Abstract
The cold shock domain is among the most evolutionarily conserved nucleic acid binding domains from prokaryotes to higher eukaryotes, including plants. Although eukaryotic cold shock domain proteins have been extensively studied as transcriptional and post-transcriptional regulators during various developmental processes, their functional roles in plants remains poorly understood. In this study, AtCSP3 (At2g17870), which is one of four Arabidopsis thaliana c old s hock domain proteins (AtCSPs), was functionally characterized. Quantitative RT-PCR analysis confirmed high expression of AtCSP3 in reproductive and meristematic tissues. A homozygous atcsp3 loss-of-function mutant exhibits an overall reduced seedling size, stunted and orbicular rosette leaves, reduced petiole length, and curled leaf blades. Palisade mesophyll cells are smaller and more circular in atcsp3 leaves. Cell size analysis indicated that the reduced size of the circular mesophyll cells appears to be generated by a reduction of cell length along the leaf-length axis, resulting in an orbicular leaf shape. It was also determined that leaf cell expansion is impaired for lateral leaf development in the atcsp3 loss-of-function mutant, but leaf cell proliferation is not affected. AtCSP3 loss-of-function resulted in a dramatic reduction of LNG1 transcript, a gene that is involved in two-dimensional leaf polarity regulation. Transient subcellular localization of AtCSP3 in onion epidermal cells confirmed a nucleocytoplasmic localization pattern. Collectively, these data suggest that AtCSP3 is functionally linked to the regulation of leaf length by affecting LNG1 transcript accumulation during leaf development. A putative function of AtCSP3 as an RNA binding protein is also discussed in relation to leaf development.
Key words: Arabidopsis, cold shock domain proteins, leaf development.
Introduction
The cold shock domain (CSD) is regarded as one of the most conserved protein domains that is widespread from bacteria to mammals. The CSD contains two consensus RNA binding motifs (RNP-1 and RNP-2) which facilitate functions such as nucleic acid binding, RNA chaperone activity, post-transcriptional regulation, and transcription regulation (Graumann and Marahiel, 1998; Sommerville, 1999; Horn et al., 2007). In eukaryotes, cold shock domain proteins contain various N-terminal and C-terminal auxiliary domains in addition to the CSD. The well studied human cold shock domain protein (Y-box binding protein-1, YB-1) performs pleiotropic roles in gene transcriptional regulation, DNA repair, and external stimuli response such as drug resistance (Kohno et al., 2003). YB-1 activation by Akt-mediated phosphorylation results in its translocation to the nucleus, leading to the regulation of cell proliferation in human ovarian cancer cells (Basaki et al., 2007). In addition, phosphorylation of YB-1 mediated by Akt-1 is regarded as the inducer for increased expression of genes related to cell proliferation and stress response by the translation of silent mRNA (Evdokimova et al., 2006).
Since Kingsley and Palis first described plant cold shock domain proteins (CSPs) (Kingsley and Palis, 1994), functional studies have been initiated in rice, wheat, and Arabidopsis. A subsequent phylogenetic analysis of plant CSPs confirmed that they are highly conserved in the Plant Kingdom (Karlson and Imai, 2003). A wheat CSP (WCSP1), two rice CSPs (OsCSP1, OsCSP2), and four Arabidopsis CSPs (AtCSPs, CSDPs) have been confirmed as functional nucleic acid binding proteins. Collective studies in plants suggest a putative functional role as RNA chaperones, although the precise biological role of cold shock domain proteins in planta remains elusive (Nakaminami et al., 2006, 2009; Fusaro et al., 2007; Kim et al., 2007, 2009; Sasaki et al., 2007; Chaikam and Karlson, 2008; Park et al., 2009; Yang and Karlson, 2011). The first functional analysis for an Arabidopsis CSP (AtGRP2/AtCSP2/CSDP2; At4g38680) by RNAi-induced gene silencing resulted in early flowering, reduced stamen number, and abnormalities during seed embryogenesis (Fusaro et al., 2007). The relationship of plant CSPs to plant development was further supported by a recent detailed investigation monitoring the entire AtCSP gene family throughout all stages of development (Nakaminami et al., 2009). Recently, two Arabidopsis cold shock domain proteins (AtCSP3; At2g17870 and AtCSP1/AtCSDP1; At4g36020) have been functionally linked to abiotic stress (Kim et al., 2009; Park et al., 2009).
Leaf organogenesis is divided into three stages: leaf initiation, establishment of leaf polarity, and cell expansion resulting in the final leaf shape (Tsukaya, 2005, 2006). In general, these stages are regulated by different gene groups (Byrne, 2005; Barkoulas et al., 2007). Leaf shape is initially established in two parts; leaf petioles and leaf blades. Arabidopsis leaf blades expand in two-dimensions which are defined as leaf-length (longitudinal) and leaf-width (lateral) axes. The ratio of the two directions is used as the criterion to determine leaf blade shape, which is further determined by the cell distribution, cell size, and their interaction in the leaf lamina. Both directions of leaf expansion are regulated by the ANGUSTIFOLIA (AN) and ROTUNDIFOLIA (ROT) genes which regulate two independent directional processes, respectively. ROTUNDIFOLIA 3 (ROT3) and ROTUNDIFOLIA 4 (ROT4) are involved in the regulation of leaf length polarity growth. A null allele of rot3 exhibits stunted leaf and floral growth and ROT3 encodes Cytochrome P450 (CYP90C1) which is a late-step regulator in the brassinosteroid biosynthesis pathway (Tsuge et al., 1996; Kim et al., 1998, 2005). Over-expression of ROT3 results in longer leaves in the longitudinal direction but is not altered in leaf width (Kim et al., 1999). An activation tagging line (rot4-1D), whose protein encodes a membrane-bound small peptide, possesses a similar leaf shape phenotype as that of rot3, but its small leaf shape is caused by a decrease in cell proliferation unlike that of ROT3 (Narita et al., 2004). Two other leaf-length regulation genes (LONGIFOLIA; LNG1 and LONGIFOLIA2; LNG2), were shown to function independently of ROT3. An activation tagged line of lng1-1D exhibited a long leaf blade phenotype along with serrated margins and other elongated tissues. Loss-of-function mutants for lng1 and lng2 exhibited a shortened length of leaf blades (Lee et al., 2006).
Regarding the regulation of leaf width, loss-of-function of ANGUSTIFOLIA (AN), SPIKE1, ANGUSTIFOLIA3 (AN3)/GRF-INTERACTING FACTOR1 (AtGIF1), and over-expression of AtHB13 (OxAtHB13) caused a similar narrow leaf shape in the leaf-width direction (Hanson et al., 2001; Folkers et al., 2002; Kim et al., 2002; Qiu et al., 2002; Kim and Kende, 2004). The AN protein, which is similar to the members of the animal CtBP protein family, arranges cortical microtubules to facilitate polar expansion of leaf cells (Folkers et al., 2002). SPIKE1 plays a role in cytoskeletal reorganization, which determines overall cell shape and tissue development (Qiu et al., 2002). Therefore, it was hypothesized that specific regulation of leaf-width is related to cytoskeleton formation which is comprised of cortical microtubules. A loss-of-function mutant of AN3/AtGIF1 and GROWTH-REGULATING FACTOR 5 (AtGRF5) showed a narrow leaf phenotype in the leaf-length direction which is caused by the defect of cell numbers in leaf blades (Kim and Kende, 2004; Horiguchi et al., 2005). AN3, a homologue of the human transcription factor SYT, accumulated in leaf primordia and this pattern was in accordance with other AtGRFs, AtGRF5 and AtGRF9, but not with AtGRF8 (Horiguchi et al., 2005). In addition, yeast two hybrid analysis identified an interaction between AN3/AtGIF1 and AtGRF5 together with AtGRF9 (Horiguchi et al., 2005). Over-expression of AN3 and AtGRF5 showed a 20–30% larger leaf size compared with the wild type, which resulted from an increase in leaf cell number (Horiguchi et al., 2005).
