Abstract
This manuscript details a general method for patterning coplanar alkylsilane monolayers using deep-ultraviolet photolithography that has broad application for high fidelity patterning of cells of varying phenotype in long-term cultures. A polyethylene glycol monolayer was formed on a silica substrate and then patterned using 193 nm light from an ArF excimer laser. The regions of photoablation were then rederivatized with (3-trimethoxysilyl propyl) diethyltriamine (DETA), yielding high contrast cytophilic islands that promoted cell adhesion and growth. Rat hippocampal neurons, motoneurons, and myoblasts were then cultured in a defined, serum-free medium on the patterned surfaces for periods in excess of 40 days. This approach has been shown to be useful as a general method for the long-term culture of multiple cell types in highly defined spatial patterns and can be used for supporting complex cocultures for creating in vitro models for biological systems.
INTRODUCTION
Surface modification and patterning have been staple techniques in bioengineering and cell biology for decades. This is due primarily to the fact that the interaction of a cell or biomolecule with a particular material is dictated by the properties of the first few nanometers of the surface of the bulk material.1 Various methods have been developed to modify the surface characteristics of bulk materials to tune the biocompatibility of a material for promoting or preventing the attachment of cells and biomolecules.2 Such methods include, but are not limited to, physical adsorption of polymers, microcontact printing, microfluidic patterning, and photolithographic pattering. Patterning cells onto various substrates has been used extensively to create cellular microarrays in which the spatial orientation of the cells with respect to one another can be tightly controlled, thereby creating a unique microenvironment for each cell. In this way individual cells can be studied in isolation, in networks, or in complex cocultures where the interaction of multiple cell types can be comparatively studied.
The basis of cell patterning lies in the interaction of membrane bound receptors with adsorbed biomolecules on the underlying substrate.3, 4, 5, 6, 7 Cells secrete adhesion proteins (fibronectin, laminin, collagen, etc.), which adsorb onto a material by physical interactions (electrostatic, van der Waals, hydrophobic interactions, etc.). The nature and extent of the adsorption are determined by the chemical characteristics of the surface. Cytophobic surfaces, those that resist cell adhesion, tend to adsorb very little protein or adsorb it in a manner such that it loses its biological activity. Cytophilic surfaces, those that promote cell adhesion and growth, tend to adsorb protein such that its biological activity is retained allowing it to bind to membrane bound receptors, such as integrins. Surfaces that resist cell adhesion generally are nonpolar and either strongly hydrophobic (such as hydrocarbon or perfluorinated surfaces) or strongly hydrophilic [such as polyethylene glycol (PEG)].8, 9, 10 Surfaces that tend to promote cell adhesion usually consist of polar, hydrophilic molecules. Aminated molecules, for example, alkylsilane (3-trimethoxysilyl propyl) diethylenetriamine (DETA) and aminopropyl triethoxysilane, are considered to be strongly cytophilic and promote the growth and differentiation of cells.
Alkylsilane self-assembled monolayers (SAMs) are a class of compounds that are used extensively for modifying the surface properties of silica substrates (silicon, glass, etc.). These compounds come in a variety of chemistries that are used for many applications and are often utilized to modulate the biological interactions of silica substrates with biological materials.11 Work has shown that it is possible to pattern the aminated alkylsilane DETA using deep-ultraviolet (DUV) photolithography.12, 13, 14, 15, 16 Using DUV, SAMs were exposed to intense ultraviolet light from an ArF excimer laser (emission wavelength 193 nm). Upon exposure the SAM underwent a photochemical reaction that cleaved the carbon-nitrogen bonds within the molecule, thereby removing the cytophilic surface coating. The ablated regions were then rederivitized with a perfluorinated silane to create a surface that resisted cell adhesion. This method was extended to produce high contrast micrometer scale patterns that were used to pattern cells into microcircuits.16 Furthermore, it was shown that the geometric cues designed into the silane patterns could induce specific polarity in neural cells and direct the outgrowth of axonal processes and the development of the dendritic field.13
Here we present a novel cell patterning method using DUV photolithography where a PEG monolayer was formed on a silica substrate and then patterned using DUV photolithography that created ablated regions suitable for rederivitization. The patterned PEG monolayers were then reacted with DETA to form cytophilic islands. This is the reverse of previously established protocols where a DETA monolayer was first patterned and then rederivatized with a cytophobic silane, such as 1,1,2,2-perfluorooctyl trichlorosilane (13F).12 This approach avoided cross reaction of the PEG silane with the DETA, as the terminal amine group can act as a nucleophile to the silane moiety of the PEG silane. The versatility of these patterned PEG-DETA substrates was then demonstrated by the culture of multiple cell types. It has been shown that these surfaces could support attachment and growth of rat hippocampal neurons, motoneurons, and muscle cells in highly constrained geometric orientations for periods in excess of 40 days in a serum-free medium. The use of serum-free medium in these experiments was necessary to promote the survival of neuronal cell types, which exist in vivo bathed in cerebrospinal fluid, which contains very little solution phase protein . This method is particularly useful for incorporating patterned cocultures with microfabricated structures and devices [electrodes, field effect transistor (FETs), microcantilevers, etc.] for creating hybrid biological systems for a wide variety of applications, including drug discovery, where uncontrolled variables such as serum are not desirable. This is particularly true for cells and tissue types that require long culture times to obtain a mature phenotype representative of that found in vivo. The ability to now create high quality patterned arrays of cells in long-term cultures in way extends the toolbox needed to create cellular microarrays and microelectromechanical systems (MEMS) devices.
EXPERTIMENTAL SETUP AND METHODOLOGY
PEG-silane preparation protocol
Substrate cleaning
Silica substrates (glass and/or silicon wafers) were cleaned using serial acid baths. Substrates were arranged in ceramic staining racks (Thomas Scientific, Swedesboro, NJ). The substrates were then immersed in a 1:1 (vol:vol) solution of methanol and concentrated HCl for at least 1 h. After 1 h the substrates were rinsed (three times) in de-ionized and transferred to a solution of concentrated sulfuric acid for at least 1 h and then the substrates were washed three times in . After this step, the rinsed substrates were boiled in for 30 min. After boiling the samples were placed in an oven for at least 3 h. The resulting surfaces were analyzed using contact angle (CA) goniometry and x-ray photoelectron spectroscopy (XPS) to verify hydrophilicity of the surfaces and the elemental composition of the surfaces, respectively. Surfaces with a CA of less than 5.0° and an elemental carbon content of approximately 5.0% or less were considered suitable for initial derivitization.
