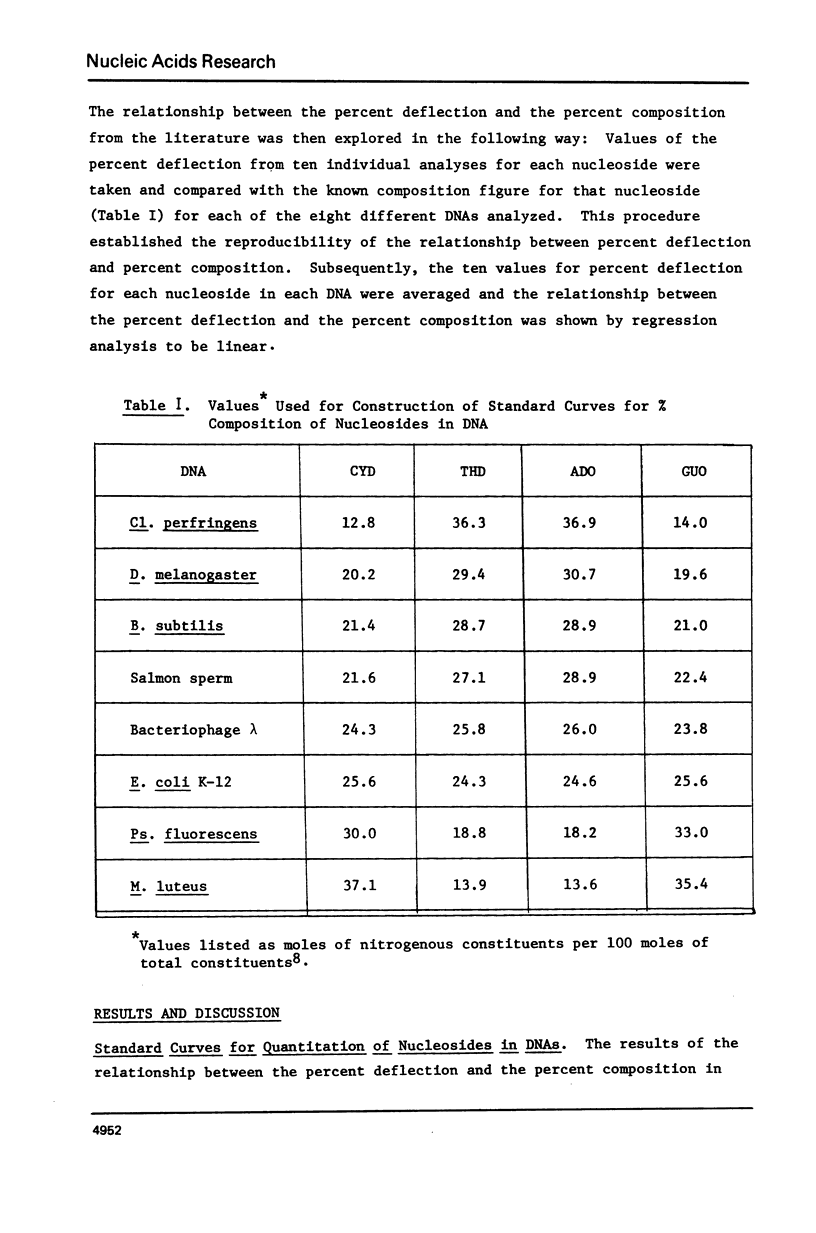
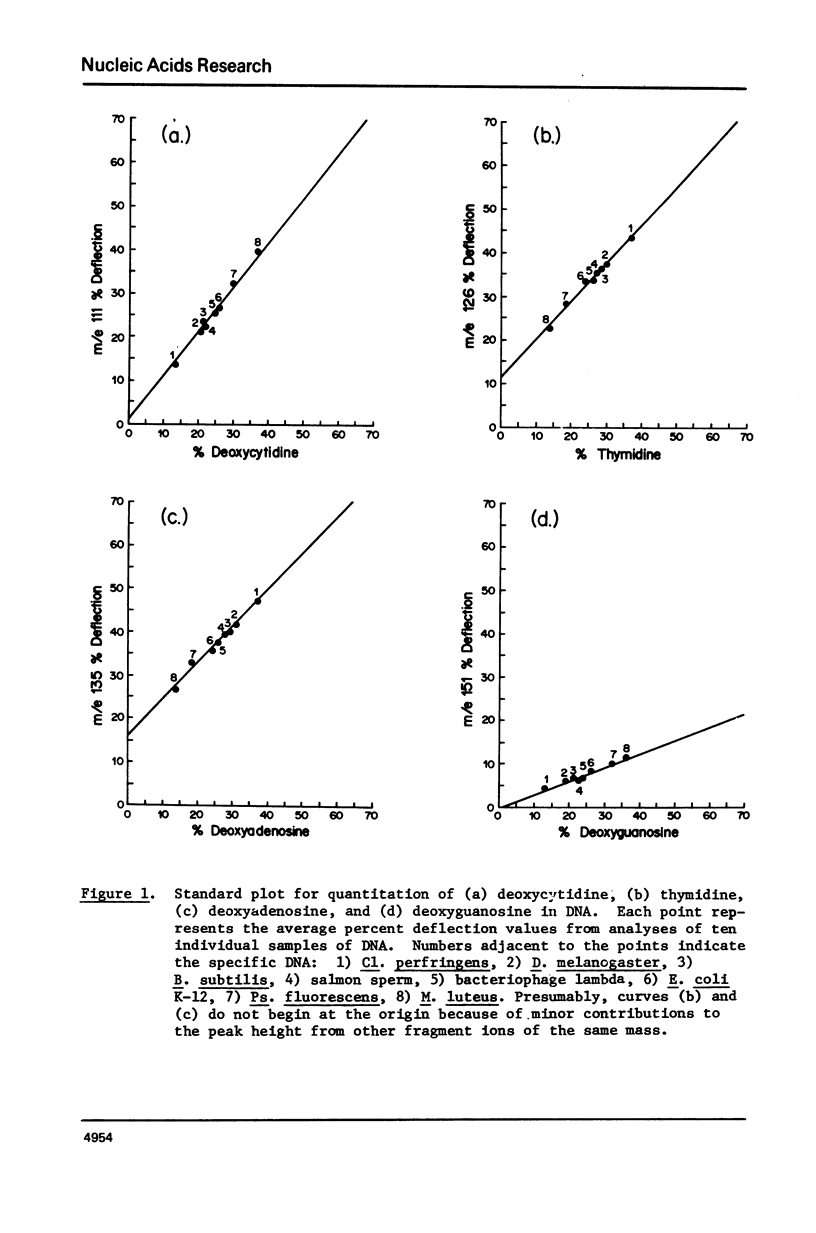
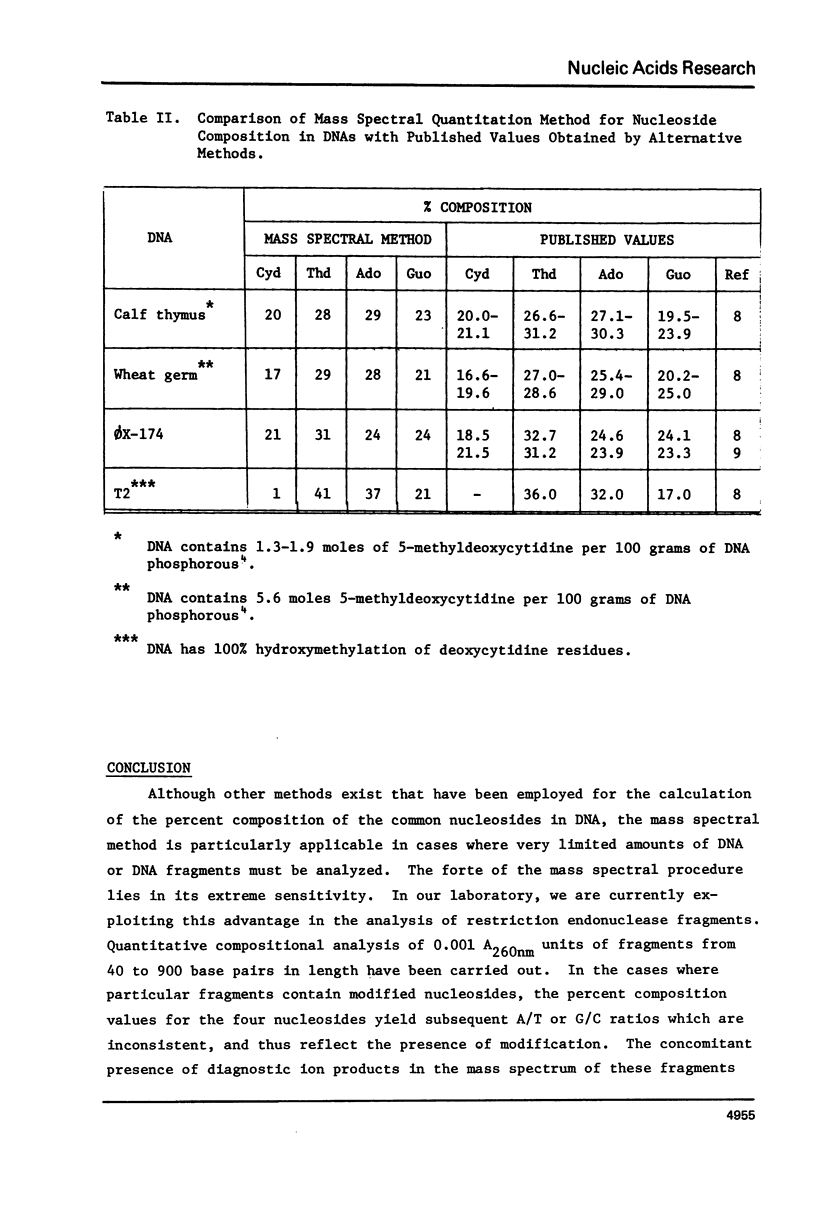
Abstract
A mass spectral method for the quantitation of the percentages of deoxyadenosine, deoxyguanosine, deoxycytidine, and thymidine in intact DNAs has been devised. Standard curves for each nucleoside have been constructed which are based upon the observation that a direct correlation exists between the heights (% deflection) of diagnostic peaks from these nucleosides in a mass spectrum and the published percent composition of specific DNAs. Analyses of DNA from Clostridiumperfringens, Micrococcusluteus, Escherichiacoli, Bacillussubtilis, Pseudomonasfluorescens, Drosophilamelanogaster, salmon sperm, and bacteriophage lambda were used to determine standard curves. The validity of the method was demonstrated by comparison of the results from the mass spectral procedure with results from the chemical analyses of the DNAs from calf thymus and wheat germ. Analysis of ØX-174 DNA yielded values consistent with the published values obtained via sequence analysis and indicated that the method is