By characterizing two independent T-DNA insertion alleles, which exhibited a small and stunted leaf shape, the first direct evidence of AtCSP3 functioning as a regulatory protein in plant growth and development is reported. In addition, the alteration in expression of a leaf development regulatory gene that is affected by loss of function of AtCSP3 during leaf development in Arabidopsis is also reported.
Materials and methods
Plant material and culture conditions
Seeds of atcsp3-1, atcsp3-2, and atcsp3-3 T-DNA insertion mutants were obtained from the Arabidopsis Biological Research Center (ABRC) with stock numbers of SALK_144972, Wisc_DsLox353G12, and SALK_022658, respectively. Col-0 wild-type seeds were purchased from Lehle Seeds (Round Rock, TX, USA). Prior to planting, seeds were stratified for 4 d at 4 °C without light. All plants were grown in Metromix 360 (Scotts Co., Marysville, OH, USA) under 16/8h and 8/16h of light/dark for long-day and short-day conditions, respectively, at 23 °C.
Quantification of transcript abundance
Experimental details which describe the transcript profiles of AtCSP3 across the stages of development and within atcsp3 mutant alleles are provided in the Supplementary material and Supplementary Table S1 at JXB online.
Morphological analysis of loss-of-function mutant of AtCSP3
Wild-type Col-0, atcsp3-2, and atcsp3-1 were grown under long-day conditions at 23 °C up to 56 DAG for morphological analyses. Representative photographs were taken among 20 different plants. For root elongation and germination tests, sterilized seeds were grown on 1× Murashige and Skoog (MS)+vitamin B5 mixture/1% sucrose/1% phytagar (Caisson Labs, North Logan, UT, USA) plates under long-day conditions. For the assessment of root elongation, seedlings were grown vertically for 5 DAG on 1× MS/1% agarose including 1% sucrose, and 5 DAG plants which had the same root length were transplanted to new plates and maintained under long-day conditions. Root elongation was determined by measuring the difference between root length at4 d after transplanting and initial root length at the time of transplanting. Statistical significance was determined with a Student’s t test analysis of atcsp3 mutant data and compared with the wild-type data.
Whole plants, aligned siliques, and leaf photographs were taken by a Nikon Coolpix 8700 digital camera (Melville, NY, Nikon). Additional photographs of small-sized tissues such as flowers and seeds were taken under a Nikon SMZ-U dissecting microscope equipped with a Nikon DXM 1200 CCD camera (Melville, NY, Nikon).
Microscopic observation and anatomic analysis
For the observation of palisade cells, leaves were fixed in Farmer’s fixative (ethanol:acetic acid 3:1 v/v) for 2–4h. Chlorophyll was completely removed by washing with 70% and 100% ethanol. To clear leaf tissue for microscopic observation, fixed leaves were soaked in 5 N NaOH at 60 °C for 2h. Nomarsky images were taken with a Nikon ECLIPSE E600 differential interference contrast (DIC) microscope equipped with a Nikon DXM 1200 CCD camera system (Melville, NY, Nikon). To observe epidermal cells on adaxial leaf surfaces, live 5th leaf tissue from a 28 DAG plant was stained with 100 µg ml–1 propidium iodide diluted in 0.1M L-arginine buffer (pH 12.4) for 2–5min. Stained leaf pictures were taken by a Zeiss Axioimager LSM 510 confocal microscope with Z-stack image generation mode. ImageJ software was used to analyse the anatomical features. Statistical significance was determined with a Student’s t test analysis of atcsp3 mutant data and compared with the wild-type data.
Transient subcellular localization of AtCSP3
The coding region for the AtCSP3 gene was amplified with primerscontaining the following restriction enzyme digestion sites; 59-TCTGTC GACATGGCGATGGAAGATCAATC-39 and 59-AGACCATGGTTTA- AGCAACCGAAGTACATT-39. Amplified PCR products were purifiedand digested with NcoI and SalI and inserted into a pre-digested sGFP(s65T) vector. DM-10 tungsten particles (Hercules, CA, Bio-Rad) were coated with 1 µg of plasmid. Particle bombardment was carried out into onion epidermal cells by using a Biolistic® PDS-1000 particle bombardment system (Hercules, CA, Bio-Rad) using the following conditions: 25 inches of Hg vacuum, 1000 psi rupture disk, and 12cm target distance. Bombarded tissue was incubated overnight at 20 °C in the dark. Images were obtained by a Zeiss Axioimager LSM-501 confocal microscope and analyzed by LSM image analysis software (German, Carl Zeiss AG).
Results
Tissue-specific expression of AtCSP3 andisolation of atcsp3 mutant alleles
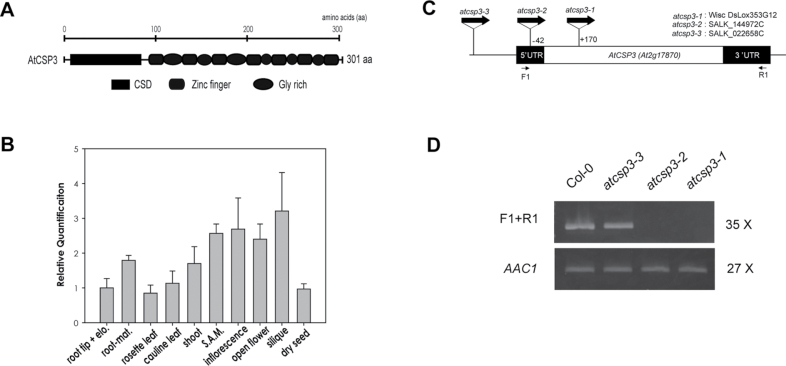
The AtCSP3 (At2g17870) genomic sequence contains a single exon with a complete open reading frame of 906 base pairs encoding a protein of 301 amino acids (Fig. 1A). The AtCSP3 protein contains a well-conserved N-terminal cold shock domain (CSD) and seven C-terminal CCHC retroviral-like zinc finger motifs interspersed by glycine-rich regions. Quantitative real-time PCR (qRT-PCR) confirmed the expression of AtCSP3 transcripts in all vegetative and reproductive tissues (Fig. 1B). In root tissue, AtCSP3 is highly expressed in the maturation area compared with the elongation zone and root tips. In aerial tissues, AtCSP3 transcripts are highly expressed in tissues with active growth and cell division such as shoot apices, inflorescences, and developing siliques. Relative mRNA accumulation levels for AtCSP3 were lower in leaves and dry seeds. Taken together, the qRT-PCR results indicated that the AtCSP3 gene is expressed in multiple tissues but shows elevated expression in meristematic and actively dividing tissues.
Fig. 1.
Tissue-specific gene expression and T-DNA insertion alleles of AtCSP3. (A) Domain architecture of the AtCSP3 protein. (B) qRT-PCR tissue specific analysis of AtCSP3 mRNA expression. Actin2 was used as an internal control for normalization. All data points for individual AtCSPs were calibrated with the respective normalized value of cDNA from root tips. The data shown represent the average and standard deviation of three replicates (elon. and mat. indicate root elongation and maturation zones; S.A.M. designates shoot apical meristem). (C) Genetic map of T-DNA insertion positions within the AtCSP3 locus. The white box represents the AtCSP3 exon and black boxes represent 59 and 39 UTRs. D) Semi-quantitative RT-PCR analysis of AtCSP3 expression of wild-type (Col-0) and AtCSP3 T-DNA insertion alleles. PCR was performed for the described cycle numbers with a flanking region primer. Selected primer regions were marked in (C). Arabidopsis actin 1 (AAc1) was used as an internal control. This figure is a representative image from three replicate reactions.
