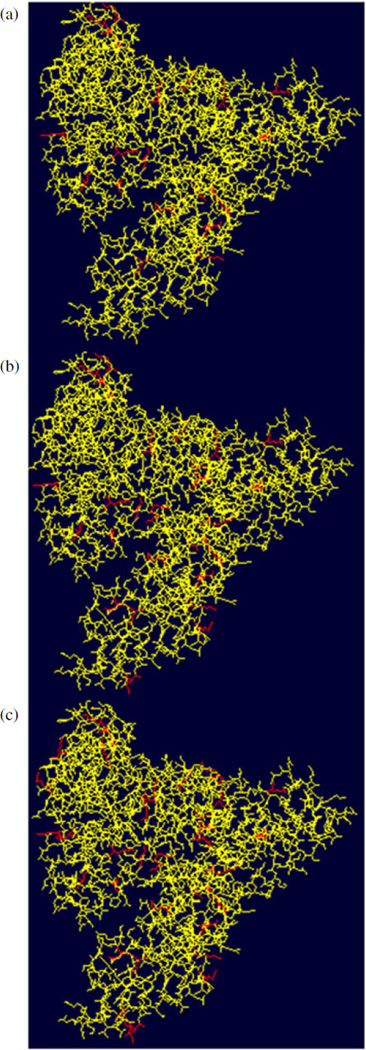
To sum it up, a total of 20 glycation sites were identified during the LC-ESI-QqTOF-MS/MS analysis of the tryptic and GluC V8 digests of the neoglycoconjugate with a hapten:BSA ratio of 4.3:1 (Fig. 4(a)) for total sequence coverage of 89%. Figure 5(a) displays the 3D structure of the BSA represented using the Swiss-Pdb Viewer software, where the glycated lysine residues are highlighted in red.[19,20] A quick glance at Fig. 5(a) allowed us to conclude that the glycated lysine residues are located on the outer surface of the protein.
Figure 4.
Neoglycoconjugate model sequences where the glycation sites are indicated by an asterix (red = identified on tryptic digests, blue = identified on GluC V8 digests and red and underlined = identified on both tryptic and GluC V8 digests) for the neoglycoconjugates with a hpaten-BSA ratio of (a) 4.3:1. (b) 6.6:1 and (c) 13.2:1.
Figure 5.
3D-structure of the neoglycoconjugate model vaccines. The glycated lysine residues are highlighted in red (Swiss-Pdb Viewer software) for the neoglycoconjugates with a hapten:BSA ratio of (a) 4.3:1, (b) 6.6:1 and (c) 13.2:1. This figure is available in colour online at wileyonlinelibrary.com/journal/jms
Mass spectrometry determination of the glycation sites on the neoglycoconjugate possessing a hapten: BSA ratio of 6.6:1
The LC-ESI-QqTOF-MS/MS analysis of the tryptic digests of the neoglycoconjugate with a hapten:BSA ratio of 6.6:1 allowed identification of 16 glycation sites on through sequencing of the observed glycopeptides (Table 1).
Figure 1(c) displays the following example of the glycopeptide which was identified during the CID-MS/MS analysis of the tryptic digests of the neoglycoconjugate with a hapten:BSA ratio of 6.6:1: VTK*CCTESLVNR (Lys 498) at m/z 990.4813 (+2).
The neoglycoconjugate with a hapten:BSA ratio of 6.6:1 was also digested with the GluC V8 endoproteinase and subsequently analyzed by LC-ESI-QqTOF-MS/MS. The glycated peptides were thus discovered after the manual sequencing of the peptides which were suspected to be glycated (same mass than a ‘peptide + carbohydrate linker’) and listed in Table 3.
The LC-ESI-QqTOF-MS/MS analysis of the GluC V8 digests of the neoglycoconjugate with a hapten:BSA ratio of 6.6:1 allowed us to identify 14 glycation sites. Moreover, the combination of the identified glycation sites from both the tryptic and GluC V8 digests gives us a total of 27 glycation sites with total sequence coverage of 90% (Fig. 4(b)).
In addition, the Fig. 5(b) which shows the 3D structure of the BSA with the glycated lysine residues highlighted in red enabled us to conclude that the glycated lysine residues are located on the outer surface of the BSA.
Mass spectrometry determination of the glycation sites on the neoglycoconjugate possessing a hapten: BSA ratio of 13.2:1
The neoglycoconjugate with a hapten:BSA ratio of 13.2:1 was also digested with trypsin and LC-ESI-QqTOF-MS/MS analysis of the digests allowed identification of 17 glycopeptides (Table 1), corresponding to 17 glycation sites.