applicable to both single and double-stranded DNAs. Results from T2 DNA, which contains no cytidine, exhibited artificially high values for adenosine, guanosine and thymidine with concomitant alteration in the A/T and G/C molar ratios. Such skewed results are useful in predicting the presence of modified nucleosides. The extreme sensitivity of the method has been exploited in the analysis of subnanogram quantities of restriction endonuclease fragments from DNA.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armbruster M. A., Wiebers J. L. Identification and location of protecting groups on the nucleotide components of chemically synthesized oligodeoxyribonucleotides by mass spectral analysis. Anal Biochem. 1977 Dec;83(2):570–579. doi: 10.1016/0003-2697(77)90060-4. [DOI] [PubMed] [Google Scholar]
- Burgar D. R., Perone S. P., Wiebers J. L. Factor analysis of the mass spectra of oligodeoxyribonucleotides. Anal Chem. 1977 Aug;49(9):1444–1446. doi: 10.1021/ac50017a037. [DOI] [PubMed] [Google Scholar]
- Burgard D. R., Perone S. P., Wiebers J. L. Sequence analysis of oligodeoxyribonucleotides by mass spectrometry. 2. Application of computerized pattern recognition to sequence determination of di-, tri-, and tetranucleotides. Biochemistry. 1977 Mar 22;16(6):1051–1057. doi: 10.1021/bi00625a004. [DOI] [PubMed] [Google Scholar]
- Sanger F., Air G. M., Barrell B. G., Brown N. L., Coulson A. R., Fiddes C. A., Hutchison C. A., Slocombe P. M., Smith M. Nucleotide sequence of bacteriophage phi X174 DNA. Nature. 1977 Feb 24;265(5596):687–695. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- Wiebers J. L. Detection and identification of minor nucleotides in intact deoxyribonucleic acids by mass spectrometry. Nucleic Acids Res. 1976 Nov;3(11):2959–2970. doi: 10.1093/nar/3.11.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebers J. L. Sequence analysis of oligodeoxyribonucleotides by mass spectrometry. Anal Biochem. 1973 Feb;51(2):542–556. doi: 10.1016/0003-2697(73)90511-3. [DOI] [PubMed] [Google Scholar]
- Wiebers J. L., Shapiro J. A. Sequence analysis of oligodeoxyribonucleotides by mass spectrometry. 1. Dinucleoside monophosphates. Biochemistry. 1977 Mar 22;16(6):1044–1044. doi: 10.1021/bi00625a003. [DOI] [PubMed] [Google Scholar]