The SIGnAL T-DNA Express website identified two SALK lines (SALK_144972C and SALK_022658C) and a Wisconsin line (WISC DsLox353G12) containing putative T-DNA insertions within the AtCSP3 locus (http://signal.salk.edu/cgi-bin/tdnaexpress). Homozygous lines for each accession were confirmed by genotyping individual plants for several generations (data not shown). DNA sequence analysis of genotyping results confirmed that SALK_022658C contains a T-DNA insertion in the promoter region far upstream from the initial codon of AtCSP3. However, SALK_144972C and Wisc DsLox 353G12 have insertions at –42 into the 59 UTR and +170 in the coding region, respectively. WiscDsLox353G12 is denoted as atcsp3-1, SALK_144972C as atcsp3-2, and SALK_022658C as atcsp3-3 (Fig. 1C).
To confirm the disruption of the AtCSP3 gene transcript in the T-DNA insertion mutants, semi-quantitative RT-PCR was performed using a forward and reverse gene-specific primer pair for the full sequence of AtCSP3 (Fig. 1C). AtCSP3 transcript was not detected in atcsp3-1 and atcsp3-2 but a full-length transcript was identified in atcsp3-3 (Fig. 1D). Therefore, two T-DNA insertion alleles were identified as loss-of-function mutant alleles disrupting AtCSP3 transcripts.
Phenotypic analysis of atcsp3 T-DNA insertion mutant alleles
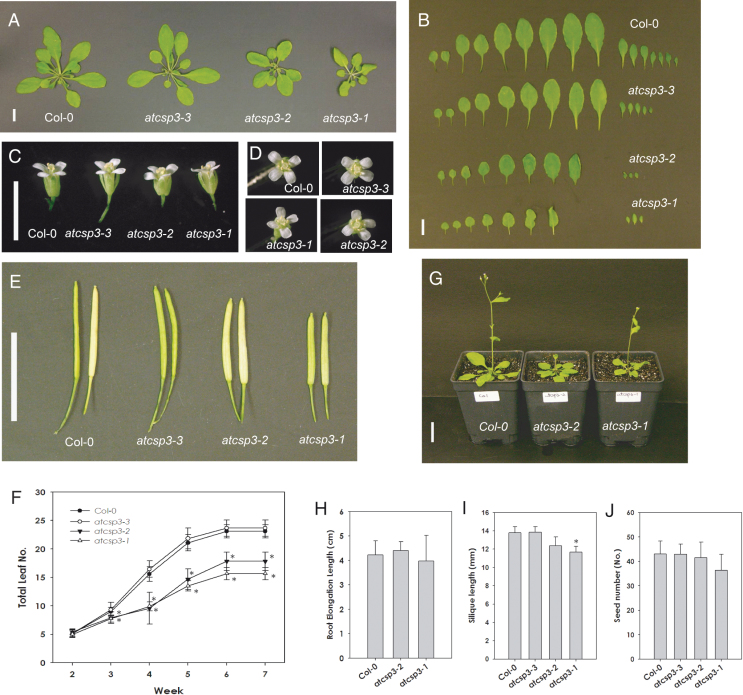
To observe the effects of the T-DNA insertion, the phenotype of the mutants was monitored from germination through to flowering and seed maturation. Under long-day conditions, none of the mutants showed germination defects as determined by the uniform emergence of radicles (data not shown). Primary root elongation was also measured on MS plates under long-day conditions and no differences were observed among the mutant alleles (Fig. 2H).
Fig. 2.
Morphological analysis of atcsp3 loss-of-function mutants. (A) Vegetative growth of the wild type (Col-0) and atcsp3 insertion lines at 28 DAG grown under long-day conditions (16/8h light/dark) at 23 °C (bar=1cm). (B) Total number of leaves of the wild type and atcsp3 insertion mutants. Rosette leaves were taken from the same plant shown in (A) and leaves were aligned starting from the first leaf from the cotyledons (Bar=1cm). (C) Comparison of flower size. Opened flowers were collected from primary bolts at 35 DAG (Bar=0.5cm). (D) Vertical views of flowers. (E) Comparison of fully matured siliques collected at 48 DAG (Bar=1cm). (F) Total leaves were counted from 14–49 DAG. Total leaves were numbered starting from the first leaf next to the cotyledons exclusive of cauline leaves (n=16). All data show the average of total leaf number and error bars indicate standard deviation. (G) 28 DAG plant size of wild-type Col-0 and atcsp3 loss-of-function mutants grown under long day conditions (Bar=1cm). (H) Root elongation measurement of the wild type and atcsp3 loss-of-function mutants (n=20). (I) Silique length comparison at 48 DAG (n=20). (J) Seed count number. Seeds were collected from the same silique that was used for silique length measurement (n=20). All plots in (F), (H), (I), and (J) show an average measured value and error bars represent standard deviation. An asterisk indicates a P-value <0.05.
During the later stages of vegetative development, atcsp3-1 and atcsp3-2 mutants exhibited smaller and a reduced number of leaves relative to the wild type but atcsp3-3 did not (Fig. 2A). Total leaf number from 28 DAG plants was compared with all alleles by counting all leaves after removing the primary shoot. Rosette leaf numbers of atcsp3-2 and atcsp3-1, including younger leaves close to the shoot apex, were fewer than the wild type (Fig. 2B, F). We also observed abnormally rounded leaf blades and shortened petioles in the atcsp3-2 and atcsp3-1 mutants. By contrast, atcsp3-3 leaves had an indistinguishable shape relative to wild-type plants. In addition to the aforementioned abnormalities in leaf shape, atcsp3-1 leaves exhibited a curly phenotype but other alleles did not exhibit a similar phenotype (Fig. 2B).
To determine if the mutant alleles showed any defect in the transition from vegetative to reproductive growth, flowering time was investigated by counting the days until the emergence of primary inflorescences (~1cm in height). Seedlings of atcsp3 mutant alleles started to flower at approximately 22 DAG and were similar to the wild type (Table 1). The similarities in the initiation of flowering time confirmed that the differences in rosette growth were not due to a delay in developmental progression (see Supplementary Fig. S1 at JXB online). The height of fully grown atcsp3-2 and atcsp3-1 plants was approximately 7cm shorter than the wild type (Table 1). Measurements of fresh weight at 42 DAG also indicated that the growth of atcsp3-2 and atcsp3-1 plants is clearly reduced relative to the wild type and atcsp3-3 (data not shown).
Table 1.
Analysis of flowering time and seedling size
| Genotype | Flowering time (DAG) | Height (cm) | Fresh weight (mg) | Sample size (n) |
| Col-0 | 22.5 ± 1 | 38.88 ± 1.28 | 395.5 ± 14.5 | 14 |
| atcsp3-3 | 22.5 ± 0.90 | 38.58 ± 1.62 | 371.5 ± 17.8 | 16 |
| atcsp3-2 | 23.25 ± 0.97 | 31.83 ± 2.53 | 256.5 ± 26.6 | 14 |
| atcsp3-1 | 22.58 ± 0.79 | 31.33 ± 2.31 | 189.6 ± 13.7 | 12 |
Among the reproductive tissues, atcsp3-1 and atcsp3-2 flowers were slightly shorter in length relative to wild-type plants. However, no abnormalities in floral organ shapes were observed in any alleles (Fig. 2C). Regarding the numbers of floral organ tissues, all alleles contained six stamens, two carpels, four petals, and four sepals (Fig. 2D). Siliques of atcsp3-1 and atcsp3-2 were shorter than thoset of the wild type (Fig. 2E, I). Trends in both atcsp3-2 and atcsp3-1 mutants revealed slightly reduced seed numbers relative to the wild type and atcsp3-3 (Fig. 2J). In summary, morphological analyses of atcsp3 T-DNA insertion alleles revealed that atcsp3-2 and atcsp3-1 mutants produce smaller plants with rounded leaf blades, shortened petioles, and shortened siliques.