Figure 1(d) shows the following example of identified glycopeptide, which was only discovered on the neoglycoconjugate with a hapten:BSA ratio of 13.2:1: FK*DLGEEHFK (Lys 36) at m/z 881.9491 (+2).
Finally, the analysis by LC-ESI-QqTOF-MS/MS of the GluC V8 digests permitted identification of 20 glycopeptides (Table 3). Moreover, we also observed glycopeptides carrying two carbohydrate-linker residues: VSRSLGK*VGTRCCTK*PE (Lys 455, Lys 463) at m/z 987.8313 (+3) and KK*FWGK*YLYE (Lys 156, 160) at m/z 1194.5626 (+2).
It is important to note in this rationale that we have identified glycopeptides which contained two diagnostic glycation sites. Unquestionably, two glycopeptides with the following peptide sequence VSRSLGKVGTRCCTKPE were detected containing one glycation site on the Lys 455, VSRSLGK*VGTRCCTKPE (Lys 455) at m/z 816.7483 (+3), and also with two glycation sites on the Lys 455 and Lys 463 residues: VSRSLGK*VGTRCCTK*PE (Lys 455, Lys 463) at m/z 987.8313 (+3). This observation supports the fact that our sample is composed of glycoforms. Figure 3(d) displays the spectra obtained during the CID-MS/MS analysis of the glycated peptide VSRSLGK*VGTRCCTK*PE (Lys 455, Lys 463) at m/z 987.8313 (+3), while the observed product ions and their mass-to-charge ratios are listed in Table 5.
Table 5.
LC-ESI-QqTOF-MS/MS analysis of the glycated peptide VSRSLGK*VGTRCCTK*PE (Lys 455, Lys 463) at m/z 987.8313 (+3)
| Molecular ion | Calculated | Experimental | Deviation | |
|---|---|---|---|---|
| m/z | m/z | Da | ||
| [b14 − B]+ | 1814.9000 | 1814.8000 | 0.1000 | |
| [b11 − B]+ | 1393.7910 | 1393.8366 | −0.0456 | |
| [y9 − B]+ | 1360.5973 | 1360.5399 | 0.0574 | |
| Y2+ | 1350.6700 | 1350.6416 | 0.0284 | |
| [b10 − B]+ | 1237.6899 | 1237.6267 | 0.0632 | |
| [Y − B]2+ | 1220.1091 | 1220.0900 | 0.0191 | |
| [y7 − B]+ | 1202.5282 | 1202.6109 | −0.0827 | |
| [b9 − B]+ | 1136.6422 | 1136.6248 | 0.0174 | |
| [b8 − B]+ | 1079.6208 | 1079.5776 | 0.0432 | |
| [b7 − B]+ | 980.5524 | 980.5371 | 0.0153 | |
| [b14 − B]2+ | 907.9539 | 907.9442 | 0.0097 | |
| [b12 − B]2+ | 777.4148 | 777.4165 | −0.0018 | |
| [b11 − B]2+ | 697.3994 | 697.3653 | 0.0341 | |
| [CTK* − B]+ | 642.2910 | 642.2561 | 0.0349 | |
| [y3 − B]+ | 625.3181 | 625.3205 | −0.0024 | |
| 600.3469 | 600.3531 | −0.0062 | ||
| 543.3255 | 543.3561 | −0.0306 | ||
| [K*VG − B]+ | 537.3026 | 537.2682 | 0.0344 | |
| [TK* − B]+ | 482.2604 | 482.2529 | 0.0075 | |
| [GK* − B]+ | 438.2342 | 438.2285 | 0.0057 | |
| 430.2414 | 430.2185 | 0.0229 | ||
| [K* − B]+ | 381.2127 | 381.1820 | 0.0307 | |
| 343.2094 | 343.2214 | −0.0120 | ||
| 336.1918 | 336.1890 | 0.0028 | ||
| B+ | 262.1290 | 262.1259 | 0.0031 | |
| [B − H2O]+ | 244.1179 | 244.1156 | 0.0023 | |
| [B − 2H2O]+ | 226.1079 | 226.1058 | 0.0021 | |
| 222.1237 | 222.1170 | 0.0067 | ||
| 187.1083 | 187.1118 | −0.0035 | ||
| [2,5A − 2H2O]+ | 152.0706 | 152.0673 | 0.0033 | |
| [y1 − H2]+ | 146.0453 | 146.0758 | −0.0305 | |
| [B − C4H6O3]+ | 142.0863 | 142.0837 | 0.0026 | |
| [2,5A − CH4O3]+ | 124.0757 | 124.0733 | 0.0024 | |