PEG-silane coating procedure
Silica substrates were coated with a PEG-terminated silane by a modified protocol from Papra et al.15 Dry toluene was prepared by distillation over metallic sodium to remove any water or other contaminants. The toluene was refluxed for a minimum of 2 h prior to collection. The first toluene fraction was discarded to ensure a minimum of contamination. Toluene was collected in clean Pyrex bottles that had been placed in an oven for at least 2 h to drive off any water adsorbed to the surface of the glass. The remaining air in the Pyrex bottle was replaced with dry ultrapure nitrogen to ensure a minimum of gaseous oxygen in the reaction mixture. The toluene was then placed in the antechamber of an MBraun glovebox (MBraun, Stratham, NH). The chamber was evacuated and refilled with dry nitrogen three times before bringing the toluene into the main chamber. Preventing pre-exposure to gaseous oxygen and water vapor was a necessary precaution to prevent the premature polymerization of the monomeric silane. Oxygen and water can act as a nucleophile to a silane moiety, thus initiating polymerization in the solution phase rather than at the surface of the substrate. The PEG silane used for these experiments 2-[methoxypoly(ethyleneoxy)propyl]trimethoxysilane (PEGSi) (Gelest, Tullytown, PA) was added to the toluene to a final concentration of 0.1% by volume. The PEGSi-toluene solution was removed from the glovebox and brought into a chemical fume hood. Concentrated HCl was added to a final volume of 0.08% (0.8 ml HCl/L) and the solution briefly stirred. The cleaned silica substrates were removed from the oven and allowed to cool to room temperature before incubation in the PEGSi-toluene solution for 1 h at room temperature. The reaction vessel was loosely covered to prevent excessive exposure. After 1 h the samples were removed and rinsed in serial washes of toluene (one time), ethanol (two times), and (one time). The washed samples were blown dry under a stream of ultrapure nitrogen and were used immediately or stored in a desiccator until needed.
Deep-UV photolithography of PEG-silane monolayers
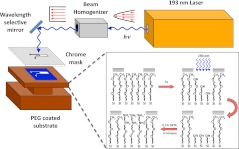
PEGSi modified silica substrates were patterned using DUV photolithography. The samples were patterned in a photolithography system of our own design (Fig. 1), which was based on a mask aligner, 193 nm ArF excimer laser (Lambda Physik, Santa Clara, CA) with an in-line beam homogenizer. A beam homogenizer was necessary for patterning of the monolayers as light emitted from the laser has a parabolic intensity profile (Fig. 1) that yields uneven ablation. The beam homogenizer refocuses the laser such that it yields a top-hat profile with an even distribution of light intensity across the ablation area. Samples were placed on the stage of the mask aligner under a chrome plated photomask, which contained the pattern to be ablated. The masks were written in dark-field polarity such that the areas corresponding to the ablated pattern were transparent and the remaining areas were opaque. When necessary, the substrate was precision aligned using the aligner stage to ensure micrometer precision placement of the pattern. The substrate was then brought into contact with the mask and a vacuum applied between the stage and mask to ensure a hard contact. A hard contact was used to minimize the gap between the substrate and mask to ensure a high contrast pattern with minimal edge effects due to refraction of the laser light. The substrates were then exposed to 193 nm ultraviolet laser light for 15–120 s with a pulse intensity of 200 mJ/pulse and a frequency of 10 Hz. After ablation the samples were removed from the aligner stage and stored for subsequent processing.
Figure 1.
(Color online) DUV photolithography system and proposed reaction scheme. Light emitted from the ArF laser has a parabolic profile that is passed through a beam homogenizer to produce a beam with a top-hat intensity profile. Using a wavelength selective mirror, the homogenized light is then reflected onto the sample, which has been placed under a patterned chrome photomask. The exposed regions of the PEGSi monolayer are then photoablated by the incident laser light leaving a substrate suitable for reaction with another silane, in this case DETA.
Backfill of patterned PEGSi monolayers with a DETA-silane
After ablation, the patterned PEGSi substrates were backfilled with DETA. Fresh distilled toluene was transferred into a Pyrex bottle that had been dried in an oven to remove excess surface water. Dry nitrogen was used to replace the air in the remaining volume of the bottle to minimize free oxygen. The bottle was sealed and transferred into an MBraun glovebox. DETA was added to the toluene to a final concentration of 0.1% (vol:vol). The DETA-toluene solution was removed from the glovebox and transferred to a Pyrex beaker, and the samples were immersed in the solution. To drive the reaction forward the solution was gently heated to no more than . Optimal reaction time was analyzed for these conditions by incubating the samples for 10, 20, and 30 min . After reaction with DETA the samples were allowed to cool to room temperature, washed three times with dry toluene, and heated to for additional 30 min in fresh toluene. The resulting samples were analyzed by XPS and contact angle goniometry.
Characterization of unpatterned and patterned silane monolayers
Contact angle goniometry analysis
The surface contact angle of the modified substrates was measured by contact angle goniometry. The samples were placed on the stage of a Ramé Hart (Netcong, NJ) contact angle goniometer. The sample stage was raised to close proximity with the outlet nozzle of the drop-dispensing pump. A drop of reagent grade ultrapure water was placed on the surface of the sample. A side-on image of the water drop on the surface was taken using an imaging video charge-coupled device camera and processed with DROPIMAGE ADVANCE software (Ramé Hart, Netcong, NJ). A normal contact angle for PEGSi was considered to be , and for DETA.
X-ray photoelectron spectroscopy analysis
XPS was performed on unpatterned and patterned samples to verify the relative amounts of the elements comprising the different monolayers. All analyses were performed on a Fisons ESCALab 220-XL with a monochromatic single-anode x-ray source. Samples were first introduced into the preplock chamber, which was then closed and evacuated for at least 30 min to a vacuum of less than by a roughing pump. The samples were then transferred into the main chamber. After introduction, the main chamber was allowed to settle to a pressure of less than . Elemental analysis was performed for identification of all relevant constituents of the silanes used according to their characteristic binding energies. A survey scan from 10 to 1200 eV was first performed to identify the major elemental constituents of the samples using a step increment of 1 eV, dwell time of 100 ms, and pass energy of 50 eV. The strong line peaks were analyzed using high resolution scans for Si 2p (103.3 eV) C 1s (284.5 eV), N 1s (402.5 eV), and O 1s (533.0 eV). High resolution scans were performed over a minimum spectral width of of the elemental center binding energy, a step increment of 0.1 eV, a dwell time of 20 ms, and pass energy of 50 eV. Relative elemental composition of the various elements was measured by calculating the area under the curves for the different elements and normalizing those areas by the sensitivity factor of the particular element, and the transmission function of the instrument. Relative atom percents were calculated from normalized peak areas.