Leaf index measurement and orbicular leaf blade shape of atcsp3 mutant alleles
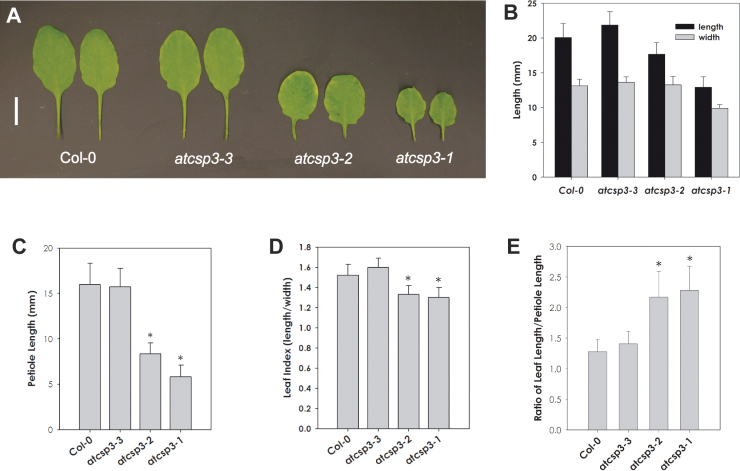
As shown in Fig. 2, both atcsp3 loss-of-function mutant alleles exhibited stunted and orbicular shaped leaves. To characterize the leaf shape of atcsp3 T-DNA insertion mutants, the length and width of the 5th leaf from 28 DAG seedlings were measured. In wild-type plants, the 5th leaf is typically elliptical in shape and contains a long petiole (Fig. 3A). In contrast, leaves in atcsp3-2 and atcsp3-1 mutants were rounded and orbicular with clear reductions in leaf blade and petiole lengths (Fig. 3B, C). The atcsp3-1 mutant exhibited the most severely stunted and rounded leaf phenotype (Fig. 3A, B). To characterize leaf shape better, leaf index was calculated by measuring the length and width of the leaf blade area (Fig. 3D). Wild-type Col-0 and atcsp3-3 plants had 1.52±0.11 and 1.60±0.09 leaf index values, respectively. In the atcsp3-2 and atcsp3-1 mutant alleles, the leaf index values decreased to 1.33±0.09 and 1.30±0.10, respectively. This reduction in leaf index values directly correlates with the increased roundness in leaf shape for these mutant alleles. Using a similar approach, the ratio of leaf blade length to petiole length (Fig. 3E) was quantified and it was confirmed that the atcsp3-2 and atcsp3-1 mutants have significantly reduced petioles relative to leaf blade length. Taken together, atcsp3-2 and atcsp3-1 leaves were orbicular in shape which resulted from a reduction of leaf length and a rounded leaf base. Since the atcsp3-3 mutation did not result in a reduction of transcript of AtCSP3 (Fig. 1D) and in the impairment of normal phenotype, atcsp3-3 was eliminated from further characterization due to its resemblance to wild-type plants.
Fig. 3.
atcsp3 loss-of-function mutants have short petioles and small-sized orbicular leaves. (A) Comparison of leaf shape of 5th leaves from 28 DAG plant (Bar=1cm). (B) Two-dimensional measurement of leaf blade length. Leaf length was measured in the leaf-length (longitudinal) and leaf-width (lateral) directions (n=20). (C) Measurement of petiole length (n=20). (D) Comparison of leaf index. Each leaf index was determined by the ratio of length to width in a leaf blade (n=20). A value close to 1.0 is indicative of increased leaf roundness. (E) The ratio of leaf blade length to petiole length (n=20). Note that the petioles of atcsp3-2 and atcsp3-1 are significantly shorter compared with leaf blade length. An asterisk indicates a P-value <0.05. (This figure is available in colour at JXB online.)
Histological characterization of atcsp3 mutants
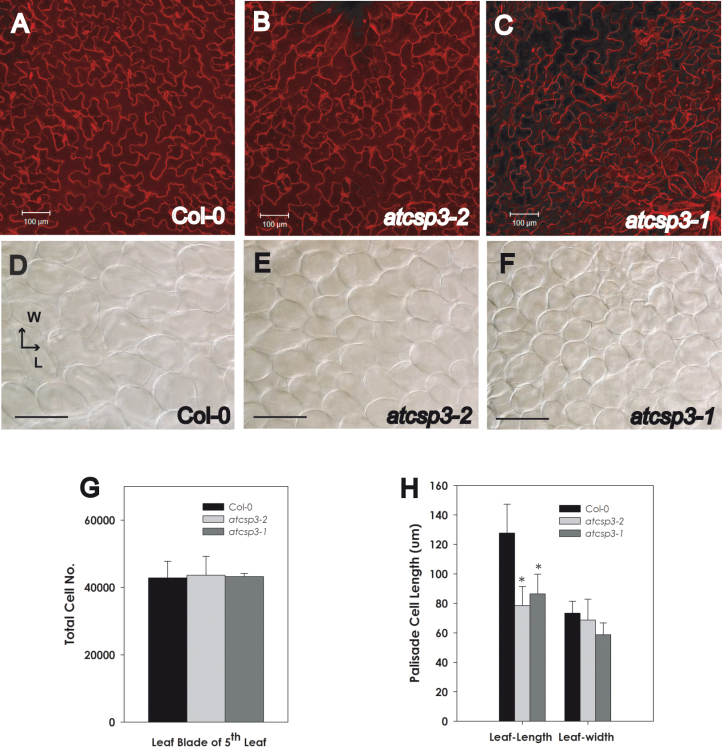
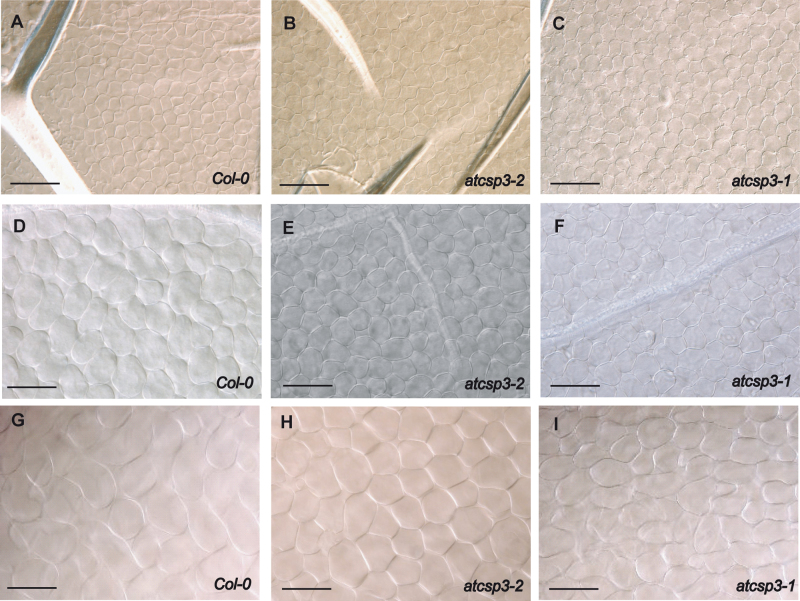
To determine why the shape of leaf blades was altered in atcsp3 mutants, epidermal and palisade mesophyll cell size and shape in the middle portion of 28 DAG 5th leaves were observed. Adaxial epidermal cells in atcsp3-2 and atcsp3-1 mutants were more densely arranged, while those of Col-0 wild type were more evenly aligned (Fig. 4A, B, C). Palisade cells in the atcsp3 mutants were smaller than that of the wild type. Similar to epidermal cells, the distribution of palisade cells in atcsp3 mutants was more condensed relative to the wild type (Fig. 4D, E, F). In relative comparison with the wild type, palisade cells within both mutants generally appeared more circular in shape. To understand why atcsp3 mutants have orbicular shaped leaves, the number and size of palisade cells were measured from microscopic images. Palisade cell numbers were quantified within subset regions of the micrographs. In order to estimate total cell numbers within an entire leaf area, cell counts within subset regions were multiplied by the ratio between the size of the microscopic image and the overall size of the examined leaves. Using this approach, the estimated total cell numbers were similar between atcsp3 mutants and the wild type (Fig. 4G).