In the product ion scan of the precursor ion at m/z 987.8313 (+3), we observed the product ion Y2+ at m/z 1350.6416 formed by the loss of the carbohydrate moiety from one of the glycated lysine residues. The product ion [Y − B]2+ at m/z 1220.0900 was given rise to by the loss of the carbohydrate portions from the two glycated lysine residues. It is important to mention that the following series of diagnostic product ions: B+ at m/z 262.1259, [B − H2O]+ at m/z 244.1156, [B − 2H2O]+ at m/z 226.1058, [2,5A − 2H2O]+ at m/z 152.0673, [B − C4H6O3]+ at m/z 142.0837, [2,5A − CH4O3]+ at m/z 124.0733, [C17H26N3O4]+ at m/z 336.1890 and [C11H16N3O2]+ at m/z 222.1170 were formed specifically from both the carbohydrate portions and the spacer moieties of the glycopeptide precursor ion. Furthermore, we have observed during this CID-MS/MS analysis ‘proper’ peptide product ions that were identified as follows: [b14 − B]+ at m/z 1814.8000, [b11 − B]+ at m/z 1393.8366, [y9 − B]+ at m/z 1360.5399, [b10 − B]+ at m/z 1237.6267, [y7 − B]+ at m/z 1202.6109, [b9 − B]+ at m/z 1136.6248, [b8 − B]+ at m/z 1079.5776, [b7 − B]+ at m/z 980.5371, [b14 − B]2+ at m/z 907.9442, [b12 − B]2+ at m/z 777.4165, [b11 − B]2+ at m/z 697.3653, [CTK* − B]+ at m/z 642.2561, [y3 − B]+ at m/z 625.3205, [K*VG − B]+ at m/z 537.2682, [TK* − B]+ at m/z 482.2529, [GK* − B]+ at m/z 438.2285 and [K* − B]+ at m/z 381.1820. All of these product ions were formed by the loss of the carbohydrate moiety exclusively, which allowed us to localize unambiguously the two different glycation sites.
Thus, the CID-MS/MS sequencing of the previously mentioned glycopeptides allowed us to identify 22 glycation sites. Moreover, the combination of the CID-MS/MS analysis of the tryptic and GluC V8 digests enabled us to discover a total of 33 glycation sites with total sequence coverage of 88% (Fig. 4(c)). As previously observed for the neoglycoconjugates with hapten:BSA ratios of 4.3:1 and 6.6:1, the glycated lysine residues for the hapten-BSA glycoconjugate with a ratio of 13.2:1 are positioned on the outer surface of the protein (Fig. 5(c)).
CONCLUSION
The LC-ES-QqTOF-MS/MS analysis of the tryptic and GluC V8 digests allowed identification of a higher number of glycation sites than previously reported for the neoglycoconjugates with a hapten:BSA ratio of 4.3:1, 6.6:1 and 13.2:1.[12] As expected, the number of glycation sites in the neoglycoconjugates increases with the hapten:BSA ratio. Indeed, 20 glycation sites were observed for the neoglycoconjugate with a hapten:BSA ratio of 4.3:1, 27 for the neoglycoconjugate with a hapten:BSA ratio of 6.6:1 and 33 for the neoglycoconjugate with a hapten:BSA ratio of 13.2:1. Moreover, it was found that the glycated lysine residues are situated on the outer surface of the BSA.
It is also interesting to note that most of the observed glycation sites were at positions similar to those observed for the Bacillus anthracis neoglycoconjugate vaccine.[15]
Footnotes
The publisher acknowledges that the United States and Canadian Governments retain the right to publish or reproduce the published form of this work, or allow others to do so, for government purposes.