Palladium-catalyzed metallization of patterned silane monolayers
Patterned samples were visualized using a palladium-catalyzed copper reduction reaction, modified from Ref. 17 that specifically deposits metallic copper on regions containing the amine-terminated silane DETA. The patterned substrates were immersed in a solution containing 0.8 mM palladium chloride and 0.6 mM NaCl for 10 min. The substrates were then rinsed three times in . A solution of 1 part dimethylamiobutyrate and 4 parts was prepared, and the samples were immersed for 10 min. The samples were again washed in , and then immersed in 10 ml of a solution of copper (II) sulfate with of formaldehyde (37.2%) until copper deposition was observed. Metallized patterns were imaged and analyzed using light microscopy.
Cell culture
Embryonic skeletal muscle
Skeletal muscle was dissected from the hind limb thighs of rat pups at embryonic day 18 (Charles River Laboratories, Wilmington, MA) according to a previously published protocol,18 with some modification. Tissue samples were collected in a sterile 15 ml centrifuge tube containing 1 ml of calcium and magnesium free phosphate buffered saline. Tissue samples were enzymatically disassociated using 3 ml of 0.05% trypsin-ethylenetriaminetetraacetic acid (EDTA) (Invitrogen, Carlsbad, CA) solution for 60 min in a water bath with agitation of 100 rpm. After 60 min, the trypsin solution was removed and 6 ml of L15 media (Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum (FBS) was added to terminate the trypsin action. The tissue was then mechanically triturated using a sterile narrow bore Pasteur pipette, allowed to settle for 3 min, and transferred to a 15 ml centrifuge tube. This was repeated three times. The dissociated tissue was centrifuged at 300 g for 10 min at on 6 ml of a 4% (wt/vol) cushion of bovine serum albumin (BSA). The pellet was resuspended in 10 ml FBS and plated on uncoated 100 mm Petri dishes for 20–30 min (depending on the amount of tissue) to allow contaminating fibroblasts to settle out. After 20–30 min the supernatant was layered on 6 ml of a 4% BSA cushion and then centrifuged at 300 g for 10 min at . The pellet was resuspended in 1.5 ml of medium.
Purified myocytes were plated at a density of 500–800 cells per square millimeter onto the modified coverslips. Myocytes were allowed to attach for 1 h after which time 3 ml of culture medium [neurobasal media containing B-27 (Invitrogen, Carlsbad, CA), Glutamax (Invitrogen, Carlsbad, CA), and pencillin/streptavidin] was added. Cultures were maintained in a incubator (relative humidity, 85%) and the culture medium was changed every 4 days.
Embryonic motoneuron
Spinal motoneurons were purified from ventral cords of embryonic day 14 (E14) rat pups. Briefly, rats were euthanized by asphyxiation. Ventral spinal cells from the embryo were collected in cold Hibernate E (BrainBits, Springfield, IL, SA)/GlutaMAX/antibiotic-antimycotic/B27 (Invitrogen, Carlsbad, CA). The cells were dissociated with 0.05% trypsin-EDTA (Invitrogen, Carlsbad, CA) treatment. The dissociated cells were layered over a 4 ml step gradient [Optipep diluted 1:1 (vol/vol) with Hibernate E/GlutaMAX/antibiotic-antimycotic/B27 and then made to 15%, 20%, 25%, and 35% (vol:vol) in Hibernate E/GlutaMAX/antibiotic-antimycotic/B27] followed by centrifugation for 15 min, using 800 g, at . This was a modification from the previously described protocols due to nonavailability of metrizamide.19, 20, 21 After centrifugation, four bands of cells were obtained, the topmost of which contained the motoneurons. These cells were further purified by immunopanning. The motoneurons were selected using the immune interaction between the motoneurons and the MAB192 antibody (1:2 dilution, ICN Biomedicals, Akron, OH) coated on the dishes.20, 22 The antibody recognized the low affinity nerve growth factor (NGF) receptor that is only expressed by ventral motoneurons at this age.23
Embryonic hippocampal neuron
Rat pups, at embryonic day 18, were dissected from timed pregnant rats that were euthanized using asphyxiation. Embryos were collected in ice cold Hibernate E/B27/Glutamax™/antibiotic-antimycotic. The hippocampi were isolated from the embryonic brain and collected in a tube containing 1 ml of Hibernate E/B27/Glutamax™/antibiotic-antimycotic. The embryonic hippocampal neurons were obtained by triturating the tissue using a Pasteur pipette. The 1 ml cell suspension was layered over a 4 ml step gradient [Optipep diluted 1:1 (vol:vol) with Hibernate E/GlutaMAX/antibiotic-antimycotic/B27 and then made to 15%, 20%, 25% and 35% (v/v) in Hibernate E/GlutaMAX™/antibiotic-antimycotic/B27] followed by centrifugation for 15 min, using 800g, at . This additional step enabled the removal of debris that arises during dissection from the damaged cells. After centrifugation, one strong band of cells was obtained at the top. The pyramidal hippocampal neurons constituted this band and were identified due to their large somas. The cells were resuspended in culture medium (neurobasal/B27/Glutamax/antibiotic-antimycotic) and plated at a density of 75 cells/mm.2, 13, 24, 25, 26, 27, 28 Half of the medium was changed after every 3–4 days.