Fig. 4.
Alteration of cell size in atcsp3 loss-of-function mutants. Anatomical analysis was performed with 5th leaves of 28 DAG plants. Images of individual mutant alleles were taken from the same position in the middle part of an adaxial leaf blade. (A–C) Propidium iodide staining images of epidermal cells of wild-type Col-0, atcsp3-2, and atcsp3-1. (D–F) Nomarsky images of palisade mesophyll cells of wild-type Col-0, atcs3-2, and atcsp3-1 (Bar=100 µm). Note that loss-of-function atcsp3 mutants have small cell sizes relative to the wild type. (G) Number of palisade cells on leaf blades. Total cell number was determined by counting the number of cells per unit area which is multiplied by the ratio between the original leaf area and observed leaf area (n=6). (H) Cell length of individual palisade cells in relation to leaf-length and width (n=30 from three different leaves). Leaf-length and leaf-width direction are marked in (D). All data in graphs are average values with error bars indicating standard deviation. An asterisk indicates a P-value <0.05.
A comparative analysis was also performed to confirm if the individual shape and size of leaf cells is altered in the mutants and could be correlated with the orbicular leaf shape mutation. To determine individual palisade parenchyma cell size, palisade cell length was measured along two-dimensions. Palisade cell length along the leaf-length axis was significantly reduced in atcsp3-1 and atcsp3-2 mutants, whereas a non-significant trend of reduced cell length along the leaf-width axis was observed (Fig. 4H). Taken together, the results of these cell dimension analyses indicate that the orbicular leaf shape of atcsp3-2 and atcsp3-1 likely results from impairment of leaf cell expansion in the leaf-length direction.
Relationship of AtCSP3 to leaf cell expansion
Leaf morphogenesis proceeds in two separate stages including a cell division and an expansion phase. In general, cell proliferation by cell division occurs during early leaf development, whereas cell expansion continues until leaves mature into full size. Abnormalities in leaf morphology can result from a disorder in either of the two or during both stages.
To determine the stage where leaf development is impaired in the atcsp3 loss-of-function mutants, palisade cell shape was monitored at three different time points during leaf development. Cell number and cell size were not altered until 14 DAG when the leaf primordia expanded into adult leaves after cell proliferation in 5th leaf blades (Fig. 5A, B, C). Differences in cell size were observed at 21 DAG, where the cells complete the transition from the cell proliferation to the cell expansion stage (Fig. 5D, E, F). Also, the distance between individual cells in atcsp3-2 and atcsp3-1 was less than the wild type, resulting in a stacked and condensed cell layer appearance. These data suggested that morphological differences in cell shape and size in atcsp3-2 and atcsp3-1 are not manifested until the end of cell proliferation. This pattern was sustained through an evaluation period of 35 DAG (see Supplementary Fig. S2 at JXB online). Thus, AtCSP3 appears to affect leaf morphology at the leaf cell expansion stage through an alteration of cell size in the leaf length direction.
Fig. 5.
Comparison of palisade cell size during leaf development. DIC images were taken from the middle part of 5th leaves at different time points from wild-type Col-0, atcsp3-2, and atcsp3-1. Upper row (A), (B), and (C) are 14 DAG 5th leaf palisade cells from the adaxial surface of leaf blades (Bar=20 µm). Middle row (D), (E), and (F) are 21 DAG 5th leaves, and bottom row (G), (H), and (I) show 28 DAG 5th leaves. Scale bars indicate 100 µm.
Expression of leaf development related genes in atcsp3 mutants
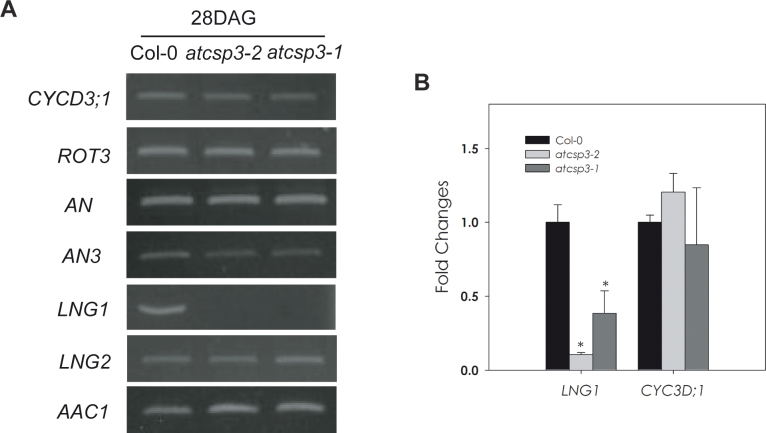
Morphological and anatomical analyses of atcsp3 loss-of-function mutants suggest that AtCSP3 functions in leaf cell expansion as a determinant of leaf shape. Therefore, there was a need to determine if the expression of known genes involved with leaf development are affected by the atcsp3 loss-of-function mutation. To address this question, a targeted expression analysis of several genes that are involved with cell polarity was performed. Specifically, expression patterns of ROT3, LNG1, and LNG2, which are leaf-length direction regulators, and AN and AN3, which are leaf-width direction regulators, were monitored. The expression of a cell division regulator (CYCD3;1) was also monitored as a means to assess if an alteration of cell division could be related to the altered leaf shape in atcsp3 mutants. Interestingly, transcript levels of LNG1 were clearly reduced in atcsp3-2 and atcsp3-1 at 28 DAG (Fig. 6A). LNG2 is a functionally redundant gene of LNG1, however, LNG2 gene expression in atcsp3-2 and atcsp3-1 did not show the same pattern of gene expression as LNG1. As reported by Kim et al., LNG1 and LNG2 exert additive effects on leaf cell expansion. Therefore, it is important to note that there was an observed reduction of LNG2 transcript from an independent microarray experiment comparing the atcsp3-1 mutant background relative to wild-type plants (data not shown). ROT3, which is another leaf-length specific regulator, was not altered relative to the wild type. Cell division and cell proliferation regulator genes such as CYCD3;1, AN3, and AN were not altered in atcsp3 mutants. By confirming similar levels of CYCD3;1 across mutants and the wild type with qRT-PCR analysis, it was concluded that the reduction of LNG1 transcripts was not related to alterations of cell division in atcsp3 mutant alleles (Fig. 6B). Future leaf development expression analyses on a transcriptome-wide level are warranted and are necessary to completely understand the role of AtCSP3 on a molecular level.
Fig. 6.
Gene expression of leaf shape determinant genes. (A) Total RNA was extracted from the 5th leaves of 28 DAG plants. With the exception of AAc1 amplification, all PCR reactions were performed for 30 cycles with the described primer sets that can be found in Supplementary Table S2 at JXB online. AAc1 was used as an internal control and was amplified for 26 cycles. This figure contains representative results that were obtained among three replicates for each individual gene. (B) Comparative quantification of LNG1 and CYCD3;1 mRNA at 28 DAG. Quantitative real-time PCR was performed to compare the levels of both gene transcripts. Actin2 was used as an internal control for normalization. Fold change was determined by relative comparison to the expression of individual genes in the wild type. The average of three replicates was graphed with error bars indicating standard deviation. An asterisk indicates a P-value <0.05.