REFERENCES
- 1.Jones C. Vaccines based on the cell surface carbohydrates of pathogenic bacteria. Ann. Brazil. Acad. Sci. 2005;77:293. doi: 10.1590/s0001-37652005000200009. [DOI] [PubMed] [Google Scholar]
- 2.Costello CE, Contado-Miller JM, Cipollo JF. A glycomics platform for the analysis of permethylated oligosaccharide alditols. J. Am. Soc. Mass Spectrom. 2007;18:1799. doi: 10.1016/j.jasms.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Domon B, Costello CE. Structure elucidation of glycosphingolipids and gangliosides using high-performance tandem mass spectrometry. Biochemistry. 1988;27:1534. doi: 10.1021/bi00405a021. [DOI] [PubMed] [Google Scholar]
- 4.Domon B, Costello CE. A systematic nomenclature for carbohydrate fragmentationsin FAB-MS/MS spectra of glycoconjugates. Glycoconj. J. 1988;5:397. [Google Scholar]
- 5.Carr SA, Huddleston MJ, Bean MF. Selective identification and differentiation of N- and O-linked oligosaccharides in glycoproteins by liquid chromatography-mass spectrometry. Protein Sci. 1993;2:183–196. doi: 10.1002/pro.5560020207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rouse JC, Vath JE. On-the-probe sample cleanup strategies for glycoprotein-released carbohydrates prior to matrix-assisted laser desorption-ionization time-of-flight spectrometry. Anal. Biochem. 1996;238:82–92. doi: 10.1006/abio.1996.0255. [DOI] [PubMed] [Google Scholar]
- 7.Scanlan CN, Pantophlet R, Wormald MR, Saphire EO, Stanfield R, Wilson IA, Katinger H, Dwek RA, Rudd PM, Burton DR. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of α → 2 mannose residues on the outer face of gp120. J. Virol. 2002;76:7306–7321. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harvey DJ. N-(2-Diethylamino)Ethyl-4-Aminobenzamide derivative for high sensitivity mass spectrometric detection and structure determination of N-linked carbohydrates. Rapid Commun. Mass Spectrom. 2000;14:862–871. doi: 10.1002/(SICI)1097-0231(20000530)14:10<862::AID-RCM957>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 9.Mechref Y, Novotny MV. Structural investigations of glycoconjugates at high sensitivity. Chem. Rev. 2002;102:321–369. doi: 10.1021/cr0103017. [DOI] [PubMed] [Google Scholar]
- 10.Huddleston MJ, Bean MF, Carr SA. Collisional fragmentation of glycopeptides by electrospray ionization LC/MS and LC/MS/MS: methods for selective detection of glycopeptides in protein digests. Anal. Chem. 1993;65:877–884. doi: 10.1021/ac00055a009. [DOI] [PubMed] [Google Scholar]
- 11.Dell A, Morris HR. Glycoprotein structure determination by mass spectrometry. Science. 2001;291:2351–2356. doi: 10.1126/science.1058890. [DOI] [PubMed] [Google Scholar]
- 12.Jahouh F, Saksena R, Aiello D, Napoli A, Sindona G, Kovac P, Banoub JH. Determination of the glycation sites in neoglycoconjugates from the terminal monosaccharide antigen of the O-PS of Vibrio cholerae O1, serotype Ogawa, and BSA revealed by matrix-assisted laser desorption-ionization tandem mass spectrometry. J. Mass Spectrom. 2010;45:1148–1159. doi: 10.1002/jms.1796. [DOI] [PubMed] [Google Scholar]
- 13.Karas M, Krüger R. Ion ormation in MALDI: the cluster ionization mechanism. Chem. Rev. 2003;103:427–440. doi: 10.1021/cr010376a. [DOI] [PubMed] [Google Scholar]
- 14.Knochenmuss R, Vertes A. Time-delayed 2-pulse studies of MALDI matrix ionization mechanisms. J. Phys. Chem. B. 2000;104:5406–5410. [Google Scholar]
- 15.Jahouh F, Hou SJ, Kováč P, Banoub JH. Determination of the glycation sites of Bacillus anthracis neoglycoconjugate vaccine by MALDI-TOF/TOF-CID-MS/MS and LC-ESI-QqTOF-tandem mass spectrometry. J. Mass Spectrom. 2011;46:993. doi: 10.1002/jms.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saksena R, Chernyak A, Karavanov A, Kováč P. Conjugating low molecular mass carbohydrates to proteins. 1. Monitoring the progress of conjugation. Meth. Enzymol. 2003;362:125. doi: 10.1016/S0076-6879(03)01010-3. [DOI] [PubMed] [Google Scholar]
- 17.Roepstorff P, Fohlman J. Proposal for a common nomenclature for sequence ions in mass spectra of peptides. Biol. Mass Spectrom. 1984;11:601. doi: 10.1002/bms.1200111109. [DOI] [PubMed] [Google Scholar]
- 18.Johnson RS, Martin SA, Biemann K, Stults JT, Watson JT. Novel fragmentation process of peptides by collision-induced decomposition in a tandem mass spectrometer: differentiation of leucine and isoleucine. Anal. Chem. 1987;59:2621. doi: 10.1021/ac00148a019. [DOI] [PubMed] [Google Scholar]
- 19.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL Workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 20.Peitsch MC. Protein modeling by E-mail. Biol. Technol. 1995;13:658. [Google Scholar]