RESULTS AND DISCUSSION
Contact angle measurements
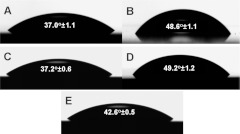
Contact angle measurements were performed on samples from all reaction conditions: control PEGSi monolayer, control DETA monolayer, and DETA backfilled onto unablated and ablated PEGSi monolayers, as well as PEGSi backfilled onto a control DETA surface. The results are seen in Fig. 2a. For the control PEGSi monolayers, the contact angle was measured to be , which is consistent with a PEGSi monolayer. The control DETA samples also exhibited a typical contact angle of . Figure 2c indicates the contact angle for DETA backfilled onto an unablated PEGSi monolayer. While the reaction conditions were identical to those of a plain clean glass immersed in the same reaction mixture, the contact angle, , did not show a significant degree of change from the control PEGSi sample. This was due to the presence of the methoxy-terminal group of the PEGSi protecting the monolayer from reaction with the DETA silane. The methoxy-terminal group was a poor nucleophile compared to the hydroxylated surface of the clean glass coverslips; therefore little to no reaction takes place on the PEGSi monolayer. Conversely, when the PEGSi was reacted onto a DETA surface the resulting contact angle and XPS data indicated evidence of PEGSi reacting onto the unprotected DETA forming a hybrid surface with a contact angle of [Fig. 2e], which is intermediate to and significantly different from that of PEGSi and DETA . Figure 2d shows the contact angle resulting for DETA reacted with a PEGSi monolayer ablated by DUV photolithography for 45 s. The contact angle for this set of conditions, , is statistically indistinguishable from that of the control DETA sample. This demonstrates that the PEGSi monolayer is ablated sufficiently to leave a surface suitable for formation of a normal DETA monolayer. Contact angles for the ablated PEGSi samples were .
Figure 2.
Contact angle measurements and droplet visualizations for (a) PEGSi control, (b) DETA control, (c) DETA backfilled into unablated PEGSi control, (d) DETA backfilled into ablated PEGSi, and (e) PEGSi backfilled into a DETA control.
XPS analysis
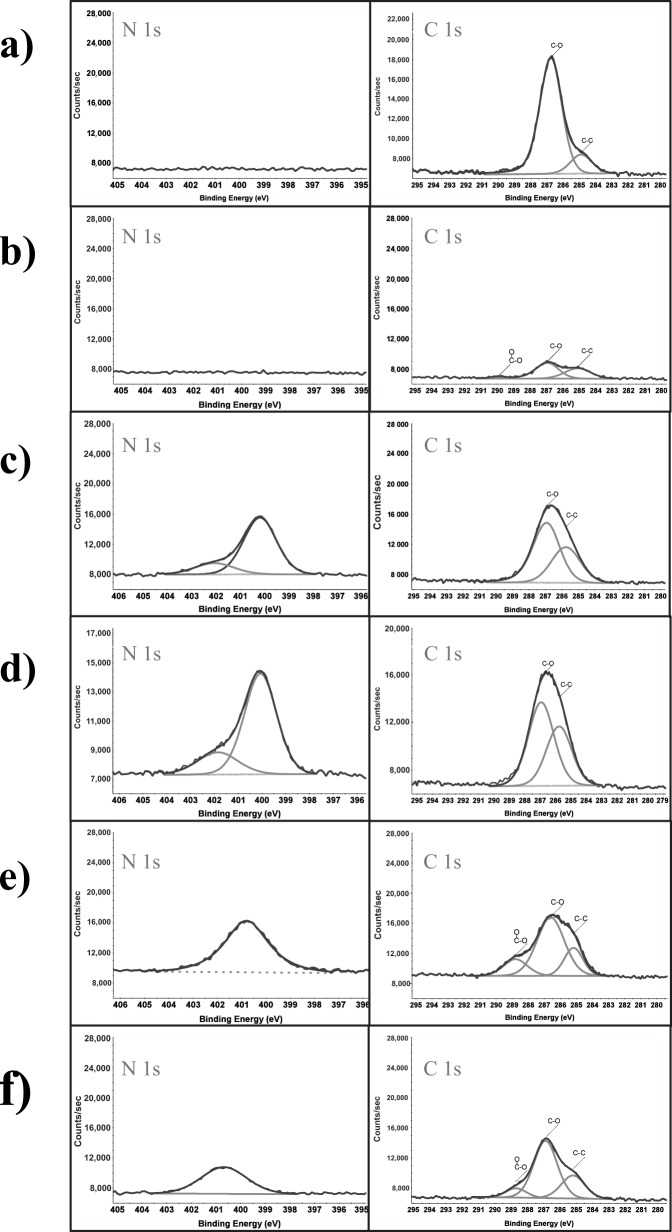
Analysis by XPS was performed to verify the chemical composition of the various surfaces tested in this study. Figure 3 indicates the representative surface compositions after optimization experiments described in Fig. 4, for PEGSi removal, and Fig. 5, for DETA incorporation. Figure 3a indicates a representative spectrum (survey spectrum, and high resolution spectra for C 1s and N 1s peaks) for a PEG monolayer. The characteristic peak for this surface is the C 1s peak. In Fig. 3a it can be seen that the C 1s spectrum is comprised of two partially overlapping peaks. The smaller peak, at , is characteristic of an aliphatic carbon peak, which corresponds to the three carbon spacer between the silane and the ethylene glycol units. The larger peak, at , corresponds to a carbon bound to an oxygen or nitrogen group. This is expected as the PEGSi used for this study is a 6–9mer of ethyleneglycol groups. The ratio of these two peaks is approximately 6:1 indicating the proportion of aliphatic to ether linkages expected from the PEGSi, Table 1.
Figure 3.
Representative high resolution N 1s and C 1s spectra for (a) PEGSi monolayer, (b) photoablated PEGSi, (c) DETA monolayer formed on ablated PEGSi surface, (d) DETA monolayer formed on clean Si, (e) mixed monolayer formed by reacting PEGSi onto a control DETA monolayer, and (f) DETA reacted onto unablated PEGSi monolayer.