Subcellular localization of AtCSP3 in onion epidermal cells
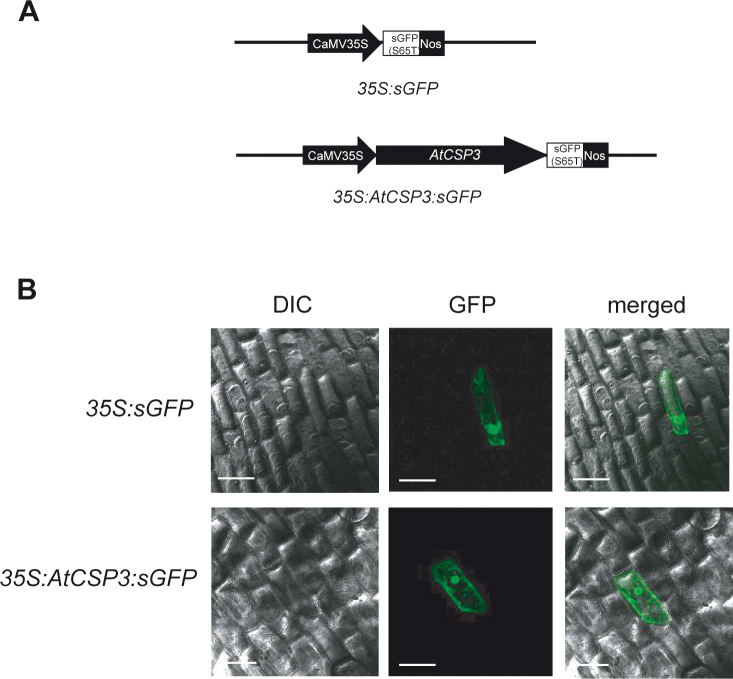
To determine the subcellular localization of the AtCSP3 full-length protein, its complete coding sequence was fused to the N-terminus of sGFP(S65T) which was driven by a CaMV 35S promoter (Fig. 7A). The sGFP(S65T) and 35S:AtCSP3:sGFP plasmids were bombarded into onion epidermal cells with a particle bombardment delivery system and cells that were transiently expressing GFP were visualized with confocal microscopy. Cells transformed with sGFP(S65T) alone showed both nuclear and cytosolic fluorescence (Fig. 7B). Consistent with a previous report with transgenic lines over-expressing an AtCSP3-GFP fusion protein (Kim et al., 2009), transiently expressed 35S:AtCSP3:sGFP fusion proteins fluoresced in both the cytosol and nucleus; confirming that that AtCSP3 is a nucleocytosolic protein.
Fig. 7.
AtCSP3 localizes in the nucleus and cytosol in onion epidermal cells. (A) Schematic representation of 35S:sGFP(S65T) and 35S:AtCSP3:sGFP vectors. The coding region of AtCSP3 was amplified by high fidelity PCR and sub-cloned into the sGFP(S65T) vector for transient expression via a particle bombardment system. (B) Transient analysis of subcellular localization for AtCSP3 in onion epidermal cells (Bar=100 µm). Note the nucleocytoplasmic localization of AtCSP3.
Discussion
AtCSP3 expression is enriched in reproductive tissues
The four AtCSPs are highly similar at the amino acid sequence level (Karlson and Imai, 2003) and have recently been characterized in relation to floral and silique development (Nakaminami et al., 2009). AtGRP2/CSDP2/AtCSP2 was characterized at a biochemical level (Sasaki et al., 2007) and RNAi mutational analysis linked its function to plant development (Fusaro et al., 2007). To begin elucidating the function of AtCSP3 in relation to plant development, its expression patterns were studied in different tissues with qRT-PCR analysis. Similar to AtGRP2/CSDP2/AtCSP2 expression patterns, AtCSP3 also accumulates in tissues like shoot apices, inflorescence meristems, open flowers, and siliques compared with mature rosette leaves and stem tissue (Fig. 1).
AtCSP3 loss-of-function generates stunted and orbicular leaf morphology
Independent T-DNA insertion alleles were selected for functional characterization and were confirmed by semi-quantitative PCR analysis. The atcsp3-2 and atcsp3-1 mutant alleles have T-DNA insertions in the 59 UTR and the cold shock domain, respectively (Fig. 1). Although a previously published report did not document any morphological alteration in an atcsp3 loss-of-function mutant under their experimental conditions (Kim et al., 2009), two independent T-DNA insertion mutations in the AtCSP3 gene clearly resulted in smaller statured plants, reduced leaf and petiole size, and an orbicular leaf shape under our experimental testing conditions (Fig. 2). The most prominent characteristic for the loss-of-function mutants is the abnormal orbicular leaf shape that is accompanied by short petioles (Fig. 3). Comparative phenotypic analyses of the independent atcsp3 T-DNA insertion mutants revealed atcsp3-1 as a strong allele and atcsp3-2 as a weak allele. As previously mentioned, AtGRP2/CSDP2/AtCSP2 RNAi mutants showed defects in flowering time, flower organogenesis, and seed development (Fusaro et al., 2007). Since AtGRP2/CSDP2/AtCSP2 and AtCSP3 exhibit high homology in their cold shock domain regions, it was necessary to confirm whether atcsp3 mutants also exhibit developmental related defects as described for AtGRP2/CSDP2/AtCSP2 down-regulated plants. With the exception of flowers with shorter lengths, atcsp3 loss-of-function mutants did not exhibit significant abnormalities in flower morphology or flowering time. Taken together, our observations suggest that AtCSPs likely have diversified roles in Arabidopsis since mutant atcsp3 phenotypes did not overlap with those previously described for the AtGRP2/CSDP2/AtCSP2 gene.
Stunted and orbicular leaf shape in atcsp3 loss-of-function mutants is affected by altered cell size
atcsp3 loss-of-function mutants exhibited an abnormal orbicular leaf morphology which has been previously investigated in different Arabidopsis mutant alleles with genetic and histochemical methods (Tsukaya et al., 2006). In general, lateral leaf blade morphology is determined by the harmonious control of two-dimensional proliferation and expansion of leaf cells. Investigations of loss-of-function rot3, rot4, an, and an3 mutant plants described a functional relationship between cell proliferation and cell expansion. As previously described, the small and rounded leaf morphology of rot3 and rot4 is generated in the leaf-length direction by the alteration of cell proliferation and cell expansion, respectively (Tsuge et al., 1996; Narita et al., 2004). The rounded leaf shape of the atcsp3 loss-of-function mutants is reflective of the rot3 and rot4 mutant phenotype (Tsuge et al., 1996; Narita et al., 2004). Even though an and an3 gene mutants exhibited different leaf shapes than rot3 and rot4 mutants, they were also implicated in the polarity-specific regulation of leaf shape (Tsuge et al., 1996; Horiguchi et al., 2005). Therefore, the molecular mechanisms controlling cell proliferation and cell elongation during leaf blade morphogenesis appear to be complex. Image analysis of the wild type and atcsp3 loss-of-function mutants indicated that the total cell number in atcsp3 leaf blades is not affected (Fig. 4G), whereas cell size was reduced in the leaf-length axis relative to the wild type (Fig. 4H). Regarding the progression in stages of development, developmental phase transitions occurred at similar time points in all lines (Table 1). Thus, the small size and stunted leaf shape of atcsp3 mutant did not correlate to differences in growth stages between mutant and wild-type plants. Instead, the orbicular mutant leaf shape in atcsp3 mutants appears to be a consequence of the reduced cell size in the leaf length direction (Fig. 3).