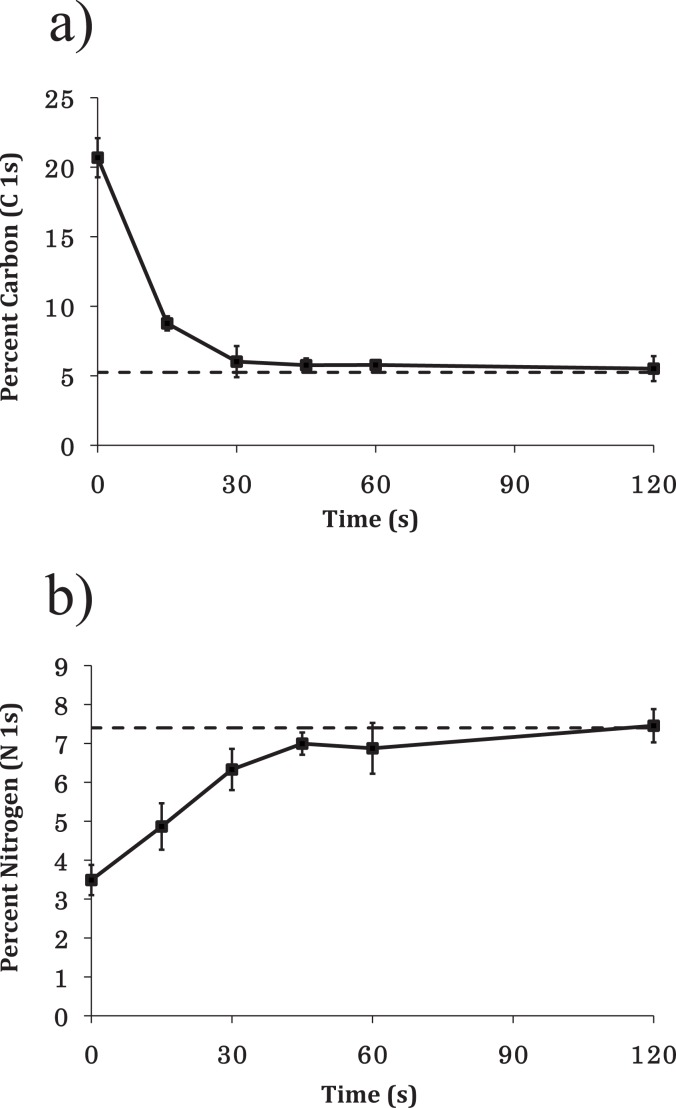
Figure 4.
(a) Atomic percent of C 1s signal vs ablation time. After 30 s atomic percent of C 1s was comparable to that of clean Si (dashed line). (b) Atomic percent of N 1s signal vs ablation time. Ablated samples were reacted with 0.1% DETA for 30 min to form a monolayer of DETA in the ablated regions. The graph shows nitrogen content (from N 1s high resolution spectra) vs ablation time. The dashed line indicates nitrogen content measured from control DETA samples.
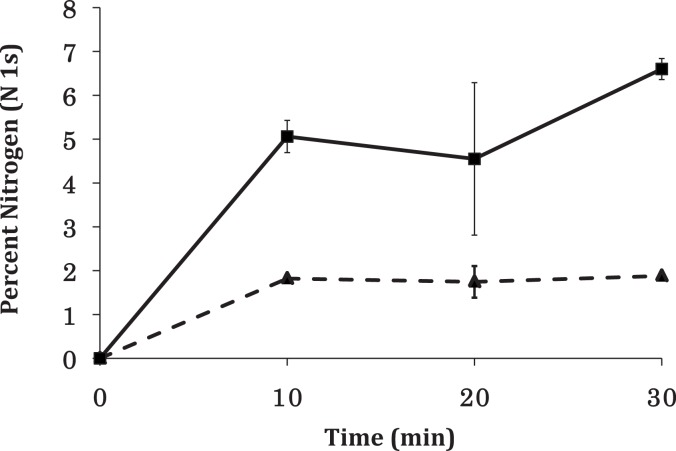
Figure 5.
Nitrogen content as incorporated into unablated vs ablated PEGSi monolayers as a function of reaction time. After 30 min reaction time, nitrogen content in the ablated PEGSi samples (solid line) reached values suitable for use in cell culture. Nitrogen content on unablated PEGSi monolayers (dashed line), however, reached a maximum of at about 10 min, which ensured that longer reaction times could be used to achieve an optimal DETA monolayer while not sacrificing pattern contrast.
Table 1.
Relative content and chemical states of carbon and nitrogen on silane surfaces. Surface types include clean silicon (Si), PEGSi, ablated PEGSi monolayer (ablPEGSi), DETA, PEGSi reacted onto DETA monolayer (DETA-PEGSi), DETA reacted onto an ablated PEGSi monolayer (ablPEGSi-DETA), and DETA reacted onto an unablated PEGSi monolayer (PEGSi-DETA).
| Si(%) | PEGSi(%) | ablPEGSi(%) | DETA-PEGSi(%) | ablPEGSi-DETA(%) | PEGSi-DETA(%) | DETA(%) | |
|---|---|---|---|---|---|---|---|
| N 1s | |||||||
| C–O/C–N | |||||||
| C–C | |||||||
| COO |
A time course study was then performed to determine the optimal ablation time for the PEGSi monolayer. Figure 4a shows the atom percent values for the C 1s spectra as a function of ablation time. It can be seen that at time zero the total carbon content was . After only 30 s of ablation time the carbon content was reduced to values comparable to those of clean silicon (dummy point at 240 s). After 30 s of ablation little change was seen in the carbon content of the samples tested. Figure 3b shows representative XPS spectra of a PEGSi coated silicon wafer ablated for 45 s by DUV photolithography. It should be noted that in these spectra the C–O component of the C 1s spectrum was drastically reduced compared to the C–C component [Fig. 3b]. This is evidence that the C–O bond of the PEGSi is photolabile at 193 nm and was possibly the site of reaction during photoablation.
The ablated PEGSi samples were then reacted with DETA to determine which ablation times yield optimal backfill of DETA into the patterned PEGSi monolayer. Figure 4b shows the percent nitrogen incorporation content versus ablation time. As seen in Fig. 4b, at 30 s of ablation time, the nitrogen content values were consistent with those of a normal DETA monolayer (between 7% and 8%). Beyond 30 s nitrogen content values remained consistent with those found in a normal DETA monolayer.
Figure 3c shows XPS spectra for a control DETA surface. The C 1s component of the XPS spectrum shows two components at 285.5 and 286.7 eV, which correspond to aliphatic carbon (C–C) and carbon in an amine (C–N) bonded environment, respectively. The primary component of the carbon signal comes from the C–N component, which was expected as five of the seven carbon atoms in DETA are bonded to nitrogen. The ratio of the two peaks reflected this (Table 1).
Figure 3d shows XPS spectra of DETA backfilled into an ablated PEGSi monolayer. The DETA monolayer deposited onto the ablated PEGSi surface was indistinguishable from that of the control DETA deposited onto a clean glass with some slight differences. The analysis of the N 1s spectrum indicated that the amount of nitrogen present was 7.4% (Table 1). This atomic percent has been found to be an ideal nitrogen content for cell culture experiments. The C 1s spectrum shows that, as with the control sample, the primary component of the C 1s signal was centered at 286.7 eV, which corresponds to an amine-bonded carbon. An aliphatic component was present at 285.5 eV and a small component at 288.9 eV, which corresponds to carbon in a highly oxidized state such as an amide or carboxylic acid group. This component of the signal may be due to oxidized carbon residue resulting from the ablation reacting with the amine containing DETA. The precise nature of carbons in this chemical state cannot be deduced from the data presented.
For the culture of cells on patterned PEGSi-DETA surfaces it was important to verify a high level of contrast between the PEGSi and DETA portions of the pattern. Therefore, it was important that a minimal amount of DETA should be incorporated into the PEGSi portion of the pattern. If DETA were able to freely react with the PEGSi monolayer, an overlayer of DETA would be formed on the PEGSi and effectively eliminate the surface contrast and hence the cytophobicity of the PEGSi surface. Also, it was necessary to optimize the reaction time to ensure DETA monolayer formation on the ablated regions while minimizing incorporation into the PEGSi regions. Figure 5 shows the nitrogen content versus reaction time as measured by XPS on control PEGSi samples as well as samples that had been ablated for 45 s. The gray trace shows the nitrogen content of DETA backfilled into ablated samples. Here it can be seen that after 30 min of ablation time the nitrogen content reached a level suitable for cell culture. Samples incubated for 20 min or less, while showing substantial DETA incorporation, were slightly lower than what we have found is optimal for culture. Conversely it can be seen in the black trace that after 10 min of reaction time, the relative nitrogen content incorporated into a control PEGSi sample does not change. DETA incorporation into a PEGSi monolayer reached its maximum at or before 10 min of reaction time. This was important as longer reaction times could be used to optimize DETA coating on ablated samples while not sacrificing incorporation of DETA into the PEGSi coating.
Figure 3e illustrates representative XPS spectra of PEGSi reacted onto a control DETA surface. Samples reacted with DETA, then PEGSi, yielded surface properties intermediate to those of either PEGSi or DETA alone. The high resolution spectrum of the C 1s region indicated that the dominant peak was found at 286.9 eV, which corresponds to either the ether or amine group found in PEGSi and DETA surfaces, respectively. However, the ratio of the ether/amine peak to that of the aliphatic carbon was intermediate to that of PEGSi and DETA. This suggests that this set of conditions yielded a mixed monolayer that was neither completely PEGSi nor DETA. As previously stated, the contact angle measurements supported this conclusion, as the contact angle was intermediate to that of pure PEGSi or DETA surfaces. It is likely that this is due to the PEGSi reacting directly to the free terminal amine of DETA, as it is a good nucleophile or could possibly be due to silanol sites unreacted during the initial DETA modification. These data also serve as further evidence that the methoxy-terminus of the PEGSi effectively protected it from a reaction with the DETA silane during backfill.
In order to test whether the nitrogen detected on the unablated PEGSi samples [Fig. 3f] was due to covalently linked or physically adsorbed DETA, samples were sonicated for 30 min in dry toluene to attempt to remove physically adsorbed silane. XPS analysis indicated that after sonication the measured nitrogen content was consistent to that in unsonicated samples (data not shown). This suggests that the DETA was covalently linked to the surface. It is possible that the DETA molecule could have reacted to the ether oxygen of PEG, or to defect sites on the substrate. However, a precise mechanism cannot be proposed from the data presented here.
As noted in Section IIIA, the contact angle of an unablated PEG surface reacted with DETA does not significantly differ from that of a control PEG surface. XPS analysis was performed to compare spectra from control PEGSi monolayers to that of unablated PEGSi monolayers reacted with DETA for 30 min at [Fig. 3f]. It can be seen from both the survey and high resolution spectra that only a very small amount of nitrogen can be detected . This evidence lends support to the hypothesis that the methoxy-terminated PEGSi effectively protected the PEG monolayer from reaction with the DETA silane allowing the formation of high contrast alkylsilane monolayer patterns.
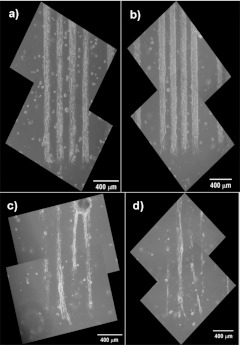
Metallization
Various types of cells were cultured on the PEGSi-DETA surfaces to verify the usefulness of this method for cell patterning; however, the patterns were first verified using a metallization technique. Figure 6 shows the results for both metallization and culture of different cell types on the PEGSi-DETA patterns. Figures 6a, 6c, 6e indicate palladium-catalyzed metallization of DETA, which in these images the metalized pattern corresponds to the dark regions of the picture, except in panel (e), which was imaged using a different filter set. The pattern in Fig. 6a consists of a series of wide lines used for culturing embryonic skeletal muscle. Figure 6c consists of an array of squares and Fig. 6e is a pattern designed specifically for creating two-cell networks of hippocampal neurons.29 The metallization results confirmed that patterns fabricated from the combination of PEGSi and DETA engineer high contrast patterns that were suitable for cell culture. Since the copper reduction reaction only occurs on regions containing the amine-terminated DETA, it can be seen that the DETA was confined primarily to regions where the PEGSi had been ablated. This result was in agreement with the XPS analysis and contact angle measurements. A faint amount of copper deposition was seen on the PEGSi regions of the DETA backfilled patterns, which indicated that a small amount of nitrogen was present in the PEGSi regions. This also was in agreement with XPS analysis (Fig. 3), which indicated that a low level of nitrogen signal in unablated PEGSi samples reacted with DETA. It was also possible that this low-level copper deposition could be due to reduced copper precipitating out onto the background in a nonspecific manner. For this reason, metallization experiments were performed on unablated PEGSi surfaces (results not shown), which confirmed that no copper deposition was seen on native PEGSi surfaces. Thus, it can be concluded that a small, yet detectable, amount of DETA was incorporated into the PEGSi regions. Both XPS and contact angle analysis indicated that the amount of DETA incorporated into the PEGSi regions was not sufficient to alter the surface properties from cytophobic to cytophilic, since the nitrogen content was well below that which is considered necessary for cell culture, and the contact angle was not significantly different from control PEGSi surfaces. Cell culture experiments confirmed that these surfaces were suitable for cell patterning.
Figure 6.