Leaf cell proliferation is primarily determined during the early leaf generation stage and partially during two-dimensional lateral leaf growth (Donnelly et al., 1999). As shown in Fig. 5, in the early stages of leaf development, wild type and atcsp3 loss-of-function mutants have similar palisade cell numbers, size, and shape. In contrast, cell expansion during the later stages of leaf generation differs between the wild type and atcsp3 loss-of-function mutants (Fig. 5). These microscopic observations support the hypothesis that AtCSP3 primarily impacts leaf development in the later stages of leaf development through an alteration of cell expansion.
atcsp3 loss-of-function mutation altersLNG1 gene expression
Leaf shape and size determination is affected by cell differentiation on adaxial and abaxial leaf surfaces during the early stages of leaf morphogenesis and two-dimensional leaf cell expansion (Barkoulas et al., 2007). Complicated temporal and spatial networks of multiple genes play important roles as determinants of leaf morphology. An incomplete transition of leaf development from the shoot apical meristem results in abnormal leaf morphology with deformation in symmetry, polarity, and flatness (Long et al., 1996; Waites et al., 1998; Timmermans et al., 1999; Ori et al., 2000; Byrne et al., 2000, 2002, 2003; Benkova et al., 2003; Heisler et al., 2005). The atcsp3 loss-of-function mutant did not exhibit phenotypic abnormalities during the leaf initiation stage. Our semi-quantitative RT-PCR data for SHOOT MERISTEMLESS (STM), ASYMMETRIC LEAVES1 (AS1), CUC1 and CUC2 (CUP-SHAPED COTYLEDON 1 and 2), and PIN-FORMED 1 (PIN1) genes using shoot apex and early leaf tissues did not identify any alteration in expression patterns (data not shown). These observations support the hypothesis that AtCSP3 primarily functions during lateral leaf development. Our histological observations, which determined that the abnormal leaf cell shape and size of atcsp3 are initiated during lateral leaf expansion, are in good accordance with this scenario (Fig. 5).
Expression analysis of a cell division marker gene (CYC3D;1) at 28 DAG did not reveal abnormalities as a result of atcsp3 loss-of-function. Therefore, AtCSP3 appears to affect LNG1 mRNA abundance independent of cell division regulation via an unknown mechanism. Observations of leaf phenotypes from the atcsp3 mutants are in good accordance to published results from functional studies of LNG1 (Lee et al., 2006). Specifically, a correlation of reduced LNG1 expression to a reduction in leaf-length was observed. LNG1 is a regulator of leaf-length specific cell expansion that functions independently of ROT3 and does not regulate expression of AN (Lee et al., 2006). Our RT-PCR data for ROT3 and AN genes in atcsp3 mutants (Fig. 6) are similar to those previously published in a study characterizing a lng1 loss-of-function mutant; suggesting that AtCSP3 does not function in relation to ROT3 and AN during two-dimensional leaf expansion (Lee et al., 2006). Over-expression of LNG2, a homologue of LNG1, results in a narrow and long leaf phenotype. In addition, an lng1-3 lng2-1 double mutant has an additive effect in leaf-length cell expansion. As revealed by semi-quantitative RT-PCR, LNG1 expression was majorly altered in the atcsp3 mutants with no apparent effect on LNG2 expression. However, independent data obtained from a comparative microarray study between the atcsp3-1 mutant and the wild type revealed a minor reduction in LNG2 transcript (data not shown). Thus, it is hypothesized that AtCSP3 primarily affects LNG1 gene function, contributing to a reduction in cell length along the leaf-length direction and there may be additive contribution from LNG2 due to a marginal reduction in transcripts. Although this targeted investigation of a sub-set of leaf-development related genes identified a predominant change in LNG1 expression, additional transcriptome analyses may reveal additional development-related genes that are affected by the disruption of AtCSP3 function and may also function to regulate cell expansion in the atcsp3 mutants.
To date, all plant CSPs tested including OsCSP1, OsCSP2, AtGRP2/CSDP2/AtCSP2, AtCSDP1, AtCSP3, and WCSP1 exhibit nucleic acid binding activity (Karlson et al., 2002; Nakaminami et al., 2006; Fusaro et al., 2007; Chaikam and Karlson, 2008; Kim et al., 2009; Park et al., 2009). AtCSP3 melts nucleic acids and complements a bacterial mutant lacking four endogenous cold shock proteins, supporting the hypothesis that AtCSP3 functions as an RNA chaperone in plants (Kim et al., 2009; Park et al., 2009). Our transient subcellular localization experiment confirmed that AtCSP3 is a nucleocytoplasmic protein (Fig. 7), consistent with a previous report of stably transformed AtCSP3:GFP fusion protein in Arabidopsis (Kim et al., 2009). In humans, the RNA binding activity of the YB-1 cold shock domain protein activates the translation of silent RNA for genes relating to cell proliferation, malignant transformation, and stress response (Evdokimova et al., 2006). In Chlamydomonas, the NAB1 cold shock domain protein also regulates LHCBM mRNA stabilization, resulting in the induction of LHCBM protein translation (Mussgnug et al., 2005). Importantly, biochemical analysis of recombinant AtCSP3 protein confirmed that it possesses nucleic acid chaperone activity which induces the unwinding of double-stranded DNA and RNA (Kim et al., 2009). Therefore, it is reasonable to consider that AtCSP3 may exert a similar role and positively affect LNG1 transcripts during leaf cell expansion during the generation of lateral leaves.
Supplementary Material
Acknowledgements
The authors would like to thank Dr Karen Martin at the West Virginia University Microscopy Imaging Center for assistance with confocal microscopy. The authors also acknowledge Dr Carina Barth and Dr Simeon Kotchoni for morphological analysis and Drs Chaikam, Panaccione, Doelling, and Yao for careful reading of this manuscript. This work was supported by a National Science Foundation grant (IBN-0416945) to DK. West Virginia Agriculture and Forestry Experiment Station Scientific Article No. 3135.