Metalized PEGSi-DETA patterns and resulting cell cultures with varying cell types. [(a), (c) and (e)] Metallization results for three different patterns [(a) lines, (b) squares, and (c) two-cell circuit pattern]. [(b), (d), and (f)] Rat cells cultured on PEGSi-DETA patterns [(b) embryonic skeletal muscle, (d) embryonic motoneuron, and (f) hippocampal neurons].
Cell culture
Multiple cell types were cultured on the PEGSi-DETA surface to verify that this method is broadly applicable to patterning cells on two-dimensional substrates. Figures 6b, 6d, 6f illustrate phase contrast microscopy images of live rat embryonic skeletal muscle [16 days in vitro (DIV)], motorneurons (10 DIV), and hippocampal neurons (19 DIV), respectively, while still in culture. In all cases the cells were confined to the DETA regions of the patterns and adhered to the patterns for the duration of the culture. Figure 6b shows embryonic skeletal muscle cultured on wide lines of DETA. It can be seen that the cells adhered and differentiated into myotubes (syncitium of myoblast that precedes myofibril formation) on the DETA regions of the lines. Myotube assembly was anisotropic with myotubes orienting themselves along the long axis of the pattern. This behavior has been demonstrated before using mouse skeletal muscle cell lines on microcontact printing patterned surfaces30 or on microfabricated cantilevers.31, 32 The myocytes then fused and differentiated into functional myotubes that exhibited spontaneous contraction as well as contraction under electrical stimulation (data not shown).
Embryonic motorneurons were also cultured on the patterned PEGSi-DETA surfaces. Figure 6d shows phase contrast images of motorneurons cultured for 10 days on surfaces patterned with an array of DETA squares. It can be seen that it was primarily the cell bodies that adhered to the square pattern, while processes were seen to extend across the PEGSi regions to contact cells on other squares. It was common to see cells extend processes onto the PEGSi while the motoneuron cell bodies remained attached to the DETA regions. This indicates that while the PEG repels adhesion of the cell soma, it is not completely repulsive to the attachment of neuronal processes. Figure 6f indicates embryonic hippocampal neurons cultured on PEGSi-DETA patterns designed for creating two-cell circuits. The patterns consisted of two circular regions ( diameter) intended for adhesion of the cell bodies. lines that extend in an arc from one somal adhesion site to another connected these circular regions, similar to Ravenscroft et al.16 and Molnar et al.29 The dashed lines extending radially from the somal adhesion sites further surrounded the somal adhesion sites. This geometry served to induce a specific polarity in the neurons similar to Stenger et al.,13 such that they would extend a dominant process (which would ultimately become the axon), and contact the soma of the opposing cell. The image in Fig. 6f illustrates cells adhered to the pattern shown in Fig. 6e. Cell bodies were confined to the circular somal adhesion sites that extended axonal processes that followed the line’s paths between the two-cell bodies.
Figure 7 indicates results from embryonic skeletal myoblasts cultured on PEGSi-DETA patterns over 41 days. Embryonic skeletal myoblasts were cultured on wide lines of DETA patterned on PEGSi surfaces. After 5 days the myoblasts fused and differentiated into myotubes. Figures 7a, 7b, 7c, 7d demonstrate myotubes formed on the DETA lines after 9, 16, 26, and 41 days in culture, respectively. It can be seen that after 26 days in culture the number of myotubes adhering to the patterns was reduced when compared to earlier culture times. This was due in part to the functional myotubes spontaneously contracting on the substrate and detaching. Despite this, a significant number of myotubes remained attached to the patterns for the duration of the experiment. The detachment of myotubes is not a desired outcome for the fabrication of hybrid devices. Ideally it should be possible to maintain a high density of myotubes in geometrically constrained patterns; however, for the purposes of these experiments, cell survival and pattern fidelity were more critical than myotube density. It was also observed that the areas between the DETA lines remained largely free of migrating cells and cell debris, and similar results were obtained in neuronal cell cultures. This long-term maintenance of pattern contrast and fidelity will prove critical in the application of this technique for creating long-term patterned cultures that can be used to create in vitro models for the study of cellular development, cell-cell interactions, and cellular response to exogenously applied compounds (growth factors, drugs, toxins, etc.).
Figure 7.
Skeletal muscle on patterned PEG-DETA surface remained confined to the DETA regions of the pattern up to 41 days on wide lines. (a) 9 days in culture, (b) 16 days in culture, (c) 26 days in culture, and (d) 41 days in culture. Although many of the myotubes at a later time pulled off the surface due to spontaneous contraction, the remaining myotubes were still confined to the patterns.
CONCLUSION
A novel method for patterning cells with PEGSi and DETA alkylsilane monolayers was developed. PEG-terminated monolayers were used as the cytophobic surface to prevent cell adhesion. The PEGSi monolayers were then patterned using deep-UV photolithography according to previously developed methods.14, 16, 13, 12 The patterned surfaces were then reacted with the amine-terminated silane DETA to create cell-adhesive islands in the nonadhesive PEGSi regions. Patterned and unpatterned surfaces were analyzed by XPS, contact angle goniometry, and palladium-catalyzed copper reduction metallization. This technique was then used to pattern various cell types from rat (motoneuron, hippocampal neuron, and skeletal muscle). Furthermore it was shown that cells were confined to the patterns for time periods in excess of 40 days.
Cell culture experiments indicated that this technique created surfaces suitable for long-term patterning of a wide variety of cell types. Culture results for hippocampal neurons, motorneurons, and skeletal muscle were presented. The applicability of this technique to a broad range of cell types make it ideal for creating complex cocultures with multiple cell types, and could be utilized to create in vitro model systems for in vivo biological circuits such as neuronal networks and neuromuscular junction constructs. Fidelity to geometric patterns is particularly important for these types of biological systems, as they often require weeks to months in culture to develop phenotypes that are representative of cells and tissues in vivo.33, 34 Furthermore, interrogation and post hoc analysis of complex cellular interactions in unpatterned cultures are difficult and often impractical. In this manner in vitro test beds could be created that control the spatial orientation of the cells in question on a well-defined substrate and in a highly controlled culture environment and then ultimately incorporated with microfluidic and solid-state electronic devices for further interrogation and experimentation.
ACKNOWLEDGMENT
The authors thank the National Institute of Health for providing the funds for this project through Grant No. R01-NS050452.
References
- Ratner B., Hoffman A., Schoen F., and Lemons J., Biomaterials Science: An Introduction to Materials in Medicine (Elsevier Academic, Amsterdam, 2004). [Google Scholar]
- Folch A. and Toner M., Annu. Rev. Biomed. Eng. 2, 227 (2000). 10.1146/annurev.bioeng.2.1.227 [DOI] [PubMed] [Google Scholar]
- Keselowsky B. G., Collard D. M., and Garcia A. J., J. Biomed. Mater. Res. Part A 66A, 247 (2003). 10.1002/jbm.a.10537 [DOI] [PubMed] [Google Scholar]
- Capadona J. R., Collard D. M., and Garcia A. J., Langmuir 19, 1847 (2003). 10.1021/la026244+ [DOI] [Google Scholar]
- Michael K. E., Vernekar V. N., Keselowsky B. G., Meredith J. C., Latour R. A., and Garcia A. J., Langmuir 19, 8033 (2003). 10.1021/la034810a [DOI] [Google Scholar]
- Keselowsky B. G., Collard D. M., and Garcia A. J., Proc. Natl. Acad. Sci. U.S.A. 102, 5953 (2005). 10.1073/pnas.0407356102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Chen S., Giachelli C. M., Ratner B. D., and Jiang S., J. Biomed. Mater. Res. Part A 74A, 23 (2005). 10.1002/jbm.a.30221 [DOI] [PubMed] [Google Scholar]
- Prime K. L. and Whitesides G. M., Science 252, 1164 (1991). 10.1126/science.252.5009.1164 [DOI] [PubMed] [Google Scholar]
- Prime K. L. and Whitesides G. M., J. Am. Chem. Soc. 115, 10714 (1993). 10.1021/ja00076a032 [DOI] [Google Scholar]
- Cooper E., Wiggs R., Hutt D. A., Parker L., Leggett G. J., and Parker T. L., J. Mater. Chem. 7, 435 (1997). 10.1039/a607204f [DOI] [Google Scholar]
- Blawas A. S. and Reichert W. M., Biomaterials 19, 595 (1998). 10.1016/S0142-9612(97)00218-4 [DOI] [PubMed] [Google Scholar]
- Stenger D. A., Georger J. H., Dulcey C. S., Hickman J. J., Rudolph A. S., Nielsen T. B., McCort S. M., and Calvert J. M., J. Am. Chem. Soc. 114, 8435 (1992). 10.1021/ja00048a013 [DOI] [Google Scholar]
- Stenger D. A. et al. , J. Neurosci. Methods 82, 167 (1998). 10.1016/S0165-0270(98)00047-8 [DOI] [PubMed] [Google Scholar]
- Hickman J. J., Bhatia S. K., Quong J. N., Shoen P., Stenger D. A., Pike C. J., and Cotman C. W., J. Vac. Sci. Technol. A 12, 607 (1994). 10.1116/1.578844 [DOI] [Google Scholar]
- Papra A., Gadegaard N., and Larsen N. B., Langmuir 17, 1457 (2001). 10.1021/la000609d [DOI] [Google Scholar]
- Ravenscroft M. S. et al. , J. Am. Chem. Soc. 120, 12169 (1998). 10.1021/ja973669n [DOI] [Google Scholar]
- Kind H., Bittner A. M., Cavalleri O., Kern K., and Greber T., J. Phys. Chem. B 102, 7582 (1998). 10.1021/jp981684o [DOI] [Google Scholar]
- Das M., Wilson K., Molnar P., and Hickman J. J., Nat. Protoc. 2, 1795 (2007). 10.1038/nprot.2007.229 [DOI] [PubMed] [Google Scholar]
- Das M., Bharqava N., Gregory C., Riedel L., Molnar P., and Hickman J. J., In Vitro Cell. Dev. Biol.: Anim. 41, 343 (2005). [DOI] [PubMed] [Google Scholar]
- Das M., Molnar P., Devaraj H., Poeta M., and Hickman J. J., Biotechnol. Prog. 19, 1756 (2003). 10.1021/bp034076l [DOI] [PubMed] [Google Scholar]
- Schnaar R. I. and Schaffner A. E., J. Neurosci. 1, 204 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettling C. et al. , J. Neurosci. 15, 3128 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q. and E. M.Johnson, Jr., J. Neurosci. 8, 3481 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner A. E., Barker J. L., Stenger D. A., and Hickman J. J., J. Neurosci. Methods 62, 111 (1995). 10.1016/0165-0270(95)00063-1 [DOI] [PubMed] [Google Scholar]
- Brewer G. J., J. Neurosci. Res. 42, 674 (1995). 10.1002/jnr.490420510 [DOI] [PubMed] [Google Scholar]
- Brewer G. J., Exp. Neurol. 159, 237 (1999). 10.1006/exnr.1999.7123 [DOI] [PubMed] [Google Scholar]
- Brewer G. J., Torricelli J. R., Evege E. K., and Price P. J., J. Neurosci. Res. 35, 567 (1993). 10.1002/jnr.490350513 [DOI] [PubMed] [Google Scholar]
- Stenger D. A., Pike C. J., Hickman J. J., and Cotman C. W., Brain Res. 630, 136 (1993). 10.1016/0006-8993(93)90651-3 [DOI] [PubMed] [Google Scholar]
- Molnar P., Kang J., Bhargava N., Das M., and Hickman J. J., in Patch-Clamp Methods and Protocols, edited by Molnar P. and Hickman J. J. (Humana, New York, 2007), Vol. 403, pp. 165–173. 10.1007/978-1-59745-529-9_10 [DOI] [Google Scholar]
- Tourovskaia A., Figueroa-Masot X., and Folch A., Nat. Protoc. 1, 1092 (2006). 10.1038/nprot.2006.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K., Das M., Wahl K. J., Colton R. J., and Hickman J. J., PLoS ONE 5, e11042 (2010). 10.1371/journal.pone.0011042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K., Molnar P., and Hickman J., Lab Chip 7, 920 (2007). 10.1039/b617939h [DOI] [PubMed] [Google Scholar]
- Potter S. M. and DeMarse T. B., J. Neurosci. Methods 110, 17 (2001). 10.1016/S0165-0270(01)00412-5 [DOI] [PubMed] [Google Scholar]
- Tourovskaia A., Figueroa-Masot X., and Folch A., Lab Chip 5, 14 (2005). 10.1039/b405719h [DOI] [PubMed] [Google Scholar]