References
- Barkoulas M, Galinha C, Grigg SP, Tsiantis M. 2007. From genes to shape: regulatory interactions in leaf development. Current Opinion in Plant Biology 10 660– 666 [DOI] [PubMed] [Google Scholar]
- Basaki Y, Hosoi F, Oda Y, et al. 2007. Akt-dependent nuclear localization of Y-box-binding protein 1 in acquisition of malignant characteristics by human ovarian cancer cells. Oncogene 26 2736– 746 [DOI] [PubMed] [Google Scholar]
- Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, Friml J. 2003. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115 591–– 602 [DOI] [PubMed] [Google Scholar]
- Byrne ME. 2005. Networks in leaf development. Current Opinion in Plant Biology 8 59– 66 [DOI] [PubMed] [Google Scholar]
- Byrne ME, Barley R, Curtis M, Arroyo JM, Dunham M, Hudson A, Martienssen RA. 2000. Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408 967– 971 [DOI] [PubMed] [Google Scholar]
- Byrne ME, Groover AT, Fontana JR, Martienssen RA. 2003. Phyllotactic pattern and stem cell fate are determined by the Arabidopsis homeobox gene BELLRINGER. Development 130 3941– 3950 [DOI] [PubMed] [Google Scholar]
- Byrne ME, Simorowski J, Martienssen RA. 2002. ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129 1957– 1965 [DOI] [PubMed] [Google Scholar]
- Chaikam V, Karlson D. 2008. Functional characterization of two cold shock domain proteins from Oryza sativa. Plant, Cell and Environment 31 995– 1006 [DOI] [PubMed] [Google Scholar]
- Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG. 1999. Cell cycling and cell enlargement in developing leaves of Arabidopsis. Developmental Biology 215 407– 419 [DOI] [PubMed] [Google Scholar]
- Evdokimova V, Ruzanov P, Anglesio MS, Sorokin AV, Ovchinnikov LP, Buckley J, Triche TJ, Sonenberg N, Sorensen PH. 2006. Akt-mediated YB-1 phosphorylation activates translation of silent mRNA species. Molecular and Cellular Biology 26 277– 292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkers U, Kirik V, Schobinger U, et al. 2002. The cell morphogenesis gene ANGUSTIFOLIA encodes a CtBP/BARS-like protein and is involved in the control of the microtubule cytoskeleton. EMBO Journal 21 1280– 1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusaro AF, Bocca SN, Ramos RL, et al. 2007. AtGRP2, a cold-induced nucleo-cytoplasmic RNA-binding protein, has a role in flower and seed development. Planta 225 1339– 1351 [DOI] [PubMed] [Google Scholar]
- Graumann PL, Marahiel MA. 1998. A superfamily of proteins that contain the cold-shock domain. Trends in Biochemical Science 23 286– 290 [DOI] [PubMed] [Google Scholar]
- Hanson J, Johannesson H, Engstrom P. 2001. Sugar-dependent alterations in cotyledon and leaf development in transgenic plants expressing the HDZhdip gene ATHB13. Plant Molecular Biology 45 247– 262 [DOI] [PubMed] [Google Scholar]
- Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, Meyerowitz EM. 2005. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Current Biology 15 1899– 1911 [DOI] [PubMed] [Google Scholar]
- Horiguchi G, Kim GT, Tsukaya H. 2005. The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. The Plant Journal 43 68– 78 [DOI] [PubMed] [Google Scholar]
- Horn G, Hofweber R, Kremer W, Kalbitzer HR. 2007. Structure and function of bacterial cold shock proteins. Cell and Molecular Life Sciences 64 1457– 1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlson D, Imai R. 2003. Conservation of the cold shock domain protein family in plants. Plant Physiology 131 12– 15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlson D, Nakaminami K, Toyomasu T, Imai R. 2002. A cold-regulated nucleic acid-binding protein of winter wheat shares a domain with bacterial cold shock proteins. Journal of Biological Chemistry 277 35248– 35256 [DOI] [PubMed] [Google Scholar]
- Kim GT, Fujioka S, Kozuka T, Tax FE, Takatsuto S, Yoshida S, Tsukaya H. 2005. CYP90C1 and CYP90D1 are involved in different steps in the brassinosteroid biosynthesis pathway in Arabidopsis thaliana. The Plant Journal 41 710– 721 [DOI] [PubMed] [Google Scholar]
- Kim GT, Shoda K, Tsuge T, Cho KH, Uchimiya H, Yokoyama R, Nishitani K, Tsukaya H. 2002. The ANGUSTIFOLIA gene of Arabidopsis, a plant CtBP gene, regulates leaf-cell expansion, the arrangement of cortical microtubules in leaf cells and expression of a gene involved in cell-wall formation. EMBO Journal 21 1267– 1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim GT, Tsukaya H, Saito Y, Uchimiya H. 1999. Changes in the shapes of leaves and flowers upon overexpression of cytochrome P450 in Arabidopsis. Proceedings of the National Academy of Sciences, USA 96 9433– 9437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim GT, Tsukaya H, Uchimiya H. 1998. The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Genes and Development 12 2381– 2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Kende H. 2004. A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proceedings of the National Academy of Sciences, USA 101 13374– 13379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Park SJ, Kwak KJ, Kim YO, Kim JY, Song J, Jang B, Jung CH, Kang H. 2007. Cold shock domain proteins and glycine-rich RNA-binding proteins from Arabidopsis thaliana can promote the cold adaptation process in Escherichia coli. Nucleic Acids Research 35 506– 516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MH, Sasaki K, Imai R. 2009. Cold Shock Domain Protein 3 regulates freezing tolerance in Arabidopsis thaliana. Journal of Biological Chemistry 284 23454– 23460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley PD, Palis J. 1994. GRP2 proteins contain both CCHC zinc fingers and a cold shock domain. The Plant Cell 6 1522– 1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno K, Izumi H, Uchiumi T, Ashizuka M, Kuwano M. 2003. The pleiotropic functions of the Y-box-binding protein, YB-1. Bioessays 25 691– 698 [DOI] [PubMed] [Google Scholar]
- Lee YK, Kim GT, Kim IJ, Park J, Kwak SS, Choi G, Chung WI. 2006. LONGIFOLIA1 and LONGIFOLIA2, two homologous genes, regulate longitudinal cell elongation in Arabidopsis. Development 133 4305– 4314 [DOI] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK. 1996. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379 66– 69 [DOI] [PubMed] [Google Scholar]
- Mussgnug JH, Wobbe L, Elles I, et al. 2005. NAB1 is an RNA binding protein involved in the light-regulated differential expression of the light-harvesting antenna of Chlamydomonas reinhardtii. The Plant Cell 17 3409– 3421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaminami K, Hill K, Perry SE, Sentoku N, Long JA, Karlson DT. 2009. Arabidopsis cold shock domain proteins: relationships to floral and silique development. Journal of Experimental Botany 60 1047– 1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaminami K, Karlson DT, Imai R. 2006. Functional conservation of cold shock domains in bacteria and higher plants. Proceedings of the National Academy of Sciences, USA 103 10122– 10127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita NN, Moore S, Horiguchi G, Kubo M, Demura T, Fukuda H, Goodrich J, Tsukaya H. 2004. Overexpression of a novel small peptide ROTUNDIFOLIA4 decreases cell proliferation and alters leaf shape in Arabidopsis thaliana. The Plant Journal 38 699– 713 [DOI] [PubMed] [Google Scholar]
- Ori N, Eshed Y, Chuck G, Bowman JL, Hake S. 2000. Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127 5523– 5532 [DOI] [PubMed] [Google Scholar]
- Park SJ, Kwak KJ, Oh TR, Kim YO, Kang H. 2009. Cold shock domain proteins affect seed germination and growth of Arabidopsis thaliana under abiotic stress conditions. Plant and Cell Physiology 50 869– 878 [DOI] [PubMed] [Google Scholar]
- Qiu JL, Jilk R, Marks MD, Szymanski DB. 2002. The Arabidopsis SPIKE1 gene is required for normal cell shape control and tissue development. The Plant Cell 14 101– 118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Kim MH, Imai R. 2007. Arabidopsis COLD SHOCK DOMAIN PROTEIN2 is a RNA chaperone that is regulated by cold and developmental signals. Biochemical and Biophysical Research Communications 364 633– 638 [DOI] [PubMed] [Google Scholar]
- Sommerville J. 1999. Activities of cold-shock domain proteins in translation control. Bioessays 21 319– 325 [DOI] [PubMed] [Google Scholar]
- Timmermans MC, Hudson A, Becraft PW, Nelson T. 1999. ROUGH SHEATH2: a Myb protein that represses knox homeobox genes in maize lateral organ primordia. Science 284 151– 153 [DOI] [PubMed] [Google Scholar]
- Tsuge T, Tsukaya H, Uchimiya H. 1996. Two independent and polarized processes of cell elongation regulate leaf blade expansion in Arabidopsis thaliana (L.) Heynh. Development 122 1589– 1600 [DOI] [PubMed] [Google Scholar]
- Tsukaya H. 2005. Leaf shape: genetic controls and environmental factors. International Journal of Developmental Biology 49 547– 555 [DOI] [PubMed] [Google Scholar]
- Tsukaya H. 2006. Mechanism of leaf-shape determination. Annual Review of Plant Biology 57 477– 496 [DOI] [PubMed] [Google Scholar]
- Tsukaya H, Imaichi R, Yokoyama J. 2006. Leaf-shape variation of Paederia foetidain Japan: reexamination of the small, narrow leaf form from Miyajima Island. Journal of Plant Research 119 303– 308 [DOI] [PubMed] [Google Scholar]
- Waites R, Selvadurai HR, Oliver IR, Hudson A. 1998. The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell 93 779– 789 [DOI] [PubMed] [Google Scholar]
- Yang Y, Karlson DT. 2011. Over-expression of AtCSP4 affects late stages of embryo development in Arabidopsis. Journal of Experimental Botany 62 2079– 2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.